Abstract
The expression of most herpes simplex virus type 1 (HSV-1) immediate-early (IE) and early (E) genes decreases late in productive infection. IE and E promoters contain various binding sites for cellular activators, including sites for Sp1, upstream of the TATA box, while late gene promoters generally lack such sites. To address the possibility that Sp1 function may be altered during the course of infection, the modification state and activity of Sp1 were investigated as a function of infection. Sp1 was quantitatively phosphorylated in HSV-1-infected cells without a significant change in abundance. The kinetics of accumulation of phosphorylated Sp1 immediately preceded the decline in E gene (thymidine kinase gene [tk]) mRNA abundance. Phosphorylation of Sp1 required ICP4; however, the proportion of phosphorylated Sp1 was reduced during infection in the presence of phosphonoacetic acid or in the absence of ICP27. While the DNA binding activity of Sp1 was not greatly affected by phosphorylation, the ability of phosphorylated Sp1 isolated from HSV-infected cells to activate transcription in vitro was decreased. These studies suggest that modification of Sp1 may contribute to the decrease of IE and E gene expression late in infection.
During productive infection by herpes simplex virus type 1 (HSV-1), viral genes from three major kinetic classes are sequentially transcribed by host RNA polymerase II machinery (1, 9, 23, 28). The activated expression of immediate-early (IE) genes, which is required for the subsequent cascade of gene expression, is achieved immediately after infection by the function of the virion transcription activator VP16 (3, 5). IE gene promoters also contain multiple binding sites for the transcription factor Sp1. Following the expression of IE proteins, the expression of early (E) genes begins, and it is maximally induced by the IE transcription activator ICP4 (25, 40, 55). Expression of most IE and E genes is subsequently attenuated, and with viral DNA replication, late (L) genes are expressed (14, 24, 55). Even though both E and L genes are highly activated by ICP4 and transcribed by the same transcription machinery, only the E genes are efficiently shut off later in infection. The mechanisms involved in attenuating IE and E gene expression late in infection remain unclear.
The transcription of each HSV-1 gene is primarily determined by the promoter of each gene. The promoters of each kinetic class have their own characteristic structure, possessing a TATA box as a common element (54, 56). Within IE and E promoters, there are binding sites for viral and/or cellular transcription activators upstream of the TATA box, such as VP16/Oct-1, Sp1, NF-1, etc. (5, 8, 27). Among these binding sites, one of the most frequently found in IE and E promoters is that for Sp1. In contrast, most of the L promoters do not have transcription activator binding sites upstream of the TATA box.
Sp1 (promoter specificity protein 1) is a 105-kDa prototype transcription activator (29, 41). It is a fairly ubiquitous and versatile protein, essential for many different cellular functions, such as cell cycle regulation and chromatin remodeling (32, 52). Sp1 has been implicated in the efficient transcription of many cellular and viral genes (32, 52).
Sp1 binds with high affinity to GC- or GT-rich promoter elements through its C-terminal three zinc finger motifs (Cys-2-His-2) (10, 15, 29). The N-terminal activation domain is glutamine rich and interacts with the Drosophila TAFII110/human TAFII130 (10, 22, 53). Subsequent in vitro studies demonstrated the mechanism of activation of Sp1. The TAFII110 subunit of Drosophila TFIID or its human homologue, TAFII130, interacts directly with Sp1 and serves as an essential cofactor for Sp1 activation (22, 53). More recently, a mediator complex called cofactor required for Sp1 (CRSP) was purified as an additional cofactor for Sp1 activity (45, 46).
Sp1 is subject to two different forms of posttranslational modifications, glycosylation and phosphorylation. Sp1 is present in multiple O-glycosylated forms and is phosphorylated by several different cellular kinases in different biological situations with different functional consequences (4, 52). Both modifications occur mainly on the N-terminal region of Sp1. Changes in O glycosylation have been shown to alter the stability of Sp1 in vivo and its interaction with other factors (21, 44). Phosphorylation has been implicated in changes in DNA binding affinity and transcriptional activation of Sp1 (2, 7, 20, 33, 38, 43).
A variety of cellular proteins including transcription factors are modified during infection during with viruses, providing a mechanism for controlling viral and cellular gene expression. Sp1 has been reported to be modified or changed in abundance in virus-infected cells. Human cytomegalovirus and simian virus 40 infection cause upregulation of Sp1 (47, 58). Human immunodeficiency virus Tat protein was reported to induce phosphorylation of Sp1 by DNA protein kinase, resulting in increased activity (7).
The studies described herein examine the effects of HSV-1 infection on Sp1 and the potential consequences of these effects for viral gene expression. It was found that Sp1 was quantitatively phosphorylated during HSV-1 infection and that the time when Sp1 was phosphorylated coincided with the decrease in E gene expression. Additionally, phosphorylation of Sp1 reduced the ability of Sp1 to activate transcription in vitro. These results suggest that phosphorylation of Sp1 during HSV-1 infection may contribute to the reduced transcription of IE and E genes late after infection.
MATERIALS AND METHODS
Viruses and cells.
Wild-type (wt) virus (KOS) was propagated and subjected to titer determination on Vero cells. IE deletion mutants d120, d106, d109, and 5dl1.2 were maintained and subjected to titer determination on appropriate complementing cell lines as previously described (12, 34, 48, 49).
Western blot analysis.
Vero cells were infected with KOS or an IE deletion mutant at a multiplicity of infection of 10. At different times postinfection (p.i.), cells were lysed directly into sodium dodecyl sulfate (SDS) sample buffer and then resolved on an SDS-8% polyacrylamide gel. The proteins were transferred to a polyvinylidene difluoride membrane. Nonspecific sites on the membrane were blocked with 5% milk in TBST (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Tween 20) and incubated with the Sp1 primary antibody (750:1 dilution in 5% milk; Santa Cruz). After several washing steps, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody. Detection was done using a chemiluminescence agent (Roche) and exposure to Hyperfilm (Amersham). For the infection in the presence of phosphonoacetic acid (PAA) (Lancaster Synthesis, Eastgate, England), medium was supplemented with 400 μg of PAA/ml during adsorption and incubation.
Electrophoretic mobility shift assay.
An Sp1 binding site oligonucleotide (5′-ATTGCATCGGGGCGGGGCGAGC-3′) (Promega) was end labeled with 32P using polynucleotide kinase (Roche). The labeled probe was purified and incubated with the cell lysate prepared from KOS-infected Vero cells for 30 min at 4°C. Whole-cell lysate was prepared as described previously (25). Binding reactions were carried out in buffer containing 50 mM HEPES (pH 7.6), 200 mM NaCl, 0.05 mM EDTA, 0.25% Nonidet P-40, 0.5 mM dithiothreitol (DTT), and 5% glycerol. Each binding reaction contained 2 μl (∼10 μg) of whole-cell lysate, 150 ng of poly(dI-dC), and 3.5 fM (∼20,000 cpm) labeled probe. Binding reaction samples were resolved on 5% nondenaturing polyacrylamide gels, which were then dried and exposed to film for autoradiography. For supershift reactions, 4 μl of Sp1 antibody was added after all reaction components described above were mixed. As a control, 1 μl of purified baculovirus-produced Sp1 (Promega) was included in the reaction instead of whole-cell lysate.
Purification of Sp1.
Sp1 was isolated from HeLa cells as previously described (26, 30). To prepare infected-cell Sp1, 6 liters of HeLa cells (∼3 × 109) were infected with KOS at a multiplicity of infection of 10. HeLa cells (3 × 109) were pelleted by centrifugation and then were incubated with 3 × 1010 PFU of KOS in 300 ml of Dulbecco's modified Eagle medium for 1 h at 37°C with stirring. Minimal essential medium modified for suspension cultures (S-MEM; Gibco BRL, Grand Island, N.Y.) was then added to give a final volume of 3 liters, and the infection was allow to proceed for a further 12 h. The phosphatase inhibitors sodium orthovanadate (400 μM), sodium fluoride (50 mM), and sodium pyrophosphate (15 mM) were added during purification of Sp1 from infected cells.
Purification of general transcription factors and RNA polymerase.
The transcription factors TFIIA, TFIIE, TFIIF, TFIIH, and TFIID and RNA polymerase II were prepared from HeLa cell nuclear extracts by sequential column fractionation as described previously (13, 18). The final AB fraction contains TFIIA, the CB fraction contains TFIIE, TFIIF, or TFIIH, and CC and the DB contain polymerase and TFIID, respectively. The concentrations of transcription factors were optimized to produce detectable basal and significant activated transcription. Recombinant human TFIIB (rTFIIB) was expressed and purified from Escherichia coli as described previously (19, 51).
In vitro transcription and primer extension.
The plasmids p4/LSWT and pTP4/LSWT were used as templates for in vitro transcription reactions (18). Supercoiled DNA templates (100 ng) were incubated with a mixture of transcription factors in the presence or absence of Sp1. The mixture of transcription factors contained 1 μl of CB fraction (TFIIE, TFIIF, or TFIIH), 2 μl of CC fraction (polymerase), 1 μl of rTFIIB, and 1 μl of DB fraction (TFIID). The final concentrations of components in reaction buffer were 40 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid-KOH (pH 7.9), 60 mM KCl, 12% glycerol, 8.3 mM MgCl2, 0.6 mM ribonucleoside triphosphates, 0.3 mM DTT, and 12 U of RNasin (Promega). The reaction was carried out in a final volume of 30 μl for 1 h at 30°C and stopped by addition of 70 μl of stop solution (150 mM sodium acetate [pH 5.3], 15 mM EDTA [pH 8.0], 150-μg/ml tRNA). The transcription products were then phenol extracted and ethanol precipitated.
For primer extension analysis, in vitro-synthesized RNA was annealed to 3 ng of 32P-labeled primer (5′-AGGGGTACGAAGCCA TACGCGCTTCTACAAGGCGCT-3′) complementary to thymidine kinase gene (tk) sequences from +90 to +125 in buffer containing 10 mM Tris-HCl (pH 7.5), 250 mM KCl, and 1 mM EDTA (pH 8.0) in a final volume of 10 μl. The annealed products were reverse transcribed using 300 U of Moloney murine leukemia virus reverse transcriptase (Gibco/BRL) in a reaction mixture containing 50 mM Tris-HCl (pH 7.5), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 1 mM deoxynucleoside triphosphates, 12 U of RNasin, and 50 μg of actinomycin D. The reverse-transcription reaction was carried out in a final volume of 40 μl for 1 h at 42°C and stopped by addition of 60 μl of stop solution (1 M ammonium acetate, 20 mM EDTA). After phenol extraction and ethanol precipitation, the final products were resolved on 6% denaturing polyacrylamide gels, which were then dried and exposed to Hyperfilm (Amersham) for autoradiography. The radioactive signals were also quantified using a phosphoimaging system (Storm 840; Molecular Dynamics).
RESULTS
The promoters of HSV-1 IE and E genes contain Sp1 binding sites upstream of the TATA box. In contrast, Sp1 binding sites are not found in true L promoters (54). After viral DNA replication, IE and E gene transcription declines and L gene expression predominates. Considering that the abundance, modification state, or activity of cellular transcription factors can be modified as a consequence of viral infection, we entertained the hypothesis that Sp1, which activates IE and E promoters, is altered with respect to its activity and that this may contribute to the decrease in IE and E gene expression as infection proceeds.
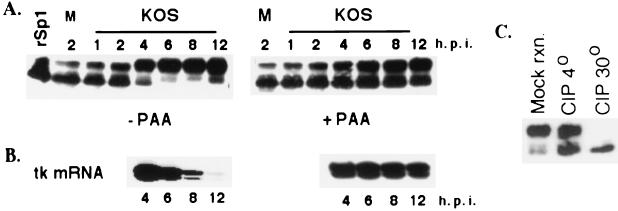
To examine the abundance and modification state of Sp1 as a function of HSV infection, Vero cells were infected with wt HSV (strain KOS) and samples for Western blot analysis were obtained at different time points after infection. The Western blot probed with Sp1 antibody showed that there was no large change in the total amount of Sp1 throughout the infection (Fig. 1A). Sp1 was present in two major forms with different electrophoretic mobilities as reported previously (30). Early in infection, most of the Sp1 was present as a faster-migrating form. However, the majority of Sp1 was modified to a slower-migrating form as infection proceeded. At approximately 4 h p.i., the slower-migrating form became the major species, and by 6 h p.i., the faster-migrating form of Sp1 was almost completely absent (Fig. 1A). When the kinetics of modification of Sp1 were compared to the kinetics of accumulation of an E gene (tk) mRNA, the modification of Sp1 to the slower-migrating form occurred slightly prior to the decrease in the level of accumulated tk message (Fig. 1B).
FIG. 1.
Modification of Sp1 during HSV-1 infection. (A) Western blot analysis of cell extracts prepared from Vero cells infected with KOS in the presence and absence of PAA. At indicated times after infection, the infected cells were harvested directly into the SDS sample buffer, resolved on an SDS-8% acrylamide gel, and transferred to a polyvinylidene difluoride membrane. The membrane was immunoblotted with Sp1-specific antibody. In the first lane, purified baculovirus-produced Sp1 (rSp1) was run alongside the samples as a control. M designates the sample from the mock infection. (B) Primer extension analysis using total RNA from KOS-infected cells in the presence and absence of PAA. RNA was isolated at the indicated times, and 6 μg of total RNA from each sample was used for primer extension reaction using a tk-specific primer. The products were resolved on a 6% sequencing gel, dried, and exposed to film for autoradiography. (C) Phosphatase treatment of infected cell Sp1. Lysate from Vero cells infected with HSV for 8 h (∼100 μg) was incubated in a reaction mixture containing 10 mM (each) sodium fluoride and sodium phosphate at 30o (mock reaction [mock rxn.]), 10 U of CIP at 4o (CIP 4o), and 10 U of CIP at 30o (CIP 30o). All reactions were performed for 10 min in 50 mM Tris-HCl (pH 7.5)-1.0 mM MgCl2. Following the reactions, the samples were subjected to Western blot analysis as described for panel A.
One characteristic of most E gene expression is that it often decreases after DNA replication. When viral DNA replication was blocked by the addition of PAA, tk mRNA abundance persisted at a greater level than in the absence of PAA (Fig. 1B). Also shown in Fig. 1A is that while there was an increase in the amount of the slower-migrating form of Sp1 as infection proceeded in the presence of PAA, the amount of the faster-migrating form of Sp1 persisted in greater quantities than that in the absence of PAA. Thus, the presence of the faster-migrating form of Sp1 correlates with increased tk mRNA abundance.
It has been shown that Sp1 is posttranslationally modified by glycosylation and phosphorylation on its Ser and Thr residues (4). These modifications are indirectly or directly involved in regulating the activity of Sp1 (2, 7, 20, 21, 33, 38, 43, 44). To determine if the slower migration of Sp1 as a function of infection was due to phosphorylation, lysate from Vero cells infected for 8 h with KOS was treated with calf intestinal alkaline phosphatase (CIP). Within 10 min of incubation at 30°C, the slower-migrating form of Sp1 was completely changed to the faster-migrating form (Fig. 1C). Incubation at 4°C resulted in less than complete conversion. The mock reaction consisted of incubating the lysate at 30o in the presence of phosphatase inhibitors and in the absence of CIP. The mobilities of the Sp1 forms in this sample resembled those of the 8-h sample in Fig. 1A. These results suggest that Sp1 is phosphorylated during HSV-1 infection.
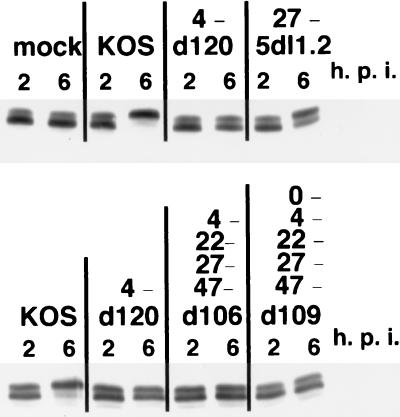
To investigate the genetic requirements for the phosphorylation of Sp1, the modification state of Sp1 was studied as a function of infection with several IE mutants (d120, d109, d106, and 5dl1.2). These viruses possess mutations in different combinations of IE genes (12, 34, 48, 49). The modification state of Sp1 in cells infected with d120, d106, and d109 was similar to that in uninfected cells (Fig. 2). A common feature of these mutants is that they do not produce ICP4. In cells infected with the ICP27 deletion mutant, 5dl1.2, a notable amount of Sp1 was converted to the slower-migrating form (Fig. 2). The pattern of Sp1 phosphorylation in 5dl1.2-infected cells was similar to that seen in KOS-infected cells in the presence of PAA (Fig. 1 and 2) in that there was partial conversion to the phosphorylated form. Unlike d120, d106, and d109, ICP4 is expressed in 5dl1.2-infected cells. As a consequence, E genes and some leaky L genes are expressed in this background (34); however, DNA replication is severely reduced. This is similar to the gene expression profile of cells infected with wt HSV in the presence of PAA (11, 31). These results suggest that ICP4 is directly or indirectly involved in the phosphorylation of Sp1. It is possible that a viral or cellular gene induced by ICP4 is also involved in the modification of Sp1. Additionally, DNA replication may be required for the maximum modification of Sp1 seen during productive infection.
FIG. 2.
Effect of IE gene mutations on the phosphorylation of Sp1. Vero cells were infected with indicated viruses, lysed at 2 and 6 h after infection, and then subjected to Western blot analysis as described for Fig. 1. The IE genes not expressed from each virus are shown.
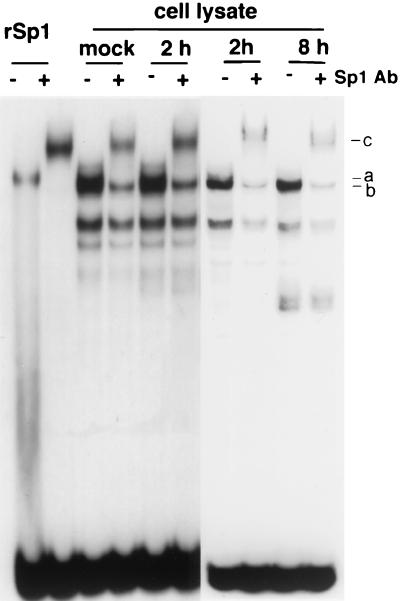
Sp1 can be phosphorylated by variety of cellular kinases, such as protein kinase A (PKA), DNA-dependant protein kinase (DNA-PK), and casein kinase II (CKII), and its activity is affected by several different mechanisms (2, 7, 20, 33, 38, 43). In some cases phosphorylation of Sp1 results in a change in its DNA binding affinity (2, 20, 33, 43). Therefore, we compared the abilities of infected-cell Sp1 and uninfected-cell Sp1 to bind to DNA in mobility shift assays. Whole-cell lysates were prepared from uninfected and KOS-infected Vero cells at 2 and 8 h p.i. Purified baculovirus-produced Sp1 was used as a control. Purified Sp1 showed a single DNA-protein complex (a), which was supershifted in the presence of an Sp1-specific antibody (c) (Fig. 3). Several DNA-protein complexes were seen by using samples from uninfected cells. One of these (a) had the same mobility as that seen with the purified Sp1 and was supershifted by the addition of antibody (c). Another complex migrating closely with the a complex (b) was not supershifted by the antibody. With the infected-cell lysates, there were no apparent differences in the amounts of Sp1-DNA complexes formed between the mock and 2-h-p.i. samples, or 2- and 8-h-p.i. samples. These results suggest that phosphorylation of Sp1 during HSV-1 infection does not cause significant changes in the DNA binding ability of Sp1 (Fig. 3). This was also confirmed indirectly during purification of Sp1 using DNA affinity chromatography. Both forms of Sp1 were purified to very similar yields (Fig. 4A).
FIG. 3.
DNA binding of uninfected and infected cell Sp1. Electrophoretic mobility shift assay using KOS-infected Vero cell lysates. Cell lysates were prepared from uninfected and infected cells at the indicated times p.i. The cell lysate was incubated with a 32P-labeled oligonucleotide containing an Sp1 binding site in the presence of nonspecific competitor poly(dI-dC). For supershifts, Sp1 antibody was added where indicated as described in Materials and Methods. Purified baculovirus-produced Sp1 was used as a control. The bands were resolved on a nondenaturing 5% polyacrylamide gel. The complexes labeled a, b, and c are described in the text.
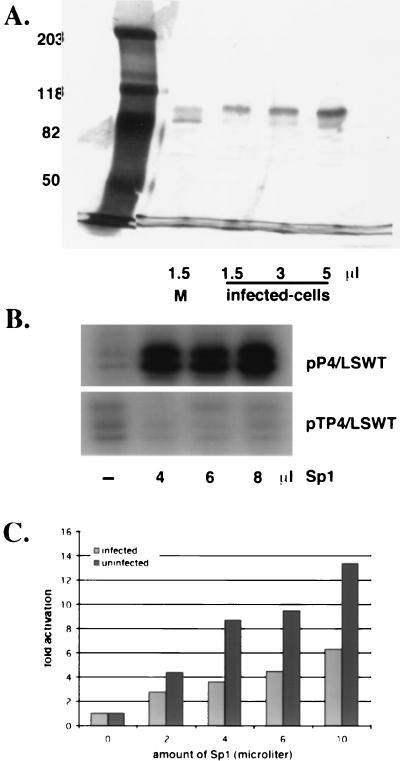
FIG. 4.
Activity of purified Sp1. (A) Silver-stained gel of purified uninfected-cell (M) and infected-cell Sp1. Nuclear extracts were prepared from 6 liters of KOS-infected (12 h) and uninfected HeLa cells. Sp1 was purified from the nuclear extracts by passage over wheat germ agglutinin and Sp1-specific DNA affinity columns as described in Materials and Methods. The indicated volumes of purified Sp1 were resolved on an SDS-8% polyacrylamide gel and subsequently silver stained. (B) Transcriptional activity of Sp1 from uninfected cells on templates containing the wt ICP4 promoter (pP4/LSWT) or one with the Sp1 binding sites deleted (pTP4/LSWT). In vitro transcription assays were performed in the presence of the indicated amounts of Sp1. Primer extension was conducted as described in Materials and Methods. (C) Transcriptional activity of infected-cell Sp1. In vitro transcription assays were performed on the wt template (pP4/LSWT) in the presence of increasing amounts of Sp1 purified from infected and uninfected cells. Plotted are the averages of the fold-induction ratios relative to the reaction that did not contain Sp1 from three different experiments.
To more directly investigate the effect of phosphorylation on Sp1 activity, Sp1 was purified from uninfected and infected HeLa cells. Using wheat germ agglutinin and DNA affinity columns (26, 30), both forms of Sp1 were purified with similar yields and to near-homogeneity (Fig. 4A). The majority of the uninfected-cell Sp1 was of the higher-mobility species, while most of the infected-cell Sp1 was comprised of the lower-mobility species. While it is difficult to determine the absolute concentration of Sp1 in these preparations, the silver-stained gel of Fig. 4A demonstrates that the two preparations of Sp1 were of similar concentrations.
To test the abilities of the two forms of Sp1 to activate a promoter containing Sp1 binding sites, the ICP4 promoter was used in reconstituted in vitro transcription reactions with and without purified Sp1 from infected and uninfected cells. The ICP4 promoter was used because it contains Sp1 binding sites and because the tk promoter is not sufficiently active in this system to produce unambiguous results. The specificity of activation was first established by adding uninfected-cell Sp1 to reactions, comparing templates driven by the entire ICP4 promoter and by an ICP4 promoter with a deletion for the upstream region. As shown in Fig. 4B, the purified Sp1 activated transcription from the ICP4 promoter. When assayed on a similar template with the Sp1 binding sites deleted (pTP4/LSWT), activation of transcription by Sp1 was not observed (Fig. 4B). These results demonstrated that the purified Sp1 was functional and specific.
Subsequently, both the infected- and uninfected-Sp1 preparations were tested at different concentrations for the ability to activate the intact ICP4 promoter (Fig. 4C). Figure 4C represents data averaged from three experiments. At all the concentrations of Sp1 tested, the uninfected-cell Sp1 was more efficient in activation of the ICP4 promoter than the infected-cell Sp1. Only after the addition of 10 μl of infected-cell Sp1 did the level of activation exceed that seen with 2 μl of uninfected-cell Sp1. The level of activation with the infected-cell Sp1 may be expected to increase at the higher concentrations if the low level of high-mobility Sp1 in this preparation (Fig. 4A) has the same activity as uninfected-cell Sp1. Therefore, we conclude that the form of Sp1 phosphorylated during infection is less functional for activation in vitro.
DISCUSSION
The results of this study show that the cellular upstream activator Sp1 is phosphorylated during infection with kinetics that coincide with the reduced expression of an E gene, tk. Furthermore, while the abundance and DNA binding ability of Sp1 was not greatly affected by infection, the activity of infected-cell Sp1, as determined by in vitro transcription using Sp1 purified from uninfected and infected cells, was less than that of uninfected-cell Sp1. Therefore, the reduced transcription of IE and E genes later in infection may in part be due to the virus-induced phosphorylation of Sp1.
Glycosylation and phosphorylation of Sp1 can affect the stability or activity of the protein (2, 7, 20, 21, 33, 38, 43, 44). A variety of cellular kinases, such as PKA, DNA-PK, and CKII, have been shown to phosphorylate Sp1 on Ser/Thr residues in the N-terminal region of the protein (2, 7, 20, 33, 38, 43). Phosphorylation of Sp1 by PKA has been shown to increase both its DNA binding and transcriptional activities (43). Human immunodeficiency virus Tat protein augments phosphorylation of Sp1 by DNA-PK, and this also increases the activity of Sp1 (7). In contrast, phosphorylation of Sp1 by CKII results in a decrease in DNA binding activity and thus possibly in transactivator function (2, 33).
At present we do not know the identity of the kinase that phosphorylates Sp1 during HSV infection. From the studies conducted with the mutant viruses it appears that ICP4 is necessary for the phosphorylation of Sp1. While purified ICP4 preparations have been shown to possess kinase activity (57), the studies at present do not distinguish between a direct role for ICP4 or a viral pathway requiring ICP4 for the expression of one of its components. Additionally, the expression of ICP4 is not sufficient for the maximum level of Sp1 phosphorylation seen beyond 4 h p.i. with wt virus. While infection-induced phosphorylation of Sp1 was seen in the absence of ICP27 and during infection with wt virus in the presence of PAA (Fig. 1 and 2), it was not as robust as during productive infection with wt virus. ICP27, like ICP4, may promote the expression of one or more proteins involved in the phosphorylation of Sp1. However, it is unlikely that ICP27 is sufficient, since infection by d120 did not result in the mobility change of Sp1 (Fig. 2). Collectively the data suggest it is possible that the product of an E or leaky-L gene may be involved. Additionally, an ICP4-dependant viral mechanism resulting in the activation of a cellular kinase may also be involved.
ICP4 is also known to negatively regulate the transcription of its own mRNA by binding to a specific sequence near the transcription start site (12, 35, 37). Not all IE genes possess such binding sites near their mRNA start sites; however, the expression of ICP4 results in the reduced abundance of all IE transcripts (14, 42). It is possible that this reflects the requirement for ICP4 for the phosphorylation of Sp1.
The results from these studies showed that the phosphorylated form of Sp1 generated during HSV-1 infection has a lower activity in transcription activation than uninfected-cell Sp1 (Fig. 4C). Regulation of Sp1-dependent transcription can be affected by changes in abundance, DNA binding affinity, stability, or the stability to interact with other proteins involved in the mechanism of Sp1 activation. As shown in Fig. 1 and 4, neither the abundance nor DNA binding ability of Sp1 was significantly affected by phosphorylation. It has been shown that Sp1 interacts with various transcription factors, such as TAFII130, TATA-binding protein, nuclear protein P74, RelA, YY1, and TAF55 (6, 16, 36, 39, 50, 53). Among these, TAFII130 has been shown to serve as a cofactor of Sp1 by directly interacting with Sp1 in vitro (17, 22, 53). The mammalian mediator, CRSP, is also involved in activation of Sp1 (45, 46). It is possible that the phosphorylation of Sp1 during infection affects interactions between Sp1 and one or more of these factors. The negative effect of phosphorylation on Sp1 activation of the ICP4 promoter (Fig. 4C) may not completely explain the reduced transcription of E and IE genes. Multiple mechanisms involving changes in the abundance and activities of other activators and coactivators may be involved in the attenuation of IE and E gene transcription late in infection.
During productive HSV infection, the viral genes are expressed such that those possessing binding sites for transcription factors, including Sp1, in their upstream promoter regions are expressed first. The expression of most of these genes is eventually attenuated and followed by the abundant transcription from HSV L promoters, which are often simply TATA boxes and initiator elements. We propose that the reduced transcription of many IE and E genes is in part due to the reduced activity of Sp1 resulting from the phosphorylation of the protein, as a consequence of an activity requiring ICP4.
Acknowledgments
We thank Lorna Samaniego and Susan Zabierowski for helpful discussions and comments on the manuscript.
This work was supported by the NIH grant AI30612.
REFERENCES
- 1.Alwine, J. C., W. L. Steinhart, and C. W. Hill. 1974. Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology 60:302-307. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 272:13489-13495. [DOI] [PubMed] [Google Scholar]
- 3.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and Kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, C. M., and R. G. Roeder. 1995. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267:531-536. [DOI] [PubMed] [Google Scholar]
- 7.Chun, R. F., O. J. Semmes, C. Neuveut, and K. T. Jeang. 1998. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 72:2615-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coen, D. M., S. P. Weinheimer, and S. L. McKnight. 1986. A genetic approach to promoter recognition during trans induction of viral gene expression. Science 234:53-59. [DOI] [PubMed] [Google Scholar]
- 9.Costanzo, F., G. Campadelli-Fiume, L. Foa-Tomasi, and E. Cassai. 1977. Evidence that herpes simplex virus DNA is transcribed by cellular RNA polymerase B. J. Virol. 21:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 11.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857-868. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon, R. A., and P. A. Schaffer. 1980. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein VP175. J. Virol. 36:189-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dynan, W. S., and R. Tjian. 1983. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35:79-87. [DOI] [PubMed] [Google Scholar]
- 16.Emili, A., J. Greenblatt, and C. J. Ingles. 1994. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol. Cell. Biol. 14:1582-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill, G., E. Pascal, Z. H. Tseng, and R. Tjian. 1994. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 91:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu, B., R. Rivera-Gonzalez, C. A. Smith, and N. A. DeLuca. 1993. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proc. Natl. Acad. Sci. USA 90:9528-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha, I., W. S. Lane, and D. Reinberg. 1991. Cloning of a human gene encoding the general transcription initiation factor IIB. Nature 352:689-695. [DOI] [PubMed] [Google Scholar]
- 20.Haidweger, E., M. Novy, and H. Rotheneder. 2001. Modulation of Sp1 activity by a cyclin A/CDK complex. J. Mol. Biol. 306:201-212. [DOI] [PubMed] [Google Scholar]
- 21.Han, I., and J. E. Kudlow. 1997. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 17:2550-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoey, T., R. O. Weinzierl, G. Gill, J. L. Chen, B. D. Dynlacht, and R. Tjian. 1993. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell 72:247-260. [DOI] [PubMed] [Google Scholar]
- 23.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 72:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imbalzano, A. N., A. A. Shepard, and N. A. DeLuca. 1990. Functional relevance of specific interactions between herpes simplex virus type 1 ICP4 and sequences from the promoter-regulatory domain of the viral thymidine kinase gene. J. Virol. 64:2620-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, S. P., and R. Tjian. 1989. Purification and analysis of RNA polymerase II transcription factors by using wheat germ agglutinin affinity chromatography. Proc. Natl. Acad. Sci. USA 86:1781-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, K. A., K. R. Yamamoto, and R. Tjian. 1985. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell 42:559-572. [DOI] [PubMed] [Google Scholar]
- 28.Jones, P. C., and B. Roizman. 1979. Regulation of herpesvirus macromolecular synthesis. VIII. The transcription program consists of three phases during which both extent of transcription and accumulation of RNA in the cytoplasm are regulated. J. Virol. 31:299-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadonaga, J. T., K. R. Carner, F. R. Masiarz, and R. Tjian. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079-1090. [DOI] [PubMed] [Google Scholar]
- 30.Kadonaga, J. T., and R. Tjian. 1986. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA 83:5889-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lania, L., B. Majello, and P. DeLuca. 1997. Transcriptional regulation by the Sp family proteins. Int. J. Biochem. Cell Biol. 29:1313-1323. [DOI] [PubMed] [Google Scholar]
- 33.Leggett, R. W., S. A. Armstrong, D. Barry, and C. R. Mueller. 1995. Sp1 is phosphorylated and its DNA binding activity down-regulated upon terminal differentiation of the liver. J. Biol. Chem. 270:25879-25884. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller, M. T. 1987. Binding of the herpes simplex virus immediate-early gene product ICP4 to its own transcription start site. J. Virol. 61:858-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murata, Y., H. G. Kim, K. T. Rogers, A. J. Udvadia, and J. M. Horowitz. 1994. Negative regulation of Sp1 trans-activation is correlated with the binding of cellular proteins to the amino terminus of the Sp1 trans-activation domain. J. Biol. Chem. 269:20674-20681. [PubMed] [Google Scholar]
- 37.O'Hare, P., and G. S. Hayward. 1985. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. J. Virol. 56:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal, S., K. P. Claffey, H. T. Cohen, and D. Mukhopadhyay. 1998. Activation of Sp1-mediated vascular permeability factor/vascular endothelial growth factor transcription requires specific interaction with protein kinase C zeta. J. Biol. Chem. 273:26277-26280. [DOI] [PubMed] [Google Scholar]
- 39.Perkins, N. D., A. B. Agranoff, E. Pascal, and G. J. Nabel. 1994. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol. Cell. Biol. 14:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persson, R. H., S. Bacchetti, and J. R. Smiley. 1985. Cells that constitutively express the herpes simplex virus immediate-early protein ICP4 allow efficient activation of viral delayed-early genes in trans. J. Virol. 54:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philipsen, S., and G. Suske. 1999. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston, C. M. 1979. Abnormal properties of an immediate early polypeptide in cells infected with the herpes simplex virus type 1 mutant tsK. J. Virol. 32:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohlff, C., S. Ahmad, F. Borellini, J. Lei, and R. I. Glazer. 1997. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J. Biol. Chem. 272:21137-21141. [DOI] [PubMed] [Google Scholar]
- 44.Roos, M. D., K. Su, J. R. Baker, and J. E. Kudlow. 1997. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol. Cell. Biol. 17:6472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu, S., and R. Tjian. 1999. Purification of transcription cofactor complex CRSP. Proc. Natl. Acad. Sci. USA 96:7137-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 47.Saffer, J. D., S. P. Jackson, and S. J. Thurston. 1990. SV40 stimulates expression of the transacting factor Sp1 at the mRNA level. Genes Dev. 4:659-666. [DOI] [PubMed] [Google Scholar]
- 48.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samaniego, L. A., N. Wu, and N. A. DeLuca. 1997. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seto, E., B. Lewis, and T. Shenk. 1993. Interaction between transcription factors Sp1 and YY1. Nature 365:462-464. [DOI] [PubMed] [Google Scholar]
- 51.Smith, C. A., P. Bates, R. Rivera-Gonzalez, B. Gu, and N. A. DeLuca. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 67:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suske, G. 1999. The Sp family of transcription factors. Gene 238:291-300. [DOI] [PubMed] [Google Scholar]
- 53.Tanese, N., D. Saluja, M. F. Vassallo, J. L. Chen, and A. Admon. 1996. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc. Natl. Acad. Sci. USA 93:13611-13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner, E. K., J. F. Guzowski, and J. Singh. 1995. Transcription of the herpes simplex virus genome during productive and latent infection. Prog. Nucleic Acid Res. Mol. Biol. 51:123-165. [DOI] [PubMed] [Google Scholar]
- 55.Watson, R. J., and J. B. Clements. 1980. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature 285:329-330. [DOI] [PubMed] [Google Scholar]
- 56.Weir, J. P. 2001. Regulation of herpes simplex virus gene expression. Gene 271:117-130. [DOI] [PubMed] [Google Scholar]
- 57.Xia, K., N. A. DeLuca, and D. M. Knipe. 1996. Analysis of phosphorylation sites of herpes simplex virus type 1 ICP4. J. Virol. 70:1061-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yurochko, A. D., E. S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E. S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J. Virol. 71:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]