Abstract
Epstein-Barr virus (EBV) classically infects and transforms B lymphocytes in vitro, yielding lymphoblastoid cell lines (LCLs). In contrast to other herpesviruses, EBV is not described as an infectious agent for monocytes. However, recent papers described in vitro infection of monocytes leading to abortive or transient viral expression. In the present study, we report the characterization of E1, a monocytic cell line infected and transformed by EBV. This cell line was derived from an LCL by a drastic electroporation and selection of neomycin-resistant cells, unfavorable to B-cell outgrowth. E1 expressed surface molecules of monocytic lineage (CD14, major histocompatibility complex class II, and CD80) and the c-fms gene, a highly specific marker for the monocytic lineage. This cell line is able to phagocytose and secrete proinflammatory monokines tumor necrosis factor alpha, interleukin-6 (IL-6), and IL-8. E1 cells are tumorigenic after injection in nude mice, and a monocytic cell line obtained from one of these tumors (TE1) displayed immunophenotype and functional properties similar to those of E1. We detected the presence of the EBV genome in both cell lines, as well as expression of the EBNA-1 and LMP-1, but not EBNA-2, viral genes, characteristic of a type II latency. LMP-1 influences the phenotype of these monocytic cell lines, as demonstrated by down-regulation of cell proliferation and membrane intercellular adhesion molecule 1 expression due to an LMP-1 antisense strategy. This is the first description of a latently infected human monocytic cell line and the first direct demonstration of an instrumental role for LMP-1 in the proliferation of EBV-transformed cell lines expressing a type II latency.
The Epstein-Barr virus (EBV), a member of the herpesvirus family, infects over 90% of healthy adults. EBV classically infects B cells, causing a benign disease, acute infectious mononucleosis, but also malignant diseases such as Burkitt's lymphoma and other B-lymphoproliferative disorders in patients with severe immunodeficiency. EBV can also infect epithelial cells and is associated with undifferentiated nasopharyngeal carcinoma (NPC) (43). More recently, many reports have shown that EBV can be associated with other pathologies, including Hodgkin's disease (HD) (4), lymphoproliferative disorders of T cells such as peripheral T-cell lymphoma in immunocompetent hosts (6), and gastric, breast, and hepatocellular adenocarcinomas (3, 21). In all the reported cases, the virus displays mainly a latency program of infection with a restricted pattern of gene expression, which can be classified in three types. Type I latency, where only EBNA-1 is expressed, is found in Burkitt's lymphoma. Coexpression of EBNA-1 and latent membrane proteins LMP-1 and LMP-2 is characteristic of a type II latency found in NPC, HD, and T-cell lymphomas, and type III latency with expression of the five EBNAs and three LMPs is found in lymphomas of immunodeficient patients.
In vitro, EBV is classically associated with the infection and transformation of quiescent B lymphocytes to yield lymphoblastoid cell lines (LCLs), and LCLs are to date a unique cellular model to study establishment and maintenance of a type III viral latency by EBV. However except for LCLs, in vitro cellular models of physiologically relevant EBV targets are currently lacking. This is particularly the case for cellular models of infection and transformation for studying a type II expression program, which is commonly found in EBV-associated malignancies. In this respect, we previously showed isolation and establishment of EBV-infected T-cell lines expressing a type II latency program after in vitro infection of peripheral blood mononuclear cells (PBMCs) by using drastic electroporation and selection of neoresistant cells unfavorable to B-cell outgrowth (17, 37).
In recent studies, in vitro target cells for EBV have been shown to be more diversified than first expected. This virus can indeed infect neutrophils (30), follicular dendritic cells (33), and astrocytic cell lines (36). Whereas other human herpesviruses such as cytomegalovirus (CMV) (20), herpes simplex virus (11), and human herpesvirus 6 (29) have been found to target macrophages, only rare studies describe infection of cells of monocytic origin by EBV (42, 47, 48). In an old report, the viral genome was detected in monoblast or early monocytic cell lines obtained from bone marrow of children with a defect in myelopoiesis (42). Another study reports six EBV-infected macrophage cultures derived from various clinical samples from adult and child patients (48). In these nonestablished cultures, latent and replicative EBV genes are expressed. More recently, the ability of EBV to infect and replicate in fresh monocytes was demonstrated, and in this case, no cellular transformation was observed (47). Here, we describe the establishment and characterization of an EBV-infected and transformed monocytic cell line (E1) obtained in the course of the in vitro infection and electroporation and selection process used to yield the T-cell lines previously characterized (17). This cell line is tumorigenic in immunodeficient mice, and a second monocytic cell line, termed TE1, was obtained from a tumor induced by injection of E1 in a nude mouse. Both cell lines expressed viral genes characteristic of type II latency, and expression of LMP-1 was instrumental for their proliferation.
MATERIALS AND METHODS
Isolation of the E1 and TE1 EBV-infected monocytic cell lines.
The isolation protocol used was previously reported and allowed EBV-infected T cells to be obtained from EBV LCLs (17). Briefly, EBV LCLs incubated with an expression vector encoding resistance to neomycin (pSV-neoR) were electroporated at 250 V and 1,200 μF for 60 ms. Following transfection, cells were plated in 24-well plates and cultured 4 to 5 weeks with selection drug G-418 (1 mg/ml). These experimental conditions were far too stringent for B cells, which rapidly died. Few slow-growing living cells were obtained; the cells grew in permanent cell lines about 2 months after the beginning of the experiment. Analysis of these cells reveals that the majority were T cells (17). However, one established cell line (E1) turned out to display a non-T- and non-B-cell phenotype.
To test the transformed phenotype of E1 cells, a tumorigenicity assay was performed with nude mice (data not shown). To this end, 106 cells per mouse were subcutaneously injected into 10 animals. Fifty percent of the animals developed tumors after 3 to 4 weeks. A cell line termed TE1 was derived from one tumor and was further analyzed in parallel with E1 cells.
Cells and culture conditions.
PBL25 (PBMCs), EBV-transformed B-EBVneo (LCL), NC5, and TC cells (T-cell lines) were described elsewhere (17, 37) and were derived from the same donor as E1 and TE1 cells. The human Burkitt's lymphoma DG75 (EBV negative), Kas, and Rafa B-LCL (EBV positive), monocytic HL60, and U937 cell lines, human Jurkat T cells, and the marmoset B95.8 B-cell line were also used in this study. All cells were propagated in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM l-glutamine, 1% nonessential amino acids, 1 mM sodium pyruvate, and gentamicin (50 μg/ml) (Gibco). The medium for E1 and TE1 cells contained also transferrin (10 μg/ml; Sigma) and bovine insulin (10 μg/ml; Organon) during the first three passages. These two cell lines were passed three times per week at a 1:4 ratio.
Immunofluorescence analysis by cytometry.
For detection of cell surface antigens, monocytic cell lines E1 and TE1 (106 cells) were stained with monoclonal antibodies directly labeled with phycoerythrin (PE) or fluorescein isothiocyanate (FITC) as described by the suppliers or in a two-step immunofluorescence assay for unlabeled antibodies. Antibodies used are listed in Table 3.
TABLE 3.
Cell surface phenotype of monocytic cell lines E1 and TE1
| Marker | Resulta for:
|
Clone | Isotype | Supplier | Fluorochrome | |
|---|---|---|---|---|---|---|
| E1 | TE1 | |||||
| CD3 | − | − | UCHT1 | IgG1 | Immunotech | FITC |
| CD2 | − | − | 39C1.5 | IgG2a | Immunotech | FITC |
| CD4 | +w | +w | 13B8.2 | IgG1 | Immunotech | FITC |
| CD8 | − | − | B9.11 | IgG1 | Immunotech | FITC |
| CD25 | − | − | ACT-1 | IgG1 | Dako | FITC |
| CD28 | − | − | CD28.2 | IgG1 | Immunotech | FITC |
| HLA-DR | + | + | B8.12.2 | IgG2b | Immunotech | FITC |
| κ chain | − | − | Polyclonal | Polyclonal | Dako | FITC |
| λ chain | − | − | Polyclonal | Polyclonal | Dako | PE |
| CD19 | − | − | J4.119 | IgG1 | Immunotech | PE |
| CD20 | − | +w | B9E9(HRC20) | IgG2a | Immunotech | FITC |
| CD10 | − | − | ALB2 | IgG2a | Immunotech | FITC |
| CD21 | +w | +w | BL13 | IgG1 | Immunotech | FITC |
| CD22 | − | − | 4KB128 | IgG1 | Dako | FITC |
| CD14 | + | + | RMO52 | IgG2a | Immunotech | FITC |
| CD80 | + | + | MAB104 | IgG1 | Immunotech | None |
| CD86 | − | − | 2331(FUN-1) | IgG1 | Pharmingen | None |
| CD1a | − | − | BL6 | IgG1 | Immunotech | None |
| CD16 | − | − | 3G8 | IgG1 | Immunotech | FITC |
| CD34 | − | − | 8G12 | IgG1 | Becton Dickinson | PE |
| CD54 | + | + | 84H10 | IgG1 | Immunotech | PE |
−, negative; +, positive; +w, weakly positive. Comparisons are to results for the control antibody.
For detection of LMP-1, cells were fixed by 4% paraformaldehyde for 30 min at room temperature and after two washes were permeabilized by Triton X-100 (0.25% [vol/vol] in phosphate-buffered saline [PBS]). After two washes, cells were then incubated for 45 min with anti-LMP-1 (CS 1-4) (Novocastra) at 4°C, and binding of anti-LMP-1 antibodies was revealed by incubation with a rabbit anti-mouse immunoglobulin (Ig) conjugated to PE.
After being washed with PBS, cells were analyzed on an EPICS-XL cytometer (Coulter).
DNA extraction.
Viral DNA for analytic PCR was extracted according to a modified Hirt procedure (8). Briefly, cells were heated at 65°C for 15 min and then lysed in 0.1% sodium dodecyl sulfate at 65°C for 20 min. NaCl was added to 1 M. Samples were kept at 4°C overnight and then centrifuged. The supernatants were extracted with phenol-chloroform-isoamyl alcohol (24:1) and incubated with 2 volumes of ethanol at −20°C for precipitation.
RNA extraction, Northern blotting, and reverse transcription.
Total RNA was extracted with RNAzol according to the supplier instructions (Bioprobes). A total of 1 μg of RNA in 9 μl of milli-Q water was mixed with 0.5 μl of primer (0.5 μg of oligo[dT]), 0.1 μl of RNasin (40 U/μl; Promega), and 1.4 μl of milli-Q water. The samples were heated at 70°C for 5 min and then allowed to cool down at room temperature. Then, samples were mixed with a solution containing 0.1 μl of RNasin (40 U/μl), 5 μl of 5× reverse transcriptase (RT) buffer (250 mM Tris-HCl [pH 8.3], 300 mM KCl, 15 mM MgCl2), 1 μl of dithiothreitol (0.1 M), 5 μl of deoxynucleotide triphosphates (10 mM), 1.9 μl of milli-Q water, and 1 μl of Moloney murine leukemia virus (MMLV) RT (200 U; Gibco). This mixture was incubated at 37°C for 45 min. Samples were heat inactivated at 95°C for 5 min, and after rapid cooling 100 U of MMLV RT in 2 μl was added and a second cycle of reverse transcription was performed.
For Northern blot analysis of c-myc, total RNAs were prepared from subconfluent cells by the isothiocyanate-CsCl gradient method (45). Denatured RNA samples (20 μg/well) were fractionated on a formaldehyde-1.2% agarose gel, transferred to a Hybond C Extra filter (Amersham), and analyzed by Northern blot hybridization. RNA amounts were quantified by hybridizing a [α-32P]dCTP GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe. Hybridizations were performed under stringent conditions with a myc-specific PCR product labeled with [α-32P]dCTP.
PCR analysis.
Fifty nanograms of cDNA was used as a template for PCR amplification. After an initial denaturation step at 94°C for 2 min, samples were subjected to 35 cycles of amplification (1 min at 55°C, 1 min at 72°C, and 1 min at 94°C). The PCR products were visualized by electrophoresis in a 2% agarose gel and ethidium bromide staining or alternatively blotted onto nylon filters (Hybond N; Amersham) and detected by Southern hybridization using a specific probe for LMP-1, EBNA-1, or EBNA-2, labeled with [α-32P]ATP (pCMV-EBNA-1 and pSG5-EBNA-2 vectors; kindly provided by E. Manet and A. Sergeant, Lyon, France).
Analysis of viral DNA sequences in infected cells was performed by PCR on 2 μl of Hirt's extracts. Specific primer pairs used in PCR experiments and the sizes of the generated fragments are listed in Table 1.
TABLE 1.
Oligomers used in PCR and for the antisense strategya
| Transcript | Oligomer | Fragment length (bp) | Sequence |
|---|---|---|---|
| LMP-1 | 5′ primerb | 408 | CAC GAC CTT GAG AGG GGC CCA |
| 3′ primerb | GCC AGA TGG TGG CAC CAA GTC | ||
| 5′ primerc | 399 | CCT TTG CTC TCA TGC TTA TAA | |
| 3′ primerc | GCC AGA TGG TGG CAC CAA GTC | ||
| AS2d | CTC TCA AGG TCG TGT TCC AT | ||
| Sc2 (scrambled)d | TTC GAC TAG ACT CCG GTT TC | ||
| EBNA-1 Qp/Cp | 5′ primerb | 421 | GTA ACT TAG GAA GCG TTT CT |
| 3′ primerb | GGT CTC CGG ACA CCA TCT CT | ||
| 5′ primerc | 80 | GGA CCT CAA AGA AGA GGG GG | |
| 3′ primerc | GCT CCT GGT CTT CCG CCT CC | ||
| EBNA-2 | 5′ primere | 177 | CTA TCT TGC GTT ACA TGG GGG ACA |
| 3′ primere | GGT CTC CGG ACA CCA TCT CT | ||
| BZLF-1 | 5′ primer | 430 | CAC CTC AAC CTG GAG ACA AT |
| 3′ primer | TGA AGC AGG CGT GGT TTC AA | ||
| Probe | GCA CAT CTG CTT CAA CAG GA | ||
| c-fms | 5′ primer | 293 | CTT TCC TAA TCC CCT TAT C |
| 3′ primer | ATT ACA GCA GTA CCA GTA TG | ||
| CSF-1 | 5′ primer | 192 | TCG GAC GCA GGC CTT GTC ATG |
| 3′ primer | GAA CAG TTG AAA GAT CCA GTG |
Oligonucleotides used in experiments involving PCR and sequencing are shown in Table 2.
For RT-PCR.
For PCR.
For antisense strategy.
For PCR and RT-PCR.
Sequencing of the LMP-1 gene after PCR amplification.
DNA extracted from all cell lines was amplified by a PCR strategy described by Mehl et al. (35). Briefly, LMP-1 PCR products were amplified by nested PCR with two sets of primers (Table 2). PCR amplification was performed in a final volume of 50 μl containing 1 μl of a 50 μM concentration of each primer, 5 μl of polymerase buffer (10×), 8 μl of a 1.25 mM solution of each deoxynucleoside triphosphate, 5 μl of dimethyl sulfoxide, 500 ng of genomic DNA, and 1.25 U of Ampli Taq gold polymerase (Perkin-Elmer). Reaction mixtures were placed in a 480 thermal cycler (Perkin-Elmer) and subjected to the following program: 35 cycles at 94°C for 1 min, 65°C (or 55°C for the nested PCR) for 1 min, and 72°C for 1 min with a pre-PCR heating step at 94°C and a final period of 10 min at 72°C to complete the reaction. PCR products were analyzed by 2% agarose gel electrophoresis and ethidium bromide staining. Each PCR product (50 ng) was sequenced with an ABI PRISM dye terminator kit (Perkin-Elmer) supplemented with 7.5 pM (each) primer. Reaction mixtures were placed in a 2400 thermal cycler (Perkin-Elmer) and subjected to the program according the manufacturer's recommendations. Sequence analysis was realized with OMIGA software. In addition to the primers already used for amplification, internal primers described by Faumont et al. (13) were used for sequencing.
TABLE 2.
Sets of oligonucleotides used for LMP-1 sequencing
| Use | Oligomers | Location | Sequence (5′-3′) | Nucleotide position |
|---|---|---|---|---|
| 5′ fragment sequencing | Outer | 8617 | GGT CCG TCG CCG GCT CCA CTC ACG AGC AGG | 168617-168646 |
| 9580 | CCA AGA AAC ACG CGT TAC TCT GAC GTA GCC | 169551-169580 | ||
| Inner | 8667 | GTT AGA GTC AGA TTC ATG GCC AGA ATC ATC G | 168667-168697 | |
| 9494 | CCT GAC ACA CTG CCC TCG AGG | 169471-169494 | ||
| 3′ fragment sequencing | Outer | 7832 | GCC TGG TAG TTG TGT TGT GCA GAG GTC | 167832-167858 |
| 8785 | CGA TTT TAA TCT GGA TGT ATT ACC ATG G | 168758-168785 | ||
| Inner | 7901 | GGC GGA GTC TGG CAA CGC CCG GGT CCT TG | 167901-167929 | |
| 8702 | GCT ACC GAT GAT TCT GGC CAT GAA TCT GAC | 168673-168702 |
The nucleotide sequences were compared with LMP-1 sequence from the prototype B95.8 strain (GenBank accession no. X66863).
Study of the phagocytic activity.
A suspension of Escherichia coli (JM 109 strain ) was incubated with a solution of FITC (1 mg/ml) overnight at 4°C and then washed twice with PBS (pH 7.2). Cells (5 × 105) were gently rotated for 3 h with labeled bacteria at 4 or 37°C. Jurkat and HL60 cells were used as negative and positive controls, respectively. After two washes with cold PBS, the internalization of labeled bacteria by phagocytosis was determined by analysis on an EPICS-XL cytometer at excitation and emission settings of 488 and 540 nm, respectively.
Determination of monokine production.
Different cell lines (HL60, U937, Jurkat, E1, and TE1) were cultured at 2 × 105 cells/ml with medium alone or with phorbol myristate acetate (PMA; 0.1 μg/ml; Sigma). Endotoxin-tested RPMI 1640 culture medium and heat-inactivated fetal bovine serum (endotoxin content, less than 0.6 U of endotoxin/ml) were purchased from Sigma. Supernatants were collected after 2, 4, 8, 24, and 40 h, or only after 40 h, of stimulation. The presence of interleukin-6 (IL-6), IL-8, and tumor necrosis factor alpha (TNF-α) was quantified by immunoenzymetric assays. TNF-α activity in the supernatant was measured by an L929 cytotoxicity bioassay.
Cell transfection and antisense experiment.
Prior to electroporation, E1 and TE1 cells were harvested and placed for 10 min in cold RPMI 1640 with 25 μM HEPES buffer (pH 7.9). Five million cells were electroporated at 255 V and 950 μF with 75 μg of tRNA plus 5 μg of pEGFP C1 (Clontech) only or with a pSV-HA LMP-1 antisense vector or with a pcDNA3 vector (Invitrogen) containing the coding sequence for mutated IκBα32/36A. The pSV-HA LMP-1-AS vector (pSV LMP-1-AS) was obtained by cloning the LMP-1 cDNA in an antisense orientation in a pSG5-derived expression plasmid (generous gift from J.-L. Baert, Lille, France). After 48 h, expression of intercellular adhesion molecule 1 (ICAM-1)/CD54 in enhanced green fluorescent protein (EGFP)-positive cells was analyzed by flow cytometry (Epics XL; Coulter) using a monoclonal antibody labeled with PE (Table 3).
An antisense oligonucleotide strategy was also performed using oligonucleotides listed in Table 1. The effect of an antisense oligodeoxynucleotide (AS2) targeted against LMP-1 was compared to that of a randomly scrambled version (SC2) used as control (34). To test the control of ICAM-1 expression by LMP-1, 20 μM AS2 or SC2 was added in culture medium each 24 h.
The effect of antisense oligonucleotides was determined by measuring the expression of ICAM-1/CD54 by flow cytometry after 48 h of culture and by measuring cell proliferation. For studying cell proliferation, cells (E1, TE1, Rafa B-EBV, or DG75) were seeded in 96-well flat-bottom microtiter plates (Costar; 40,000 cells per well) in OPTIMEM with AS2 or SC2 oligonucleotides (5 or 10 μM) and 1 μl of Lipofectamine (Gibco) to enhance oligonucleotide uptake. After 16 h, medium was replaced by RPMI 1640 supplemented with fetal calf serum in the presence of oligonucleotides. For the last 10 h of culture, 1 μCi of [methyl-3H]thymidine (Amersham)/well was added. After 48 h of culture, cells were harvested and [3H]thymidine uptake was measured in a liquid scintillation counter (MicroBetaTM TriLux). To compare proliferative responses between different cell lines, proliferation was standardized to a stimulation index calculated as follows: (mean counts per minute of triplicate cultures with AS2 or SC2 [5 or 10 μM]/mean counts per minute of triplicate cultures of the same cells without oligonucleotides) × 100.
Western blotting analysis.
Western blot analysis was performed according to techniques already published (17). Briefly, following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Euromedex), proteins were transferred onto membranes (Immobilon-P; Millipore) by electroblotting. Membranes were blocked with 0.2% casein in PBS-0.1% Tween and then incubated with the primary antibodies. Antibodies used in this study are mouse monoclonal anti-LMP-1 (S12; generous gift from P. Busson), a mouse monoclonal antiactin, a mouse monoclonal anti-BCL-2, a rabbit polyclonal anti-TRAF-2, and a rabbit polyclonal anti-CSF1-R, all purchased from Santa Cruz Biotechnology. Following a washing (in PBS-0.2% Tween), membranes were incubated with a peroxidase-conjugated rabbit anti-mouse or donkey anti-rabbit serum (Jackson Immunoresearch) and revealed with the ECL kit (Amersham) before autoradiography. For PMA stimulation, cells were stimulated with 2.5 nM PMA for 48 h before being harvested.
RESULTS
Characterization of E1, a non-T non-B cell line expressing cell surface antigens of monocytic cell lineage.
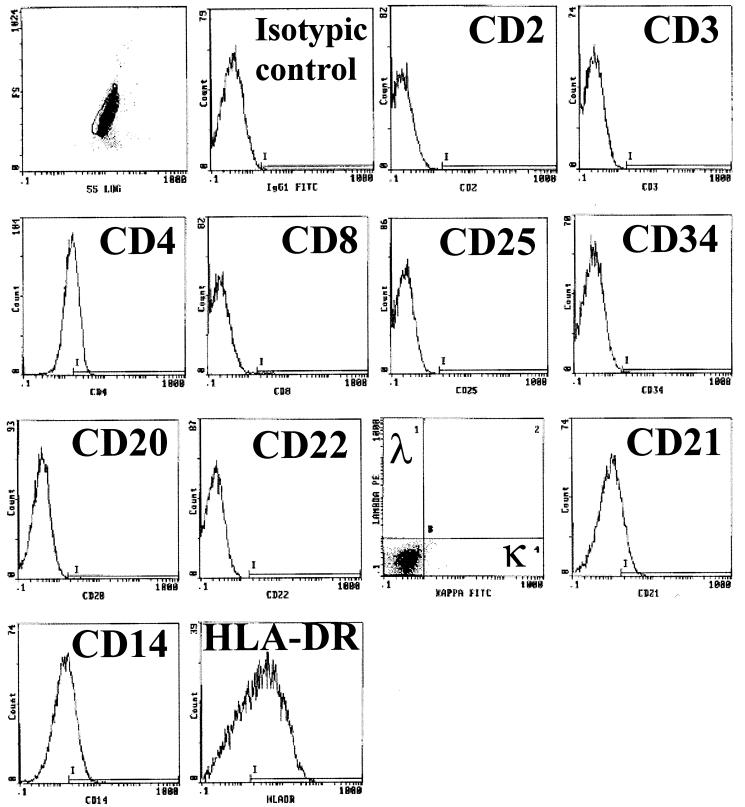
The first step to yield the EBV-transformed E1 cell line was infection of PBMCs with B95.8 supernatants. The LCL obtained was electroporated with a vector for the selectable-marker neoR gene. After 4 to 5 weeks of culture with high doses of G-418, only non-B cells remained. Conditions of electroporation and selection were very drastic for B-cell outgrowth. We analyzed the surviving non-B cells, which grew in permanent cell lines 2 months after the beginning of the experiment. As we previously reported, the majority of the cell lines obtained with this protocol were naive T cells (17). However one cell line (E1) did not display surface antigens specific for mature T cells (CD3, CD2, and CD28), and no rearrangement of T-cell receptor genes was detected. These cells were also devoid of B-cell-specific markers (CD19, CD20, light chain of IgG[κ] or IgG[λ]) or NK cells (CD16, CD56) (Fig. 1 and Table 3). On the other hand, this cell line expressed surface antigens classically found on peripheral monocytes, basically CD14 and HLA-DR, or found after activation or culture in vitro (CD80) (15). In addition, E1 was weakly positive for CD4 and CD21 expression and constitutively negative for CD86 and CD1a expression (Table 3). This pattern of surface antigen expression, except for that aspect related to CD86 (15), argues in favor of their monocytic origin. During long-term culture (over 6 months), the E1 cell line was stable, as indicated by constancy in cell marker expression. This ability to proliferate in long-term cultures is characteristic of a transformed phenotype.
FIG. 1.
Membrane markers expressed by E1 and analyzed by flow cytometry. Antibodies used for immunophenotyping are listed in Table 3.
Established cell line E1 was injected into nude mice, and tumors were obtained (data not shown), showing that this cell line readily exhibits a malignant phenotype. Cell line TE1 was derived from one of these tumors. The TE1 cell line displays the same pattern of surface marker expression as E1, except that CD20 is weakly expressed on this cell line (Table 3). Nevertheless, no expression of rearranged immunoglobulin genes was detected by nested RT-PCR for E1 and TE1 (data not shown).
E1 and TE1 express c-fms and its specific ligand.
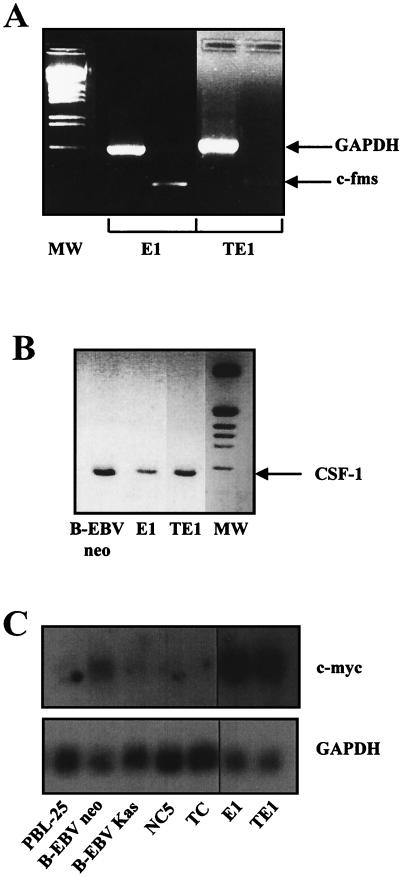
To further demonstrate the monocytic origin of E1 and TE1, we investigated by RT-PCR the expression of the receptor for macrophage colony-stimulating factor/colony-stimulating factor 1 (CSF-1) (c-fms or CSF-1 receptor), which is implicated in the differentiation of monocytic cells. This highly specific marker of the monocytic lineage (46) is expressed in both cell lines (Fig. 2A). The expression of CSF-1 was also detected by RT-PCR in both cell lines (Fig. 2B).
FIG. 2.
Detection of c-fms, CSF-1, and c-myc transcripts. Total RNA was isolated from both cells. RNA was reverse transcribed and amplified with pairs of PCR primers specific for the c-fms (A) or CSF-1 (B) gene. Specific products were visualized by ethidium bromide staining on a 1.8% agarose gel. (C) Northern blot experiments performed with total RNA from the cell lines listed and hybridized with c-myc- and GAPDH-specific probes. MW, molecular weight markers.
In addition, since the c-myc gene is induced and seems instrumental in the proliferative effects that result from the binding of CSF-1 to its receptor (7, 10), we tested expression of this proto-oncogene. Overexpression of c-myc in the E1 and TE1 cell lines was detected by Northern blotting, whereas c-myc was weakly expressed in the NC5 and TC T-cell lines (Fig. 2C).
The E1 and TE1 monocytic cell lines are infected by EBV.
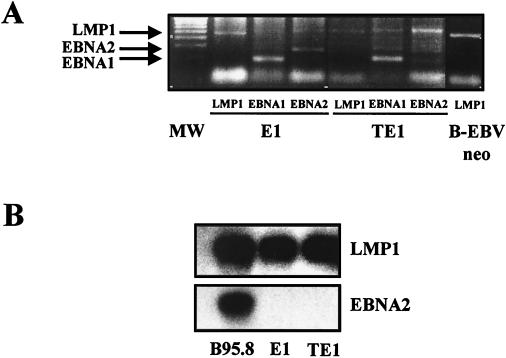
To confirm that E1 and TE1 cells were infected by EBV, the presence of viral DNA in Hirt's extracts was investigated (8). Thus, we could detect by PCR the LMP-1-, EBNA-1-, and EBNA-2-encoding genes (Fig. 3A). In addition, preparative PCR was performed to determine the sequences of the LMP-1 gene (also called BNLF-1) and its promoter on DNA extracted from various EBV-infected cells. These included the E1 and TE1 cell lines, as well as the two infected T-cell lines, NC5 and TC, derived from the same donor (17, 37). LMP-1 sequences of the corresponding B-LCL (B-EBVneo) and donor PBMCs (PBL25) were also analyzed. Except for the results for PBL25 cells, the sequences of 5′ and 3′ PCR-amplified fragments encompassing the LMP-1 coding region were identical to those obtained from the prototype B95.8 strain. Sequence analysis of LMP-1 from PBL25 showed 15 sequence variations and two deletions compared to the B95.8 strain. However, only nine of the variations, found mainly in the transmembrane region, resulted in nonconservative amino acids (Table 4). The two deletions were found in the region coding for the cytoplasmic domain of the protein. One corresponds to the 30-bp deletion that was reported as possibly associated with an increased transforming potential of the LMP-1 protein (32). The other affects the number of repeats of the 33-bp motif (4 versus 4.5 repeats in PBL25 and B95.8 sequences, respectively) (1). This result demonstrates the presence of a very divergent resident EBV strain in donor cells but confirms that the viral strain present in our monocytic cell lines is derived from the B95.8 strain. This last point was corroborated by sequence analyses performed on the LMP-1 promoter that showed a perfect colinearity between the B95.8 strain and the strain present in the in vitro-transformed cell lines (data not shown).
FIG. 3.
E1 and TE1 expressed the EBV genome. (A) Detection of the EBV genome in infected monocytes. The presence of viral DNA in Hirt's extracts was evaluated by PCR amplification of three EBV latent genes: LMP-1, EBNA-1, and EBNA-2. The corresponding PCR products (see primers in Table 2) were visualized by ethidium bromide staining on a 1.8% agarose gel. An LCL was used as a positive control for detection of the viral gene. Lane MW, 100-bp ladder. (B) Detection of viral RNA in E1 and TE1 by RT-PCR and Northern blotting experiments using primers and probes specific for a selected set of viral genes (see primers in Table 2). B95.8 cells were used as a positive control.
TABLE 4.
Substitutions and deletions in the amino acid sequence of LMP-1 identified in EBV strains infecting donor cells or derived cell lines
| Nucleotide position | B95-8 sequence
|
Mutationa in PBL25 | ||
|---|---|---|---|---|
| Codon | Nucleotide or change | Amino acid | ||
| 5′ region | ||||
| 169183 | 46 | G | Asp | A/Asn |
| 169003 | 106 | T | Phe | A/Ile |
| 168943 | 126 | A | Leu | C/Phe |
| 168934 | 129 | G | Met | T/Ile |
| 168934 | 132 | A | Arg | G/Gly |
| 168889 | 144 | T | Phe | A/Ile |
| 168871 | 150 | A | Asp | C/Ala |
| 168868 | 151 | A | Leu | C/Ile |
| 3′ region | ||||
| 168754 | 189 | A | Gln | C/Pro |
| 168265-168294 | 352-343 | 30-bp deletion | NAb | Deletion |
Comparison is to the B95-8 reference strain (GenBank accession no. X66863). The mutated nucleotide and corresponding amino acid are shown. No mutations in cell lines B-EBVneo, NC5, TC, E1, and TE1 were found.
NA, not applicable.
EBV-infected monocytic cell lines expressed viral gene products characteristic of a type II latency.
Since an EBV genome was clearly detected in E1 and TE1 cells and since both cell lines presented a transformed phenotype and oncogenic properties, we decided to evaluate the presence of specific viral mRNA transcripts in these cells. Thus, we determined the expression pattern of EBV in E1 and TE1 by performing RT-PCR and blotting experiments using primers and probes specific for a selected set of viral genes. Figure 3B shows that E1 and TE1 expressed an LMP-1 transcript with a size similar to that of the transcript found in B95.8 cells. By contrast, no expression of EBNA-2 was detected in both cells. In addition, we found that the EBNA-1 transcript is selectively expressed from the Qp promoter and that the BZLF-1 mRNA, specific for the lytic phase, was not present (data not shown). As no BZLF-1 transcript was found in these cells, we did not explore further expression of the late lytic transcripts.
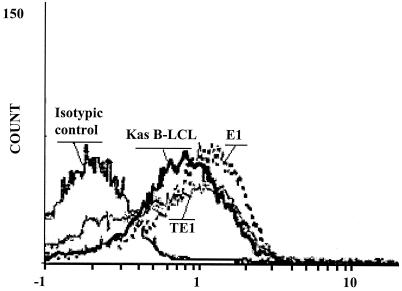
Furthermore, the LMP-1 protein product was detected by Western blotting (data not shown) and by flow cytometry analysis (Fig. 4). The expression levels of LMP-1 in E1 and TE1 are very similar to that found in Kas cells, a LCL used as a positive control. LMP-1 expression is more homogeneous in E1 than in TE1. Expression of EBNA-1 and LMP-1 together with the lack of EBNA-2 expression specifies type II latency for the virus in both cell lines.
FIG. 4.
Detection of LMP-1 protein expression in E1 and TE1 cell lines. After fixation and permeabilization, cells were first incubated with anti-LMP-1 antibodies (CS1-4) and then with an anti-mouse Ig labeled with PE. Cells were then analyzed by flow cytometry. Kas B-LCL was used as a positive control. The signal obtained with the isotypic control for E1 is indicated.
The E1 and TE1 cell lines display phagocytic activity and monokine production.
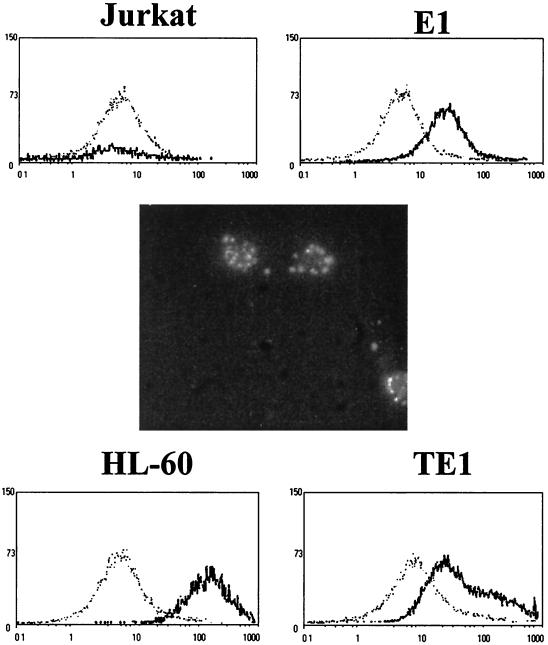
The main characteristic of monocytes is the ability to phagocytose foreign organisms in order to present antigens to T cells through class II proteins of the major histocompatibility complex. It has been reported that infection of monocytes by EBV alters their phagocytic activity (47). Figure 5 shows that E1 and TE1 cells exhibit a phagocytic activity when incubated with FITC-labeled E. coli at 37°C. After incubation at 4°C, this activity was strongly decreased to a low level comparable to that obtained at 37°C for Jurkat cells, the negative-control cell line.
FIG. 5.
Demonstration of phagocytic properties for EBV-infected monocytes E1 and TE1. E1 and TE1 were incubated with a suspension of E. coli labeled with FITC at 4 (dotted lines) and 37°C (solid lines). After two washes with cold PBS, cells were analyzed on a cytometer at 488 nm. Jurkat cells were used as a negative control, and HL60 cells were used as a positive control. In the center, phagocytosis of E. coli labeled with FITC by E1 cells at 37°C is shown by microscopic fluorescence analysis.
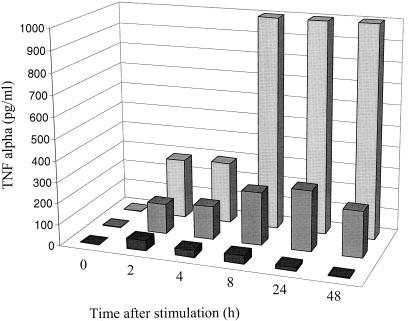
Monocytes and macrophages produce a set of cytokines, such as IL-8, IL-6, and TNF-α, which play the main role in the biological effect of these cells on the immune response, particularly during inflammation. IL-8, the prototypic CXC chemokine, is necessary for the recruitment of leukocytes in inflammatory processes. After 40 h of culture, E1 and TE1 cells produced IL-8 constitutively. This production was enhanced after stimulation by PMA. The amounts of IL-8 produced for these cell lines were higher than those produced for monocytic control cell lines HL60 and U937. IL-6 induces production of acute-phase proteins by hepatocytes. IL-6 was produced by E1 and TE1 cells mainly after activation by PMA. TNF-α plays a key role in the orchestration of inflammation. Its biological activity in L929 cells can be measured by using the cytotoxicity assay. Biologically active TNF-α is still present 40 h after stimulation of E1 and TE1 cells but not after stimulation of HL60 and U937 cell lines (Table 5). To investigate the basis of the persistence of this monokine 40 h after the activation, the kinetics of TNF-α production in E1 and TE1 cells and in a control monocytic cell line (U937) were analyzed. For U937, the maximum TNF-α production arises between 2 and 4 h after activation. For E1 and TE1, the appearance of the peak is delayed (24 h for E1) and the amounts produced by these two cell lines are higher than that produced by the U937 cell line (Fig. 6). Both these findings can explain the persistence of detectable levels of biologically active TNF-α at 40 h. As expected, control Jurkat cells produced TNF-α, IL-6, and small amounts of IL-8.
TABLE 5.
Production of three monokines (TNF-α, IL-6, and IL-8) in cell supernatants after 40 h of culture with (activated) or without (constitutive) cell activation by PMA and ionomycin
| Cell line | Expression | Levela of:
|
||
|---|---|---|---|---|
| TNF-α (U/ml) | IL-6 (pg/ml) | IL-8 (pg/ml) | ||
| Jurkat | Constitutive | < | < | 15 |
| Activated | < | < | 497 | |
| E1 | Constitutive | < | 10.2 | 356 |
| Activated | 14 | 15.4 | 20,520 | |
| TE1 | Constitutive | < | < | 780 |
| Activated | 26 | 18.4 | 24,740 | |
| U937 | Constitutive | < | 10.3 | 75 |
| Activated | < | 36.4 | 5,098 | |
| HL60 | Constitutive | < | < | 261 |
| Activated | < | < | 8,053 | |
IL-6 and IL-8 levels were evaluated by enzyme immunoassay, and TNF-α levels were determined by a cytoxicity test against L929.<, lower than the sensitivity threshold of the test.
FIG. 6.
Kinetics of TNF-α production in culture supernatants of E1 (dark gray bars) and TE1 (light gray bars) after stimulation by PMA (0.1 μg/ml). The TNF-α level was evaluated with an enzyme-linked immunosorbent assay. The U937 cell line (black bars) was used as positive cell control.
Antisense to LMP-1 down-regulates E1 and TE1 cell proliferation as well as expression of adhesion molecule ICAM-1 and the BCL-2 antiapoptotic protein
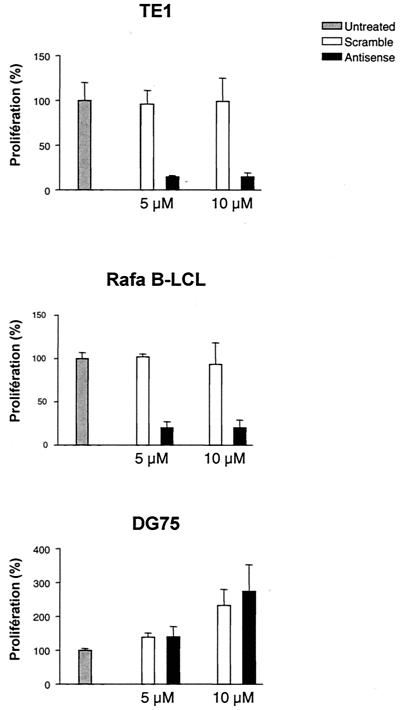
Since LMP-1 was demonstrated to display oncogenic properties (2, 50), we then wanted to know whether LMP-1 played a role in the immortalized and transformed phenotype of these cell lines. As a first approach, we investigated the proliferative rate of cells treated with LMP-1 antisense oligonucleotides. To this end, we designed an antisense oligodeoxynucleotide (AS2) corresponding to the first 20 nucleotides of the LMP-1 mRNA coding sequence (34). A randomly scrambled version of the LMP-1 antisense oligodeoxynucleotide (SC2) was used as a control (Fig. 7A). To show the efficacy of antisense oligonucleotides, the effects on the expression of LMP-1 were determined by immunoblotting using the S12 anti-LMP-1 monoclonal antibody. LMP-1 expression was significantly down-regulated in TE1 (Fig. 8B) and Rafa B-LCL (not shown) cells treated with the antisense oligodeoxynucleotide (AS2) compared to expression in control cells. As shown in Fig. 7, inhibiting LMP-1 reduced by 80% the [3H]thymidine incorporation of TE1 antisense oligodeoxynucleotide-treated cells. The 5 μM dose was as efficient as 10 μM, indicating that 5 μM is already a plateau dose. A weaker effect was observed with the E1 cell line (data not shown). As already reported by others (25), the LMP-1 antisense oligonucleotide also significantly reduced the proliferative rate of LCLs but did not selectively affect the proliferation of DG75, a Burkitt lymphoma cell line negative for EBV. A cytotoxic effect of LMP-1 antisense oligonucleotides can be ruled out because of the increase of DG 75 cell proliferation and the noncytotoxic effect observed for all the cell lines tested with scrambled oligonucleotides. This result indicates that LMP-1 is essential for the proliferation of E1 and TE1, pinpointing the critical role played by EBV infection in the phenotype and the properties of these cells.
FIG. 7.
Inhibition of proliferation of EBV-positive cells by an LMP-1 antisense oligonucleotide. Proliferative responses were evaluated by measuring [3H]thymidine uptake after 48 h of cell culture. Cell lines TE1, Rafa B-LCL, and DG 75 were cultured alone or with oligonucleotide AS2 (▪) or SC2 (□) at two concentrations (5 and 10 μM). Results were expressed as percentages compared with that for the untreated control. The proliferative rate of untreated cells was arbitrarily defined as 100%. The values represent the means of triplicate assays ± standard deviations.
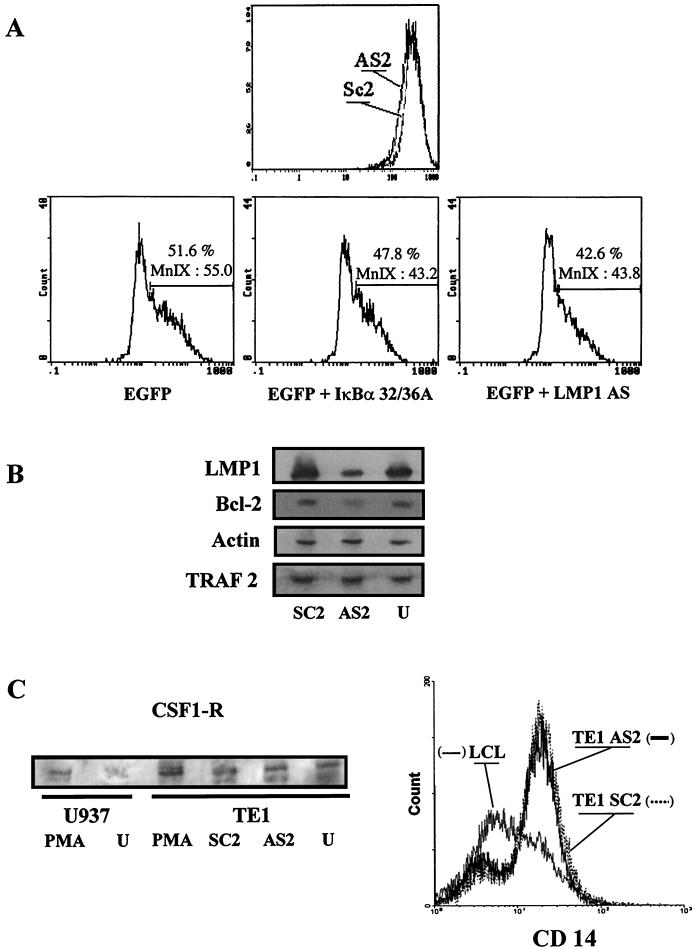
FIG. 8.
Effect of LMP-1 inhibition on expression of different markers in E1 or TE1 cells. (A) Effect on ICAM-1/CD54 expression on E1 cells analyzed by flow cytometry. (Top) The LMP-1 antisense oligonucleotide (AS2) down-regulated membrane expression of LMP-1 molecular target ICAM-1 on E1 cells. Scramble oligomers were used as the control (SC2). (Bottom) The same results were obtained with cells cotransfected with pEGFP-C1 and pSV LMP-1-AS or with pcDNA3 containing the coding sequence for mutated IκBα32/36A. The percentage of ICAM-1-expressing cells and the density of expression (MnI X) in these conditions are indicated. (B) Western blot analysis of several markers after LMP-1 inhibition experiments with TE1 cells. Immunoblots were probed with the S12 monoclonal antibody (LMP-1) or antibodies specific for TRAF-2, BCL-2, and actin proteins. Detection was performed after antisense oligonucleotide (AS2) or scramble (SC2) treatment, and results were compared to those for untreated cells (U). (C) Western blot and cytometry analyses of monocytic markers. (Left) Detection of CSF1-R after AS2, SC2, or PMA treatment in TE1 cells. U937 cells untreated or stimulated by PMA were used as the control for CSF1-R induction. (Right) Flow cytometry profile for CD14 labeling after treatment of TE1 cells. Rafa B-LCL is shown as a negative control.
Numerous reports using LCLs or transient expression of LMP-1 in noninfected cells stress the role of this receptor-like protein in induction of genes involved in cellular proliferation and/or activation (12). This is the case for ICAM-1, a gene encoding an adhesion molecule (CD54) induced or up-regulated by LMP-1 partly through induction of a NF-κB pathway (9). Thus, we have investigated by flow cytometry analysis modulation of ICAM-1 expression by LMP-1 in E1 and TE1 cells using the antisense strategy. As shown in Fig. 8A, we found that addition of AS2 to the culture medium reproducibly down-regulated ICAM-1 expression compared to addition of the scrambled control (SC2). The apparently modest down-regulation of ICAM-1 after antisense oligonucleotide treatment is relevant of LMP-1-independent constitutive expression of this protein in TE1 cells as well as in LCLs.
Similar results were obtained with antisense vectors in cotransfection experiments. A down-regulation of ICAM-1 expression was obtained for EGFP-positive cells after cotransfection with an EGFP-expressing vector and a plasmid encoding an antisense transcript of LMP-1 (pSV LMP-1-AS) or a mutated form of IκBα (Fig. 8A).
In LCLs, down-regulation of prototypic antiapoptotic protein BCL-2 was tested as a good indicator of loss of LMP-1 expression (24). We thus investigated the effects of antisense oligonucleotide treatment on expression of BCL-2, as well as TRAF-1 and TRAF-2, well-known mediators of the signaling properties of LMP-1. Whereas BCL-2 expression was reduced both in TE1 cells (Fig. 8B) and LCLs (data not shown), TRAF-2 was not affected. TRAF1, which is an LMP-1 target gene in LCLs and other cell lines (8) (data not shown), was not detected in TE1 cells (data not shown).
We wanted to check the consequences of LMP-1 inhibition on expression of monocytic markers by TE1. Using, respectively, flow cytometry and Western blot analysis, we did not detect any modulation of CD14 and CSF-1-R expression after LMP-1 antisense oligodeoxynucleotide treatment (Fig. 8C). As a control for the identity of the band detected with the anti-CSF-1-R antibody, we showed that it was induced after PMA treatment, further emphasizing a macrophage differentiation property shared with the U937 monocytic cell line (Fig. 8C). Finally, constitutive IL-8 production by TE1 was not affected by the antisense experiments (data not shown). Thus, these cells kept some functional or housekeeping characteristics in spite of loss of LMP-1-dependent proliferative properties.
DISCUSSION
In this work, we established and characterized two EBV-transformed monocytic cell lines. These cell lines, E1 and TE1, display a transformed and malignant phenotype as shown by their capacity to grow in long-term cultures and to develop tumors in nude mice. The E1 cell line was isolated from PBMCs infected in vitro, and TE1 was derived from a tumor induced by E1 cells in an immunodeficient mouse. Immunofluorescence, biochemical, and functional analyses confirmed their monocytic origin. The cells were infected by the B95.8 EBV strain used for infection as ascertained by sequencing the LMP-1 gene. Expression of EBNA-1 and LMP-1 at the RNA and/or protein levels was documented by RT-PCR and immunofluorescence analysis, respectively. Importantly, inactivation of LMP-1 in these cells severely hampered their proliferative properties, suggesting that EBV gene expression plays a significant role in their transformed phenotype.
In addition, this is the first report of a type II latency established in monocytic cell lines infected by EBV. Long-term cultures of macrophages obtained from biological samples have been described (48). All of them expressed latent genes, as assessed by detection of EBNA-2 and LMP-1 proteins, and some expressed one or more genes of the lytic phase such as BZLF-1. However, unlike the E1 and TE1 cell lines, none of these cultures were established, nor did they induce tumors in nude mice. Importantly, the E1 and TE1 cells expressed LMP-1 and EBNA-1 but not EBNA-2 or lytic-phase protein BZLF-1. This pattern corresponds to a type II latency, previously reported for NPC, T-cell lymphoma, and HD, as well as in the in vitro-infected T-cell lines that we previously characterized (17).
Recently, it has been found that EBV can infect fresh human monocytes, in which a replicative cycle is observed, without viral persistence and without installation of latency. In addition, those EBV-infected monocytes have reduced phagocytic activity and TNF-α production (47). This inhibitory effect on TNF-α production seems to be triggered by the virus without penetration into the cells (16). These processes could reflect a strategy developed by the virus to escape immune system control in vivo. In contrast, the protocol we used, with a 5-week selective phase, could favor the appearance of infected cells in a latent phase rather than in a lytic phase. In addition, both E1 and TE1 display phagocytic activity and secrete high levels of proinflammatory cytokines such as TNF-α. Therefore, the viral status in the respective monocytic cell lines obtained may dictate the functional properties of the cells.
Some human hemophagocytic syndromes are related to EBV infection (22, 26, 27). Related to the pathogenesis of this EBV-associated syndrome, enhanced phagocytosis and secretion of TNF-α by monocytes/macrophages (31, 40) seemingly result from an activation of their functions by EBV-infected T cells. We show here that, consequent to their infection by EBV, monocyte/macrophage cell lines display abnormally high TNF-α production, reminiscent of the effects of hemophagocytic syndromes. Therefore, our results argue for a careful reexamination of EBV infection of macrophages in hemophagocytic syndromes, which was not precisely investigated.
The two monocytic cell lines described here expressed a nonmutated LMP-1 protein. To assess LMP-1 functionality in E1 and TE1 cells, expression of ICAM-1, an LMP-1 target molecule, was examined. After 48 h of cell culture with an antisense LMP-1 oligonucleotide or with a plasmid encoding an antisense LMP-1 transcript, down-regulation of ICAM-1, compared to what was found for control cells, was observed in both cases. This result indicates that, in E1 and TE1 cells, LMP-1 is functional and can modulate cellular genes. As expected (5, 14), a dominant-negative IκBα32/36A, which inhibits the NF-κB pathway, affected ICAM-1 expression, suggesting that ICAM-1 down-regulation following introduction of a LMP-1 antisense oligodeoxynucleotide partially relied on the NF-κB pathway.
The critical proliferative and antiapoptotic roles played by LMP-1 in various cellular models including LCLs is well established (23, 24, 25). Though more controversial, some reports show a correlation between LMP-1 expression and tumorigenicity in different cell types (28, 38, 44). Both the monocyte E1 cells and the NC5 T-cell line we previously described (17, 37) expressed LMP-1 and are tumorigenic after injection into immunodeficient mice. The fact that this tumorigenic property is not shared by LCLs seems not related to a difference in the levels of LMP-1 expression, since B95.8-transformed E1 cells and the Kas B-LCL expressed similar amounts of the LMP-1 protein. In line with this observation, we did not detect any mutation in the LMP-1 promoter region of the EBV strain infecting these cell lines. LMP-1 inactivation using an antisense strategy suggests an instrumental role for LMP-1 in the proliferative status of the infected monocytes. Indeed, the results clearly demonstrate that cells with reduced LMP-1 expression were greatly impaired in their proliferation. Moreover, the fact that BCL-2 was down-regulated after antisense oligonucleotide treatment suggests a role for LMP-1 in TE1 cell survival, as is the case for LCLs (24).
The role of LMP-1 in establishment and maintenance of the proliferative status of LCLs expressing type III latency is well known (43). Lack of relevant experimental models hampered investigation of this role in other EBV-infected cell lines expressing LMP-1. Recently however, Noguchi et al. showed that proliferation of two LMP-1-positive NK lymphoma cell lines was insensitive to the LMP-1 antisense strategy used in this work (39). Thus, to our knowledge, this is the first report of the instrumental role of LMP-1 in maintaining proliferation of EBV-infected cells expressing a type II latency.
Of note, whereas ectopic expression of c-myc renders proliferation of LCLs independent of LMP-1 (41), c-myc overexpression did not alleviate the LMP-1 dependence of the growth of EBV-transformed monocytes. This key regulator of cell proliferation could be targeted by the binding of CSF-1 to its cognate receptor, expressed by both E1 and TE1 cells. Whether this results from a possible autocrine effect of CSF-1 produced by these cells and what could be the role of LMP-1 in this case remain to be addressed. As a preliminary result, we showed that, like other monocytic markers (CD14 and IL-8 secretion), CSF-1-R expression was not affected by LMP-1 inhibition.
A unique infected monocyte cell line was isolated concomitantly with four T-cell lines. This observation suggests that infection and transformation of monocytes by EBV are exceptional events. Nevertheless, direct infection of T cells by EBV is also very rare, as demonstrated recently with a novel green fluorescent protein-expressing EBV (49). Thus, the existence of very few T cells and even fewer monocytes in an overwhelming majority of B cells could explain the isolation of four T-cell lines and of a unique monocytic cell line infected by EBV in our model. In agreement with this hypothesis, we were unable to detect CD14-expressing cells in LCLs, due probably to the paucity of monocytic cells in these cell lines (data not shown). This impeded the screening of additional infected clones by classical isolation processes. Infection of T cells and monocytes and their immortalization occur in exceptional cases, possibly influenced by the isolation protocol.
In B cells, the initial event for the entry of EBV is the interaction of the viral outer envelope glycoproteins gp350 and gp220 with CD21, the main receptor for EBV. In contrast, the path for entry of EBV in monocytes is not known. E1 and TE1 weakly expressed CD21 in a small percentage of cells and also HLA-DR molecules, known as CD21 coreceptors (18, 49). Alternatively, apoptotic bodies derived from EBV-carrying B lymphocytes can be a source of viral transfer to phagocytosing cells such as monocytes/macrophages (19). Thus, the massive apoptosis triggered by the electroporation and selection processes used to yield E1 cells could favor this kind of entry. Finally, cellular fusion of an infected B or T cell with a monocytic cell was ruled out, since no rearrangement of Ig or T-cell receptor genes was detected.
In conclusion, our T and monocytic cell lines can serve as tools to elucidate this infection and transformation processes and represent unique in vitro models to study EBV-infected cells displaying a type II latency.
Acknowledgments
E.M. and E.A. contributed equally to this work.
We thank Philippe Gosset for help in performing monokine production assays, Jean Luc Baert, Jean Feuillard, Alain Sergeant, Evelyne Manet for reagents, and Véronique Fafeur for critical reading of the manuscript.
This work was supported by the Association pour la Recherche sur le Cancer (ARC; grants 9615 and 5455; fellowship for E.A.), CNRS, Lille II, Institut Pasteur de Lille, and INSERM.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Baichwal, V. R., and B. Sudgen. 1988. Transformation of Balb/3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 12:1-9. [PubMed] [Google Scholar]
- 3.Baumforth, K. R. N., L. S. Young, K. J. Flavell, C. Constandinou, and P. G. Murray. 1999. The Epstein-Barr virus and its association with human cancers. Mol. Pathol. 52:307-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brousset, P., F. Meggetto, S. Chittal, F. Bibeau, J. Arnaud, B. Rubin, and G. Delsol. 1993. Assessment of the methods for the detection of Epstein-Barr nucleic acids and related gene products in Hodgkin's disease. Lab. Investig. 69:483-490. [PubMed] [Google Scholar]
- 5.Cahir-McFarland, E. D., D. M. Davidson, S. L. Schauer, J. Duong, and E. Kieff 2000. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 97:6055-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. L., R. H. Sadler, D. M. Walling, I. J. Su, H.-C. Hsieh, and N. Raab-Traub. 1993. Epstein-Barr virus (EBV) gene expression in EBV-positive peripheral T-cell lymphomas. J. Virol. 67:6303-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, M., D. Wang, and M. F. Roussel. 1999. Expression of c-Myc in response to colony-stimulating factor requires mitogen-activated protein kinase kinase-1. J. Biol. Chem. 274:6553-6558. [DOI] [PubMed] [Google Scholar]
- 8.Chinsky, J., and R. Soeiro. 1981. Fv-1 host restriction of Friend leukemia virus: analysis of unintegrated proviral DNA. J. Virol. 40:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devergne, O., E. D. Cahir-McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey, A., H. She, L. Kim, A. Boruch, D. L. Guris, K. Carlberg, S. M. Sebti, D. T. Woodley, A. Imamoto, and W. Li. 2000. Colony-stimulating factor-1 receptor utilizes multiple signalling pathways to induce cyclin D2 expression. Mol. Biol. Cell 11:3835-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domke-Opitz, I., and H. Kirchmer. 1990. Stimulation of macrophages by endotoxin results in the reactivation of a persistent herpes simplex infection. Scand. J. Immunol. 32:69-75. [DOI] [PubMed] [Google Scholar]
- 12.Eliopoulos, A. G., and A. B. Rickinson. 1998. LMP1 masquerades as an active receptor. Curr. Biol. 8:R196-R198. [DOI] [PubMed] [Google Scholar]
- 13.Faumont, N., T. Al Saati, P. Brousset, C. Offer, G. Delsol, and F. Meggetto. 2001. Demonstration by single cell PCR that Reed-Sternberg cells as bystander B lymphocytes are infected by different Epstein-Barr virus in Hodgkin's disease. J. Gen. Virol. 82:1169-1174. [DOI] [PubMed] [Google Scholar]
- 14.Feuillard, J., M. Schuhmacher, S. Kohanna, M. Asso-Bonnet, F. Ledeur, R. Joubert-Caron, P. Bissières, A. Polack, G. W. Bornkamn, and M. Raphaël. 2000. Inducible loss of NF-κB activity is associated with apoptosis and Bcl-2 down-regulation in Epstein-Barr virus-transformed B lymphocytes. Blood 95:2068-2075. [PubMed] [Google Scholar]
- 15.Fleischer, J., E. Soeth, N. Reiling, E. Grage-Griebenow, H. D. Flad, and M. Ernst. 1996. Differential expression and function of CD80 (B7-1) and CD86 (B7-2) on human peripheral blood monocytes. Immunology 89:592-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosselin, J., L. Flamand, M. D'Addario, J. Hiscott, I. Stephanescu, D. V. Ablashi, R. C. Gallo, and J. Menezes. 1992. Modulatory effects of Epstein-Barr, herpes simplex and human herpes-6 viral infection and co-infection on cytokine synthesis. J. Immunol. 149:181-187. [PubMed] [Google Scholar]
- 17.Groux, H., F. Cottrez, C. Montpellier, B. Quatannens, J. Coll, D. Stehelin, and C. Auriault. 1997. Isolation and characterization of transformed human T-cell lines infected by Epstein-Barr virus. Blood 89:4521-4530. [PubMed] [Google Scholar]
- 18.Haan, K. M., W. W. Kwok, R. Longnecker, and P. Speck. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmgren, L., A. Szeles, E. Rajnavölgyi, J. Folkman, G. Klein, I. Ernberg, and K. I. Falk. 1999. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood 93:3956-3963. [PubMed] [Google Scholar]
- 20.Ibanez, C. E., R. Schrier, P. Ghazal, P. Wiley, and J. A. Nelson. 1991. Human cytomegalovirus productively infects primary differentiated macrophages. J. Virol. 65:6581-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawa, K 2000. Epstein-Barr virus-associated diseases in humans. Int. J. Hematol. 71:108-117. [PubMed] [Google Scholar]
- 22.Kawaguchi, H., T. Miyashita, H. Herbst, G. Niedobitek, M. Asada, M. Tsuchida, R. Hanada, A. Kinoshita, M. Sakurai, N. Kobayashi, et al. 1993. Epstein-Barr virus-infected T lymphocytes in Epstein-Barr virus-associated hemophagocytic syndrome. J. Clin. Investig. 92:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawanishi, M. 1997. Expression of Epstein-Barr virus latent membrane protein 1 protects Jurkat T cells from apoptosis induced by serum deprivation. Virology 228:244-250. [DOI] [PubMed] [Google Scholar]
- 24.Kenney, J. L., M. E. Guinness, T. Curiel, and J. Lacy. 1998. Antisense to the Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP-1) suppresses LMP-1 and Bcl-2 expression and promotes apoptosis in EBV-immortalized B-cells. Blood 92:1721-1727. [PubMed] [Google Scholar]
- 25.Kenney, J. L., M. E. Guinness, M. Reiss, and J. Lacy. 2001. Antisense to the Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP1) sensitizes EBV-immortalized B-cells to transforming growth factor-beta and chemotherapeutic agents. Int. J. Cancer 91:89-98. [DOI] [PubMed] [Google Scholar]
- 26.Kikuta, H. 1995. Epstein-Barr virus associated hemophagocytic syndrome. Leukoc. Lymphoma 16:425-429. [DOI] [PubMed] [Google Scholar]
- 27.Kikuta, H., Y. Sakiyama, S. Matsumoto, T. Oh-Ishi, T. Nakano, T. Nagashima, T. Oka, T. Hironaka, and K. Hirai. 1993. Fatal Epstein-Barr virus associated hemophagocytic syndrome. Blood 82:3259-3264. [PubMed] [Google Scholar]
- 28.Kim, K.-R., T. Yoshizaki, H. Miyamori, K. Hasegawa, T. Horikawa, M. Furukawa, S. Harada, M. Seiki, and H. Sato. 2000. Transformation of Madin-Darby canine kidney (MDCK) epithelial cells by Epstein-Barr virus latent membrane protein 1 (LMP1) induces expression of Ets1 and invasive growth. Oncogene 19:1764-1771. [DOI] [PubMed] [Google Scholar]
- 29.Kondo, K., T. Kondo, T. Okuno, M. Takahashi, and K. Yamanishi. 1991. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 72:1401-1408. [DOI] [PubMed] [Google Scholar]
- 30.Larochelle, B., L. Flamand, P. Gourde, D. Beauchamp, and J. Gosselin. 1998. Epstein-Barr virus infects and induces apoptosis in human neutrophils. Blood 92:291-299. [PubMed] [Google Scholar]
- 31.Lay, J. D., C. J. Tsao, J. Y. Chen, M. E. Kadin, and I. J. Su. 1997. Upregulation of tumor necrosis factor-alpha gene by Epstein-Barr virus and activation of macrophages in Epstein-Barr virus-infected T cells in the pathogenesis of hemophagocytic syndrome. J. Clin. Investig. 100:1969-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, S. N., Y. S. Chang, and S. T. Liu. 1996. Effect of a 10-amino acid deletion on the oncogenic activity of latent membrane protein 1 of Epstein-Barr virus. Oncogene 12:2129-2135. [PubMed] [Google Scholar]
- 33.Lindhout, E., A. Lakeman, M. L. C. M. Mevissen, and C. de Groot. 1994. Functionally active Epstein-Barr virus-transformed follicular dendritic cell-like cell lines. J. Exp. Med. 179:1173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattia, E., S. Chichiarelli, T. Hickish, A. Gaeta, C. Mancini, D. Cunningham, and J. van Renswoude. 1997. Inhibition of in vitro proliferation of Epstein-Barr virus infected B cells by an antisense oligodeoxynucleotide targeted against EBV latent membrane protein LMP1. Oncogene 15:489-493. [DOI] [PubMed] [Google Scholar]
- 35.Mehl, A. M., N. Fischer, M. Rowe, F. Hartmann, H. Daus, L. Trumper, M. Pfreundschuh, N. Muller-Lantzsch, and F. A. Grasser. 1998. Isolation and analysis of two strongly transforming isoforms of the Epstein-Barr virus (EBV)-encoded latent membrane protein-1 (LMP1) from a single Hodgkin's lymphoma. Int. J. Cancer 76:194-200. [DOI] [PubMed] [Google Scholar]
- 36.Menet, A., C. Speth, C. Larcher, W. M. Prodinger, M. G. Schwendinger, P. Chain, M. Jäger, F. Schwarzmann, H. Recheis, M. Fontaine, and M. P. Dierich. 1999. Epstein-Barr virus infection of human astrocyte cell line. J. Virol. 73:7722-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montpellier, C., P. Crépieux, B. Quatannens, B. Delobel, M. F. Croquette, D. Stehelin, C. Auriault, H. Groux, and J. Coll. 1997. Homologous T and B cells immortalized in vitro by the Epstein-Barr virus exhibit differential genetic and functional features. Int. J. Oncol. 11:87-96. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson, L. J., P. Hopwood, I. Johannessen, J. R. Salisbury, J. Codd, D. Thorley-Lawson, and D. H. Crawford. 1997. Epstein-Barr virus latent membrane protein does not inhibit differentiation and induces tumorigenicity of human epithelial cells. Oncogene 15:275-283. [DOI] [PubMed] [Google Scholar]
- 39.Noguchi, T., K. Ikeda, K. Yamamoto, I. Yoshida, A. Ashiba, J. Tsuchiyama, K. Shinagawa, T. Yoshino, M. Takata, and M. Harada. 2001. Antisense oligodeoxynucleotides to latent membrane protein 1 induce growth inhibition, apoptosis and Bcl-2 suppression in Epstein-Barr virus (EBV)-transformed B-lymphoblastoid cells, but not in EBV-positive natural killer cell lymphoma cells. Br. J. Haematol. 114:84-92. [DOI] [PubMed] [Google Scholar]
- 40.Ohga, S., A. Matsuzaki, M. Nishizaki, T. Nagashima, T. Kai, M. Suda, and K. Ueda. 1993. Inflammatory cytokines in virus hemophagocytic syndrome. Am. J. Pediatr. Hematol. Oncol. 15:291-298. [PubMed] [Google Scholar]
- 41.Polack, A., K. Hortnagel, A. Pajic, B. Christoph, B. Baier, M. Falk, J. Mautner, C. Geltinger, G. W. Bornkamm, and B. Kemples. 1996. c-myc activation renders proliferation of Epstein-Barr virus (EBV)-transformed cells independent of EBV nuclear antigen 2 and latent membrane protein 1. Proc. Natl. Acad. Sci. USA 93:10411-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Revoltella, R. P., E. Vigneti, A. Fruscalzo, M. Park, G. Ragona, G. Rocchi, and E. Calef. 1989. Epstein-Barr virus DNA sequences in precursor monocyte-macrophage cell lines established from the bone marrow of children with maturation defects of haematopoiesis. J. Gen. Virol. 70:1203-1215. [DOI] [PubMed] [Google Scholar]
- 43.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 44.Roberts, M. L., and N. R. Cooper. 1998. Activation of a Ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology 240:93-99. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 7.19-7.22. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 46.Sariban, E., K. Imamura, M. Sherman, V. Rothwell, P. Pantazis, and D. Kufe. 1989. Downregulation of c-fms gene expression in human monocytes treated with phorbol esters and colony-stimulating factor 1. Blood 74:123-129. [PubMed] [Google Scholar]
- 47.Savard, M., C. Bélanger, M. Tardif, P. Gourde, L. Flamand, and J. Gosselin. 2000. Infection of primary human monocytes by Epstein-Barr virus. J. Virol. 74:2612-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimakage, M., M. Kimura, S. Yanoma, M. Ibe, S. Yokota, G. Tsujino, T. Kozuka, T. Dezawa, S. Tamura, A. Ohshima, M. Yutsudo, and A. Hakura. 1999. Expression of latent and replicative-infection genes of Epstein-Barr virus in macrophage. Arch. Virol. 144:157-166. [DOI] [PubMed] [Google Scholar]
- 49.Speck, P., and R. Longnecker. 1999. Epstein-Barr virus (EBV) infection visualized by EGFP expression demonstrates dependence on known mediators of EBV entry. Arch. Virol. 144:1123-1137. [DOI] [PubMed] [Google Scholar]
- 50.Wang, D., D. Liebowitz, and E. Kieff. 1985. An Epstein-Barr virus membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]