Abstract
Cowpea mosaic virus (CPMV) replication induces an extensive proliferation of endoplasmic reticulum (ER) membranes, leading to the formation of small membranous vesicles where viral RNA replication takes place. Using fluorescent in situ hybridization, we found that early in the infection of cowpea protoplasts, CPMV plus-strand RNA accumulates at numerous distinct subcellular sites distributed randomly throughout the cytoplasm which rapidly coalesce into a large body located in the center of the cell, often near the nucleus. The combined use of immunostaining and a green fluorescent protein ER marker revealed that during the course of an infection, CPMV RNA colocalizes with the 110-kDa viral polymerase and other replication proteins and is always found in close association with proliferated ER membranes, indicating that these sites correspond to the membranous site of viral replication. Experiments with the cytoskeleton inhibitors oryzalin and latrunculin B point to a role of actin and not tubulin in establishing the large central structure. The induction of ER membrane proliferations in CPMV-infected protoplasts did not coincide with increased levels of BiP mRNA, indicating that the unfolded-protein response is not involved in this process.
Infection with positive-stranded RNA viruses often causes extensive membrane rearrangements in the host cell, establishing a distinct compartment where viral RNA synthesis occurs. The viral replication complexes are associated with these membranes, which can originate from different intracellular membranes including the late and early endomembrane system (21, 22, 27, 29, 30, 33). Despite the central role of such a virus-induced membranous compartment in the replicative cycle, the cellular components involved in the formation of this compartment are largely unknown.
Cowpea mosaic virus (CPMV), a bipartite positive-stranded RNA virus, is the type member of the comoviruses, which bear strong resemblance to animal picornaviruses in both the gene organization and the amino acid sequence of replication proteins (1, 14). Both RNA1 and RNA2 are translated into large polyproteins, which are proteolytically cleaved into the different cleavage products by the 24-kDa proteinase (24K) (Fig. 1). The proteins encoded by RNA1 are necessary and sufficient for replication, whereas RNA2 codes for the capsid proteins and the movement protein. The RNA1-encoded 87-kDa protein (87K) contains a domain specific to RNA-dependent RNA polymerases; however, a 110-kDa protein (110K; 87K plus 24K) is the only viral protein present in highly purified, RNA-dependent RNA polymerase preparations capable of elongating nascent RNA chains, suggesting that fusion to 24K is required for replicase activity (13).
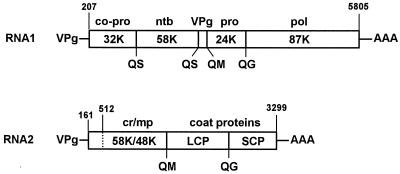
FIG. 1.
Genetic organization of the CPMV genome. Open reading frames in the RNA molecules are indicated by open bars. The nucleotide positions of the start and stop codons are shown above the open reading frame, and the cleavage sites in the polyproteins are shown below. The 110K (24K plus 87K) intermediate processing product is the full-length polymerase (13). Abbreviations: co-pro, cofactor for proteinase; ntb, nucleotide-binding protein; pro, proteinase; pol, core polymerase; cr, cofactor for RNA2 replication; mp, movement protein; LCP, large coat protein; SCP, small coat protein.
Upon infection of cowpea plants with CPMV, a typical cytopathic structure is formed, often adjacent to the nucleus, consisting of an amorphous matrix of electron-dense material that is traversed by rays of small membranous vesicles (12). Autoradiography in conjunction with electron microscopy on sections of CPMV-infected leaves treated with [3H]uridine revealed that CPMV RNA replication was closely associated with the membranous vesicles (12). Additional support for that view came from analyzing different fractions of homogenates of CPMV-infected leaves in which the double-stranded, replicative form of CPMV RNA was present mainly in the microsomal fraction (2). Also, the viral RNA-dependent RNA polymerase activity was found in the crude membrane fraction of CPMV-infected leaves (13, 39). However, as observed by electron microscopy, the bulk of the replication proteins in CPMV-infected cells were immunolocalized not to the vesicles but to the adjacent electron-dense structures, suggesting that only a small part is present in active replication complexes (37).
The membranous vesicles induced upon CPMV infection may originate from the endoplasmic reticulum (ER). Through the use of transgenic Nicotiana benthamiana plants expressing the green fluorescent protein (GFP) targeted to the lumen of the ER, it was demonstrated that CPMV infection leads to a strong proliferation of ER membranes and that these membranes are associated with the viral cytopathic structure (9). For poliovirus, the ER has also been suggested to serve as source for virally induced membranous vesicles, although immunoisolated vesicles were found to contain marker proteins of both the ER and the late endomembrane system (29). It has been proposed that the small membranous vesicles in CPMV-infected cells are the result of the unfolded-protein response (9). This response can occur after overcrowding of ER membranes and results in a proliferation of ER membranes and the upregulation of ER chaperones like protein disulphide isomerase and the lumenal binding protein (BiP) (for a review, see reference 16).
In this study, the intracellular distribution of CPMV RNA during virus infection was visualized by fluorescent in situ hybridization (FISH). The combined use of FISH and immunofluorescence detection of viral proteins allowed us to determine the spatial relationship of CPMV RNA accumulation and the accumulation of CPMV proteins involved in replication and encapsidation and to establish the role of cellular components in the formation of the cytopathic structure. Furthermore, we tested whether the unfolded-protein response was involved in the proliferation of ER membranes in CPMV-infected cells by monitoring the level of BiP mRNA accumulation.
MATERIALS AND METHODS
Plasmids.
Plasmid pTB1552(+) was used as a template to produce the fluorescein-labeled probes used in the in situ hybridization experiments. To create pTB1552(+), the SstI-BamHI fragment of the coding region of RNA1 (comprising nucleotides [nt] 2305 to 3857) was released from pTB1G and subcloned into the plasmid vector Bluescript SK(+) (Stratagene, Inc.). The construction of pUC-mGFP5-ER, which contains the plant-optimized GFP5 with an N-terminal Arabidopsis thaliana basic chitinase signal sequence and a C-terminal HDEL ER retention signal under the control of a CaMV35S promoter, was described previously (9). pMON talin-YFP (generously provided by Gerard van der Krogt, Wageningen University, Wageningen, The Netherlands) contains the actin-binding domain of Dictyostelium talin fused to yellow fluorescent protein (YFP) (27a). The construction of a fusion of GFP with the tubulin-binding part of mammalian MAP4 (GFP-MBD) (generously provided by Richard Cyr, The Pennsylvania State University, University Park, Pa.) has been described previously (23). To determine BiP mRNA levels, plasmid pBLP2 (generously provided by Jürgen Denecke, University of Leeds, Leeds, United Kingdom) was used, which contains tobacco BiP (10).
Transfection of cowpea protoplasts and immunofluorescent labeling.
Cowpea (Vigna unguiculata L.) mesophyll protoplasts were prepared and transfected by polyethylene glycol-mediated transformation as described previously (32). Protoplasts were harvested at different time points postinfection for immunofluorescent staining. One volume of fixing solution (4% paraformaldehyde, 0.1% glutaraldehyde, 0.25 M mannitol, 50 mM sodium phosphate; pH 6) was added to the protoplast suspension. After incubation for 15 min, the liquid was removed, replaced with fixing solution, and allowed to incubate for another 30 min. The cells were washed three times with phosphate-buffered saline (PBS) and spotted on polylysine-coated microscope slides. The protoplasts were permeabilized with a 0.5% Triton X-100 solution in PBS for 10 min. In the case that the protoplasts were used for in situ hybridization, the slides were immersed in cold methanol for 10 min to reduce background staining of the chlorophyll. This step was omitted when the GFP organellar markers were used, since this step abolishes GFP fluorescence. Nonspecific antibody binding was reduced by incubation for 10 min in blocking solution (5% bovine serum albumin in PBS). Subsequently, the protoplasts were incubated for 1 h with dilutions of the primary anti-48K/58K (38), anti-CPMV (37), anti-32K (15), or anti-110K (31) serum in blocking solution. After three washes with PBS, the protoplasts were incubated with goat antirabbit antibodies conjugated to Cy3 (Sigma) for another hour. After two washes with PBS, the cells were either mounted with cover slides by using Citifluor or prepared for in situ hybridization.
In situ hybridization.
In situ hybridization was performed essentially as described previously (24), with a minor modification. After acetylation, the dehydration step was omitted. The fluorescent probe that recognizes CPMV plus-strand RNA was obtained by in vitro transcription of pTB1552(+) linearized with SpeI in the presence of fluorescein-12-UTP (Roche Diagnostics GmbH) according to the manufacturer's recommendation.
Fluorescence microscopy.
A Zeiss LSM 510 confocal microscope was used to obtain images. Optical sections were made at 1-μm intervals, and projections of serial optical sections were obtained by using the software provided by the manufacturer. GFP and fluorescein fluorescence were observed with standard settings (excitation wavelength, 488 nm; emission band pass filter, 505 to 550 nm). Cy3 fluorescence was detected with the following settings: excitation wavelength, 543 nm; emission band pass filter, 560 to 615 nm. In experiments of dual localization, both fluorophores were scanned independently to reduce the possibility of crossover between the cannels. Furthermore, single immunodetection controls verified the absence of fluorescence crossover.
Northern blotting.
At various times postinfection, protoplasts were harvested and centrifuged at low speed (600 × g). Total RNA was isolated from the cell pellet by using Trizol reagent (Gibco BRL) according to the instructions of the manufacturer. Standard procedures were followed for denaturing the RNA with glyoxal, electrophoresing in a 1% agarose gel, and blotting to a GeneScreen membrane (NEN Research Products). The blots were hybridized with a 32P-labeled probe prepared by random-primer labeling of an EcoRI fragment of pBLP2 to detect BiP mRNA.
RESULTS
Localization of CPMV RNA in infected protoplasts.
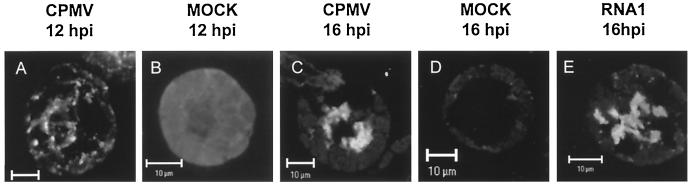
The intracellular accumulation of CPMV RNA during the infection of cowpea protoplasts was visualized by FISH by using a plus-strand-specific probe corresponding to nt 2894 to 3857 of CPMV RNA1. The probe was labeled by in vitro transcription in the presence of fluorescein isothiocyanate-UTP. The earliest time at which viral proteins and infectious viral particles can be detected in cowpea protoplasts by immunofluorescence and by a local lesion assay, respectively, is 12 h postinfection (hpi). Between 12 and 24 hpi, the number of infected cells and the production of infectious particles increase rapidly, reaching a maximum at 36 hpi (18). This implies that viral RNA synthesis peaks between 12 and 24 hpi in these cells. Cowpea protoplasts were infected with CPMV RNA1 and RNA2, collected at 16 hpi, and hybridized with the probe. The fluorescent signals were measured with a focal depth of 1 μm by confocal microscopy. The majority of the plus-strand RNA was localized in a large, irregularly shaped body that was often located near the nucleus (Fig. 2C). Similar localization patterns were observed with protoplasts harvested at 24 hpi (data not shown) and 36 hpi (Fig. 3B ). To assess background staining of the fluorescent probe, mock-infected protoplasts were subjected to FISH and the fluorescent signals were measured with settings of the confocal microscope identical to those used for the CPMV-infected sample at 16 hpi. No significant background staining was observed (Fig. 2D).
FIG. 2.
Intracellular distribution of plus-strand viral RNA in protoplasts infected with CPMV RNA. Protoplasts were collected at the indicated time points after infection with CPMV RNA1 and RNA2 (CPMV), CPMV RNA1 (RNA1), or water (mock) and hybridized with a fluorescein-RNA probe that recognized plus-strand CPMV RNA1. Fluorescent signals were visualized by confocal microscopy measuring optical sections with a focal depth of 1 μm. (A) At 12 hpi, viral RNA was localized in multiple small bodies dispersed over the cytoplasm. (C and E) At 16 hpi, viral RNA was observed in one or several large, amorphous bodies of fluorescence located in the center of protoplasts infected with CPMV (C) or with CPMV RNA1 alone (E). (B and D) Background fluorescence was evaluated in mock-infected protoplasts observed with identical settings at 12 hpi (B) and 16 hpi (D). Bars, 10 μm.
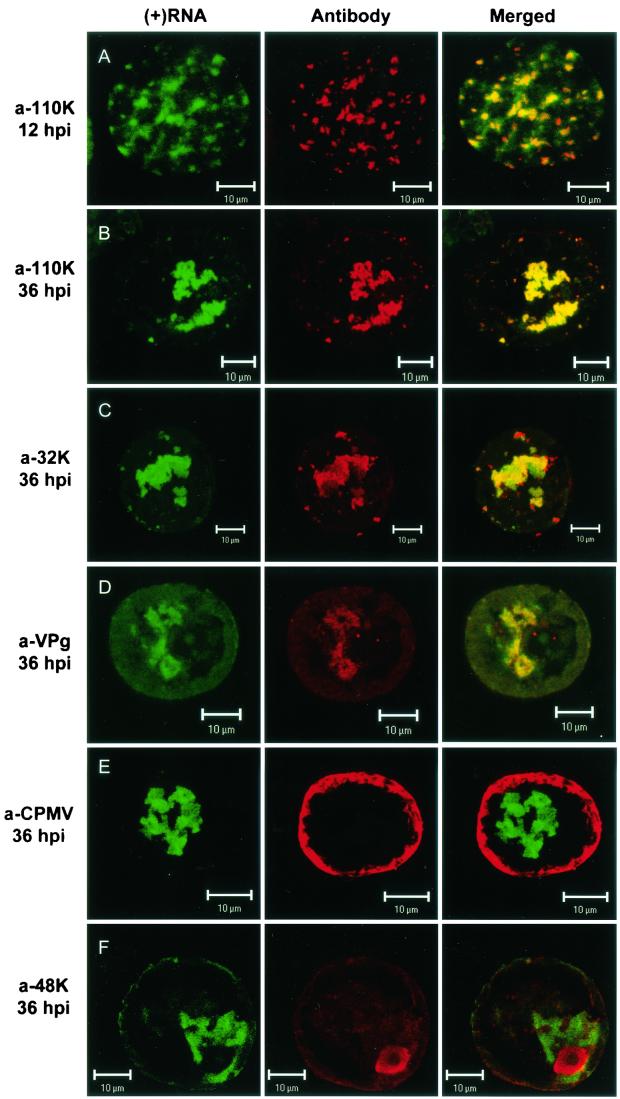
FIG.3.
Localization of plus-strand viral RNA with respect to different viral proteins in CPMV-infected protoplasts. Protoplasts were collected at the indicated time points after infection with CPMV RNA, hybridized with a fluorescein-RNA probe that recognized plus-strand CPMV RNA1 (green signal), and immunostained with the indicated antisera (red signal). (A and B) At 12 and 36 hpi, the 110K polymerase was localized almost exclusively in sites where viral RNA accumulated. Colocalization of the two signals is shown in the merged images as yellow. (C) The 32K cofactor for the proteinase also colocalized substantially with viral RNA. (D) The majority of the proteins recognized by anti-VPg (a-VPg) serum colocalized with viral RNA. (E) Viral particles localized in the periphery of the cell and did not colocalize with the sites of viral RNA accumulation. (F) The 58K protein recognized by the anti-48K/58K serum localized mainly in the nucleus and did not colocalize with viral RNA. Bars, 10 μm.
To determine the localization of plus-strand RNA early in infection, protoplasts were harvested at 12 hpi. At this time, the plus-strand RNA labeling was still weak and signals higher than the background signal could be observed only in a small percentage of the protoplasts (typically 5%). Approximately half of these protoplasts displayed the accumulation of plus-strand RNA in a large irregular body located near the nucleus, as was observed at later times. In the other half, the fluorescence pattern differed markedly and plus-strand RNA was observed in multiple smaller bodies scattered over the cytoplasm (Fig. 2A). At later time points (14 to 36 hpi), this fluorescence pattern was no longer observed, suggesting that the smaller bodies are a preliminary stage preceding the large juxtanuclear amorphous structure. Again the background staining was measured in mock-infected protoplasts with confocal settings similar to those used for CPMV-infected protoplasts at 12 hpi. The fluorescent background staining was low and diffusely spread over the cytoplasm (Fig. 2B).
In order to visualize the accumulation of minus-strand RNA, a minus-strand-specific probe was prepared corresponding to nt 2305 to 3857 of RNA1. With this probe, a fluorescent signal above the background signal could not be observed at 12, 16, or 36 hpi (data not shown), even when the samples were denatured thermally at 65°C prior to hybridization, a treatment reported to be necessary for the detection of poliovirus minus-strand RNA (7). In addition, with probes spanning different regions of RNA1 or RNA2, no specific signal corresponding to minus-strand RNA was observed (data not shown). This suggests that in CPMV-infected protoplasts, minus-strand RNA accumulates much less than does plus-strand RNA, a suggestion in line with earlier findings obtained in Northern hybridization studies (11).
Taken together, these results suggest that plus-strand CPMV RNA starts to accumulate at multiple sites early in infection, sites which quickly coalesce into a large, irregularly shaped body located juxtanuclearly.
Combined localization of plus-strand RNA and CPMV proteins.
The accumulation of plus-strand viral RNA in a large amorphous structure resembles the immunofluorescence pattern observed previously with the use of antibodies against the viral replication proteins (9, 32). On the ultrastructural level, the replication proteins accumulate in electron-dense material found in close proximity to the small membranous vesicles that are the site of RNA synthesis and presumably of RNA accumulation. To examine the spatial relationship between the site of viral plus-strand RNA accumulation and the location of CPMV proteins involved in replication, CPMV-infected protoplasts were immunostained with different antibodies prior to in situ hybridization. The distribution of the viral proteins recognized by antiserum raised against the 110K polymerase and visualized with Cy3-conjugated secondary antibody (Fig. 3B; red) was coincident with that of viral plus-strand RNA (Fig. 3B; green) at 36 hpi, as is clearly visible in the digitally superimposed image (Fig. 3B; merged), where green and red signals that coincide together produce a yellow signal. In addition, at 12 hpi, when the large amorphous structure in the center of the cell was not yet formed, the majority of the plus-strand RNA colocalized with 110K (Fig. 3A). Through the use of antibodies raised against the RNA1-encoded VPg and 32K, a similar colocalization was found (Fig. 3C and D) at 36 hpi, although with slightly more variation in the intensity of the red and green fluorescence than was observed with the anti-110K serum. At present we do not know whether these differences reflect real differences in the localization of minor portions of the replication proteins recognized by anti-32K and anti-VPg serum and the viral RNA.
After synthesis, plus-strand RNA is encapsidated in viral particles. To investigate the spatial relationship between plus-strand RNA visualized by FISH and the particles, immunolabeling was performed with an antibody raised against purified virus particles. As shown in Fig. 3E, the majority of the viral particles accumulated at the periphery of the cell at 36 hpi, whereas plus-strand RNA was found centrally, near the nucleus. It is clear from the merged image that colocalization of viral RNA and viral particles was not observed, which is somewhat surprising, because the viral particles contained plus-strand RNA. This suggests that encapsidated plus-strand RNA does not hybridize to the probe under these experimental conditions.
With anti-48K/58K serum that recognized both the 48K movement protein (MP) and the co-C-terminal 58K protein, the majority of the anti-48K/58K signal was present in the nucleus at 36 hpi whereas only a faint signal was present diffusely in the cytoplasm, which only partly overlapped with the fluorescently labeled viral RNA (Fig. 3F). The signal in the nucleus is in agreement with the results of earlier studies localizing 58K, an RNA2-encoded protein essential for RNA2 replication, in the nucleus (38). The faint signal in the cytoplasm could be either MP or 58K. The lack of colocalization of plus-strand RNA with 58K, MP, or the capsid proteins may suggest that these RNA2-encoded proteins play no role in the establishment of the sites of viral RNA accumulation. This suggestion was strengthened by the observation that the localization of viral RNA in cowpea protoplasts infected with RNA1 alone was similar to the localization observed in cells infected with both RNA1 and RNA2 (Fig. 2E).
Effects of cytoskeletal inhibitors on the establishment of the site of replication.
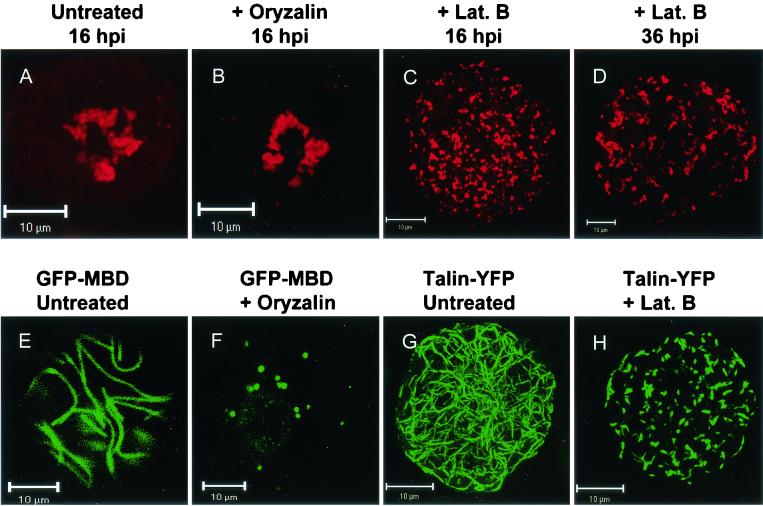
As shown above, both viral RNA and the 110K polymerase colocalize in numerous small bodies early in infection, which later in infection coalesce into a large, irregularly shaped body. The possible involvement of the plant cytoskeleton in the formation of this structure was tested by using the cytoskeletal inhibitors latrunculin B and oryzalin. The highly specific toxin latrunculin B, isolated from a red sea sponge, has been shown to effectively depolymerize actin filaments in eukaryotic cells (reference 3 and references therein). Oryzalin is an herbicide which strongly binds to plant tubulin monomers, thereby stimulating the complete disassembly of the microtubular network (reference 4 and references therein). Protoplasts were infected with CPMV and then divided into three equal portions. One sample was left untreated, whereas either oryzalin (10 μM) or latrunculin B (20 μM) was added to the incubation medium of the other two samples directly after infection. The protoplasts were collected at 16 and 36 hpi and prepared for immunofluorescent staining with antibodies raised against the 110K polymerase. The percentage of infected protoplasts did not differ in the treated and untreated samples. In the untreated and oryzalin-treated protoplasts, the polymerase was present in the large juxtanuclear body at 16 hpi (Fig. 4A and B) and at 36 hpi (data not shown). The pattern of distribution of the polymerase in latrunculin B-treated protoplasts was strikingly different. Fluorescence was present in numerous small bodies that were randomly scattered throughout the cytoplasm (Fig. 4C). A similar pattern was observed at 36 hpi (Fig. 4D). The pattern of multiple smaller bodies resembled that of polymerase distribution observed early in infection, suggesting that latrunculin B arrests the movement of these bodies to a juxtanuclear position.
FIG. 4.
Disruption of the actin but not the microtubular network induced changes in the localization of the viral 110K polymerase in CPMV-infected protoplasts. Immediately after infection, protoplasts were treated with oryzalin (10 μM) (B) or latrunculin B (20 μM) (C and D) or left untreated (A) and harvested at the indicated time points, followed by immunostaining with anti-110K serum. In untreated (A) and oryzalin-treated (B) protoplasts, the polymerase was present in a central large amorphous structure at 16 hpi. In cells treated with latrunculin B, the polymerase was present in numerous small spots scattered throughout the cytoplasm at both 16 hpi (C) and 36 hpi (D). Protoplasts transiently expressing GFP-MBD to visualize the microtubules (E and F) or Talin-YFP to visualize the actin cytoskeleton (G and H) show that oryzalin (F) and latrunculin B (H) were active at the concentration used at 16 h posttransfection. Lat. B, latrunculin B. Bars, 10 μm.
In parallel experiments, it was verified that both inhibitors were active at the concentrations used. For this purpose, the cowpea protoplasts were transfected with either a plant expression vector encoding GFP-MBD (23) or an expression vector encoding a fusion of the YFP with the actin-binding part of the Dictyostelium talin gene (Talin-YFP) (20). In protoplasts treated with oryzalin, the typical microtubular network labeled by GFP-MBD was rigorously disturbed, as was observed in live protoplasts 16 and 36 h posttransfection by confocal microscopy (Fig. 4E and F and data not shown). In addition, the actin cytoskeleton labeled with Talin-YFP was severely disturbed upon treatment with latrunculin B, and only very small fluorescent strands that were not interconnected remained (Fig. 4G and H).
Taken together, these data indicate that integrity of the actin filaments but not of the microtubular cytoskeleton is required for the formation of the juxtanuclear large shapeless body.
CPMV-induced membrane proliferation.
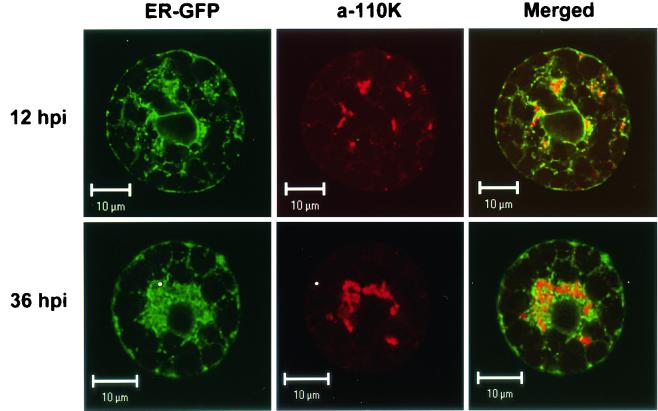
CPMV infection resulted in the formation of proliferated ER membranes that surrounded and traversed the site of RNA replication stained with anti-110K serum at 36 hpi (Fig. 5, bottom row) (9). To determine whether the proliferated ER membranes were already formed early in infection, when the replication complexes are present and dispersed over the cytoplasm, cowpea protoplasts were transfected with CPMV RNA together with pUC-mGFP5-ER, a plant expression vector that encodes the GFP targeted to the lumen of the ER (17). The protoplasts were harvested at 12 hpi and prepared for immunostaining with anti-110K serum. It should be noted that mGFP5-ER remains fluorescent during immunostaining procedures and that the GFP fluorescence could be separated from the fluorescence of Cy3, which was used for staining the viral polymerase. As shown in Fig. 5 (top row), the smaller bodies stained with anti-110K serum were closely associated with proliferated ER membranes.
FIG. 5.
Dual localization of ER-targeted GFP (ER-GFP) and the viral 110K polymerase (a-110K) in CPMV-infected protoplasts. Cells were fixed at 12 hpi (top row) and 36 hpi (bottom row) and processed for immunofluorescence by using antibodies raised against the 110K polymerase. ER-GFP retained its fluorescence throughout the procedure. Proliferated ER membranes surrounded and traversed the sites of 110K accumulation both early (12 hpi) and late (36 hpi) in infection. Bars, 10 μm.
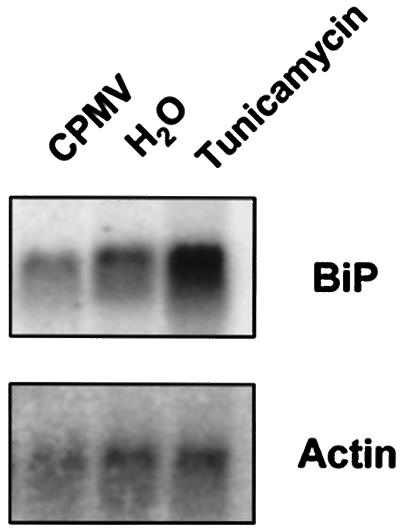
Previously it has been suggested that the ER membrane proliferation might occur as a result of the unfolded-protein response triggered by CPMV infection (9). In the present study, we tested this hypothesis in cowpea protoplasts by monitoring the mRNA levels of the ER lumenal binding protein BiP, which has been reported to be strongly upregulated in yeast, mammalian, and plant cells during the unfolded-protein response (16, 19). Northern blot analysis with a tobacco BiP probe revealed that the level of BiP mRNA in CPMV-infected protoplasts was not elevated compared to that in uninfected protoplasts at 36 hpi, when the CPMV-induced membrane proliferation was at its maximum (Fig. 6). Similar results were obtained at earlier time points (data not shown). As a control to ensure that the unfolded-protein response could be induced in our cowpea protoplast system, tunicamycin, a drug that inhibits N-glycosylation of ER membrane proteins and potently elicits the unfolded-protein response, was used. Treatment of the protoplasts with this drug led to a sharp increase in BiP mRNA levels (Fig. 6). These results suggest that the ER membrane proliferations associated with CPMV replication are probably not due to the unfolded-protein response.
FIG. 6.
Effect of CPMV infection on BiP mRNA levels in cowpea protoplasts. Cowpea protoplasts were infected with CPMV (CPMV) or uninfected (H2O and Tunicamycin). Prior to the harvesting of the protoplasts (36 hpi), the sample designated Tunicamycin was treated with tunicamycin (20 μg/ml) for 2.5 h. Total RNA was extracted, separated on an agarose gel, and blotted on a nylon membrane. The blots were probed for BiP, and an actin probe (Actin) was used as a control for loading differences. CPMV infection (CPMV) did not lead to an increase in BiP mRNA levels compared to those of the uninfected protoplasts (H2O). Treatment with tunicamycin (Tunicamycin) resulted in upregulation of BiP expression.
DISCUSSION
The use of FISH and immunolocalization allowed us to better define the subcellular sites of viral RNA replication during the course of CPMV infection. Early in infection, the fluorescent probe that recognizes plus-strand RNA was present at numerous subcellular sites distributed over the cytoplasm that quickly coalesced into a large structure located in the center of the cell. Strikingly, almost no fluorescence was present in the cytoplasm at the periphery of the cells, which was the site where the viral particles were shown to accumulate, indicating that only unencapsidated plus-strand RNA hybridizes with the fluorescent probe. It has been shown previously that under conditions that leave viral RNA unprotected by coat protein, the genomic RNA is unstable (11), suggesting that in CPMV-infected cells, the large majority of plus-strand viral RNA is present in viral particles whereas only a small proportion is present as unencapsidated RNA. However, we observed no clear differences in the plus-strand RNA1 signal in protoplasts infected with RNA1 alone and in protoplasts coinfected with RNA2, which encodes the capsid proteins. This further strengthened the notion that when FISH is used, encapsidated viral RNA is not detected. Thus, the signal observed with FISH probably corresponds to recently synthesized RNA, implying a direct correlation of the FISH signal with the site of viral replication. This is in agreement with the observation that the FISH signal colocalized almost perfectly with the viral 110K polymerase during infection. The subcellular sites where 110K and the viral RNA were found to colocalize are believed to correspond, on the ultrastructural level, with the cytopathic structure consisting of electron-dense material and clusters of small membranous vesicles. We were unable to determine whether the 110K polymerase and the viral RNA were associated specifically with vesicles and/or the electron-dense material due to the limited resolution of fluorescence microscopy compared to electron microscopy.
The numerous sites where the viral RNA and the 110K colocalized early in infection were closely associated with proliferated ER membranes, supporting the view that ER membranes act as a source for the small membranous vesicles. This is in agreement with earlier observations of live N. benthamiana cells with ER-GFP, in which multiple, highly mobile bodies consisting of proliferated ER were observed in newly infected cells (9). In addition, for poliovirus, it has been shown that vesiculation of intracellular membranes starts early in infection at multiple sites in the cytoplasm of infected cells (7). In CPMV-infected protoplasts, the smaller bodies containing viral RNA could be detected from 12 hpi onwards but they rapidly (within 2 h) coalesced into a large, irregularly shaped body located in the center of the cell, often near the nucleus. Previous experiments with a local lesion assay and the detection of viral proteins have indicated that the majority of viral RNA synthesis occurs between 12 and 24 hpi (18), which suggests that the formation of this cytopathic structure early in infection is important to promote efficient RNA replication. Consistent with this are findings that in cells infected with poliovirus or tobacco mosaic virus (TMV), the formation of a large juxtanuclear structure containing viral RNA, nonstructural proteins, and ER membranes precedes bulk RNA synthesis (7, 24). Many positive-stranded RNA viruses induce the formation of a membranous compartment in the cytoplasm where viral replication occurs, and it has been proposed that such compartmentalization increases the local concentrations of virus-encoded proteins and viral RNA (8).
Based on the effects of latrunculin B and oryzalin on the distribution of the viral polymerase, we suggest that in CPMV-infected protoplasts, intracellular trafficking of replication complexes to the large juxtanuclear structure occurs via association with the actin cytoskeleton and not the microtubular network. Actin-based movement in plant cells has also been reported recently for individual Golgi stacks in live plant cells by using GFP constructs specifically targeted to the Golgi apparatus (6, 26). In contrast to CPMV, for the movement of TMV replication complexes to a juxtanuclear position early in infection, the integrity of both the actin and the microtubular cytoskeleton is required, as was demonstrated by using cytochalasin D and oryzalin (24).
After the formation of the large cytopathic structure, unencapsidated viral RNA remained in the center of the cell and was not redistributed to the periphery of the cell. In contrast, viral particles did accumulate in the periphery of the cell, presumably to be transported to the neighboring cell via plasmodesmata. The observation that almost no capsid material was found in the central structure suggests that after encapsidation, the viral particles rapidly spread to the periphery of the cell for intercellular transport. The 48K MP did not colocalize with viral RNA, and the distribution of viral RNA was similar in protoplasts infected with either RNA1 and RNA2 or RNA1 alone, suggesting that the 48K MP does not influence the distribution of viral RNA. In contrast, the TMV 30K MP colocalized with viral RNA at all points during infection and was essential for the establishment of the large body near the nucleus observed in the middle stages of infection (24). Furthermore, at late stages of infection, viral RNA was dispersed over the cytoplasm and at the periphery of the cell (24). These differences between CPMV and TMV might reflect the different mechanisms these viruses use for their cell-to-cell movement, since TMV RNA spreads from cell to cell as a ribonucleoprotein complex of TMV RNA and 30K MP (for a review, see reference 5) whereas CPMV RNA is encapsidated before intercellular transport and moves from cell to cell as viral particles through tubules formed by the MP in plasmodesmata (34-36).
CPMV infection induces an extensive rearrangement of intracellular membranes, but the cellular mechanism underlying this vesiculation is unclear. Based on the observation that poliovirus-induced vesicles share cytological characteristics (a double membrane and cytosolic content) with autophagic vacuoles that are formed in noninfected cells in response to nitrogen or amino acid starvation, autophagy has been proposed as a mechanism of induction of the vesicles (29, 30). On the other hand, CPMV-induced vesicles do not possess similar features, which makes it unlikely that autophagy is involved for CPMV. Another mechanism that has been proposed for poliovirus suggests that poliovirus infection interferes with ER-to-Golgi transport, leading to the accumulation of membranous vesicles (25). This view is supported by a recent study that showed that the poliovirus-induced vesicles bud from the ER and colocalize with the COPII components Sec13 and Sec31, suggesting that poliovirus-induced vesicles are homologous to the vesicles of the anterograde membrane transport pathway (28). The extensive ER proliferation observed in CPMV-infected cells has not been reported for poliovirus-infected cells, which may suggest that CPMV uses a different cellular mechanism to induce vesiculation. CPMV infection did not result in an increase of the level of BiP mRNA in protoplasts, which is a marker for the unfolded-protein response. The reliability of this marker was tested with tunicamycin, which elicited a clear response in protoplasts. Since in yeast, animal, and plant cells upregulation of BiP is a hallmark in the unfolded-protein response (16, 19), it is unlikely that this stress response is responsible for the ER membrane proliferations. Further studies will be necessary to unravel the molecular pathways leading to CPMV-induced ER proliferation and vesiculation.
Acknowledgments
We thank Jürgen Denecke, Richard Cyr, and Gerard van der Krogt for generously providing the biological material indicated in Materials and Methods. We gratefully acknowledge Jeroen Pouwels for useful comments and technical assistance.
This work was supported by The Netherlands Foundation of Chemical Research with financial aid from The Netherlands Organization for Scientific Research.
REFERENCES
- 1.Argos, P., G. Kamer, M. J. Nicklin, and E. Wimmer. 1984. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant comovirus suggest common ancestry of these virus families. Nucleic Acids Res. 12:7251-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assink, A. M., H. Swaans, and A. Van Kammen. 1973. The localization of virus-specific double-stranded RNA of cowpea mosaic virus in subcellular fractions of infected Vigna leaves. Virology 53:384-391. [DOI] [PubMed] [Google Scholar]
- 3.Baluska, F., J. Jasik, H. G. Edelmann, T. Salajova, and D. Volkmann. 2001. Latrunculin B-induced plant dwarfism: plant cell elongation is F-actin-dependent. Dev. Biol. 231:113-124. [DOI] [PubMed] [Google Scholar]
- 4.Baskin, T. I., J. E. Wilson, A. Cork, and R. E. Williamson. 1994. Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 35:935-942. [PubMed] [Google Scholar]
- 5.Beachy, R. N., and M. Heinlein. 2000. Role of P30 in replication and spread of TMV. Traffic 1:540-544. [DOI] [PubMed] [Google Scholar]
- 6.Boevink, P., K. Oparka, S. Santa Cruz, B. Martin, A. Betteridge, and C. Hawes. 1998. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15:441-447. [DOI] [PubMed] [Google Scholar]
- 7.Bolten, R., D. Egger, R. Gosert, G. Schaub, L. Landmann, and K. Bienz. 1998. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent in situ hybridization. J. Virol. 72:8578-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, K. W. 1999. Replication of tobacco mosaic virus RNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:613-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carette, J. E., M. Stuiver, J. Van Lent, J. Wellink, and A. Van Kammen. 2000. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74:6556-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denecke, J., M. H. Goldman, J. Demolder, J. Seurinck, and J. Botterman. 1991. The tobacco luminal binding protein is encoded by a multigene family. Plant Cell 3:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Varennes, A., and A. J. Maule. 1985. Independent replication of cowpea mosaic virus bottom component RNA: in vivo instability of the viral RNAs. Virology 144:495-501. [DOI] [PubMed] [Google Scholar]
- 12.De Zoeten, G. A., A. M. Assink, and A. Van Kammen. 1974. Association of cowpea mosaic virus-induced double-stranded RNA with a cytopathological structure in infected cells. Virology 59:341-355. [DOI] [PubMed] [Google Scholar]
- 13.Eggen, R., A. Kaan, R. Goldbach, and A. Van Kammen. 1988. Cowpea mosaic virus RNA replication in crude membrane fractions from infected cowpea and Chenopodium armaranticolor. J. Gen. Virol. 69:2711-2720. [Google Scholar]
- 14.Franssen, H., J. Leunissen, R. Goldbach, G. Lomonossoff, and D. Zimmern. 1984. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 3:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franssen, H., M. Moerman, G. Rezelman, and R. Goldbach. 1984. Evidence that the 32,000-Dalton protein encoded by bottom-component RNA of cowpea mosaic virus is a proteolytic processing enzyme. J. Virol. 50:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hampton, R. Y. 2000. ER stress response: getting the UPR hand on misfolded proteins. Curr. Biol. 10:R518-R521. [DOI] [PubMed] [Google Scholar]
- 17.Haseloff, J., K. R. Siemering, D. C. Prasher, and S. Hodge. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hibi, T., G. Rezelman, and A. Van Kammen. 1975. Infection of cowpea mesophyll protoplasts with cowpea mosaic virus. Virology 64:308-318. [DOI] [PubMed] [Google Scholar]
- 19.Jelitto-Van Dooren, E. P., S. Vidal, and J. Denecke. 1999. Anticipating endoplasmic reticulum stress. A novel early response before pathogenesis-related gene induction. Plant Cell 11:1935-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kost, B., P. Spielhofer, and N. H. Chua. 1998. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16:393-401. [DOI] [PubMed] [Google Scholar]
- 21.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marc, J., C. L. Granger, J. Brincat, D. D. Fisher, T. Kao, A. G. McCubbin, and R. J. Cyr. 1998. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10:1927-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mas, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maynell, L. A., K. Kirkegaard, and M. W. Klymkowsky. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nebenfuhr, A., L. A. Gallagher, T. G. Dunahay, J. A. Frohlick, A. M. Mazurkiewicz, J. B. Meehl, and L. A. Staehelin. 1999. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121:1127-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Pouwels, J., G. N. van der Krogt, J. van Lent, T. Bisseling, and J. Wellink. The cytoskeleton and the secretory pathway are not involved in targeting the cowpea mosaic virus movement protein to the cell periphery. Virology, in press. [DOI] [PubMed]
- 28.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Bokhoven, H., J. W. van Lent, R. Custers, J. M. Vlak, J. Wellink, and A. van Kammen. 1992. Synthesis of the complete 200K polyprotein encoded by cowpea mosaic virus B-RNA in insect cells. J. Gen. Virol. 73:2775-2784. [DOI] [PubMed] [Google Scholar]
- 32.van Bokhoven, H., J. Verver, J. Wellink, and A. van Kammen. 1993. Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J. Gen. Virol. 74:2233-2241. [DOI] [PubMed] [Google Scholar]
- 33.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Lent, J., J. Wellink, and R. Goldbach. 1990. Evidence for the involvement of the 58K and 48K proteins in intracellular movement of cowpea mosaic virus. J. Gen. Virol. 71:219-223. [Google Scholar]
- 35.Verver, J., J. Wellink, J. Van Lent, K. Gopinath, and A. Van Kammen. 1998. Studies on the movement of cowpea mosaic virus using the jellyfish green fluorescent protein. Virology 242:22-27. [DOI] [PubMed] [Google Scholar]
- 36.Wellink, J., and A. van Kammen. 1989. Cell-to-cell transport of cowpea mosaic virus requires both the 58K/48K proteins and the capsid proteins. J. Gen. Virol. 70:2279-2286. [Google Scholar]
- 37.Wellink, J., J. Van Lent, and R. Goldbach. 1988. Detection of viral proteins in cytopathic structures in cowpea protoplasts infected with cowpea mosaic virus. J. Gen. Virol. 69:751-755. [Google Scholar]
- 38.Wellink, J., J. W. van Lent, J. Verver, T. Sijen, R. W. Goldbach, and A. van Kammen. 1993. The cowpea mosaic virus M RNA-encoded 48-kilodalton protein is responsible for induction of tubular structures in protoplasts. J. Virol. 67:3660-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zabel, P., H. Weenen-Swaans, and A. van Kammen. 1974. In vitro replication of cowpea mosaic virus RNA: I. Isolation and properties of the membrane-bound replicase. J. Virol. 14:1049-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]