Abstract
We show here that the DNA helicase activity of the parvoviral initiator protein NS1 is highly directional, binding to the single strand at a recessed 5′ end and displacing the other strand while progressing in a 3′-to-5′ direction on the bound strand. NS1 and a cellular site-specific DNA binding factor, PIF, also known as glucocorticoid modulating element binding protein, bind to the left-end minimal replication origin of minute virus of mice, forming a ternary complex. In this complex, NS1 is activated to nick one DNA strand, becoming covalently attached to the 5′ end of the nick in the process and providing a 3′ OH for priming DNA synthesis. In this situation, the helicase activity of NS1 did not displace the nicked strand, but the origin duplex was distorted by the NS1-PIF complex, as assayed by its sensitivity to KMnO4 oxidation, and a stretch of about 14 nucleotides on both strands of the nicked origin underwent limited unwinding. Addition of Escherichia coli single-stranded DNA binding protein (SSB) did not lead to further unwinding. However, addition of recombinant human single-stranded DNA binding protein (RPA) to the initiation reaction catalyzed extensive unwinding of the nicked origin, suggesting that RPA may be required to form a functional replication fork. Accordingly, the unwinding mediated by NS1 and RPA promoted processive leading-strand synthesis catalyzed by recombinant human DNA polymerase δ, PCNA, and RFC, using the minimal left-end origin cloned in a plasmid as a template. The requirement for RPA, rather than SSB, in the unwinding reaction indicated that specific NS1-RPA protein interactions were formed. NS1 was tested by enzyme-linked immunosorbent assay for binding to two- or three-subunit RPA complexes expressed from recombinant baculoviruses. NS1 efficiently bound each of the baculovirus-expressed complexes, indicating that the small subunit of RPA is not involved in specific NS1 binding. No NS1 interactions were observed with E. coli SSB or other proteins included as controls.
Parvoviruses infect a broad range of invertebrate and vertebrate species, including humans (18). At the molecular level, this group of viruses encapsidates a small linear single-stranded DNA genome of about 5 kb bracketed by short unique palindromic sequences which base pair to form hairpin duplexes (1). The viral replication strategy resembles rolling-circle replication (RCR) using unidirectional leading-strand DNA synthesis and probably evolved from prokaryotic RCR replicons (21, 34, 48). As in RCR systems, a viral protein, in this case NS1, serves as a site-specific DNA binding protein, which binds the viral origin and initiates replication by introducing a single-stranded nick close to the core recognition site (13, 15, 16). This reaction leaves NS1 covalently attached to the 5′ end at the nick site via a phosphotyrosine bond and generates a free 3′ hydroxyl group of the nucleotide at the cleavage site, which serves as a primer for DNA synthesis (44). The linear genome replicates through a series of concatameric intermediates by a mechanism dubbed rolling-hairpin replication, in which a replication fork is flipped back and forth along the genome by rearrangement of the terminal palindromes. The steps in this process are primarily controlled by NS1, and sequential DNA synthesis is mediated by host replication factors present in the S phase of the cell cycle. In this process, the palindromic sequences play a key role in replication because they contain the viral origins, which are not only essential for replication initiation but also necessary for resolution of replicative intermediates, breaking them down to unit length genome duplexes (1-3, 16, 17, 19, 20, 53).
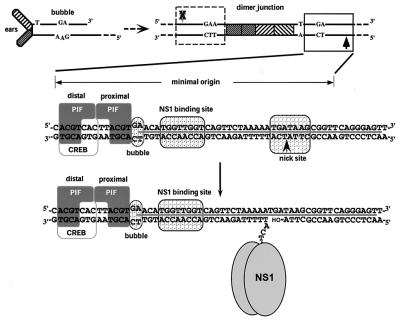
As depicted in Fig. 1, the left-end palindrome of the single-stranded minute virus of mice (MVM) genome can be folded to form a Y-shaped hairpin structure containing a mismatch in the stem, designated the “bubble” sequence, where a GA dinucleotide opposes a GAA trinucleotide. In addition, this palindrome contains enhancer elements for the initiating viral promoter and provides a terminal base pair with a free 3′ hydroxyl group, termed oriLH (for hairpin origin), which primes conversion of the genome into a monomeric duplex form by a cellular DNA polymerase(s) (3, 4). In such monomeric duplex intermediates, however, the turnaround form of the left-end palindrome cannot function as an active origin, due to the presence of the bubble mismatch. After being replicated to the dimer intermediate, the extended hairpin forms a palindromic double-stranded sequence termed the dimer junction, because it bridges two unit length head-to-head duplex genomes. In the dimer junction, the nucleotides of the bubble sequence now occur, as duplex DNA, on each side of an axis of symmetry, creating the GAA and TC arms, where the two potential origin sequences (Fig. 1) differ in length by a single nucleotide. During replication, the dimer junction is resolved into two structures, an “extended” palindromic form, and a “turnaround” form that recreates the left-end hairpin. The extra nucleotide in the GAA arm renders this origin inactive and restricts replication initiation to the TC arm of the dimer junction, causing the resolution process to be predominantly asymmetric (17, 20).
FIG. 1.
Formation and organization of MVM oriL. In the upper left is the structure of the left-end hairpin (oriLH) showing the 3′ hydroxyl group used for priming replication and the mismatched bubble sequence as present in the parental single-stranded viral genome. In the upper right is the organization of the left-end hairpin sequences within the duplex dimer junction generated by replication through the hairpin. The hatched boxes represent the palindromic sequences that were originally folded to give the ears of the hairpin form. The boxed sequence (expanded below) represents the minimum active replication origin on the outboard arm, oriLTC, while the sequence in the dashed box represents the corresponding origin on the inboard arm, oriLGAA, which is inactive. The arrows denote the potential nick sites on each side of the junction, solid for active and X-ed out shaded for inactive. The middle diagram illustrates the sequence of oriLTC, showing the different elements involved in replication. The PIF binding site (see the text) overlaps a consensus binding site for the CREB/ATF family of host transcription factors. The two tetranucleotide motifs bound by PIF are labeled “distal” and “proximal” to indicate their positions relative to the NS1 binding site. The other boxes indicate sequences involved in the bubble dinucleotide (or trinucleotide) spacer element, the ACCA2 NS1 binding site, and the nick site, a specific sequence required for nicking and covalent attachment of NS1. The heavy line between the DNA strands indicates sequences protected by NS1 from DNase I digestion. The position of the nick reflects the new determination described in this report. The bottom diagram shows the nicking and covalent attachment of an oligomer of NS1 via a phosphotyrosine bond. The nicking reaction liberates a 3′ hydroxyl group, which primes DNA synthesis mediated by host cell DNA polymerases.
Mapping of the minimal sequences necessary for replication initiation of the TC arm has revealed that the origin is approximately 50 bp long and contains three distinct recognition elements (20). NS1 binds site specifically to the (ACCA)2 repeat motif in an ATP-dependent manner, forming a large complex that protects sequences surrounding the recognition site, including the nick site, from DNase I digestion (15). However, while NS1 can bind by itself, it cannot nick the origin but requires the cooperation of a cellular factor, called parvovirus initiation factor (PIF), also known as glucocorticoid modulating element binding protein (10, 11). We have recently described the purification, cloning, and expression of PIF, which is a widely expressed site-specific DNA binding heterodimer consisting of two related polypeptides, p79 and p96 (12). Recombinant baculovirus- or HeLa-derived PIF coordinately binds two copies of a tetranucleotide, ACGT, located at one end of the origin, across the bubble sequence from the NS1 binding site. PIF and NS1 can each bind the TC and GAA arms equally well by themselves, and the discrimination between the two forms of the origin sequence is mediated by the cooperative binding of PIF and NS1 across the TC, but not the GAA, bubble sequence. On the TC arm, therefore, PIF and NS1 form a high-affinity ternary complex capable of nicking and subsequently initiating replication at this origin (13).
While it has not yet been possible to reconstitute replication of any full-length parvovirus genomes using only purified cellular components, discrete steps of replication for MVM have been reconstituted in vitro using as templates recombinant NS1, fractionated human cell extracts, and the minimal TC origin cloned in plasmids (20, 43). Replication from such templates appears to employ a unidirectional leading-strand synthesis mechanism. In vitro reconstitution of replication has led to the identification of proliferating cellular nuclear antigen (PCNA), single-stranded DNA binding protein (RPA), and unidentified cellular DNA polymerases as necessary for replication (10). By analogy to simian virus 40 (SV40) replication, the requirement for PCNA and RPA points to a requirement for the components involved in classical leading-strand synthesis, which should also require replication factor C (RFC), the clamp-loading factor. RFC recognizes the primer-template junction, loads PCNA onto the template to form the sliding clamp that stabilizes DNA polymerase δ (Pol δ) interaction with its template, and promotes processive chain elongation (49-52).
In this report, we demonstrate that leading-strand synthesis initiated from the minimal origin can be entirely reconstituted with purified recombinant factors of human origin. While NS1 and PIF are necessary for replication initiation, we have found that both unwinding of the origin and replication fork formation are dependent on a specific intermolecular interaction between NS1 and RPA. We also show that catalysis of processive leading-strand synthesis can be reconstituted on a PIF-NS1-initiated origin by addition of recombinant RFC, PCNA, RPA, and Pol δ. On the basis of the presented data, we propose a molecular model of the parvovirus replication fork.
MATERIALS AND METHODS
Plasmids.
The minimal origin templates used for replication, nicking, and KMnO4 assays were pL1-2 TC and pL1-2 GAA, which contain MVM oriLTC and oriLGAA, respectively (20).
Recombinant proteins.
The generation of recombinant baculoviruses expressing p96 and p79 subunits of PIF, amino-terminal glutathione-S-transferase (GST), and His (hexahistidine)-tagged MVM NS1 and the purification of the recombinant products have been described previously (9, 12). Baculoviruses encoding an amino-terminal His-tagged form of the major subunit of human single-stranded DNA binding protein (His-RPA70) and the two minor subunits (RPA34 and RPA12) were generous gifts from C. Umbrecht and T. Kelly, Johns Hopkins University Medical School, Baltimore, Md. Functional recombinant RPA was expressed by coinfection of insect Sf9 cells and purified as previously described (10, 14). The five-subunit complex of human RFC was expressed and purified from insect cells coinfected with baculoviruses, encoding the individual subunits His-p140, p40, p38, p37, and p36, which were a generous gift from J. Hurwitz, Memorial Sloan-Kettering Cancer Center, New York, N.Y. Human PCNA was expressed in Escherichia coli strain BL21(DE3)(pT7/PCNA), kindly provided by B. Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y., and purified as described by Fien and Stillman (26).
The open reading frames of the putative four subunits of human DNA Pol δ (31, 35, 37, 47), p125, p66, p50, and p12, were amplified by PCR from 50 ng of HeLa cDNA using Klentaq polymerase mix (Clontech, Palo Alto, Calif.) and the following primer sets designed from the relevant sequences deposited in GenBank: p125, 5′-G G G A T G G A T G G C A A G C G G C G G C C A G G-3′ and 5′-C C A T G G G A T G C T T G C A A G G T C A C C A G G-3′; p66, 5′-A C C A T G G C G G A C C A G C T T T A T C T G G-3′ and 5′-C C A A G T C T C T G A T C T A C C A G A G A T G G-3′; p50, 5′-G C C A T G T T T T C T G A G C A G G C T G C C C-3′ and 5′-C A G C C T C C A T C T G G G C C T C T C T G G T C-3′; and p12, 5′-G C C A T G G G C C G G A A G C G G C T C A T C A C T G-3′ and 5′-T C T T A C G T G G T G C C T C A T A G G G G A T A G A G-3′. The cycling parameters were 94°C for 60 s, 36 cycles at 94°C for 30 s and 67°C for 5 to 3 min, and finally 10 min at 67°C. The PCR products were cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.), verified by dideoxy sequencing, excised by EcoRI digestion, and subcloned into the EcoRI site of the baculovirus transfer vector pVL1393 or pAcHLT-A (BD Biosciences, Brøndby, Denmark) for production of untagged or histidine-tagged recombinant products, respectively. Recombinant baculoviruses were generated by cotransfection of the appropriate baculovirus transfer vector containing the desired gene and Bsu36I linearized Bakpak6 baculovirus DNA (Clontech) into insect Sf9 cells. The Pol δ holoenzyme was expressed by coinfection of insect High Five cells with recombinant baculoviruses expressing p125, His-tagged p66, p50, and p12 using multiplicities of infection of 10, 8, 4, and 8, respectively. The cells were incubated at 28°C, harvested 40 to 44 h postinfection, washed twice in phosphate-buffered saline (PBS), resuspended in 25 mM HEPES-KOH (pH 7.6)-5 mM KCl-1.5 mM MgCl2, and lysed by Dounce homogenization. After 20 min of incubation, the lysate was adjusted to 450 mM NaCl and further incubated for 30 min. The lysate was cleared by centrifugation for 30 min at 20,000 × g, and the supernatant was loaded onto an Ni2+ metal chelate agarose (Qiagen GmbH, Hilden, Germany) column (0.7 ml of resin per 109 cells), equilibrated in buffer A (25 mM HEPES-KOH [pH 7.6], 0.5 mM MgCl2, 0.5 mM dithiothreitol [DTT], and 10% glycerol) adjusted to 450 mM NaCl. The column was washed with the same buffer and eluted with buffer A adjusted to 100 mM NaCl and 100 mM imidazole. The eluted fractions were flash frozen in liquid N2 and stored at −80°C. Selected fractions were further fractionated by fast protein liquid chromatography using a Superdex 200 HR 10/30 fast protein liquid chromatography gel filtration column (Pharmacia, Uppsala, Sweden) equilibrated in buffer A adjusted to 200 mM NaCl. The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and handled as described above. All procedures were performed on ice or at 4°C in the presence of complete EDTA-free protease inhibitor (Boehringer, Mannheim, Germany). Single-stranded DNA binding protein from E. coli was purchased from Pharmacia.
Helicase assays.
The substrate for the standard helicase assay was prepared essentially as previously described (38). Briefly, a 341-bp HaeIII restriction fragment of M13mp18 replicative-form DNA (Pharmacia) was denatured and annealed to M13mp18(+) single-stranded DNA. The annealed partial duplex was labeled at the 3′ end by incubation with [32P]dCTP and Sequenase. Unincorporated nucleotides and unannealed HaeIII fragment DNA were separated by SizeSep 400 spin column chromatography (Pharmacia). For preparation of the substrate for determination of helicase directionality, the unlabeled HaeIII fragment annealed to the M13 template was digested with ClaI. This restriction enzyme cleaves once within the duplex region linearizing the annealed M13 template, generating two duplex regions 200 and 141 bp long with a 2-nucleotide 5′ overhang. The free 3′ end of each of these duplexes was labeled with [32P]dCTP in the presence of dGTP using Sequenase and purified by spin column chromatography as described above. The two substrates were incubated at 37°C for 30 min in buffer A, adjusted to 150 (for GST-NS1) or 25 (for SV40 T antigen [TAg]) mM NaCl, 4 mM MgCl2, 4 mM ATP, 1 mM DTT, and 50 μg of bovine serum albumin (BSA) per ml in the presence of different amounts of purified GST-NS1 or SV40 large TAg, respectively, as described in the legend to Fig. 2. The samples were analyzed by electrophoresis through 6% acrylamide gels. The gels were fixed in 10% acetic acid, dried, and exposed for autoradiography.
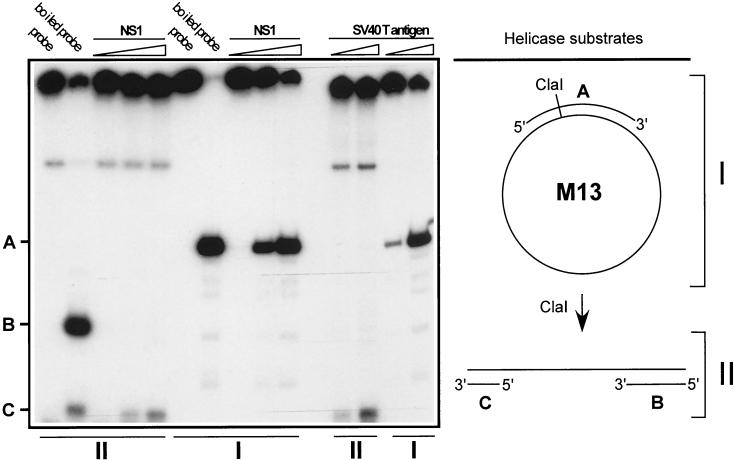
FIG. 2.
Determination of NS1 helicase directionality. The preparation and 3′-end labeling of helicase substrates were done as described in Materials and Methods. Lanes I represent the circular helicase substrate before ClaI digestion, diagrammed on the upper right, which serve as a positive control. Lanes II contained the ClaI-digested helicase substrate, diagrammed on the lower right, used to determine directionality. As indicated at the top of the gel, the different helicase substrates were incubated with increasing amounts of GST-NS1 (10, 30, or 90 ng) or purified SV40 large TAg (25 and 75 ng). Helicase assays were carried out as described in Materials and Methods. A, 341-base HaeIII fragment derived from double-stranded M13 DNA; B and C, linear substrate with 143- (C) and 202-bp (B) duplexes connected by a stretch of single-stranded M13 DNA in which each of the 3′ ends is labeled (directional substrate).
Nicking and unwinding assays.
Nicking assays were performed essentially as described previously (13). Briefly, purified recombinant PIF heterodimer was incubated for 20 to 30 min at 37°C in the presence of ATP, various amounts of GST-NS1 as specified in the figure legends, and a 32P-labeled DNA fragment containing the minimal left-end origin region of MVM. For nicking coupled with unwinding of the template, increasing amounts of RPA or E. coli single-stranded binding protein (SSB), as indicated in each figure legend, were titrated into the nicking reactions. The reactions were terminated by adding an equal volume of 25 mM Tris-HCl (pH 8.0), 5 mM EDTA, 100 mM NaCl, 1% SDS, and 10% glycerol and incubating the mixture at 60°C for 15 min. The samples were analyzed by electrophoresis through 6% acrylamide gels equilibrated in 0.2% SDS. The gels were fixed in 10% acetic acid, dried, and exposed for autoradiography.
KMnO4 assays.
KMnO4 assay substrates were separately labeled with 32P at the 3′ end of each strand as previously described (11). The assays were performed in the presence of purified GST-NS1, recombinant PIF, and ATP, as indicated in the figure legends. After 7 min of incubation, the reaction mixtures were adjusted to 3 mM KMnO4 and incubated for a further 3 min. The reactions were terminated by addition of two volumes of ice-cold ethanol, and the precipitates were collected by centrifugation. The samples were resuspended in 100 μl of 1 M piperidine and incubated for 30 min at 90°C. The piperidine was then removed by multiple rounds of lyophilization. For mapping of the NS1 nick site, nicking assay substrates labeled with 32P on one strand at the 5′ end were incubated in the presence of purified GST-NS1, recombinant PIF, and ATP as indicated in the figure legends. After 30 min of incubation, the samples were incubated with proteinase K at 50°C overnight, phenol-chloroform extracted, and ethanol precipitated. For alignment of residues oxidized by KMnO4 and the nick site, the DNA probes used for the assays were also chemically cleaved at G and GA residues according to the method of Maxam and Gilbert (40). Samples were analyzed by electrophoresis through 6% denaturing polyacrylamide gels, fixed, dried, and exposed for autoradiography.
Replication assays.
For M13 primer extension assays, 50 ng of singly primed M13mp18 single-stranded DNA was incubated for 1 h at 37°C with different combinations of RPA, SSB, PCNA, RFC, and Pol δ holoenzyme, as detailed in the figure legends, in the replication buffer described below. The reactions were terminated by addition of 100 μl of 10 mM Tris-HCl-5 mM EDTA-0.5% SDS and incubated for 1 h with 50 μg of proteinase K. Unincorporated nucleotides in the samples were removed by G-50 spin column chromatography, and the product was extracted with phenol-chloroform before electrophoresis through 1.5% alkaline agarose gels. Replication assays were carried out in the presence of optimized amounts of recombinant PIF, His-NS1, RPA, SSB, PCNA, RFC, and Pol δ holoenzyme, as indicated in the figure legends. The assay mixtures contained 25 mM HEPES-KOH (pH 7.8), 25 mM NaCl, 7 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, 4 mM ATP, 0.1 mM dCTP, 0.1 mM dGTP, 0.1 mM dTTP, 0.04 mM dATP, 5 μCi of [32P]dATP, 40 mM creatine phosphate, 10 μg of phosphocreatine kinase, 10% glycerol, and 100 ng of the appropriate DNA template in a total volume of 30 μl. After incubation at 37°C for 2 h, the reactions were terminated by addition of 100 μl of 10 mM Tris-HCl-5 mM EDTA-0.5% SDS and incubated at 50°C for 1 h with 50 μg of proteinase K. Unincorporated nucleotides in the samples were removed by G-50 spin column chromatography, and the product was extracted with phenol-chloroform. The DNA was then linearized by digestion with HindIII prior to electrophoresis on 1% agarose gels. The gels for replication assays and M13 primer extension assays were fixed in 10% trichloroacetic acid, dried, and exposed for autoradiography. Incorporation of [α-32P]dAMP was monitored by spotting 10% of the reaction mixtures on to Whatman DE81 paper followed by three successive washes with 0.5 M Na2HPO4, one wash with H2O, and one wash with ethanol, after which incorporation was quantitated by liquid scintillation counting.
Protein-protein interaction assays.
Protein-protein interactions were analyzed by an enzyme-linked immunosorbent assay (ELISA). Maxisorp 96-well plates (Merck Eurolab, Albertslund, Denmark) were incubated overnight at 4°C with 1.5 μg of purified protein in PBS as indicated in the figure legends, washed twice in PBS, blocked overnight in PBS containing 3% BSA, and washed twice in PBS. The coated plates were then incubated with increasing amounts of purified His-NS1 or GST-NS1 fusion protein for 1 h at 4°C in PBS containing 0.1% BSA and, when appropriate, in the presence of 0.5 mM ATP and 0.5 mM MgCl2, followed by four successive washes with PBS. Bound NS1 was detected by incubation for 45 min with a 1:1,000 dilution of a rabbit polyclonal antiserum directed against the carboxyl terminus of NS1. After three washes with PBS, bound antibody was detected by incubation for 45 min with a horseradish peroxidase-coupled goat anti-rabbit immunoglobulin G conjugate (Dako, Glostrup, Denmark) diluted in PBS-0.1% BSA. The wells were washed six times with PBS and developed using o-phenylenediamine and hydrogen peroxide according to the manufacturer's instructions. Optical densities were read at 490 nm.
RESULTS
NS1 helicase activity has 3′-5′ polarity.
The intrinsic helicase activity of NS1 is believed to play a crucial role in virus replication (42, 43). Thus, examination of the directionality of the NS1 helicase action would provide important information about the mechanism of parvovirus replication fork translocation. For the measurement of helicase activity, partly duplex DNA molecules were constructed by annealing a 341-base HaeIII fragment (Fig. 2, band A) derived from double-stranded M13 DNA to plus-sense single-stranded circular M13 DNA (standard substrate). To determine the directionality of the NS1 helicase-mediated translocation, the partial duplex was further digested with ClaI and radiolabeled as described in Materials and Methods. This creates a linear substrate with 143- and 202-bp duplexes (bands C and B, respectively) connected by a stretch of single-stranded M13 DNA in which each of the 3′ends is labeled (directional substrate). The two substrates were incubated in the presence of ATP and increasing amounts of purified GST-NS1 or SV40 TAg, as shown in Fig. 2. SV40 TAg has previously been shown to possess 3′-5′ helicase activity (28) and was included as a positive control. Both NS1 and TAg were able to displace the 341-nucleotide strand from the standard template, indicating that the preparations possessed helicase activity capable of extensive unwinding. Both NS1 and TAg displaced the 143-nucleotide strand from the directional substrate, while no release of the 202-nucleotide fragment was observed. This indicates that, like TAg, NS1 does not attack blunt-ended DNA but binds to recessed 5′ ends and translocates in a 3′-5′ direction on the longer strand, as would be necessary to promote strand displacement synthesis from the 3′ OH of a nick.
NS1 helicase does not displace the nicked strand after covalent attachment.
We have previously shown that the cellular heterodimeric, site-specific binding factor PIF is required for NS1 nicking and covalent attachment to the minimal origin. While PIF and NS1 bind the TC (oriLTC) and GAA (oriLGAA) arms equally well by themselves, PIF and NS1 bind cooperatively only to oriLTC (13). On oriLTC, they form a high-affinity ternary complex that catalyzes NS1-mediated site-specific nicking and leads to replication initiation at this origin. Since covalent attachment of NS1 to the origin is a prerequisite for initiating DNA synthesis, and thereby replication fork formation, the nicking reaction was analyzed in more detail in order to determine the extent of origin unwinding mediated by the NS1 helicase after nicking. For this, a DNA fragment containing oriLTC was labeled at the 3′ ends of both strands and then incubated in the presence of NS1, PIF, and ATP. The reaction products were adjusted to 0.5% SDS and either heated at 60°C to abolish noncovalent protein-DNA interactions or boiled to separate the strands prior to analysis on SDS-polyacrylamide gels. In this assay, retarded species represent double- or single-stranded origin DNA covalently attached to NS1, depending on the level of unwinding. As shown in Fig. 3A, the origin is not nicked in the absence of PIF. However, when NS1, PIF, and ATP are present, a single slowly migrating band (band A) is observed, indicating covalent attachment of NS1 to the origin. To examine whether the origin was present in a double- or single-stranded form, the products of the nicking reaction were boiled in SDS before being loaded on the gel. This changed the mobility of the nicked origin to a faster-migrating species (band B) representing NS1 covalently bound to the lower labeled strand, as depicted in the model of the nicking reaction shown in Fig. 3. Species B has been characterized previously (10) as the NS1-linked single strand, since it shows protease-sensitive mobility and is specifically immunoprecipitated by anti-NS1 antibodies. Thus, despite the helicase activity of NS1, its covalent attachment to the origin does not cause unwinding to a level that leads to dissociation of the NS1-attached DNA strand from the origin duplex.
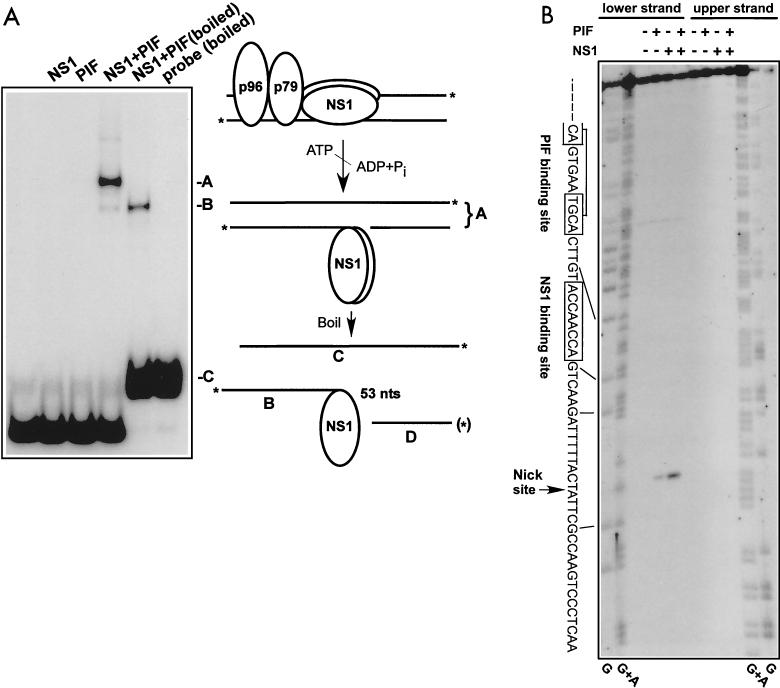
FIG. 3.
Characterization of the nick site in the minimal left-end origin. (A) A DNA fragment containing the oriLTC sequence was 32P labeled at its 3′ ends as described in Materials and Methods; incubated in the absence or presence of NS1 (100 ng), PIF (25 ng), and ATP (3 mM); and analyzed on a neutral SDS-polyacrylamide gel. Only covalent NS1-DNA complexes are significantly retarded in this gel system. For lane NS1+PIF (boiled), the nicking reaction mixture was denatured by boiling immediately before electrophoresis. A schematic model of the nicking reaction is depicted to the right of the gel autoradiograph. The asterisks indicate the positions of the 32P label at the 3′ end of each strand of the origin DNA. The letters A, B, and C correlate the DNA or DNA-protein structures produced in the reaction with bands on the gel. The fragment D labeled with an asterisk in parentheses indicates the 5′-end-labeled fragment analyzed in panel B. (B) DNA fragments containing the oriLTC sequence, separately labeled at the 5′ end of either the upper or lower strand (as depicted in panel A), were incubated with 3 mM ATP in the presence (+) or absence (−) of NS1 (100 ng) and PIF (25 ng) as indicated at the top of the gel. The samples were digested with proteinase K before analysis on a 6% denaturing polyacrylamide gel. Lanes G and G+A contain the products of G- and GA-specific chemical sequencing reactions of each labeled substrate and were used for aligning the position of the nick site. The sequence of the lower strand, depicted in panel A, is shown on the left side of the gel, and the lines indicate the position of this sequence in relation to the chemical sequencing reaction. An arrow indicates the putative nick site, representing the position of migration of fragment D in panel A. The boxed sequences show the positions of the specific NS1 and PIF binding sites.
Mapping the position of the nick in oriLTC.
The in vitro recapitulation of the nicking reaction afforded an opportunity to map the exact position of the nick within the oriLTC sequence. This has been difficult to measure with 3′-end-labeled probes, because proteinase K does not completely remove the NS1 polypeptide, and the residual oligopeptide decreases the mobility of its attached strand by a significant amount that is difficult to interpret in terms of nucleotide equivalents. To get around this problem, we separately labeled each 5′ end of the origin substrate through polynucleotide kinase and used these substrates in the nicking reaction. This approach has previously been frustrated by the presence of phosphatases in the replication extracts and PIF preparations, purified from HeLa cells, that we used in earlier assays. The present use of a system comprising only purified, overexpressed, recombinant proteins did not exhibit this drawback, and a clean result was obtained, as shown in Fig. 3B. Following nicking of a substrate labeled at the 5′ end of the lower strand (as depicted in Fig. 1 and 3A), a single product, equivalent to fragment D in Fig. 3A, can be seen on the sequencing gel, while none is produced from a similarly labeled upper strand. The appearance of this band is dependent on NS1 and PIF and maps the nick to a position 2 nucleotides more 3′ on the lower strand than previous estimates have placed it.
Nicking and covalent attachment of NS1 to the origin exposes a short stretch of single-stranded DNA juxtaposed to the nick site.
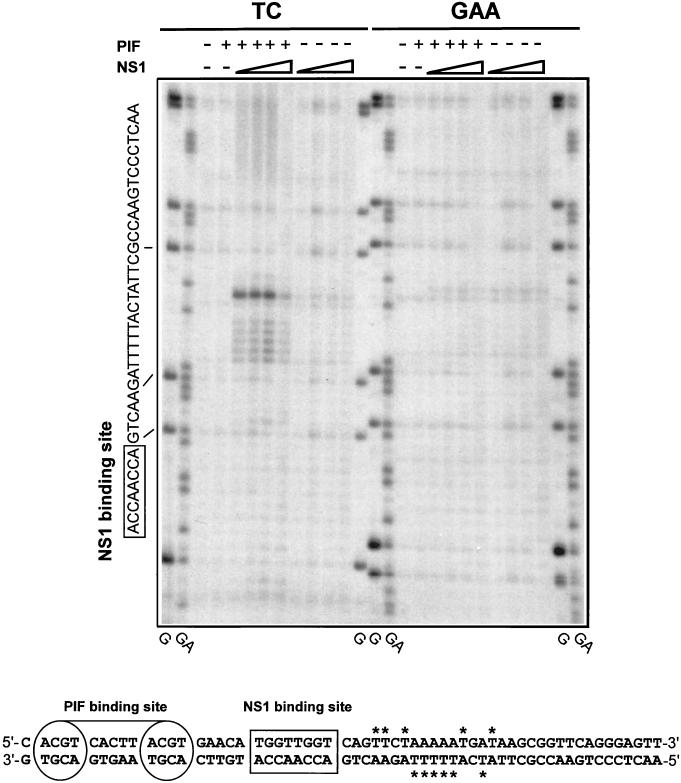
To explore the level of local unwinding and distortion caused by the nicking reaction, the presence of unpaired thymidines within the origin was probed by their susceptibility to oxidation by KMnO4 and subsequent alkaline hydrolysis. DNA fragments containing either oriLTC or oriLGAA, 3′-end labeled on the nicked strand, were incubated in the presence of PIF, NS1, and ATP as indicated in Fig. 4. It should be noted that the nicked templates were not digested with proteinase K, and thus, no strong bands should have been observed at the position of the nicked DNA. Consequently, only the DNA fragments that had been cleaved from attached NS1 by the oxidation reaction were resolved in the gel. The reaction products were analyzed on a denaturing polyacrylamide gel and aligned to G and GA sequencing reactions derived from the labeled substrate. Strong KMnO4 sensitivity was observed for oriLTC only in the presence of PIF and NS1. Even at saturating NS1 concentrations, however, the oxidized residues were limited to a maximum stretch of 14 nucleotides located between the nick site and the NS1 binding site. The most strongly oxidized residue observed was the thymidine immediately juxtaposed to the nick site identified in Fig. 3B, further supporting the revised assignment of the position of the nick. Only background oxidation was detected for oriLGAA, indicating that this sequence does not undergo unwinding or that nicking is necessary for exposure of single-stranded DNA. Similar analysis of the unnicked strand in oriLTC pinpointed the same region as sensitive to KMnO4 oxidation (data not shown). Oxidation-sensitive residues for both strands are indicated and summarized in Fig. 4. To investigate whether unwinding preceding the nicking reaction generated the exposed thymidine residues in oriLTC, nicking was abolished (data not shown) by inverting the nick site sequence by site-directed mutagenesis, generating oriLTC-INV, in which the nick site sequence was changed from TGATAAG to ACTATTC. No KMnO4 oxidation above background was detected on this modified template (data not shown), suggesting that nicking and/or covalent attachment of NS1 is necessary for exposure of single-stranded DNA in the origin and that any unwinding occurring before nicking is limited, or at least undetectable in the assay. In summary, while the ternary complex formed by NS1 and PIF on oriLTC catalyzes nicking and covalent attachment of NS1, subsequent unwinding is limited to a short sequence located between the nick site and the NS1 binding motif, despite the fact that NS1 itself is a potent helicase.
FIG. 4.
PIF and NS1 nicking of the minimal left-end origin causes local unwinding. DNA fragments containing either the oriLTC or oriLGAA sequence were 32P labeled at the 3′ end of the lower strand (Fig. 1) and incubated with 3 mM ATP in the presence (+) or absence (−) of NS1 (increasing amounts of purified GST-tagged NS1 [25, 50, 100, and 225 ng]) and PIF (50 ng of purified PIF p79-p96 complex) as indicated at the top of the gel. The samples were exposed to KMnO4 and subjected to alkaline hydrolysis before analysis on a 6% denaturing sequencing polyacrylamide gel. TC and GAA indicate the form of oriL that was used as a substrate in the left and right sets of KMnO4 assays, respectively. Lanes G and GA contain the products of G- and GA-specific chemical sequencing reactions of each labeled substrate and were used to align the positions of KMnO4-sensitive residues. The sequence of the relevant strand of the origin is shown on the left, and the lines indicate the alignment of this sequence to the chemical sequencing reaction. The boxed sequence shows the position of the specific NS1 binding site. At the bottom is a summary of the positions of KMnO4-sensitive thymidine residues (labeled with asterisks) detected in both strands of the origin.
Human RPA catalyzes unwinding of the nicked NS1-origin complex.
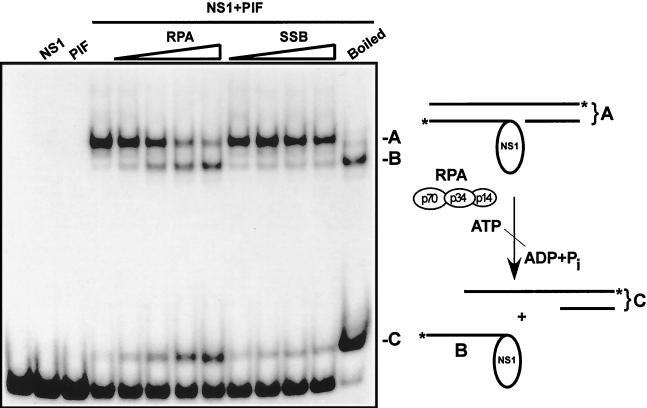
The limited unwinding of oriLTC in the presence of NS1 and PIF, described above, suggests that other cellular factors were involved in the more extensive unwinding during subsequent replication. Previous experiments have shown that RPA is necessary for in vitro replication initiated from MVM replication origins (10), making it a prime candidate for catalysis of origin unwinding. To test this possibility, human heterotrimeric RPA was expressed in insect cells using recombinant baculoviruses, purified, and added into the nicking reaction. A DNA fragment containing oriLTC was 3′-end labeled on both strands and incubated in the presence of NS1, PIF, ATP, and increasing amounts of purified RPA or E. coli SSB. Reaction products in 0.5% SDS were heated at 60°C or boiled and analyzed on SDS-polyacrylamide gels. As shown in Fig. 5, nicking of the origin is completely dependent on the presence of PIF. When NS1, PIF, and ATP are all present, a new slowly migrating band is observed, demonstrating covalent attachment of NS1 to the origin. The inclusion of increasing amounts of RPA in the reaction mixture progressively converted the nicked origin to a faster-migrating species with a mobility similar to that in the boiled nicking reaction, representing NS1 covalently bound to the labeled, nicked strand of the template. Concomitantly, a band migrating slightly more slowly than the double-stranded origin template appeared, representing the complementary part of the unwound origin. A model of this unwinding reaction is depicted in Fig. 5. Replacement of RPA with E. coli SSB in the nicking reaction did not change the mobility of the nicked template and therefore did not appear to support origin unwinding. Similarly, no unwinding of the template was detected during assay of the inactive oriLTC-INV template, described above, in the presence of NS1, PIF, and RPA (data not shown). Together, these data indicate that RPA specifically catalyzes extensive unwinding of the nicked strand that has NS1 covalently attached.
FIG. 5.
RPA catalyzes unwinding of the nicked minimal left-end origin. A DNA fragment containing the oriLTC sequence, 32P labeled at its 3′ ends, was incubated with 3 mM ATP in the presence or absence of NS1 (100 ng) and PIF (25 ng) and increasing amounts of either recombinant RPA (0.125, 0.25, 0.5, and 1 μg) or E. coli SSB (0.125, 0.25, 0.5, and 1 μg), as indicated at the top of the gel. For lane NS1+PIF (boiled), the nicking reaction mixture was boiled immediately before electrophoresis in an SDS-polyacrylamide gel. Note that only covalent NS1-DNA complexes are significantly retarded in this assay. A schematic model of the nicking-coupled unwinding reaction is depicted at the right of the gel autoradiograph. The asterisks indicate the position of the 32P label on each strand of the origin DNA.
Furthermore, we also observed that the short 5′-end-labeled fragment of the lower strand (fragment D in Fig. 3A) produced during the nicking reaction remains annealed to the intact upper strand during the reaction in which the NS1-attached strand is displaced (data not shown). That this hybrid is stable in the presence of both RPA and excess NS1 confirms the directionality of the NS1 helicase. It also indicates that the 3′ hydroxyl-bearing nucleotide produced by the nick remains base paired during the subsequent unwinding phase, available to serve as a primer for ensuing DNA synthesis.
Recombinant RFC, PCNA, Pol δ, and RPA or SSB catalyze processive replication on an M13 substrate.
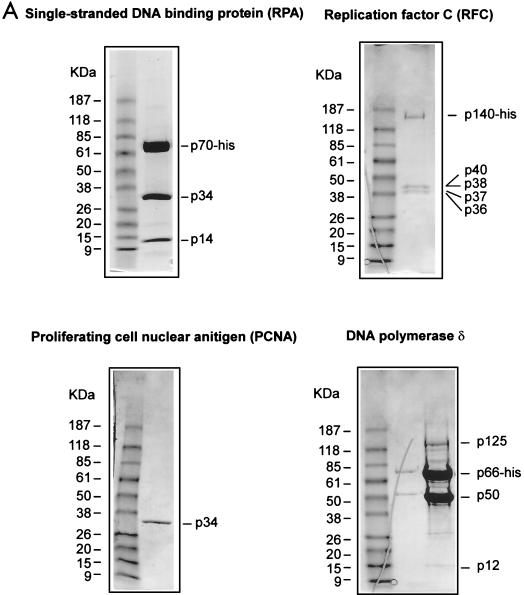
Parvoviruses employ a self-primed, continuous, unidirectional leading-strand synthesis mechanism for replication, potentially bypassing the necessity for lagging-strand synthesis. This probably reduces the requirement for DNA polymerases and accessory factors to only those components involved in leading-strand synthesis (21). Nicking and covalent attachment of NS1 to MVM origins liberates the 3′ hydroxyl groups necessary for priming DNA replication. PCNA and RPA have previously been shown to be required for in vitro MVM DNA synthesis (10), supporting a leading-strand replication mechanism. The classical components of the leading-strand synthesis machinery include PCNA, a ring-shaped homotrimeric protein forming a sliding clamp at the primer-template junction, Pol δ, RPA, and the heteropentameric multifunctional protein RFC, which loads PCNA and Pol δ onto the template. We propose that these factors, in concert with NS1, form the parvoviral replication fork. In order to examine whether the unwinding of oriLTC mediated by NS1 and RPA would allow processive unidirectional leading-strand synthesis, the leading-strand synthesis components were expressed as recombinant proteins in either insect cells or E. coli and purified as shown in Fig. 6A. SDS-PAGE analysis of the purified RPA and RFC preparations showed the expected results, comprising trimeric and pentameric complexes, respectively, each with an equimolar stoichiometry of the individual subunits, in agreement with previously reported data (8, 30). In contrast, the p50 and p66 subunits of Pol δ were clearly overrepresented in the purified polymerase complex compared to the p12 and p125 subunits. While characterization of the recombinant Pol δ complex was not our aim in these studies, we examined the effects of omitting each of the four subunits in turn (data not shown). The p125 subunit is absolutely required for activity, ruling out significant contamination with insect cell- or baculovirus-derived DNA polymerase subunits in our Pol δ preparations. We were not able to show a requirement for the p12 subunit in the M13-based leading-strand synthesis assay described here, which is surprising, since Podust and colleagues (45) have reported that addition of this newly described subunit enhances synthesis on a poly(dA)-oligo(dT) template primer by 15-fold. The low representation of both p12 and p125 may mean that the majority of the functional, p125-containing complexes in our preparations do not also contain the p12 subunit.
FIG. 6.
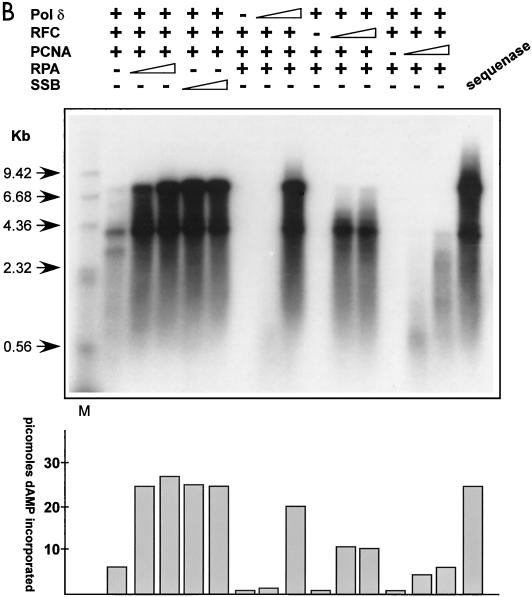
Recombinant RPA/SSB, RFC, PCNA, and Pol δ replicate primed M13 DNA. (A) SDS-PAGE analyses of the purified human recombinant RPA, RFC, PCNA, and Pol δ preparations used in replication and protein-protein interaction assays (see Materials and Methods for details). The positions of the relevant purified proteins or subunits are indicated on the right of each gel, and the positions of a standard set of molecular mass markers run in the first lane of each gel are shown on the left. The gels were stained with Coomassie brilliant blue. (B) Autoradiograph of an M13 singly primed extension assay analyzed by alkaline agarose gel electrophoresis. Singly primed M13 DNA (7.3 kb) was incubated with deoxynucleoside triphosphates, including [32P]dATP and ATP, in the presence (+) or absence (−) of titrated amounts of purified recombinant RFC, PCNA, Pol δ, and RPA or SSB. For reactions including RPA or E. coli SSB, the amounts added were 0.5 or 1.5 μg, respectively, and elsewhere + represents addition of 1.5 μg of either protein. RFC titrations were 5 and 25 ng, and elsewhere + represents 125 ng of protein added. PCNA titrations contained 10 or 25 ng, and elsewhere + indicates 100 ng of protein added. For Pol δ titrations, 5 or 25 ng was added to the assay, and elsewhere + represents 125 ng of protein added. The first lane contains a 32P-labeled, HindIII-digested λ phage DNA marker (M), while the arrows on the left indicate the sizes of individual fragments. The amount of dAMP (in picomoles) incorporated into each replication product is indicated in the histogram at the bottom.
Initially, we characterized the abilities of recombinant Pol δ, RFC, PCNA, and RPA to catalyze processive leading-strand synthesis, without the requirement for DNA unwinding, by incubating titrated amounts of the components with singly primed M13 single-stranded DNA, ATP, and deoxynucleotides. The reaction products were analyzed on alkaline agarose gels, as shown in Fig. 6B, to allow estimation of the sizes of the newly synthesized DNA strands. The extent of replication occurring in each reaction was quantitated by measuring incorporation of [32P]dAMP, using the DE81 absorption assay described in Materials and Methods. As shown in Fig. 6B, the four recombinant replication factors quantitatively and efficiently catalyzed processive leading-strand synthesis, making full-length M13 DNA (7.3 kb) comparable to that synthesized by Sequenase. Efficient DNA elongation was dependent on the presence of the predicted replication factors. If any one of the recombinant components was omitted from the reaction, only low levels of DNA synthesis were observed. Importantly for the studies of RPA-assisted, NS1-mediated unwinding described below, no strand displacement activity was observed when the extended DNA strand reached the primed site in this primed M13 DNA assay. Significantly, E. coli SSB completely substituted for RPA in this chain elongation assay, consistent with previous reports by others of the reconstitution of leading-strand synthesis by purified cellular factors (39), indicating the correct functionality of our recombinant products.
Recombinant NS1, PIF, RPA, RFC, PCNA, and Pol δ catalyze processive replication initiated from oriLTC.
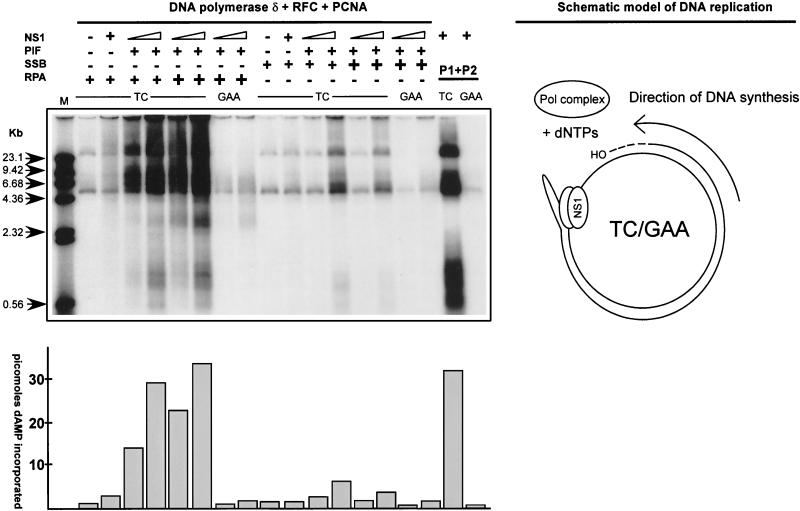
As shown in Fig. 5, RPA, but not SSB, appears to catalyze unwinding on NS1-nicked origin templates, suggesting that RPA may also exclusively permit unidirectional leading-strand synthesis on such a template. To test this possibility, plasmids pL1-2 TC, containing the active left-end origin oriLTC, and pL1-2 GAA, which contains the inactive oriLGAA and served as a negative control, were used as templates for reconstitution of replication. The plasmids were incubated with recombinant Pol δ, RFC, and PCNA, using either RPA or SSB as a single-stranded-DNA binding protein, and tested for catalysis of leading-strand synthesis controlled by PIF and NS1 in the presence of ATP and deoxynucleotides. Again, replication occurring in each reaction was quantitated by measuring total incorporation of [32P]dAMP, and reaction products were analyzed by agarose gel electrophoresis following linearization with the restriction endonuclease HindIII, which cuts once in the template plasmid. As shown in Fig. 7, only minimal replication occurred if NS1 and/or PIF was omitted from the reaction, confirming that nicking and covalent attachment of NS1 control DNA synthesis in this system. The presence of PIF, NS1, RPA, Pol δ, and accessory proteins reconstituted replication and supported abundant DNA synthesis initiated from the active minimal origin, while only low levels of replication were observed for the inactive origin. Interestingly, the deproteinized products predominantly migrate as species much larger than the template, despite HindIII digestion. This would not be the case if the displaced strand were being converted to duplex DNA rather than remaining a continuous single strand, presumably complexed with RPA. Replacement of RPA with SSB reduced replication by at least sevenfold, indicating that NS1 and SSB do not mediate fork progression at a level allowing efficient processive strand elongation. These results indicate the probability that specific NS1-RPA interactions are necessary for fork progression and the catalysis of processive leading-strand DNA synthesis.
FIG. 7.
Reconstitution of replication initiated from the minimal left-end origin. Replication reaction mixtures were assembled using combinations of purified recombinant His-tagged NS1, PIF p79-p96 complex, RPA or E. coli SSB, RFC, PCNA Pol δ, or previously described phosphocellulose fractions of 293 cell extracts as positive controls (10) as indicated above the gel. Plasmid pL1-2 TC containing the minimal active left-end origin was the replication-positive template, and pL1-2 GAA was used as a negative control. The different purified proteins were titrated to achieve optimal replication. For PIF, RFC, PCNA, and Pol δ, constant amounts of 50, 125, 100, and 100 ng, respectively, were used in the assay. The increasing amounts of NS1, as indicated at the top of the gel, were 90 and 270 ng. RPA or E. coli SSB was added (+) in 0.5- or 1.5-μg amounts. Replicated DNA was digested sequentially with proteinase K and HindIII as described in Materials and Methods and then analyzed by agarose gel electrophoresis, along with the 32P-labeled λ phage DNA markers described in the legend to Fig. 6B. The amount of dAMP (in picomoles) incorporated into each replication product is indicated in the histogram at the bottom of the autoradiograph. A schematic model of the replication reaction is at the right.
NS1 binds specifically to RPA.
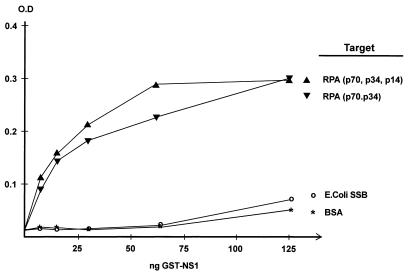
The differential specificities of RPA and SSB in origin unwinding and catalysis of replication indicate that direct molecular interactions between RPA and NS1 might be involved in coordination of fork progression. To address this question, potential NS1-RPA and NS1-SSB interactions were explored using an ELISA-based protein-protein interaction assay. Purified recombinant RPA or SSB was immobilized by coating wells in ELISA plates. Following blocking, the coated wells were incubated with increasing amounts of purified GST-NS1 in the presence of ATP. After the wells were washed, bound GST-NS1 was detected using an antibody directed against the carboxyl terminus of NS1 and a secondary horseradish peroxidase-coupled antibody. As shown in Fig. 8, the binding of GST-NS1 to RPA increased in a concentration-dependent manner and showed saturability at the highest NS1 concentrations. In contrast, no binding to SSB or BSA (the negative control) was detected. Similar results (data not shown) were obtained using His-tagged NS1, ruling out any contribution to RPA binding by the GST tag on the NS1. Moreover, purified GST protein did not bind RPA (data not shown). Thus, NS1 efficiently bound each of the baculovirus-expressed complexes, indicating that the small subunit of RPA is not involved in specific NS1 binding.
FIG. 8.
Binding of NS1 to purified RPA. Purified RPA complex comprising three (p70, p34, p14) or two (p70, p34) subunits or E. coli SSB was immobilized on ELISA wells and incubated with increasing amounts of purified GST-NS1. The different target proteins are indicated at the right. In each ELISA, bound NS1 was detected by a rabbit polyclonal antiserum directed against its carboxyl terminus, followed by horseradish peroxidase-coupled anti-rabbit immunoglobulin G antibody and a chromogenic substrate. ELISA wells coated with BSA served as negative controls. Inclusion of DNase I or micrococcal nuclease in these assays did not quantitatively or qualitatively affect the observed interactions. O.D., optical density.
DISCUSSION
The formation of eukaryotic or bacterial replication forks is a multistep process involving sequence-specific recognition of the origin, establishment of an initiator-protein complex, unwinding of the origin followed by priming of DNA synthesis, and chain elongation. A common feature of the replication fork is that DNA is synthesized in a semidiscontinuous manner, where the leading strand is polymerized continuously while the lagging strand is synthesized discontinuously by the joining of Okazaki fragments. The assembly of the replication fork is temporally ordered and orchestrated by specific intermolecular interactions between the replication factors and the origin (22). Parvoviruses employ a unidirectional leading-strand replication strategy, priming DNA synthesis either from the 3′ hydroxyl groups of terminal hairpin structures in the single-stranded viral genome or from 3′ ends generated during nicking of double-stranded viral origins by the virally encoded replicator protein NS1. This lack of a requirement for lagging-strand synthesis predicts that the formation of the parvoviral replisome is less complex than for those of the bidirectional origins of double-stranded DNA viruses or the host cell (21).
Previous studies have shown that the early events in replication initiation from the minimal left-end replication origin of MVM require the cooperative binding of the cellular sequence-specific factor PIF with NS1. PIF appears to stimulate the ATP-dependent assembly of an NS1 oligomer on the origin and might stabilize the local unwinding of the origin mediated by the ATP-driven helicase function of NS1. This limited local unwinding might expose the nick site in the single-stranded form known to be optimal for site-specific nicking and covalent attachment of NS1 (13). Accordingly, origin substrates that have been constructed to present the nick site in a constitutively single-stranded form are nicked by NS1 in a PIF- and ATP-independent manner (42).
In many cases, the binding of viral initiator proteins to the origin causes local unwinding. For example, SV40 TAg is known to cause unwinding of 8 bp within the early palindrome of the SV40 origin (5), while bovine papillomavirus (BPV) E1 distorts a region of up to 50 bp within the papillomavirus origin (27). The unwinding process is often facilitated by regions within the origin having low helical stability and thus having a tendency to unwind when placed under torsional stress (36, 41). In this study, the level of unwinding mediated by NS1 was tested in direct nicking assays in the presence of PIF. Despite the fact that NS1 has potent intrinsic helicase activity, no major unwinding of the whole origin template was observed. To examine the level of local unwinding, the origin was probed with KMnO4, and a distinct stretch of ∼14 bp on both strands, which includes an (A:T)5 tract located between the specific nick-sequence and the NS1 binding site, was found to be distorted. To discriminate between unwinding and nicking, distortion preceding nicking was examined by abolishing nicking through site-directed mutagenesis of the specific nick sequence in the origin. In the mutant, the origin should resemble the SV40 and BPV origins without the modification of the template by nicking. However, no unwinding was detected on such a template, indicating a role for the (A:T)5 tract other than providing helical instability, or alternatively, that the distortions are not detectable in the assay. Thus, unwinding appears to be not only dependent on nicking by NS1 but also limited to a short region, suggesting that covalent attachment of NS1 restricts its ability to further unwind the origin.
In this study, we have shown that NS1 and RPA play key roles in the formation of the parvoviral replication fork. While NS1 nicking and covalent attachment to the origin only causes limited local unwinding of the origin, NS1 and RPA physically interact and coordinate extensive unwinding. The level of fork migration mediated by NS1 and RPA allows further assembly of replication proteins and promotes processive leading-strand elongation catalyzed by Pol δ and accessory factors, indicating that these components could constitute a complete parvoviral replisome.
Interestingly, RPA binds DNA 10 to 20 nucleotides long, with a preference for short polypyrimidine sequences (32, 33), just as observed for the nicked origin. This suggests that the polypyrimidine tract provides an entry site for RPA and thus affords an obvious role for RPA in origin unwinding. Accordingly, the inclusion of RPA in the nicking reaction led to extensive unwinding, again with the prerequisite for covalent NS1 attachment to the origin. Our findings indicate that while NS1 may expose enough single-stranded DNA template for RPA to bind, it probably recruits RPA for this binding through direct protein-protein interaction, since the single-stranded DNA binding activity of E. coli SSB could not substitute for RPA in this unwinding reaction. Whether this specificity could be explained by an essential role for direct NS1-RPA interaction in recruitment alone or also in the unwinding reaction itself could not be deduced from the present experimental data. However, NS1-RPA interaction may modulate several functions of both proteins nonexclusively, including stimulation of NS1 activity and coordination of progressive unwinding and stabilization of the DNA template.
In our studies of the interaction between NS1 and RPA, no quantitative differences in binding to either the RPA heterotrimer or a complex comprising its two larger subunits were observed. We have found it possible to express the individual RPA subunits as soluble proteins only in bacteria, in contrast to the fully functional RPA that was expressed in insect cells. Although the level of binding was reduced compared to that of the native complex, preliminary results of interactions with the individual subunits indicate that NS1 binds preferentially to the major subunit of RPA (data not shown). Specific interaction with RPA has been demonstrated for other DNA viral initiator proteins, including SV40 TAg and BPV E1 protein, which both involve the major subunit of RPA (23, 29), indicating that this may be a major theme in viral origin unwinding.
To examine if the extent of unwinding observed allowed concomitant processive replication of the template, we sought to reconstitute leading-strand synthesis controlled by NS1 and RPA in vitro. We have previously reported the isolation of cellular factors necessary for establishment of oriL-dependent leading-strand synthesis, using extracts from human cell lines fractionated on phosphocellulose to reconstitute replication in vitro (10). This approach led us to identify PCNA, RPA, and PIF as necessary for viral DNA replication, in addition to a fraction previously shown to contain polymerases and accessory factors involved in SV40 replication (7). In the present study, we expressed the putative four-subunit DNA Pol δ complex and the clamp loader, RFC, from recombinant baculoviruses. These factors were known to be responsible for leading-strand synthesis in the SV40 system, and we tested them for reconstitution of replication in the MVM oriL-based system.
We chose to adopt a recombinant strategy rather than using factors purified from HeLa cells for several reasons. First, purification of Pol δ is known to be hampered by the loss of subunits during purification, and there is still some controversy about the numbers of subunits in the active complex. Moreover, it has recently been shown that the p66 subunit, which makes direct contact with PCNA and is necessary for processive synthesis, is preferentially lost or degraded during conventional purification (7, 31, 37). The lack of “complete” Pol δ activity may be masked in the SV40 replication system, since nearly full replication can be mediated by α-polymerase alone (25). However, the four-subunit enzyme might play a more crucial role in an exclusively unidirectional leading-strand system like that of MVM, a possibility we are currently exploring. Secondly, the purification of Pol δ and its accessory factors is tedious, and the presence of other polymerases in cell extracts complicates purification. Finally, recombinant expression of Pol δ and RFC, together with that of NS1, PIF, PCNA, and RPA, has made it possible to reconstitute replication based solely on purified recombinant polypeptides, paving the way for reconstitution of full-length genomic replication by the rolling-hairpin mechanism. Such recombinant replication machinery is subject to facile genetic manipulation and thus provides a model system for further studies of cellular or viral factors that regulate viral replication.
Initially, recombinant Pol δ, RFC, PCNA, and RPA were tested for replication activity on singly primed single-stranded DNA to test for processivity while bypassing any requirement for unwinding double-stranded DNA in front of the extending 3′ end. Despite the aberrant subunit composition of the Pol δ preparation, the recombinant factors efficiently catalyzed processive elongation of full-length circular M13 DNA. Moreover, E. coli SSB efficiently substituted for RPA, supporting another established feature of leading-strand DNA synthesis and helping to substantiate the fidelity of the recombinant system (39). When NS1, PIF, recombinant Pol δ, and accessory factors were examined using minimal left-end origin cloned in a plasmid as a template, efficient replication was observed, demonstrating the abilities of RPA and NS1 to mediate unwinding and fork progression. However, in contrast to the M13 replication assay, E. coli SSB did not support replication initiated from the minimal origin, confirming that NS1 and RPA specifically mediate fork progression and helping to rule out simple strand displacement as driving replication. The recombinant factors employed in the replication assay were also found to be the minimal set of proteins necessary for processive replication, suggesting that NS1 itself does not provide any of the functions of, for example, the polymerase accessory factors.
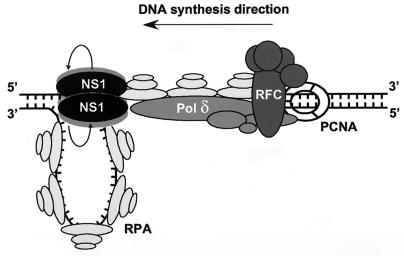
Taken together, the data suggest that parvoviral fork progression is mediated by unwinding coordinated by NS1 and RPA. The covalently attached NS1 may oligomerize with another NS1 molecule(s) and provide the helicase function necessary for unwinding. Further, we suggest that, in concert with RPA, the NS1 helicase rotates around the DNA helix, spooling off the NS1-attached strand and translocating the fork, as diagrammed in Fig. 9. Like SV40 TAg (28), NS1 is a 3′-5′ helicase, suggesting that NS1 forward migration takes place on the strand being replicated. The unwinding process appears to be independent of synthetic extension of the liberated 3′ end, since unwinding is not inhibited by Pol δ and accessory factors, at least in the absence of deoxynucleotides (J. Christensen, unpublished data). In turn, this suggests that DNA polymerization is not dependent on, or coordinated by, the advancing NS1-RPA-driven fork. However, assembly of the complete replisome may also involve interactions with the polymerase complex. Preliminary data indicate that NS1 may directly interact with RFC and assist in loading of RFC to the free 3′ end at the nick and thus indirectly load PCNA and Pol δ (J. Christensen, unpublished). The dynamics of RFC in loading the polymerase complex for extension of free 3′ ends are controversial, but recent data indicate that RFC loads PCNA and Pol δ to the primer site using RPA as a “common touchpoint.” Furthermore, these proteins stay at the template, and the 3′ end is never left free of protein during strand elongation (54), suggesting that an NS1-RFC interaction could also play a role during elongation.
FIG. 9.
Model of a minimal parvovirus replication fork. The model is based partly on the data described in this report and partly on the body of published data describing the interactions of RPA, RFC, PCNA, and Pol δ (6, 23, 24, 45, 46, 54). See the text for additional description.
In summary, NS1 and RPA specifically coordinate the progression of the parvoviral replication fork. Subsequent assembly of recombinant leading-strand DNA synthesis machinery consisting of RFC, PCNA, and DNA Pol δ efficiently catalyzed processive DNA synthesis. These factors will provide an important tool for studying the reconstitution of replication of the full-length parvovirus genome.
Acknowledgments
We thank Susan Cotmore for extensive discussions and for critically reading the manuscript and Ulla Toftegaard for excellent technical assistance.
This work was supported by grants from the Danish Center for Biotechnology (to J.C.) and by U.S. Public Health service grant AI26109 (to P.T.) from the National Institutes of Health.
REFERENCES
- 1.Astell, C. R., M. B. Chow, and D. C. Ward. 1985. Sequence analysis of the termini of virion and replicative forms of minute virus of mice DNA suggests a modified rolling hairpin model for autonomous parvovirus DNA replication. J. Virol. 54:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astell, C. R., Q. Liu, C. E. Harris, J. Brunstein, H. K. Jindal, and P. Tam. 1996. Minute virus of mice cis-acting sequences required for genome replication and the role of the trans-acting viral protein, NS-1. Prog. Nucleic Acid Res. Mol. Biol. 55:245-285. [DOI] [PubMed] [Google Scholar]
- 3.Baldauf, A. Q., K. Willwand, E. Mumtsidu, J. P. Nuesch, and J. Rommelaere. 1997. Specific initiation of replication at the right-end telomere of the closed species of minute virus of mice replicative-form DNA. J. Virol. 71:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashir, T., R. Horlein, J. Rommelaere, and K. Willwand. 2000. Cyclin A activates the DNA polymerase delta-dependent elongation machinery in vitro: a parvovirus DNA replication model. Proc. Natl. Acad. Sci. USA 97:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell J. 60:181-184. [DOI] [PubMed] [Google Scholar]
- 6.Braun, K. A., Y. Lao, Z. He, C. J. Ingles, and M. S. Wold. 1997. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry 36:8443-8454. [DOI] [PubMed] [Google Scholar]
- 7.Brush, G. S., T. J. Kelly, and B. Stillman. 1995. Identification of eukaryotic DNA replication proteins using simian virus 40 in vitro replication system. Methods Enzymol. 262:522-548. [DOI] [PubMed] [Google Scholar]
- 8.Cai, J., F. Uhlmann, E. Gibbs, H. Flores-Rozas, C. G. Lee, B. Phillips, J. Finkelstein, N. Yao, M. O'Donnell, and J. Hurwitz. 1996. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc. Natl. Acad. Sci. USA 93:12896-12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, J., S. F. Cotmore, and P. Tattersall. 1995. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 69:5422-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, J., S. F. Cotmore, and P. Tattersall. 1997. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J. Virol. 71:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, J., S. F. Cotmore, and P. Tattersall. 1997. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J. Virol. 71:5733-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, J., S. F. Cotmore, and P. Tattersall. 1999. Two new members of the emerging KDWK family of combinatorial transcription modulators bind as a heterodimer to flexibly spaced PuCGPy half-sites. Mol. Cell. Biol. 19:7741-7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, J., S. F. Cotmore, and P. Tattersall. 2001. Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur. J. Virol. 75:7009-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costello, F., N. Steenfos, K. T. Jensen, J. Christensen, E. Gottschalck, A. Holm, and B. Aasted. 1999. Epitope mapping of Aleutian mink disease parvovirus virion protein VP1 and 2. Scand. J. Immunol. 49:347-354. [DOI] [PubMed] [Google Scholar]
- 15.Cotmore, S. F., J. Christensen, J. P. Nuesch, and P. Tattersall. 1995. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J. Virol. 69:1652-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., J. P. Nuesch, and P. Tattersall. 1992. In vitro excision and replication of 5′ telomeres of minute virus of mice DNA from cloned palindromic concatemer junctions. Virology 190:365-377. [DOI] [PubMed] [Google Scholar]
- 17.Cotmore, S. F., J. P. Nuesch, and P. Tattersall. 1993. Asymmetric resolution of a parvovirus palindrome in vitro. J. Virol. 67:1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91-174. [DOI] [PubMed] [Google Scholar]
- 19.Cotmore, S. F., and P. Tattersall. 1992. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J. Virol. 66:420-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotmore, S. F., and P. Tattersall. 1994. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 13:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotmore, S. F., and P. Tattersall. 1996. Parvovirus DNA replication, p. 799-813. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.DePamphillis, M. (ed.). 1996. DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 23.Dornreiter, I., L. F. Erdile, I. U. Gilbert, D. von Winkler, T. J. Kelly, and E. Fanning. 1992. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 11:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ducoux, M., S. Urbach, G. Baldacci, U. Hubscher, S. Koundrioukoff, J. Christensen, and P. Hughes. 2001. Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta. J. Biol. Chem. 276:49258-49266. [DOI] [PubMed] [Google Scholar]
- 25.Eki, T., T. Matsumoto, Y. Murakami, and J. Hurwitz. 1992. The replication of DNA containing the simian virus 40 origin by the monopolymerase and dipolymerase systems. J. Biol. Chem. 267:7284-7294. [PubMed] [Google Scholar]
- 26.Fien, K., and B. Stillman. 1992. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell. Biol. 12:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillette, T. G., M. Lusky, and J. A. Borowiec. 1994. Induction of structural changes in the bovine papillomavirus type 1 origin of replication by the viral E1 and E2 proteins. Proc. Natl. Acad. Sci. USA 91:8846-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goetz, G. S., F. B. Dean, J. Hurwitz, and S. W. Matson. 1988. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem. 263:383-392. [PubMed] [Google Scholar]
- 29.Han, Y., Y. M. Loo, K. T. Militello, and T. Melendy. 1999. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 73:4899-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 31.Hughes, P., I. Tratner, M. Ducoux, K. Piard, and G. Baldacci. 1999. Isolation and identification of the third subunit of mammalian DNA polymerase delta by PCNA-affinity chromatography of mouse FM3A cell extracts. Nucleic Acids Res. 27:2108-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, C., B. F. Paulus, and M. S. Wold. 1994. Interactions of human replication protein A with oligonucleotides. Biochem. J. 33:14197-14206. [DOI] [PubMed] [Google Scholar]
- 33.Kim, C., R. O. Snyder, and M. S. Wold. 1992. Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 12:3050-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornberg, A., and T. A. Baker. 1991. DNA replication, 2nd ed. W. H. Freeman, New York, N.Y.
- 35.Lee, M. Y., Y. Q. Jiang, S. J. Zhang, and N. L. Toomey. 1991. Characterization of human DNA polymerase delta and its immunochemical relationships with DNA polymerase alpha and epsilon. J. Biol. Chem. 266:2423-2429. [PubMed] [Google Scholar]
- 36.Lin, S., and D. Kowalski. 1994. DNA helical instability facilitates initiation at the SV40 replication origin. J. Mol. Biol. 235:496-507. [DOI] [PubMed] [Google Scholar]
- 37.Liu, L., J. Mo, E. M. Rodriguez-Belmonte, and M. Y. Lee. 2000. Identification of a fourth subunit of mammalian DNA polymerase delta. J. Biol. Chem. 275:18739-18744. [DOI] [PubMed] [Google Scholar]
- 38.Matson, S. W. 1986. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3′ to 5′ direction. J. Biol. Chem. 261:10169-10175. [PubMed] [Google Scholar]
- 39.Matsumoto, T., T. Eki, and J. Hurwitz. 1990. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc. Natl. Acad. Sci. USA 87:9712-9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 41.Natale, D. A., R. M. Umek, and D. Kowalski. 1993. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 21:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nuesch, J. P., J. Christensen, and J. Rommelaere. 2001. Initiation of minute virus of mice DNA replication is regulated at the level of origin unwinding by atypical protein kinase C phosphorylation of NS1. J. Virol. 75:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nuesch, J. P., S. F. Cotmore, and P. Tattersall. 1992. Expression of functional parvoviral NS1 from recombinant vaccinia virus: effects of mutations in the nucleotide-binding motif. Virology 191:406-416. [DOI] [PubMed] [Google Scholar]
- 44.Nuesch, J. P., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122-135. [DOI] [PubMed] [Google Scholar]
- 45.Podust, V. N., L. S. Chang, R. Ott, G. L. Dianov, and E. Fanning. 2002. Reconstitution of human DNA polymerase δ using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem. 277:3894-3901. [DOI] [PubMed] [Google Scholar]
- 46.Shikata, K., S. Ohta, K. Yamada, C. Obuse, H. Yoshikawa, and T. Tsurimoto. 2001. The human homologue of fission yeast cdc27, p66, is a component of active human DNA polymerase delta. J. Biochem. (Tokyo) 129:699-708. [DOI] [PubMed] [Google Scholar]
- 47.Sun, Y., Y. Jiang, P. Zhang, S. J. Zhang, Y. Zhou, B. Q. Li, N. L. Toomey, and M. Y. Lee. 1997. Expression and characterization of the small subunit of human DNA polymerase delta. J. Biol. Chem. 272:13013-13018. [DOI] [PubMed] [Google Scholar]
- 48.Tattersall, P., and D. C. Ward. 1976. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature 263:106-109. [DOI] [PubMed] [Google Scholar]
- 49.Tsurimoto, T., and B. Stillman. 1991. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 266:1950-1960. [PubMed] [Google Scholar]
- 50.Tsurimoto, T., and B. Stillman. 1991. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J. Biol. Chem. 266:1961-1968. [PubMed] [Google Scholar]
- 51.Waga, S., G. Bauer, and B. Stillman. 1994. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 269:10923-10934. [PubMed] [Google Scholar]
- 52.Waga, S., and B. Stillman. 1994. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369:207-212. [DOI] [PubMed] [Google Scholar]
- 53.Willwand, K., E. Mumtsidu, G. Kuntz-Simon, and J. Rommelaere. 1998. Initiation of DNA replication at palindromic telomeres is mediated by a duplex-to-hairpin transition induced by the minute virus of mice nonstructural protein NS1. J. Biol. Chem. 273:1165-1174. [DOI] [PubMed] [Google Scholar]
- 54.Yuzhakov, A., Z. Kelman, J. Hurwitz, and M. O'Donnell. 1999. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 18:6189-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]