Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is etiologically associated with Kaposi's sarcoma and several other malignancies. The lack of an efficient infection system has impeded the understanding of KSHV-related pathogenesis. A genetic approach was used to isolate infectious KSHV. Recombinant bacteria artificial chromosome (BAC) KSHV containing hygromycin resistance and green fluorescent protein (GFP) markers was generated by homologous recombination in KSHV-infected BCBL-1 cells. Recombinant KSHV genomes from cell clones that were resistant to hygromycin, expressed GFP, and produced infectious virions after induction with tetradecanoyl phorbol acetate (TPA) were rescued in Escherichia coli and reconstituted in 293 cells. Several 293 cell lines resulting from infection with recombinant virions induced from a full-length recombinant KSHV genome, named BAC36, were obtained. BAC36 virions established stable latent infection in 293 cells, harboring 1 to 2 copies of viral genome per cell and expressing viral latent proteins, with ≈0.5% of cells undergoing spontaneous lytic replication, which is reminiscent of KSHV infection in Kaposi's sarcoma tumors. TPA treatment induced BAC36-infected 293 cell lines into productive lytic replication, expressing lytic proteins and producing virions that efficiently infected normal 293 cells with a ≈50% primary infection rate. BAC36 virions were also infectious to HeLa and E6E7-immortalized human endothelial cells. Since BAC36 can be efficiently shuttled between bacteria and mammalian cells, it is useful for KSHV genetic analysis. The feasibility of the system was illustrated through the generation of a KSHV mutant with the vIRF gene deleted. This cellular model is useful for the investigation of KSHV infection and pathogenesis.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, is a gammaherpesvirus and the proposed etiologic agent of Kaposi's sarcoma, primary effusion lymphoma, and a subset of multicentric Castleman's disease (27). While the life cycle of KSHV is similar to that of other gammaherpesviruses and consists of latent and lytic replication phases, the virology and cellular biology of KSHV infection in KSHV-related malignancies remains unclear. In Kaposi's sarcoma lesions, the majority of tumor cells are latently infected by KSHV, indicating the pathogenic role of viral latent infection (13, 17, 22, 39, 40). Nonetheless, a small number of tumor cells also undergo spontaneous viral lytic replication, which is thought to be essential for sustaining Kaposi's sarcoma lesions through a paracrine mechanism (3, 17, 30, 39, 41). The establishment of a cellular model for KSHV infection could facilitate the understanding of KSHV-related pathogenesis.
Although KSHV has been cultured by using cell lines established directly from primary effusion lymphoma (2, 6-8, 10, 16, 21, 24, 33, 43) or from peripheral blood mononuclear cells of primary effusion lymphoma patients (20), efficient KSHV primary infection remains elusive. KSHV infects a variety of human cell types, including B, T, endothelial, epithelial, fibroblast, and keratinocyte cells, and nonhuman cell types, including owl monkey kidney and baby hamster kidney fibroblast cells, in some cases resulting in the establishment of long-term cultures (9, 11, 18, 19, 23, 25, 28, 29, 31, 32, 44). Nevertheless, primary infection rates in these systems remain low (<10%). While KSHV infects umbilical vein endothelial cells by cell-to-cell contact with ≈30% primary infection efficiency (36), the transmission mechanism and sustainability of the cultures are unclear. The lack of a cellular model for efficient KSHV infection has impeded the understanding of KSHV-related pathogenesis.
Over 90 genes or open reading frames have been identified in KSHV genome. Many of them have been characterized in cell culture after cloning (27); however, their functions in KSHV infection remain unclear. Traditionally, the biologic functions of herpesviral genes could be examined by using viral mutants (12). Recombinant herpesviruses can be generated by homologous recombination in infected cells with DNA fragments carrying the alleles of mutations or assembling overlapping inserts of cosmid clones covering the entire viral genome (12), or more recently, by cloning the viral genome as a bacterial artificial chromosome (BAC) (26). The selection of recombinant viruses relies on plaque formation or immortalization of lymphoblastoid cell lines. Because of the lack of an efficient infection system, the generation of KSHV mutants has been difficult and has impeded the delineation of the functions of KSHV genes.
In this study, we have cloned KSHV genomes as BAC and selected recombinant KSHV that are highly infectious to mammalian cells. Primary infection of 293 cells with recombinant KSHV virions reached ≈50%, followed by the establishment of stable latent infection that mimicked KSHV infection in Kaposi's sarcoma tumor cells. Furthermore, recombinant KSHV genomes can be shuttled between bacteria and mammalian cells to facilitate KSHV genetic analysis.
MATERIALS AND METHODS
Construction of replacement plasmids.
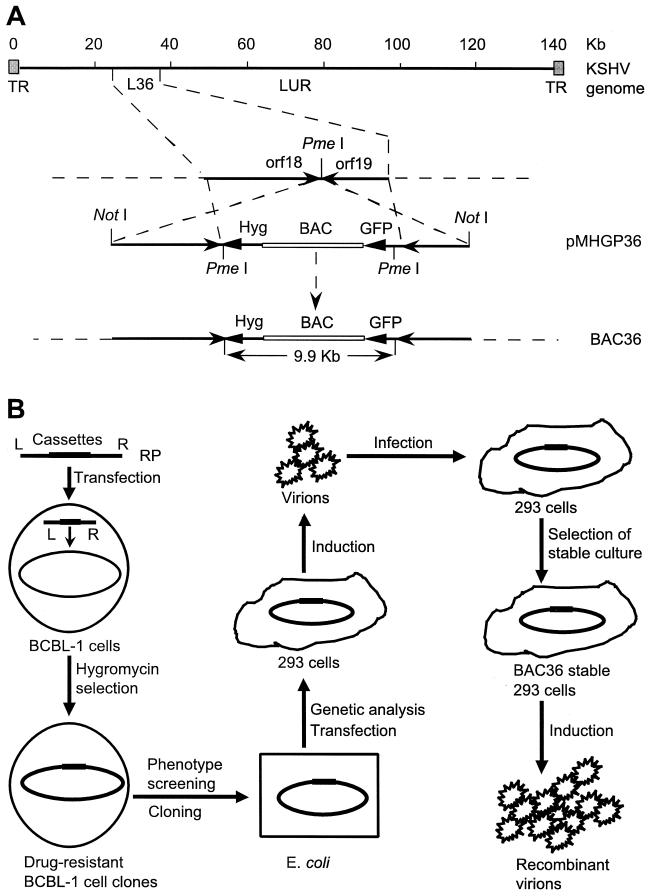
Plasmid MBO131B was generated by inserting a BamHI linker into the unique SalI site of pMBO131. A FspI/HindIII fragment from pTK-Hyg (Clontech, Palo Alto, Calif.) containing a hygromycin resistance cassette (Hyg) was inserted into the SmaI and HindIII sites of pBluescript II SK(+) to produce pSKTK-Hyg. pEGFP-C1 (Clontech) was cut with BglII and BamHI to remove the multiple cloning sites and religated to yield pEGFP-M. An AseI/MluI fragment from pGEFP-M was then Klenow blunted and cloned into the Klenow-blunted HindIII site of pSTK-Hyg to generate plasmid pHG, in which a HindIII site was recovered between Hyg and the green fluorescent protein (GFP) cassette by ligating the blunted HindIII and MluI ends. BglII and PmeI linkers were inserted into the PstI and HindIII sites of pHG, respectively, to produce plasmid pHGP. A BamHI/BglII fragment from pHGP carrying Hyg and the GFP cassette was inserted into the unique BamHI site of pBMO131B to yield plasmid pMHGP. A 12-kb NotI-excised insert from the L36 lambda phage clone (34), used as flanking sequences for homologous recombination, was self-ligated, digested with the unique cutter PmeI, and cloned into the PmeI site of pMHGP to produce replacement plasmid pMHGP36 (Fig. 1A).
FIG. 1.
Molecular cloning of infectious KSHV and establishment of a genetic manipulation system. (A) Schematic illustration of molecular cloning of KSHV genome. Replacement plasmid (RP) pMHGP36, containing the BAC vector, Hyg, GFP cassette, and the L36 locus as the flanking sequences, was integrated into the PmeI site between orf18 and orf19 in the long unique region (LUR) of the viral genome by homologous recombination in KSHV-infected BCBL-1 cells. (B) Strategy for the isolation of infectious KSHV. pMHGP36 was transfected into KSHV-infected BCBL-1 cells to generate recombinant virus. Cell clones were selected for hygromycin resistance, GFP expression, and production of infectious virions after TPA induction. Episomal DNA from selected cell clones was then prepared and transformed into E. coli to obtain recombinant KSHV BAC36. The rescued episomes were transfected into 293 cells and induced to produce infectious virions, which were used to infect normal 293 cells. BAC36-infected 293 cell lines were then established with hygromycin selection. The selected cells were induced to produce infectious recombinant virions. TR, terminal repeat.
Cell culture and induction of KSHV lytic replication.
BCBL-1 cells infected with KSHV and BC-1 cells dually infected with KSHV and Epstein-Barr virus were cultured with RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, Mo.), 50 μg of gentamicin per ml, and 2 mM glutamine. HeLa and 293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 50 μg of gentamicin, and 2 mM glutamine. E6E7-immortalized human dermal microvascular endothelial cells obtained from A. V. Moses were cultured in endothelial-SFM medium (Gibco-BRL, Gaithersburg, Md.) supplemented with 10% human serum (Sigma), 100 U of penicillin, 100 μg of streptomycin, 25 μg of endothelial cell growth supplement (Becton-Dickinson, Bedford, Mass.), 40 μg of heparin (Sigma), and 200 μg of G418 (Sigma) per ml (29). To induce KSHV lytic replication, the cultured cells were treated with 20 ng of 12-O-tetradecanoyl phorbol-13-acetate (TPA; Sigma) per ml for 2 to 4 days (21).
Cell transfection.
Plasmid DNA was prepared with standard methods by using the Qiagen kit (Qiagen Inc., Valencia, Calif.). For B-cell transfection, approximately 4 × 106 cells were pelleted by centrifugation and suspended in 400 μl of RPMI 1640 medium supplemented with 10% FBS. The cells and 20 μg of plasmid DNA were placed in 0.4-cm electroporation cuvettes, and the transfection was carried out at 250 V and 960 μF with a Gene Pulser II electroporation system (Bio-Rad Laboratories, Richmond, Calif.). Transfection of 293 cells was carried out with Lipofectamine 2000 according to the manufacturer's instructions (Gibco).
Infection of cells with recombinant KSHV virions.
Cells plated onto 3.5-cm dishes were infected with 0.45-μm-filtered cell culture supernatant containing BAC36 virions as described previously (29). GFP expression was used to monitor infection. In some experiments, infection was carried out with inoculum treated with 0.5 mg of DNase per ml for 1 h at 37°C.
Establishment of stable recombinant KSHV-infected cell lines.
pMHGP36-transfected BCBL-1 cells were seeded in 96-well plates at 5,000 cells/well and selected with medium containing 150 μg of hygromycin per ml. Stable cultures were also established from 293 and HeLa cultures infected with BAC36 virions.
Transformation of recombinant KSHV into bacteria.
Escherichia coli strain DH10B was transformed with 1 μg of episomal DNA isolated from BCBL-1 or 293 cells harboring recombinant KSHV genomes by using an alkaline lysis procedure (42). Electroporation was carried out at 1.8 kV, 100 Ω, and 25 μF in 0.1-cm cuvettes. The bacteria were grown on Luria-Bertani (LB) plates containing 15 μg of chloramphenicol per ml.
Analysis of KSHV genome.
Field inversion gel electrophoresis was performed by using a Hoefer PC 500 switch back pulse controller according to the manufacturer's instructions (Amersham Pharmacia Biotech, Piscataway, N.J.). The gel was run for 10 min at 175 V with a pulse time of 1 to 20 s, followed by another 36 h at 175 V with a pulse time of 0.8 to 1.5 s. For Southern blot hybridization, restriction enzyme-digested DNA was separated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled probe (21). Specific signals were detected with a Molecular Imager (Bio-Rad).
To determine the number of viral genomes per cell, 10 μg of DNA from approximately 5 × 105 BC-1 cells containing 2 × 107 to 4 × 107 KSHV genomes was used as a copy number control (21). Purified BAC36 DNA (15 ng) was also used as a control. β-Actin probe was used to calibrate the loading of different cell lines.
Analysis of KSHV gene expression.
To analyze KSHV gene expression by Northern blot hybridization, total RNA was isolated from uninduced and TPA-induced BCBL-1 cells, BAC36-infected 293 cells, and normal 293 cells, fractionated on a 1% formaldehyde agarose gel, transferred to nylon membranes, and hybridized with 32P-labeled riboprobes (45). Protein expression was detected by immunofluorescence assay (IFA) (20). Latent nuclear antigen (LNA) encoded by orf73 was detected with a rat anti-LNA monoclonal antibody (ABI, Baltimore, Md.) and revealed with a rabbit anti-rat immunoglobulin-tetramethylrhodamine isothiocyanate (TRITC) conjugate (Sigma). Minor capsid protein (mCP) encoded by orf65 was detected with mouse polyclonal antibodies prepared with purified mCP recombinant protein and revealed with a rabbit anti-mouse immunoglobulin-TRITC conjugate (Dako, Rothskilde, Denmark).
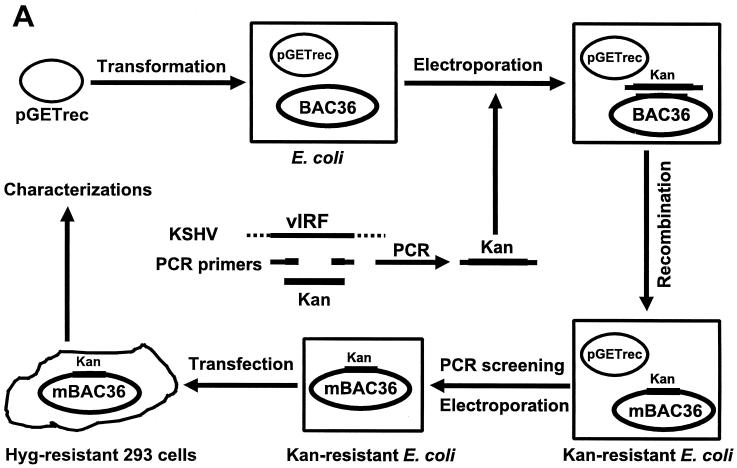
Generation of KSHV mutant with deletion of vIRF gene (orf-K9).
A PCR product of a kanamycin resistance cassette (Kan) flanking sequences from two ends of the viral interferon regulatory factor (vIRF) gene was electroporated into E. coli that had been transformed with BAC36 and plasmid pGETrec. Episomal DNA was isolated from bacterial colonies containing KSHV mutant in which the vIRF gene was replaced with Kan after a second round of transformation into E. coli to eliminate the pGETrec plasmid and analyzed further (see Fig. 7A).
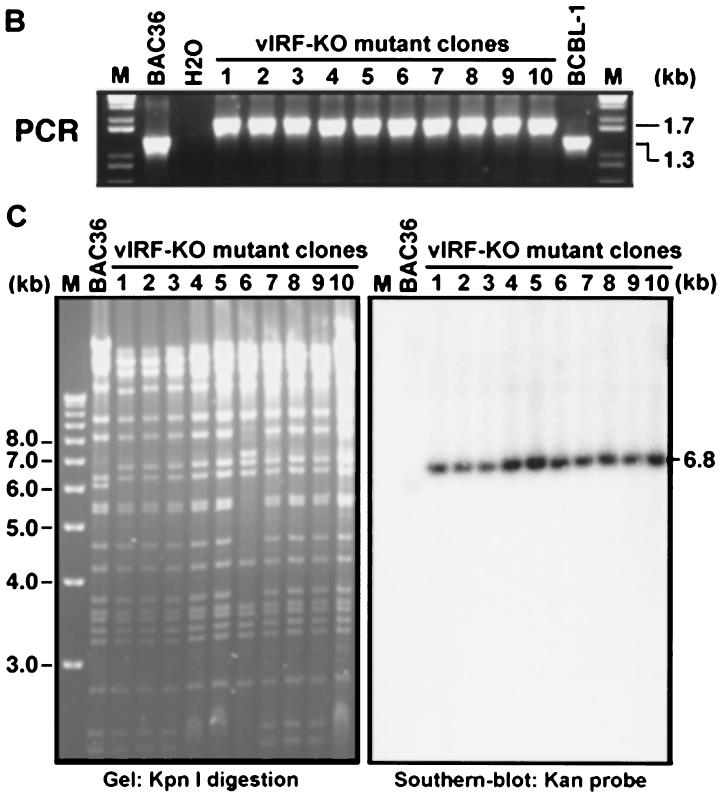
FIG. 7.
Generation of a KSHV mutant with a deletion of the vIRF gene. (A) A Kan cassette PCR product flanking sequences from outside the vIRF gene was electroporated into E. coli previously transformed with BAC36 and plasmid pGETrec. Episomal DNA isolated from bacterial colonies containing the KSHV mutant in which the vIRF gene was replaced with Kan after a second round of transformation into E. coli to eliminate the pGETrec plasmid was analyzed by PCR assay, restriction enzyme digestion, and Southern blot hybridization. The mutant virus was then reconstituted in 293 cells and used for phenotype analysis. (B) PCR assay to confirm the deletion of the vIRF gene in BAC36. BAC36 produced a band of 1.3 kb, while the mutant virus produced a band of 1.7 kb because of the size differences between the vIRF gene and Kan. (C) KpnI digestion of BAC36 and mutant virus and Southern blot hybridization with Kan as the probe.
RESULTS
Cloning of infectious KSHV as BAC in E. coli and reconstitution in 293 cells.
BAC vector sequences were inserted into the KSHV genome through transfection of a replacement plasmid into BCBL-1 cells followed by homologous recombination between the replacement plasmid and the viral genome (Fig. 1A). Since screening of recombinant KSHV could not be achieved through plaque selection or formation of lymphoblastoid cell lines, Hyg and the GFP cassette were inserted along with the BAC vector into the KSHV genome. Viral genome position 33195 (based on BC-1-derived KSHV sequences; GenBank accession no. U75698) was chosen as the insertion point because there is a unique PmeI site at the ends of two open reading frames in opposite orientations, orf18 and orf19. A lambda phage clone, L36, overlapping this region was used as the flanking sequences, and the resulting full-length recombinant KSHV was named BAC36.
A screening procedure was used to ensure the selection of cell clones harboring infectious KSHV (Fig. 1B). Cell clones that were resistant to hygromycin, expressed GFP, and could be induced to produce infectious virions after TPA treatment were selected. Out of 159 hygromycin-resistant BCBL-1 clones, 3 (cl2, cl3, and cl4) expressed GFP and produced infectious virions after TPA induction. Episomal DNA was isolated from these three cell clones and transformed into E. coli. A large number of chloramphenicol-resistant colonies were obtained, which were likely to harbor BAC36. Recombinant KSHV genomes from one of the hygromycin-resistant BCBL-1 cell clones (cl1) that expressed GFP but failed to produce infectious virions after TPA induction was also rescued in bacteria for analysis.
To reconstitute BAC36 in mammalian cells, it was transfected into 293 cells. Two days after transfection, approximately 5 to 10% of the cells expressed GFP. The cells were treated with TPA, and the supernatant was used to inoculate normal 293 cells. Approximately 1 to 5% of the cells expressed GFP 2 days after inoculation with supernatants from cells transfected with BAC36 from cl2, cl3, and cl4, an indication that these cultures were infected by BAC36 virions. As expected, no GFP expression was observed in cultures inoculated with supernatant from cells transfected with cl1 episomal DNA. After 4 to 6 weeks of hygromycin selection, all the cells became GFP positive. To ensure homogeneity, two of these cultures, each from an independent BCBL-1 clone (cl2 and cl3), were cloned further, and GFP-expressing clones were randomly picked and expanded. A total of 11 stable 293 clonal cell lines harboring BAC36 were obtained. These BAC36-infected stable clones were used for subsequent genetic and phenotypic characterizations.
BAC36 contains the full-length KSHV genome with the replacement plasmid integrated at the expected site.
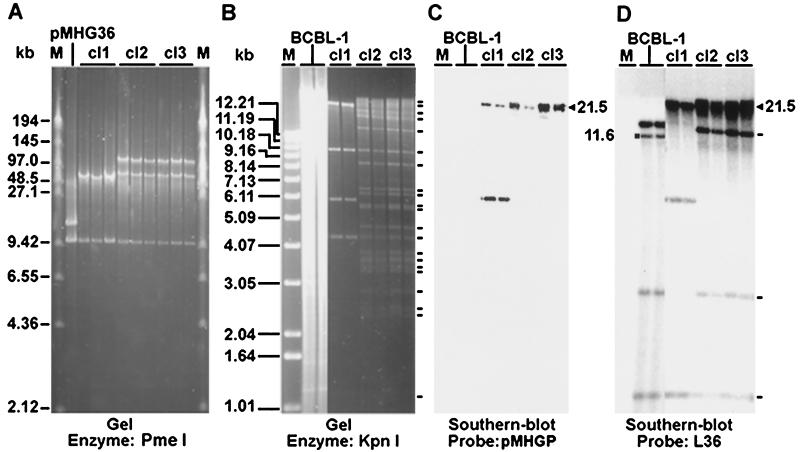
To characterize BAC36-infected 293 cell lines, the episomes were recovered after transformation into E. coli. If the recombination between pMHGP36 and KSHV genome occurred correctly, a 9.9-kb band representing the replacement plasmid and a 170-kb band representing the full-length viral genome should be observed after PmeI digestion. As expected, a band of 9.9 kb was observed in bacterial colonies from all cell lines; however, two other bands of 110 and 60 kb instead of one band of 170 kb were observed in the cl2 and cl3 genomes (Fig. 2A ), indicating the presence of another PmeI site located approximately at the genomic position of 93 kb or 143 kb in the BCBL-1 KSHV genome, which is absent in the BC-1 KSHV genome. Episomal DNA from cl1 had only one band of 50 kb besides the 9.9-kb band, suggesting that a portion of the viral genome was deleted, explaining why it was noninfectious.
FIG. 2.
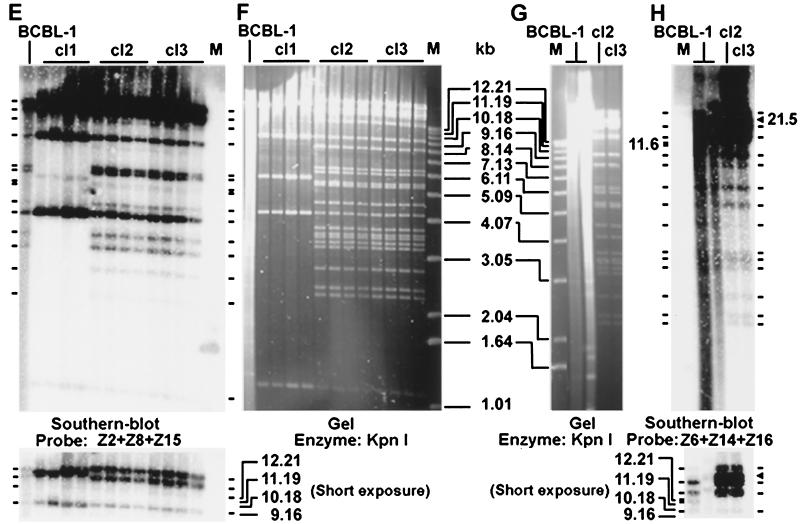
Genetic analysis of BAC36. (A) Field inversion gel electrophoresis of PmeI-digested BAC DNA. BAC DNA was isolated from chloramphenicol-resistant E. coli colonies transformed with cl1, cl2, and cl3 episomal DNA recovered from 293 cell lines. pMHGP36 was included as a control. (B, F, and G) KpnI-digested BAC DNA from bacterial colonies recovered from 293 cell lines was resolved on a 0.8% agarose gel. Episomal DNA prepared from BCBL-1 cells was used as the control. (C, D, E, and H) Southern hybridization with the following probes: pMHGP (C); L36 (D); and inserts from six cosmid clones covering the entire BC-1 KSHV genome (E and H). Restriction bands identified on gels or by Southern blot hybridization are indicated with short lines. The band marked with a square was only present in BCBL-1 cells, which was shifted to the band marked with a triangle in BAC DNA because of the insertion of the replacement plasmid. Panels C and D show the same membrane from panel B; panel E shows the membrane from panel F; panel H shows the membrane from panel G. Lanes M, size markers.
BAC36 DNA was further digested with KpnI, a multiple cutter of the KSHV genome (Fig. 2B, F, and G). Episomal DNA isolated from BCBL-1 cells was analyzed in parallel. BAC36 contained most of the bands calculated based on the available BC-1 genomic sequences; however, discrepancies were observed in several bands, which were presumably due to the insertion of the replacement plasmid and sequence variations between BC-1 and BCBL-1 KSHV genomes. Southern blot hybridization was further performed by using pMHGP, L36, and a set of cosmid clones covering the entire BC-1 KSHV genome (34) as probes. The pMHGP probe detected one band of 21.5 kb in cl2 and cl3, which was absent in BCBL-1 cells (Fig. 2B and C), indicating the integration of pMHGP in the genomes. The L36 probe also detected the 21.5 kb band in cl2 and cl3 but not in BCBL-1 cells (Fig. 2B and D); however, a band of 11.6 kb was detected in BCBL-1 cells but not in cl2 and cl3, confirming the integration of pMHGP in this band. Three other bands of 1.1, 2.7, and 14 kb were also detected in BCBL-1 cells and cl2 and cl3 (Fig. 2B and D).
All the bands detected in BCBL-1 cells by cosmid clones Z2, Z8, and Z15 were present in cl2 and cl3 (Fig. 2E and F). Similarly, all bands detected in BCBL-1 cells by cosmid clones Z6, Z14, and Z16 were also present in cl2 and cl3 except the 11.6-kb band, which was only present in BCBL-1 cells but was shifted to 21.5 kb in cl2 and cl3 (Fig. 2G and H).
To confirm the presence of the terminal repeat sequence, BAC36 DNA was digested with NotI. A band of 800 to 810 bp was observed and had staining intensity far greater than that of any other bands (data not shown), indicating its overrepresentation in the genome and hence its identity as a repeat sequence. The above results demonstrated that BAC36 contained the full-length KSHV genome with the replacement plasmid inserted into the expected genomic position.
BAC36-infected 293 cell lines maintained stable KSHV genomes and expressed viral latent transcripts and proteins.
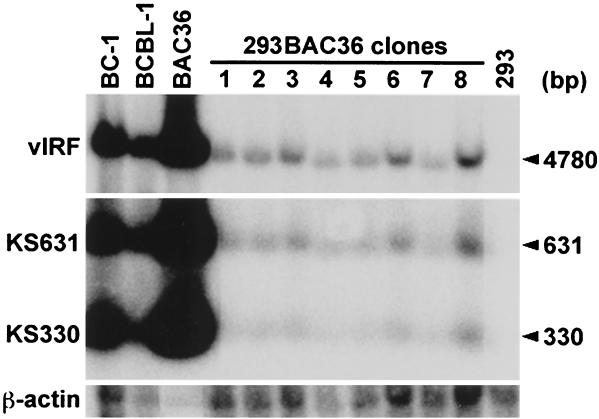
To determine KSHV genomic copy number in BAC36-infected 293 cells, Southern blot hybridization was performed on BamHI-digested DNA with KS330 (orf26), KS631 (orf75), and vIRF as probes. DNA from BC-1 cells, which were previously determined to harbor 40 to 80 KSHV genomes per cell (21), was used as a copy number control. BAC36-infected 293 cell lines were determined to harbor 1 to 2 viral genomes per cell (Fig. 3).
FIG. 3.
Determination of KSHV genomic copy number in BAC36-infected cells by Southern blot hybridization. BamHI-digested DNAs from 293 cells, BAC36-infected 293 cells, and KSHV-infected BC-1 and BCBL-1 cells were resolved in an agarose gel, transferred to a nylon membrane, and hybridized with KS330, KS631, and vIRF probes. Then 10 μg of DNA from approximately 5 × 105 BC-1 cells containing 2 × 107 to 4 × 107 KSHV genomes was used as the copy number control. Purified BAC36 DNA (15 ng) was also used as a control. β-Actin was used to calibrate any loading differences between cell lines. BAC36-infected 293 cell lines were determined to harbor 1 to 2 copies of BAC36 per cell.
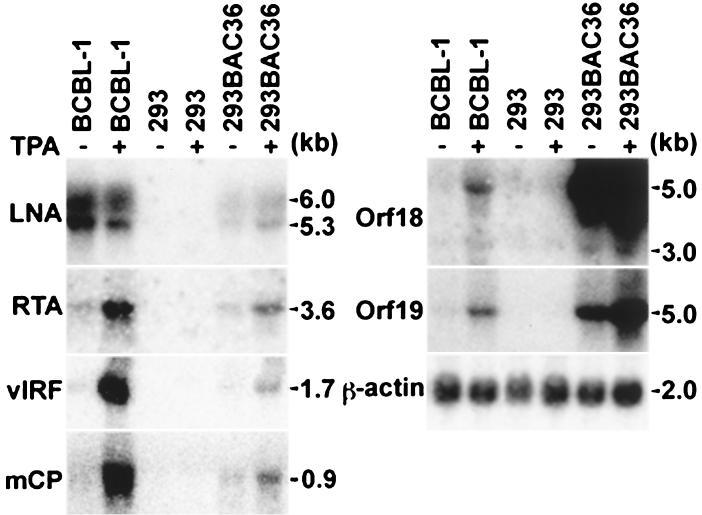
To examine KSHV gene expression, Northern blot hybridization was performed with different KSHV gene probes (Fig. 4). The LNA probe detected two transcripts of 5.3 and 6.0 kb in BAC36-infected 293 cells and BCBL-1 cells, though the latter had a stronger expression level, possibly reflecting its higher viral genomic copy number per cell. Similar to BCBL-1 cells, BAC36-infected 293 cells also weakly expressed RTA (orf50), vIRF, and mCP transcripts. Weak expression of two transcripts of 3 and 5 kb detected by the orf18 probe and one transcript of 5 kb detected by the orf19 probe was observed in BCBL-1 cells. The same orf18 and orf19 transcripts were also detected in BAC36-infected 293 cells, but at much stronger levels, indicating that insertion of the replacement plasmid has not disrupted but enhanced the expression of these transcripts, possibly because of the enhancer sequences in the replacement plasmid (Fig. 4).
FIG. 4.
Detection of KSHV gene expression in BAC36-infected cells by Northern blot hybridization. Total RNA was extracted from uninduced and TPA-induced BAC36-infected 293 cells, 293 cells, and BCBL-1 cells, resolved on agarose gels, transferred to nylon membranes, and hybridized with riboprobes of different KSHV genes. The β-actin probe was used to monitor RNA loading.
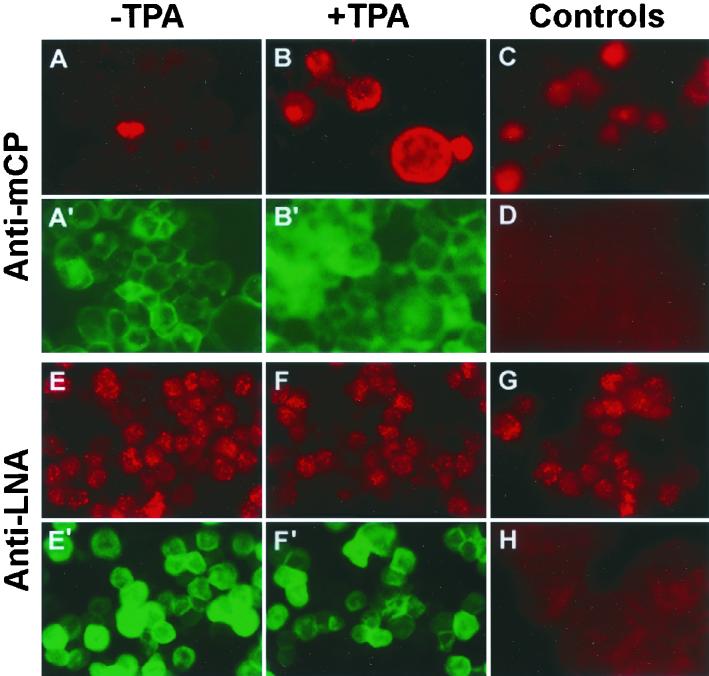
IFA showed that all BAC36-infected cells strongly expressed LNA (Fig. 5E), while less than 0.5% of cells also expressed mCP (Fig. 5A), reflecting the weak expression of RTA, vIRF, and mCP transcripts. These results suggest that the majority of the BAC36-infected cells were in the latent phase of replication, with a small number of cells undergoing spontaneous lytic replication. This situation is reminiscent of KSHV infection in Kaposi's sarcoma tumors.
FIG. 5.
Detection of KSHV gene expression by IFA. Uninduced (A, E, G, and H) and TPA-induced (B, C, D, and F) BAC36-infected 293 cells (A, B, E, and F), 293 cells (D and H), and BCBL-1 cells (C and G) were stained for LNA (E to H) or mCP (A to D) protein expression. Cells shown in panels A′, B′, E′, and F′, which were from the same fields as those shown in panels A, B, E, and F, respectively, expressed GFP, indicating the presence of BAC36.
To determine the stability of BAC36 latent infection in 293 cells, cultures were maintained for 3 months, and KSHV genomic copy number in these cells was again examined by Southern blot hybridization. All the cell lines maintained 1 to 2 KSHV genomes per cell after this period of culture. Examination of episomal DNA by restriction enzyme digestion and Southern blot hybridization after recovery in E. coli did not reveal any genomic alterations (data not shown). Northern blot hybridization and IFA did not detect any differences in viral gene expression in these cells from cells of early passages (data not shown). These results indicated that BAC36 had maintained stable latent infection in 293 cells.
BAC36-infected 293 cell lines can be induced into lytic replication, producing highly infectious virions.
BAC36-infected 293 cells were treated with TPA to induce viral lytic replication. The expression of KSHV lytic transcripts detected by the RTA, vIRF, and mCP probes was increased 18-, 25-, and 22-fold, respectively, while the expression of latent transcripts detected by the LNA probe was increased 1.8-fold (Fig. 4). In parallel, the number of mCP-positive cells detected by IFA was increased from 0.5 to 12% (Fig. 5A and B). No differences in LNA staining by IFA were observed before and after TPA induction (Fig. 5E and F). These results indicated that BAC36-infected cells could be induced into lytic replication.
To further determine whether viral lytic replication in BAC36-infected 293 cells was productive, supernatants from TPA-treated cultures were used to inoculate normal 293 cells. Approximately ≈50% of 293 cells expressed GFP 2 days after inoculation (Fig. 6). Treatment of the inoculum with DNase did not decrease the percentage of GFP-positive cells. When 293 cells were incubated with DNA from a mammalian GFP expression vector, pEGFP-C1 (10 μg/ml), no GFP expression was observed. These results indicated de novo infection of 293 cells by BAC36 virions. Examination of BAC36 genomes and gene expression also confirmed viral infection of these cells (data not shown).
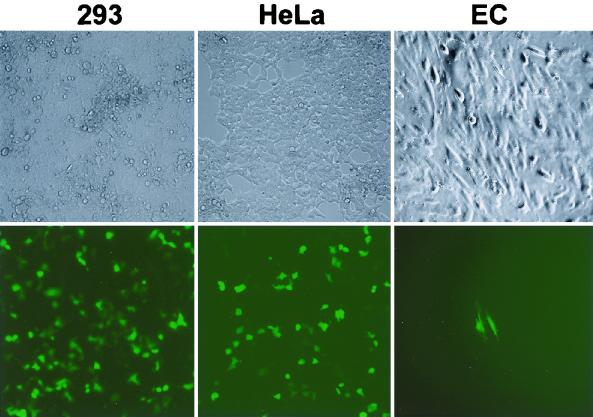
FIG. 6.
Infection of 293 cells, HeLa cells, and E6E7-immortalized human dermal microvascular endothelial cells (EC) with culture supernatant from TPA-induced, BAC36-infected 293 cells. Cells were observed 2 days postinfection with a fluorescence microscope.
BAC36 established persistent latent infection following primary infection.
To determine whether BAC36 virions could establish latent infection in 293 cells, cells infected with BAC36 virions were selected with hygromycin. All cells became GFP positive within 4 to 6 weeks. Further analysis of viral genome and gene expression did not detect any differences in these newly established cell lines from those examined above, indicating that BAC36 virions have established latent infection in these cultures. The BAC36 virus has subsequently been successfully transmitted to normal 293 cells for an additional five passages.
Infection of other cell types by BAC36 virions was also observed. Primary infection rates of 20 to 30% were achieved with HeLa cells (Fig. 6), indicating that HeLa cells can support KSHV infection. Several stable BAC36-infected HeLa cell cultures were established. Infection of E6E7-immortalized human dermal microvascular endothelial cells was observed; however, primary infection rates never exceeded 1% (Fig. 6). The number of infected cells did not increase in subsequent culture. No long-term culture of BAC36-infected endothelial cells was obtained. Infection of several B-cell lines, including Bjab and BCBL-1 cells, was carried out; however, no evidence of infection, based on GFP expression, was observed.
Generation of KSHV mutant with a deletion of the vIRF gene.
The above results demonstrated that BAC36 could be shuttled between bacterial and mammalian cells. Thus, it can be used for KSHV genetic analysis. To demonstrate the feasibility of the system, a KSHV mutant with a deletion of the vIRF gene was generated (Fig. 7 ). PCR amplification with primers immediately outside the deleted sequence region detected a band of 1.3 kb in BAC36 DNA and a band of 1.7 kb in DNA from mutant colonies, confirming the replacement of the vIRF gene by Kan in BAC36 (Fig. 7B). KpnI digestion of the episomal DNA from KSHV mutant clones also showed that they all had the restriction bands observed in BAC36 except for one band of 6.2 kb, which was shifted to 6.8 kb in the mutant virus (Fig. 7C). Southern blot hybridization with Kan as the probe showed that the 6.8-kb band was indeed the Kan cassette. DNA sequencing of this region also confirmed the replacement of the vIRF gene by Kan in the KSHV mutant.
DISCUSSION
We have isolated a recombinant KSHV BAC36 whose virions are capable of infecting 293 cells with ≈50% primary infection efficiency. Close to 100% primary infection rate was also observed when the inoculum was concentrated fivefold by centrifugation (data not shown). Furthermore, BAC36 can be serially transmitted to normal 293 cells for at least six passages. The recombinant KSHV established stable latent infection in 293 cells following primary infection. Several lines of evidence indicated that persistent infection of 293 cells by BAC36 mimicked KSHV infection of tumor cells in Kaposi's sarcoma lesions: (i) latent BAC36-infected 293 cells maintained low copy numbers (1 to 2) of viral genomes per cell (5) and (ii) the majority of BAC36-infected cells were in the latent phase of replication, with a small number of cells undergoing spontaneous lytic replication (17, 30, 39-41). This infection system will be useful for understanding the virology and cellular biology of KSHV infection.
A total of 159 hygromycin-resistant BCBL-1 cell clones harboring recombinant viruses or at least Hyg were obtained from the initial screening. Only three of them expressed GFP and produced infectious recombinant virions after TPA induction. One explanation for the low efficiency of infectious recombinant virion production is the presence of lytic replication-defective viruses in the BCBL-1 culture. In fact, we have recently isolated a lytic replication-defective virus from a BCBL-1 culture (unpublished data). It is also possible that the recombination of the replacement plasmid with KSHV genomes was inefficient and/or occurred improperly. Further analysis of these nonproductive cell clones could be enlightening in this regard.
Viral mutagenesis is a powerful tool for examining the functions of viral genes and has been widely used for other herpesviruses (12). More recently, the genomes of several herpesviruses have been cloned as BAC to facilitate their genetic analysis (1, 4, 14, 26, 35, 37, 38). A recent report describes the construction of a recombinant BAC KSHV by inserting a replacement plasmid into orf56 (15), a gene that is essential for viral replication. The recombinant KSHV underwent spontaneous but abortive lytic replication without establishing latent infection. Primary infection of recombinant virions generated through complementary orf56 expression only reached a ≈0.1% rate in 293 cells. Thus, the application of this system is limited.
In contrast, we have introduced the replacement plasmid into the locus between orf18 and orf19 and successfully cloned a highly infectious virus KSHV. Several important steps have been achieved in this study: (i) a full-length KSHV genome was cloned as a BAC with Hyg and the GFP cassette in designated positions; (ii) recombinant KSHV was successfully reconstituted in 293 cells; (iii) latent BAC36 genomes were induced into lytic replication, producing infectious virions; (iv) BAC36 virions efficiently infected 293 cells and established persistent latent infection; and (v) BAC36 genomes were easily recovered in bacteria. Together, these results have shown that BAC36 can be used as a bacterial-mammalian shuttle for KSHV genetic analysis. The feasibility of the system has been demonstrated through the generation of a KSHV mutant with the vIRF gene deleted. The introduction of Hyg and the GFP cassette into the viral genome facilitates the selection of stable cell lines infected by the recombinant virus. This feature is particularly valuable for cells that have low primary infection efficiency, such as human primary endothelial cells.
KSHV has been shown to infect and transform human primary endothelial cells (18). BAC36 virions also infected human primary endothelial cells, albeit at a very low rate. It would be important to determine whether BAC36 can transform human primary endothelial cells. Several KSHV genes have been shown to have transforming function in rat fibroblast cells (27). Genetic dissections could lead to the identification of a candidate viral gene(s) that is critical for KSHV transformation of human primary endothelial cells and provide insight into KSHV-related pathogenesis.
ADDENDUM
After revision of the manuscript, we analyzed the BAC36 genome with a KSHV microarray recently constructed in our laboratory. The results showed that the BAC36 genome contained all the KSHV-encoded genes (orfs), thus further confirming the conclusion that BAC36 contains the full-length KSHV genome.
We also succeeded in infecting human primary dermal microvascular endothelial cells with BAC36 virions at a primary infection rate of ≈50%. Two stable BAC36-infected human primary dermal microvascular endothelial cell lines have been established.
Acknowledgments
We thank Michael O'Connor at the University of Minnesota for providing plasmid pMBO131; Panayiotis A. Ioannou at Murdoch Children's Research Institute, Australia, for providing plasmid pGETrec; and Ashlee V. Moses at Oregon Health Sciences University for providing human papillomavirus E6E7-immortalized human dermal microvascular endothelial cells.
This work was supported by the Elsa U. Pardee Foundation, the Association for International Cancer Research, and NIH grant HL60604-01 to S.-J. Gao.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 3.Blasig, C., C. Zietz, B. Haar, F. Neipel, S. Esser, N. H. Brockmeyer, E. Tschachler, S. Colombini, B. Ensoli, and M. Sturzl. 1997. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J. Virol. 71:7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. D. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpes virus (KSHV) infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 6.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carbone, A., A. M. Cilia, A. Gloghini, D. Capello, T. Perin, D. Bontempo, V. Canzonieri, U. Tirelli, R. Volpe, and G. Gaidano. 2000. Primary effusion lymphoma cell lines harboring human herpesvirus type-8. Leuk. Lymphoma 36:447-456. [DOI] [PubMed] [Google Scholar]
- 8.Carbone, A., A. M. Cilia, A. Gloghini, D. Capello, M. Todesco, S. Quattrone, R. Volpe, and G. Gaidano. 1998. Establishment and characterization of Epstein-Barr virus-positive and Epstein-Barr virus-negative primary effusion lymphoma cell lines harboring human herpesvirus type-8. Br. J. Haematol. 102:1081-1089. [DOI] [PubMed] [Google Scholar]
- 9.Cerimele, F., F. Curreli, S. Ely, A. E. Friedman-Kien, E. Cesarman, and O. Flore. 2001. Kaposi's sarcoma-associated herpesvirus can productively infect primary human keratinocytes and alter their growth properties. J. Virol. 75:2435-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two AIDS-related lymphoma cell lines containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 11.Ciufo, D. M., J. S. Cannon, L. J. Poole, F. Y. Wu, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coen, D. M., and R. F. Ramig. 1996. Viral genetics, p. 101-139. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology. Lippincott-Raven, Philadelphia, Pa.
- 13.Davis, M. A., M. Sturzl, C. Blasig, A. Schreier, H.-G. Guo, M. Reitz, S. R. Opalenik, and P. J. Browning. 1997. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 89:1868-1874. [DOI] [PubMed] [Google Scholar]
- 14.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delecluse, H. J., M. Kost, R. Feederle, L. Wilson, and W. Hammerschmidt. 2001. Spontaneous activation of the lytic cycle in cells infected with a recombinant Kaposi's sarcoma-associated virus. J. Virol. 75:2921-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drexler, H. G., C. C. Uphoff, G. Gaidano, and A. Carbone. 1998. Lymphoma cell lines: in vitro models for the study of HHV-8+ primary effusion lymphomas (body cavity-based lymphomas). Leukemia 12:1507-1517. [DOI] [PubMed] [Google Scholar]
- 17.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flore, O., S. Rafii, S. Ely, J. J. O'Leary, E. M. Hyjek, and E. Cesarman. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature 394:588-592. [DOI] [PubMed] [Google Scholar]
- 19.Foreman, K. E., J. Friborg, W. Kong, C. Woffendin, P. J. Polverini, B. J. Nickoloff, and G. J. Nabel. 1997. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N. Engl. J. Med. 336:163-171. [DOI] [PubMed] [Google Scholar]
- 20.Gao, S.-J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 21.Gao, S.-J., Y.-J. Zhang, J.-H. Deng, C. S. Rabkin, O. Flore, and H. B. Jenson. 1999. Molecular polymorphism of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J. Infect. Dis. 180:1466-1476. [DOI] [PubMed] [Google Scholar]
- 22.Katano, H., Y. Sato, T. Kurata, S. Mori, and T. Sata. 1999. High expression of HHV-8-encoded ORF73 protein in spindle-shaped cells of Kaposi's sarcoma. Am. J. Pathol. 155:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kliche, S., E. Kremmer, W. Hammerschmidt, U. Koszinowski, and J. Haas. 1998. Persistent infection of Epstein-Barr virus-positive B lymphocytes by human herpesvirus 8. J. Virol. 72:8143-8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacoste, V., J. G. Judde, G. Bestett, J. Cadranel, M. Antoine, F. Valensi, E. Delabesse, E. Macintyre, and A. Gessain. 2000. Virological and molecular characterisation of a new B lymphoid cell line, established from an AIDS patient with primary effusion lymphoma, harboring both KSHV/HHV8 and Epstein-Barr virus viruses. Leuk. Lymphoma 38:401-409. [DOI] [PubMed] [Google Scholar]
- 25.Mesri, E. A., E. Cesarman, L. Arvanitakis, S. Rafii, M. A. S. Moore, D. N. Posnett, D. M. Knowles, and A. S. Asch. 1996. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J. Exp. Med. 183:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, P. S., and Y. Chang. 2001. Molecular virology of Kaposi's sarcoma-associated herpesvirus. Phil. Trans. R. Soc. Lond. B 356:499-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, P. S., S.-J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. M. Knowles, R. Garber, D. J. McGeoch, P. Pellett, and Y. Chang. 1996. Primary characterization of a herpesvirus-like agent associated with Kaposi's sarcoma. J. Virol. 70:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moses, A. V., K. N. Fish, R. Ruhl, P. P. Smith, J. G. Strussenberg, L. Zhu, B. Chandran, and J. A. Nelson. 1999. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J. Virol. 73:6892-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onstein, J. M., S. Alkan, A. Blauvelt, K. T. Jeang, M. D. Weinstein, D. Ganem, and B. Herndier. 1997. Visualization of human herpesvirus type 8 in Kaposi's sarcoma by light and transmission electron microscopy. AIDS 11:F35-F45. [DOI] [PubMed] [Google Scholar]
- 31.Panyutich, E. A., J. W. Said, and S. A. Miles. 1998. Infection of primary dermal microvascular endothelial cells by Kaposi's sarcoma-associated herpesvirus. AIDS 12:467-472. [DOI] [PubMed] [Google Scholar]
- 32.Renne, R., D. Blackbourn, D. Whitby, J. Levy, and D. Ganem. 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 34.Russo, J. J., R. A. Bohenzky, M. Chien, K. Chen, D. Maddalena, M. Yan, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of Kaposi's sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saeki, Y., T. Ichikawa, A. Saeki, E. A. Chiocca, K. Tobler, M. Ackermann, X. O. Breakefield, and C. Fraefel. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum. Gene Ther. 9:2787-2794. [DOI] [PubMed] [Google Scholar]
- 36.Sakurada, S., H. Katano, T. Sata, H. Ohkuni, T. Watanabe, and S. Mori. 2001. Effective human herpesvirus 8 infection of human umbilical vein endothelial cells by cell-mediated transmission. J. Virol. 75:7717-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher, D., B. K. Tischer, W. Fuchs, and N. Osterrieder. 2000. Reconstitution of Marek's disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 74:11088-11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staskus, K. A., W. D. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stürzl, M., C. Blasig, A. Schreier, F. Neipel, C. Hohenadl, E. Cornali, G. Ascherl, S. Esser, N. H. Brockmeyer, M. Ekman, E. E. Kaaya, E. Tschachler, and P. Biberfeld. 1997. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi's sarcoma. Int. J. Cancer 72:68-71. [DOI] [PubMed] [Google Scholar]
- 41.Stürzl, M., A. Wunderlich, G. Ascherl, C. Hohenadl, P. Monini, C. Zietz, P. J. Browning, F. Neipel, P. Biberfeld, and B. Ensoli. 1999. Human herpesvirus-8 (HHV-8) gene expression in Kaposi's sarcoma (KS) primary lesions: an in situ hybridization study. Leukemia 13:S110-S112. [DOI] [PubMed] [Google Scholar]
- 42.Sun, T. Q., D. A. Fernstermacher, and J. M. Vos. 1994. Human artificial episomal chromosomes for cloning large DNA fragments in human cells. Nat. Genet. 8:33-41. [DOI] [PubMed] [Google Scholar]
- 43.Uphoff, C. C., A. Carbone, G. Gaidano, and H. G. Drexler. 1998. HHV-8 infection is specific for cell lines derived from primary effusion (body cavity-based) lymphomas. Leukemia 12:1806-1809. [DOI] [PubMed] [Google Scholar]
- 44.Vieira, J., P. O'Hearn, L. Kimball, B. Chandran, and L. Corey. 2001. Activation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J. Virol. 75:1378-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, X. P., Y. J. Zhang, J. H. Deng, H. Y. Pan, F. C. Zhou, E. A. Montalvo, and S. J. Gao. 2001. Characterization of the promoter region of the viral interferon regulatory factor encoded by Kaposi's sarcoma-associated herpesvirus. Oncogene 20:523-530. [DOI] [PubMed] [Google Scholar]