Abstract
Herpes simplex virus type 1 (HSV-1)-based amplicon vectors have a large transgene capacity and can efficiently infect many different cell types. One disadvantage of HSV-1 vectors is their instability of transgene expression. By contrast, vectors based on adeno-associated virus (AAV) can either persist in an episomal form or integrate into the host cell genome, thereby supporting long-term gene expression. AAV expresses four rep genes, rep68, -78, -40, and -52. Of those, rep68 or rep78 are sufficient to mediate site-specific integration of the AAV DNA into the host cell genome. The major disadvantage of AAV vectors is the small transgene capacity (∼4.6 kb). In this study, we constructed HSV/AAV hybrid vectors that contained, in addition to the standard HSV-1 amplicon elements, AAV rep68, rep78, both rep68 and -78, or all four rep genes and a reporter gene that was flanked by the AAV inverted terminal repeats (ITRs). Southern blots of Hirt DNA from cells transfected with the hybrid vectors and HSV-1 helper DNA demonstrated that both the AAV elements and the HSV-1 elements were functional in the context of the hybrid vector. All hybrid vectors could be packaged into HSV-1 virions, although those containing rep sequences had lower titers than vectors that did not. Site-specific integration at AAVS1 on human chromosome 19 was directly demonstrated by PCR and sequence analysis of ITR-AAVS1 junctions in hybrid vector-transduced 293 cells. Cell clones that stably expressed the transgene for at least 12 months could easily be isolated without chemical selection. In the majority of these clones, the transgene cassette was integrated at AAVS1, and no sequences outside the ITR cassette, rep in particular, were present as determined by PCR, ITR rescue/replication assays, and Southern analysis. Some of the clones contained random integrations of the transgene cassette alone or together with sequences outside the ITR cassette. These data indicate that the long-term transgene expression observed following transduction with HSV/AAV hybrid vectors is, at least in part, supported by chromosomal integration of the transgene cassette, both randomly and site specifically.
Herpes simplex virus type 1 (HSV-1)-based amplicon vectors are bacterial plasmids that contain two HSV-1 cis elements, an origin of DNA replication (ori), and a DNA packaging/cleavage signal (pac). These two elements are sufficient to support replication of the vector DNA and packaging of the concatemeric replication products into virions in the presence of HSV-1 helper functions (34). HSV-1 helper functions can be provided by a replication-conditional helper virus (8, 20) or a replication-competent, packaging-defective HSV-1 genome cloned as either a set of cosmids (7) or a bacterial artificial chromosome (14, 29, 30, 36). As the HSV-1 ori and pac sequences are smaller than 1 kb, amplicons have the capacity to accommodate at least 150 kbp of foreign DNA, the size of the HSV-1 genome (40). Amplicons have been demonstrated to transfer a wide range of transgenes with a high efficiency into many different cells both in culture and in vivo (6, 7, 15, 25, 30, 42, 45). However, the major drawback of HSV-1-based vectors is that transgene expression is in general transient (6, 7, 16).
On the other hand, adeno-associated virus (AAV) vectors, which have a very limited transgene capacity (∼4.6 kb), have been shown to mediate long-term gene expression. The wild-type AAV genome is a linear, single-stranded DNA of 4.7 kb containing 145-base inverted terminal repeats (ITRs) at both ends that flank two clusters of genes, rep and cap (23, 35). The ITRs contain the origin of DNA replication and the packaging signal. The rep gene encodes four overlapping proteins, Rep78, -68, -52, and -40, from two different promoters, p5 and p19. ITRs and either Rep78 or Rep68 are sufficient for replication of the AAV genome in the presence of a helper virus, such as adenovirus or HSV-1. In the absence of helper virus, ITRs and either Rep78 or -68 are sufficient to mediate the integration of the AAV genome into a specific site, termed AAVS1, on chromosome 19 of human cells (18, 21, 22, 32, 38).
Recently, HSV/AAV hybrid vectors have been designed to combine the large transgene capacity of HSV-1 with the potential for site-specific integration and long-term gene expression of AAV in a single vector. In addition to HSV-1 ori and pac, HSV/AAV hybrid vectors contain the AAV rep gene and a transgene cassette that is flanked by AAV ITRs. HSV/AAV hybrid vectors showed a much higher stability of transgene expression both in culture and in vivo than that of standard amplicons, but the mechanisms responsible for this, in particular the question of whether these vectors mediate site-specific integration in AAVS1, have not been addressed (5, 6, 16). The aim of this study was to determine qualitatively whether HSV/AAV hybrid vectors can mediate site-specific integration at AAVS1.
MATERIALS AND METHODS
Cell culture.
VERO 2-2 cells (33), human 293 cells, and the Detroit 6 cell line 7374 (4), which is latently infected with wild-type AAV2, were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum.
Vector constructs.
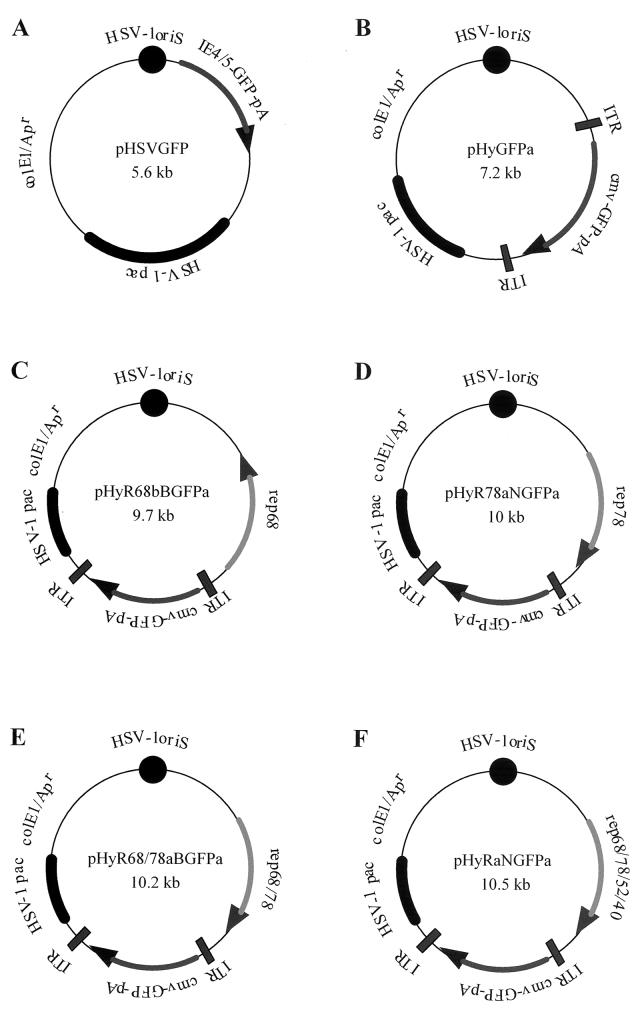
pHSVGFP (Fig. 1A) is a standard HSV-1 amplicon which expresses the gene for green fluorescent protein (GFP) from the HSV-1 immediate-early (IE) 4/5 promoter (1). pHyGFPa (Fig. 1B) is similar to pHSVGFP, except that the GFP reporter gene is controlled by the human cytomegalovirus (HCMV) IE1 enhancer/promoter and flanked by AAV ITRs. Construction of pHyGFPa required two cloning steps. First, plasmid pHSVGN (5) was cleaved with BglII and SphI and blunt end repaired with T4 DNA polymerase, and the 2-kb fragment which contains the HCMV IE1 enhancer/promoter, GFP coding sequences, and the bovine growth hormone (bGH) polyadenylation signal was ligated between the DraIII (blunt end repaired) and SnaBI sites of pAV2 (ATCC no. 37216; 19); the resulting plasmid was named pAV2GFP. Of note, pAV2 contains the entire AAV2 genome; cleavage of pAV2 with DraIII and SnaBI eliminated nucleotides 235 to 4490 of the AAV2 genome, including rep and cap coding sequences, but retained the ITRs. Second, pAV2GFP was cut with BglII, which flanks the ITR-HCMV IE1 enhancer/promoter-GFP-bGH poly(A)-ITR cassette, and the 2.3-kb fragment was inserted into the BamHI site located between the HSV-1 origin of DNA replication (oriS) and the HSV-1 DNA packaging/cleavage signal (pac) of pHSVNot (a standard HSV-1 amplicon that contains no transgene cassette; C. Fraefel, unpublished data). The resulting plasmid was named pHyGFPa. Plasmids p5R68 and p5R78, which express the rep68 and rep78 genes, respectively, from the p5 promoter were kindly provided by M. Urabe, Jichi Medical School, Tochigi, Japan (38). To construct pHyR68bBGFPa (Fig. 1C), p5R68 was cut with NotI and blunt end repaired with T4 DNA polymerase, and the 2.4-kb fragment containing rep68 and SV40 poly(A) sequences was inserted into the unique SphI site (blunt end repaired) of pHSVNot; the resulting plasmid was named pHSVR68b. Then, the 2.3-kb BglII fragment from pAV2GFP, which contains the ITR-flanked transgene cassette, was inserted into the unique BglII site of pHSVR68b. To construct pHyR78aNGFPa (Fig. 1D), p5R78 was cleaved with NotI and blunt end repaired with T4 DNA polymerase, and the 2.3-kb fragment containing rep78 and SV40 poly(A) sequences was inserted into the unique SphI site (blunt end repaired) of pHSVNot; the resulting plasmid was named pHSVR78a. Then, the 2.3-kb BglII fragment from pAV2GFP, which contains the ITR-flanked transgene cassette, was blunt end repaired and inserted into the unique NotI site (blunt end repaired) of pHSVR78a. The construction of pHyR68/78aBGFPa (Fig. 1E) included three cloning steps. First, the mutation in the splice donor of rep68 was repaired on p5R78 by replacing the SalI-XhoI fragment with the 0.9-kb SalI-XhoI fragment from pSSV9, a plasmid that contains the entire AAV genome. The resulting clone, pR68/78, has been confirmed by sequence analysis. Second, pR68/78 was cut with NotI and blunt end repaired with T4 DNA polymerase, and the 2.3-kb fragment containing rep68/78 and SV40 poly(A) sequences was inserted into the unique SphI site (blunt end repaired) of pHSVNot; the resulting plasmid was named pHSVR68/78a. Third, the 2.3-kb BglII fragment from pAV2GFP, which contains the ITR-flanked transgene cassette, was blunt end repaired and inserted into the unique NotI site (blunt end repaired) of pHSVR68/78a. The construction of pHyRaNGFPa (Fig. 1F) included several cloning steps. First, the mutations of the ATG codon of rep40 and -52 and the splice donor of rep68 were repaired on p5R78 by replacing the BglII-XhoI fragment with the 2.4-kb XbaI-EcoNI (blunt-end-repaired) fragment from pSSV9. The resulting plasmid was confirmed by sequence analysis and designated pRep. Then, pRep was cut with NotI and blunt end repaired with T4 DNA polymerase, and the 2.5-kb fragment was inserted into the unique SphI site (blunt) of pHSVNot to form pHyRa. In parallel, the 2.3-kb BglII fragment from pAV2GFP was inserted into the BamHI site of pUC18-Not, a derivative of pUC18 in which the multiple cloning site is flanked by NotI sites (12). Then, the resulting plasmid, pAV2GFP-Not, was cleaved with NotI, and the 2.3-kb fragment containing the ITR-flanked transgene cassette was inserted into the unique NotI site of pHyRa, forming pRep. All plasmids containing ITRs were cloned and maintained in Escherichia coli SURE cells (Stratagene, La Jolla, Calif.).
FIG. 1.
Schematic representation of HSV-1 amplicon (A) and HSV/AAV hybrid vectors (B to F) used in this study. See Materials and Methods for details on the construction. The HSV-1 origin of DNA replication (oriS) and DNA packaging/cleavage signal (pac), the E. coli origin of DNA replication (colE1) and ampicillin resistance gene (Apr), and the AAV ITRs and rep sequences (rep68, rep78, rep68/78, and rep68/78/52/40) are shown. The vectors contain a transgene cassette consisting of the HSV-1 IE 4/5 promoter (A) or the HCMV IE1 enhancer/promoter (B to F), the gene for GFP, and the bGH polyadenylation signal (pA). Arrows indicate the orientations of the GFP or rep genes.
Western blot analysis.
DNA transfections were performed by using the Lipofectamine procedure as described by the manufacturer (Life Technologies, Basel, Switzerland). Briefly, 106 VERO 2-2 cells were plated on 6-cm-diameter tissue culture plates. The following day, the cultures were cotransfected with 0.5 μg of vector DNA and packaging-defective HSV-1 helper DNA (2 μg of fHSVΔpacΔ27ΔKn and 0.2 μg of pEBHICP27) as described previously for the packaging of HSV-1 amplicons into virus particles (29, 30). fHSVΔpacΔ27ΔKn is a bacterial artificial chromosome that contains a packaging-defective HSV-1 genome with deletions in the pac signals and the IE2 gene; pEBHICP27 expresses the IE2 gene and complements the defect in fHSVΔpacΔ27ΔKn (29). Two days after transfection, the cells were washed with phosphate-buffered saline (PBS), lysed with 350 μl of 3× sodium dodecyl sulfate (SDS) loading buffer (150 mM Tris-HCl [pH 6.8], 6% SDS, 0.3% bromophenol blue, 30% glycerol, 300 mM β-mercaptoethanol), and boiled for 5 min. Samples (2 × 105 cell equivalents) were separated by SDS-10% polyacrylamide gel electrophoresis and then transferred onto a nitrocellulose membrane. The membrane was blocked with 5% nonfat dry milk in PBS-T (PBS containing 0.05% Tween 20) for 2 h at 37°C and then rinsed once with PBS-T and washed twice for 15 min and twice for 5 min at room temperature with TBS-T (50 mM Tris-HCl [pH 7.9], 150 mM NaCl, 0.01% Tween 20). The primary antibody (mouse anti-AAV Rep protein clone 303.9; Research Diagnostics Inc., Flanders, N.J.), which reacts with all four AAV Rep proteins, was diluted 1:25 in TBS-T containing 2% nonfat dry milk and applied to the membrane. After incubation for 1 h at room temperature, the membrane was washed twice for 15 min and twice for 5 min at room temperature with TBS-T and then incubated for 30 min with an alkaline phosphatase-conjugated rabbit anti-mouse immunoglobulin G (IgG) (Pierce, Rockford, Ill.) at 1:20,000 in TBS-T containing 5% nonfat dry milk. The membrane was washed repeatedly with TBS-T, and the bands were visualized using the substrate 5-bromo-4-chloro-3-indolyl phosphate (BCIP)-nitroblue tetrazolium according to the instructions provided by the supplier (Sigma).
Replication assays.
VERO 2-2 cells were transfected with vector DNA alone or cotransfected with vector DNA and packaging-defective HSV-1 helper DNA as described above. Two days after transfection, extrachromosomal DNA was isolated by the procedure described by Hirt (13). The DNA was digested extensively either with DpnI alone or with DpnI and a restriction endonuclease that cuts only once within the vector plasmids and was subjected to Southern analysis. After the DNA was separated on a 0.7% agarose gel and before transferring to a nylon membrane (Hybond N+; Amersham), the upper part of the gel (above 10 kb) was treated with 0.1 N HCl for 10 min to facilitate the transfer of high-Mr molecules. Hybridization with a digoxigenin-labeled GFP probe and immunological detection using an alkaline phosphatase-conjugated anti-digoxigenin antibody and chemiluminescence substrate (CDP Star) were performed as described by the supplier (Roche, Rotkreuz, Switzerland). The GFP probe was a 747-bp fragment derived from vector plasmid pHyRaNGFPa (10 ng) by PCR amplification using primers GFP1 (5′GGATCCATCGCCACCATGGTGAGCAAGGGC3′) and GFP2 (5′GTCGCGGCCGCTTTACTTGTACAGCTCGTC3′) and the PCR DIG Probe Synthesis Kit (Roche). PCR conditions were as follows. An initial denaturation step at 94°C for 4 min was followed by 35 cycles of amplification for 1 min at 94°C, 30 s at 56°C, and 1 min at 72°C. For final extension, the reaction mixture was incubated at 75°C for 10 min.
Preparation of vector stocks.
VERO 2-2 cells were transfected with vector plasmid DNA and packaging-defective HSV-1 helper DNA, vector particles were harvested 3 days after transfection, and vector titers were determined as described elsewhere (29).
Infection of human 293 cells with vector stocks and isolation of stable, GFP-positive cell clones.
Monolayers of 293 cells in 6-cm-diameter tissue culture plates were infected with stocks of pHSVGFP, pHyGFPa, or pHyRaNGFPa at a multiplicity of infection of 1 transducing unit per cell. Two days later, the cells were trypsinized and sorted by flow cytometry (FACSTAR; Becton Dickenson) at 103 GFP-positive cells per well in 96-well plates. After 2 weeks, 90% of the cells from 6 wells of the 96-well plate were pooled and expanded in T25-tissue culture flasks. The remaining cells were diluted and plated at 500 cells per plate onto 10-cm-diameter tissue culture plates to allow the formation of clones. Ten days later, GFP-positive clones were picked, expanded separately in wells of a 96-well plate, clone purified again on 10-cm-diameter tissue culture plates, and then expanded.
PCR analysis of genomic DNA.
GFP sequences were amplified by using the primers and conditions described above (replication assay) and 0.5 μg of genomic DNA as template. A 744-bp fragment within the rep coding sequence was amplified using primers rep5′ (5′GAACGCGATATCGCAGCCGCCATGCCGGG3′) and repNt3′ (5′GGATCCGAATTCACTGCTTCTCCGAGGTAATC3′). Conditions were as follows. An initial denaturation step at 94°C for 4 min was followed by 35 cycles of amplification for 2 min at 94°C, 30 s at 50°C, and 2 min at 72°C. For the final extension, the reaction mixture was incubated at 75°C for 10 min. Nested PCR to detect integration of vector sequences at AAVS1 was performed with a set of GFP- and AAVS1-specific primers, and genomic DNA was isolated from 293 cells 2 days after infection with pHyRaNGFPa. Primers GFP1 (see above) and AAVS1 1A (39) were used in the first round of amplification with 0.5 μg of genomic DNA as template. Four percent of the product was used in a second round of amplification using primers GFP4 (5′GGAGGACGGCAACATCCTGGGGCACAA3′) and AAVS2 2A (39). Conditions were as described by Tsunoda et al. (39). The product of the second PCR amplification was purified on a 1% agarose gel and cloned into plasmid pCR2.1TOPO (Invitrogen, De Schelp, The Netherlands). Eight clones were selected for sequence analysis (Microsynth, Balgach, Switzerland).
Rescue/replication of ITR-flanked transgene cassettes from GFP-positive cell clones.
Then, 106 cells from the GFP-positive 293 RaGa cell clones were plated in 6-cm-diameter tissue culture plates and, 24 h later, either mock transfected or transfected with packaging-defective HSV-1 helper DNA alone or cotransfected with packaging-defective HSV-1 helper DNA and a rep-expressing plasmid (pRep). Two days later, Hirt DNA was prepared, separated on a 0.7% agarose gel, transferred to a nylon membrane, hybridized to a digoxigenin-labeled GFP probe, and detected as described above (replication assay).
Southern analysis for colocalization of AAVS1 and GFP sequences.
Total genomic DNA was prepared from two T75-tissue culture flasks essentially as described by Sambrook et al. (31). The DNA (15 μg) was cleaved overnight with EcoRI or HindIII, separated on 0.7% agarose gels, and blotted onto nylon membranes (Hybond N+; Amersham). The membranes were prehybridized for 2 h at 68°C and then hybridized overnight at 68°C to random primed 32P-labeled probes. To detect the hybridizing bands, the membranes were exposed to X-ray film for 4 to 7 days. To rehybridize to a different probe, the membranes were stripped with 1 mM Tris-HCl (pH 8.0)-1 mM EDTA (pH 8.0)-0.1× Denhardt's reagent for 2 h at 75°C (31). The first probe was a ∼290-bp fragment within AAVS1 derived from plasmid pRVK, which contains the 3.5-kb EcoRI-KpnI fragment from AAVS1 (17), by PCR amplification using primers 79 and 80 (for nucleotide sequences and conditions, see reference 38). The second probe was the 747-bp GFP fragment derived from pHyRaNGFPa by PCR amplification using primers GFP1 and GFP2 (see above). The third probe was the 744-bp rep fragment derived from pHyRaNGFPa by PCR amplification using primers rep5′ and repNt3′ (see above).
RESULTS
Genetic elements from HSV-1 and AAV are functional in context of HSV/AAV hybrid vector.
Figure 1 is a schematic representation of the HSV-1 amplicon and the HSV/AAV hybrid vectors used in this study. All vectors contain the HSV-1 cis elements, oriS and pac, for replication of the vector DNA and packaging into virus particles in the presence of HSV-1 helper functions in mammalian cells. pHSVGFP (Fig. 1A) is a standard HSV-1 amplicon that expresses the GFP reporter gene from the HSV-1 IE 4/5 promoter. Plasmid pHyGFPa (Fig. 1B) is an HSV/AAV hybrid vector that contains a GFP reporter gene controlled by the HCMV IE1 enhancer/promoter and flanked by AAV ITRs. Plasmids pHyR68bBGFPa, pHyR78aNGFPa, pHyR68/78aBGFPa, and pHyRaNGFPa (Fig. 1C to F) are similar to pHyGFPa except that sequences for rep68, rep78, rep68/78, or all four rep genes are included. The construction of the vectors is described in detail in Materials and Methods.
We determined (i) whether the individual rep genes are expressed from the different hybrid vectors and (ii) whether the genetic elements required for replication of the two viruses are functional in the context of the hybrid vector.
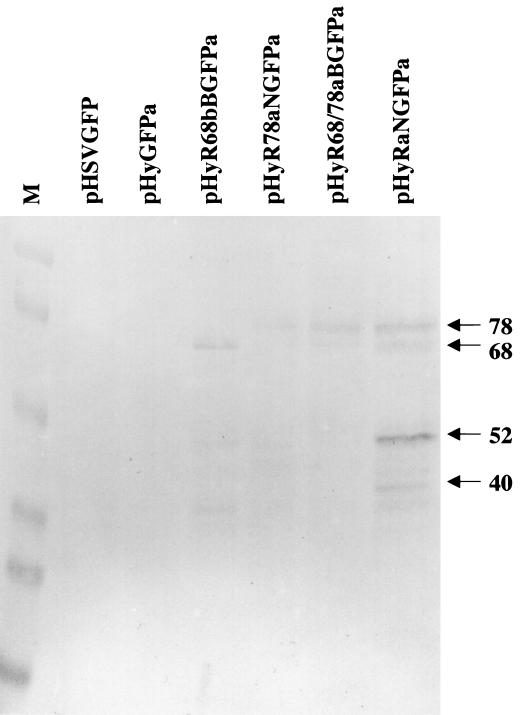
For Western analysis, VERO 2-2 cells were cotransfected with vector DNA and packaging-defective HSV-1 helper DNA. Twenty-four hours after transfection, total protein was resolved by SDS-polyacrylamide gel electrophoresis and analyzed using a monoclonal antibody that recognizes all four Rep proteins (Fig. 2). No Rep-specific bands were detected in cells transfected with pHSVGFP or pHyGFPa, which do not contain any rep sequences. In both pHyR68bBGFPa- and pHyR78aNGFPa-transfected cells, immunoreactive bands with sizes consistent with those expected for Rep68 and Rep78a, respectively, could be detected (38). The same bands were detected also in cells transfected with pHyR68/78aBGFPa, which contains the entire sequences of rep68 and rep78. Immunoreactive protein bands with sizes consistent with Rep40, -52, -68, and -78 were detected in cells transfected with pHyRaNGFPa, which contains the sequences for all four Rep proteins. This result shows that the individual rep genes were expressed from the hybrid vectors.
FIG. 2.
Western blot analysis of rep expression from different vectors. pHSVGFP, pHyGFPa, pHyR68bBGFPa, pHyR78aNGFPa, pHyR68/78aBGFPa, and pHyRaNGFPa DNA was transfected into VERO 2-2 cells, cell lysates were prepared 24 h later, and total protein was resolved by SDS-polyacrylamide gel electrophoresis and analyzed using a monoclonal antibody that recognizes all four Rep proteins as described in Materials and Methods. Lane M, molecular weight standard. Immunoreactive bands with sizes consistent with those of Rep78, -68, -52, and -40 are indicated.
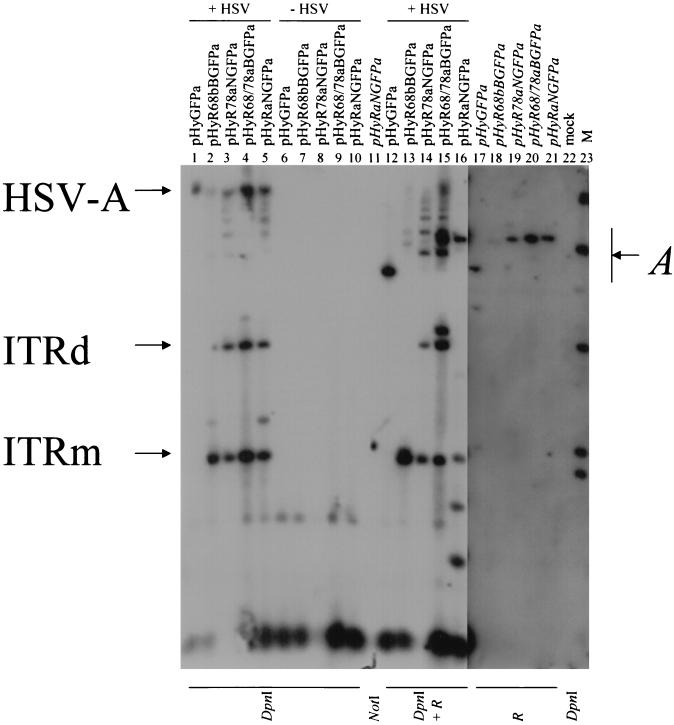
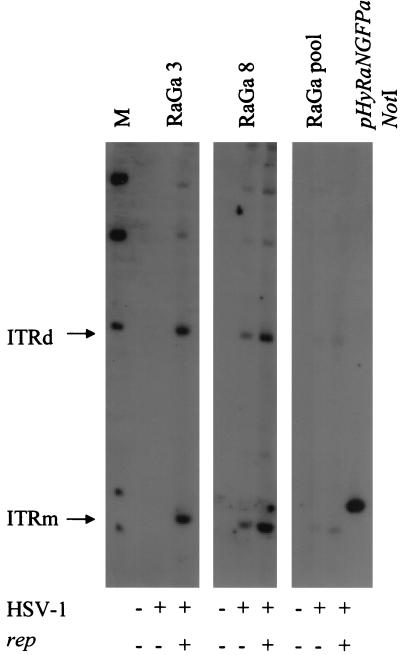
For a functional analysis of the Rep proteins and the other genetic elements required for replication of the two parent viruses, VERO 2-2 cells were transfected with vector DNA alone or cotransfected with vector DNA and packaging-defective HSV-1 helper DNA. After 2 days, Hirt DNA was prepared, digested with DpnI to cut input DNA, and analyzed by Southern blotting using a digoxigenin-labeled GFP probe. The results are shown in Fig. 3 and can be summarized as follows. All rep-containing hybrid vectors produced DpnI-resistant monomers and dimers of the ITR-flanked transgene cassette in the presence of HSV-1 helper DNA. Also, high-Mr replication products, presumably both the concatemeric products of replication from HSV-1 oriS and higher-order multimers of the ITR-flanked transgene cassette, were visible in the presence (Fig. 3, lanes 2 to 5) but not in the absence (Fig. 3, lanes 7 to 10) of HSV-1 helper DNA. In the presence of HSV-1 helper DNA, pHyGFPa, which does not contain rep sequences, also produced a high-Mr replication product but no replicative ITR intermediates (lane 1). In the presence of a rep-expressing plasmid (pRep) and HSV-1 helper DNA, pHyGFPa also produced replicative DNA intermediates of the ITR-flanked transgene cassette (not shown). No ITR intermediates were produced from pHSVGFP, which contains neither rep nor ITRs, under any condition (not shown). As a positive control for the hybridization to the GFP probe and as a size standard for the monomeric ITR-flanked transgene cassette, pHyRaNGFPa DNA isolated from E. coli was cleaved with NotI, which flanks the ITR-flanked transgene cassette in this plasmid (lane 11).
FIG. 3.
Southern analysis of AAV ITR and HSV-1 amplicon replication. VERO 2-2 cells were transfected with either vector DNA and packaging-defective HSV-1 DNA (+HSV; lanes 1 to 5) or vector DNA alone (−HSV; lanes 6 to 10). Hirt DNA was prepared 24 h later, digested with either DpnI alone (lanes 1 to 10) or DpnI and a restriction endonuclease (R) that cuts only once within the vector plasmids (lanes 12 to 16), and analyzed by Southern blot analysis using a digoxigenin-labeled GFP probe. Hirt DNA from mock-transfected cells served as a negative control (mock; lane 22). Monomeric and dimeric replication intermediates of the ITR-flanked transgene cassette (ITRm and ITRd), high-Mr replication products of the vector DNA from HSV-1 oriS (HSV-A), and monomeric or linearized vector DNA (A) are shown. Lanes 17 to 21, vector DNA was isolated from E. coli cells and digested with R; lane 21, pHyRaNGFPa DNA isolated from E. coli cells and digested with NotI served as positive control and size standard for ITRm, as NotI flanks the ITR-flanked transgene cassette in this vector (lane 11). Lane M, molecular weight standards.
To identify the high-Mr replication products observed in Fig. 3, lanes 1 to 5, the DNA samples were cleaved with DpnI and an additional restriction endonuclease that cuts only once within the vector plasmids (lanes 12 to 16). High-Mr replication products representing concatemeric HSV-1 amplicon DNA would be reduced to the size of the linearized vector plasmid isolated from E. coli (lanes 17 to 21). In addition, monomeric, dimeric, and higher-order multimeric replication intermediates of the ITR cassette would be fragmented if the recognition site for the single cutter is located within the ITR cassette but not if it is located outside the ITR cassette. For pHyGFPa, which contains no rep gene and, therefore, does not produce replication intermediates of the ITR cassette under these experimental conditions, the high-Mr replication product observed in lane 1 disappeared when the Hirt DNA was cleaved with DpnI and SphI (lane 12); instead, a band of 7.2 kb, the size of linearized pHyGFPa isolated from E. coli (lane 17). became visible. Hirt DNA from pHyR68bBGFPa-, pHyR78aNGFPa-, and pHyR68/78aBGFPa-transfected cells was digested with restriction endonucleases that have a single recognition site outside the ITR-flanked transgene cassette (NotI, BglII, and NotI, respectively). Consistent with this, the ladders of high-Mr DNAs that were predicted to be the higher-order multimeric replication intermediates of the ITR cassette remained intact (lanes 13 to 15). Additional bands of the sizes of the linearized vector DNAs isolated from E. coli (lanes 18 to 20) became visible. These bands are consistent with fragmentation of high-Mr replication products from HSV-1 oriS. Hirt DNA from pHyRaNGFPa-transfected cells was digested with DpnI and NheI, which cuts within the ITR-flanked transgene cassette (lane 16). The bands that were predicted to represent high-Mr replication products, as well as monomeric and dimeric replication intermediates of the ITR cassette, largely disappeared. In its place, a band of 10.5 kb, the size of the linearized pHyRaNGFPa plasmid DNA (lane 21), and bands of ∼2 kb and ∼1 kb, consistent with the fragments expected from NheI digestion of monomers and multimers of the ITR cassette, became prominent. Some of the bands observed (e.g., lanes 2 and 5, ∼2.3 kb; lane 16, ∼1.5 and ∼5 kb) are difficult to explain and may represent rearrangements or different configurations of the ITR cassette. Nevertheless, our conclusion from this experiment was that HSV-1 oriS as well as AAV rep genes and ITRs are functional in the context of the hybrid vectors. Similar results were obtained upon transfection of 293 cells (not shown).
HSV/AAV hybrid vectors are packaged into HSV-1 particles.
The question of whether the hybrid vectors are packageable in HSV-1 particles was addressed in the following experiment. VERO 2-2 cells were transfected with vector plasmids and packaging-defective HSV-1 helper DNA, and vector stocks were prepared and titers were determined as described in Materials and Methods. Table 1 shows the titers of the different vector stocks. All vectors that contained rep sequences produced 20- to 2,100-fold lower titers than pHSVGFP or pHyGFPa. In the presence of a rep-expressing plasmid (pRep) during packaging, the titers of pHSVGFP and pHyGFPa were also reduced by 50- to 100-fold. pHyRaNGFPa, which contains all four rep genes, produced the highest titers among the rep-containing hybrid vectors and was therefore used for the infection of 293 cells and isolation of stable, GFP-positive cell clones (see below). In conclusion, the results show that all hybrid vectors are packageable in HSV-1 particles and indicate that rep expression interferes with HSV-1 replication/packaging on a level not yet determined.
TABLE 1.
Vector titersa
| Vector | Titer (TU/ml)b |
|---|---|
| pHSVGFP | 1.2 × 107 ± 2.1 × 106 |
| pHyGFPa | 1.5 × 107 ± 3.1 × 106 |
| pHyR68bBGFPa | 7.1 × 103 ± 2.5 × 103 |
| pHyR78aNGFPa | 1.0 × 105 ± 6.7 × 104 |
| pHyR68/78aBGFPa | 5.4 × 104 ± 2.6 × 104 |
| pHyRaNGFPa | 7.5 × 105 ± 1.1 × 105 |
| pHSVGFP + pRep | 2.1 × 105 ± 1.4 × 105 |
| pHyGFPa + pRep | 1.3 × 105 ± 8.0 × 104 |
Packaging was performed as described in Materials and Methods.
Vector titers of unconcentrated cell lysates were determined on VERO 2-2 cells by using a fluorescence microscope. Results are shown as the mean ± standard deviation from at least three independent packaging experiments.
HSV/AAV hybrid vectors mediate site-specific integration at AAVS1 of human chromosome 19.
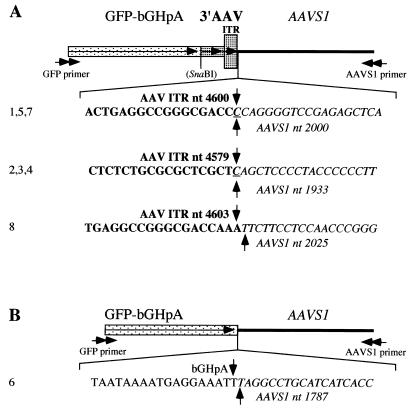
To identify site-specific integration events on the level of the nucleotide sequence, we performed PCR with a set of nested primers specific for GFP and AAVS1 on genomic DNA isolated from human 293 cells at 2 days after infection with pHyRaNGFPa. A product of ∼1 kb was cloned and a total of eight clones were sequenced (Fig. 4). Three different junction breakpoints all located within 3′ ITR and AAVS1 were identified in seven of the eight clones (Fig. 4A). In one of the clones, the crossover was between bGH polyadenylation signal (located downstream of the GFP coding sequence) and AAVS1 (Fig. 4B). All breakpoints within AAVS1 were located between nucleotides 1787 and 2025 of the sequence determined by Kotin and colleagues (17).
FIG. 4.
Sequence analysis of integration junctions between AAV ITR-flanked transgene cassette and AAVS1. The junctions were amplified by PCR using a set of nested primers specific for GFP (GFP primer) and AAVS1 (AAVS1 primer). The PCR product was cloned and a total of eight clones were sequenced. A schematic representation of the sequences and the nucleotides around the junction breakpoints in the different clones are shown. GFP, gene for GFP; bGHpA, bGH polyadenylation signal; 3′AAV, 3′ termini of the AAV genome; ITR, AAV ITR; AAVS1, AAV integration site on chromosome 19; SnaBI, site of insertion of the GFP-bGHpA cassette into the AAV genome in plasmid pAV2 (see Materials and Methods for details on the construction). Horizontal arrows indicate the directions of primers, transgene, AAV genome, and ITR; bold nucleotides represent sequences from AAV ITR; italic nucleotides represent sequences from AAVS1; vertical arrows and either bold or italic nucleotide (nt) numbers indicate the breakpoints in AAV ITR or AAVS1, respectively; underlined nucleotides are common to AAV ITR and AAVS1; numbers on the left (1 to 8) are the clone numbers. (A) The junctions in the clones are between AAV ITR and AAVS1; (B) the junction in the clone is between bGHpA sequences and AAVS1.
Characterization of stable, GFP-positive cell clones.
The goal of this experiment was to isolate (without selective pressure) and characterize stable, GFP-positive cell clones from pHyRaNGFPa-infected 293 cells. For this, 293 cells were infected with pHyRaNGFPa or, as controls, pHyGFPa or pHSVGFP at a multiplicity of infection of 1 transducing unit per cell and, 2 days later, sorted by fluorescence-activated cell sorting (FACS) to collect all GFP-positive cells in a 96-well plate (1,000 cells/well). After 2 weeks, 90% of the cells from six wells were pooled and propagated in T25-tissue culture flasks (designated 293 RaGa pool). The remaining cells were diluted and plated at 500 cells per plate onto 10-cm-diameter tissue culture plates to allow the formation of clones. Ten days later, GFP-positive clones (designated 293 RaGa) were picked, propagated separately in wells of a 96-well plate, clone purified again on 10-cm-diameter tissue culture plates, and then expanded (Fig. 5). Most of the clones remained GFP positive (100% of the cells) for at least 12 months, except for clones RaGa 2, 4, 5, and 6, which gradually lost GFP fluorescence over a period of 2 to 4 weeks. Surprisingly, no stable, GFP-positive clones were obtained from the pHSVGFP- or pHyGFPa-transduced cultures, although the latter appeared to express GFP for longer periods.
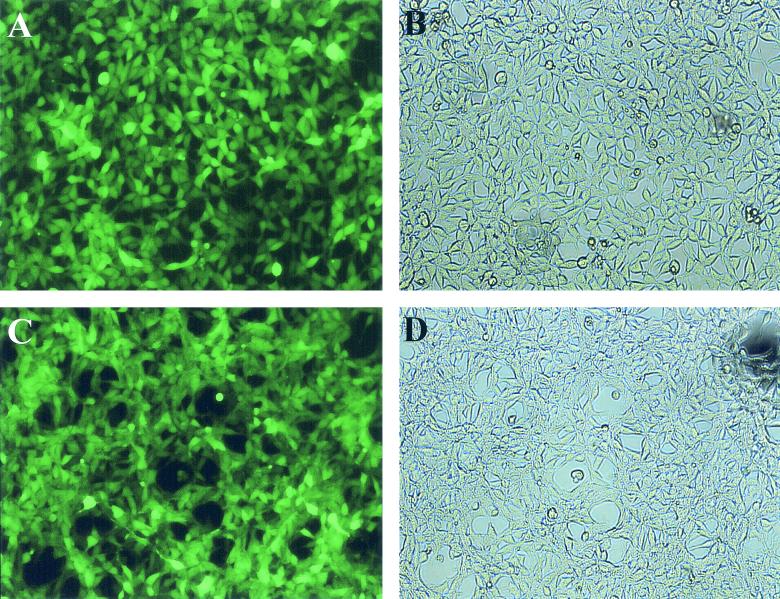
FIG. 5.
Photomicrographs showing 293 RaGa cells at 4 months after isolation from a pool of pHyRaNGFPa-infected 293 cells. (A and B) 293 RaGa clone 10; (C and D) 293 RaGa clone 12. (A and C) Standard fluorescence microscopy; (B and D) light microscopy. Original magnification, ×10.
Genomic DNA from the GFP-positive 293 RaGa clones was first analyzed by PCR for the presence of GFP and rep sequences. The 747-bp GFP fragment could be amplified in all clones except for clone 2, which had lost all GFP fluorescence at the time of analysis (not shown). Interestingly, the 744-bp rep fragment could also be amplified in some of the clones, in particular RaGa 4, 5, 6, 8, and 14, which indicates that sequences outside the ITR cassette have integrated at least in these clones (not shown). The gradual loss of GFP-fluorescent cells in clones RaGa 4, 5, and 6 may be due to the presence of functional rep genes, which may be toxic for the cell.
Next, we analyzed if the transgene cassette can be rescued and amplified from the 293 RaGa clones and if the rep sequences identified by PCR in some of the clones encode functional Rep proteins. For analysis, 106 cells from the 293 clones RaGa 3 (GFP positive and rep negative by PCR) and RaGa8 (GFP positive and rep positive by PCR) and cells from the pool (293 RaGa pool) were (i) not transfected or transfected with packaging-defective HSV-1 helper DNA alone or (ii) cotransfected with packaging-defective HSV-1 helper DNA and a rep-expressing plasmid (pRep). Hirt DNA was extracted 2 days later, separated by electrophoresis, blotted onto a nylon membrane, and hybridized to a digoxigenin-labeled GFP probe (Fig. 6). Cotransfection with HSV-1 helper DNA and pRep (HSV-1+/rep+) resulted in the rescue and replication of the transgene cassette in all three cultures. In clone RaGa8, which was rep positive by PCR, and in the 293 RaGa pool, which should have represented all different clones, but not in clone RaGa3 (rep negative by PCR), the transgene cassette was rescued also in the absence of pRep (HSV-1+/rep−). This indicates that 293 RaGa8 and the 293 RaGa pool contained functional rep genes. Clone RaGa3, which was rep negative by both PCR and rescue assay, may contain either no or disrupted rep sequences.
FIG. 6.
Southern analysis of rescue and replication of transgene cassette from 293 RaGa cells. 293 RaGa cells from clones 3 and 8 and from the pool were either transfected with packaging-defective HSV-1 DNA alone (HSV-1, +) or cotransfected with packaging-defective HSV-1 DNA and the rep-containing plasmid pRep (HSV-1, +, and rep, +), and 24 h later, Hirt DNA was prepared and analyzed by Southern blotting using a digoxigenin-labeled GFP probe. Hirt DNA isolated from mock-transfected cells served as controls (HSV-1, −, and rep, −). Monomeric and dimeric replication intermediates of the ITR-flanked transgene cassette (ITRm and ITRd) are shown. pHyRaNGFPa DNA isolated from E. coli and digested with NotI served as a positive control and as a size standard for ITRm. Lane M, molecular weight standard.
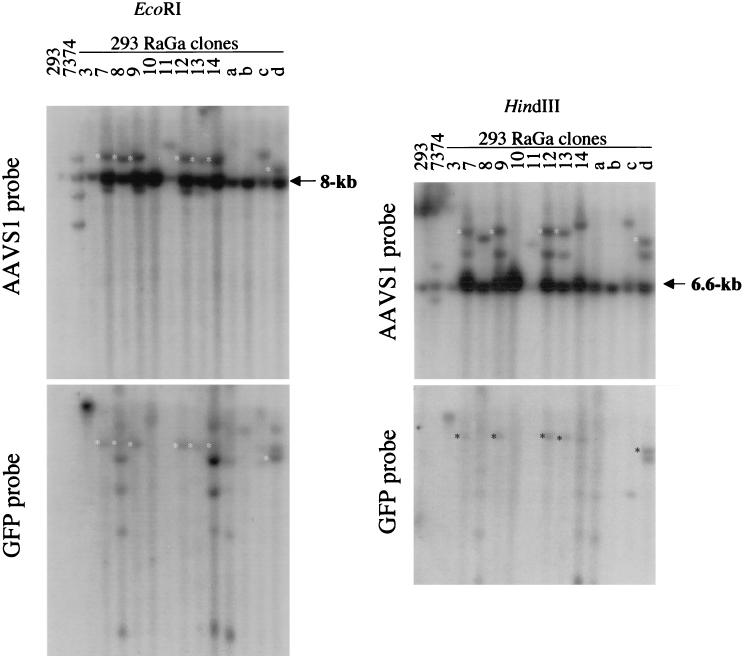
To determine whether integration by the HSV/AAV hybrid vector in the 293 RaGa clones occurred at AAVS1 or randomly, genomic DNA was digested with EcoRI, separated by electrophoresis, blotted, and hybridized to a 32P-labeled AAVS1 probe (Fig. 7A). An 8-kb band containing the AAVS1 preintegration site was detected in all lanes. AAVS1-specific rearrangements were detected in clones RaGa 7, 8, 9, 11, 12, 13, 14, c, and d and in the Detroit 6 cell line 7374, which is latently infected with wild-type AAV. Rehybridization of the same blot with a GFP probe revealed that in clones 7, 8, 9, 12, 13, 14, and d, AAVS1-rearranged bands colocalized with GFP-specific bands. GFP-specific bands that did not colocalize with AAVS1-rearranged bands (random integrations) were detected in clones RaGa 3, 8, 10, 14, a, b, c, and d. Consistent with the PCR analysis and ITR rescue experiments, DNA from RaGa 8 and the RaGa pool hybridized also with a probe specific for the rep gene which is located outside the ITR cassette (not shown). To confirm that the GFP- and AAVS1-specific EcoRI fragments in clones 7, 8, 9, 12, 13, 14, and d are colinear, a second restriction endonuclease digestion was performed with HindIII (Fig. 7B). A ∼6.6-kb HindIII band containing the AAVS1 preintegration site was detected in all lanes. GFP-specific and AAVS-1 specific HindIII bands colocalized in clones RaGa 7, 9, 12, 13, and d. In summary, these results show that HSV/AAV hybrid vectors mediate both site-specific and random integrations.
FIG. 7.
Southern analysis of 293 RaGa cell clones. Total genomic DNA was digested with EcoRI (A) or HindIII (B). Southern blots were first hybridized to the AAVS1 probe and then rehybridized to the GFP probe. Genomic DNA from parental 293 cells and from the Detroit 6 cell line 7374, which is latently infected with wild-type AAV, served as negative and positive controls, respectively. The 8- and 6.6-kb bands correspond to the EcoRI and HindIII AAVS1 preintegration fragments, respectively. ∗, colocalization of AAVS1- and GFP-specific EcoRI (A) and HindIII (B) fragments.
DISCUSSION
Helper virus-free HSV-1 amplicons can accommodate foreign DNA of any size between 1 and 150 kbp (40). As HSV-1 amplicons give room to almost unlimited combinations of genetic elements, they belong to the most versatile viral vectors. They can allow the insertion of true genomic sequences as well as cDNAs (40). Furthermore, large transcriptional regulatory sequences for cell type-specific expression (15), multiple transgenes, and genetic elements from other viruses to create hybrid vectors (16, 37, 41) may be used in a modular fashion. Optimally, HSV-1-based hybrid vectors combine the advantages of their parent vector systems while eliminating their inherent disadvantages. One so-far-unsolved problem of HSV-1 amplicons is the inefficiency of long-term transgene expression which, at least in dividing cells, may be due to loss of vector sequences. Consequently, HSV-1-based hybrid vectors have been designed to stabilize the transgene by either episomal retention and segregation or genomic integration. For example, hybrid vectors have been designed to combine specific features of HSV-1 amplicons and AAV. AAV, which has a very limited transgene capacity (∼4.6 kb), can integrate its genome into the AAVS1 locus on chromosome 19 of human cells. The viral rep gene and the ITRs are sufficient for this process. Previous reports have shown that AAV rep and ITRs included in the HSV-1 amplicon can extend transgene expression both in culture and in vivo. However, the mechanisms responsible for this and, in particular, the question of whether these vectors mediate site-specific integration at AAVS1 have not been addressed (5, 6, 16). The present study provides direct evidence that HSV/AAV hybrid vectors do mediate AAVS1-specific integration.
The HSV/AAV hybrid vectors described here contain, in addition to the standard HSV-1 amplicon elements ori and pac, the AAV rep gene and a transgene that is flanked by AAV ITRs. In the presence of HSV-1 helper functions, the hybrid vectors were replicated and packaged into HSV-1 particles and, in addition, launched the production of replicative intermediates of the ITR cassette. Rep proteins have been shown to inhibit the replication of a number of viruses, including human immunodeficiency virus type 1 (2), bovine papillomavirus type 1 (11), and adenovirus (44). This appears to be true also for HSV-1, as the titers of packaged vectors were drastically reduced when rep was provided in cis or in trans. The titers of vectors that contained no ITR cassette were also reduced in the presence of rep, indicating that rep-mediated excision of the ITR cassette was not or was not solely responsible for the reduced titers. Interestingly, the lowest titers were obtained from vectors that contained only rep78 and/or rep68, and the highest titers were obtained from a vector that contained all four rep genes.
In the absence of HSV-1 helper functions, HSV/AAV hybrid vectors mediated genomic integration and long-term gene expression following infection of 293 cells (this study) and other human cells (companion paper by Wang et al. 43). PCR and sequence analysis showed that in all but one of the amplified fragments, the junctions were between AAV ITR (nucleotides 4579 to 4603 of the AAV genome) and AAVS1 (nucleotides 1933 to 2025 of the published sequence [17]). In one fragment, the junction was between the polyadenylation signal of the GFP transgene and AAVS1 nucleotide 1787. Similar junctions have been identified in cells infected with wild-type AAV (9, 10, 17, 18, 21, 22, 32), baculovirus/AAV hybrid vectors (24), or adenovirus/AAV hybrid vectors (27). AAVS1-specific integration occurs also following transfection with AAV-based plasmids. However, while some of these reports indicate that sequences outside the ITR cassette did not integrate into AAVS1 (26), others show that the ITR cassette integrated along with the vector backbone (3, 38, 39). PCR and ITR rescue/replication experiments showed that some of the GFP-positive 293 RaGa cells described here (e.g., RaGa 8 and RaGa pool) also contained sequences outside the ITR cassette. However, Southern analysis revealed that genomic integration occurred not at AAVS1 but randomly in these cells. By contrast, sequences outside the ITR cassette were not detected in clones with AAVS1-specific integrations only; however, the number of clones analyzed was relatively small.
The goal of this study was to qualitatively determine—rather than statistically quantify—whether HSV/AAV hybrid vectors integrate at AAVS1 of human chromosome 19. It was anticipated that application of selective pressure would favor random, multiple integrations over AAVS1-specific integration of the ITR cassette (3). On the other hand, non AAVS1-specific integrations of ITR-flanked transgene sequences along with the vector backbone, rep in particular, may be underestimated, as cells positive for rep and GFP sequences appeared to be unstable (e.g., RaGa 4, 5, and 6). We chose to isolate GFP-positive clones without selective pressure. Others have shown that cells expressing rep have a lower growth rate, in particular 293 cells which constitutively express the adenovirus E1A gene, a transactivator of the p5 promoter of rep (3, 46). Interestingly, 293 RaGa clone 8 produced functional Rep proteins, as shown by ITR rescue/replication assays, and remained GFP positive for at least 12 months. Surprisingly, without selective pressure, the isolation of GFP-positive clones from cells infected with standard HSV-1 amplicon vectors or HSV/AAV hybrid vectors that did not contain rep was not successful. Therefore, we cannot answer the question whether ITRs alone (without rep) can enhance genomic integration. However, our data support that rep enhances both integration of transgene and vector sequences at random sites and integration of the ITR-flanked transgene cassette at AAVS1.
The results of this study open the possibility to analyze whether ITR-flanked transgenes with sizes that exceed the AAV genome can be inserted at and rescued from AAVS1. These experiments can be performed not only in cultured human cells but also in vivo, as transgenic animals that contain the AAVS1 locus from human chromosome 19 are available (28).
Acknowledgments
We thank E. Niederer (Swiss Federal Institute of Technology, Zurich) and R. Nunez (University of Zurich, Zurich, Switzerland) for FACS analysis, M. Urabe (Jichi Medical School, Tochigi, Japan) for providing plasmids p5R68 and p5R78, Y. Saeki (Massachusetts General Hospital, Charlestown, Mass.) for plasmids fHSVΔpacΔ27ΔKn and pEBHICP27, and X. O. Breakefield (Massachusetts General Hospital, Charlestown, Mass.) for helpful discussions.
This work was supported by the Swiss National Science Foundation, grant no. 31-58758.99 (C.F.).
REFERENCES
- 1.Aboody-Guterman, K. S., P. A. Pechan, N. G. Rainov, M. Sena-Esteves, A. Jacobs, E. Y. Snyder, P. Wild, E. Schraner, K. Tobler, X. O. Breakefield, and C. Fraefel. 1997. Green fluorescent protein as a reporter for retrovirus and helper virus- free HSV-1 amplicon vector-mediated gene transfer into neural cells in culture and in vivo. Neuroreport 8:3801-3808. [DOI] [PubMed] [Google Scholar]
- 2.Antoni, B. A., A. B. Rabson, I. L. Miller, J. P. Trempe, N. Chejanovsky, and B. J. Carter. 1991. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J. Virol. 65:396-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balague, C., M. Kalla, and W. W. Zhang. 1997. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J. Virol. 71:3299-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns, K. I., T. C. Pinkerton, G. F. Thomas, and M. D. Hoggan. 1975. Detection of adeno-associated virus (AAV)-specific nucleotide sequences in DNA isolated from latently infected Detroit 6 cells. Virology 68:556-560. [DOI] [PubMed] [Google Scholar]
- 5.Costantini, L. C., D. R. Jacoby, S. Wang, C. Fraefel, X. O. Breakefield, and O. Isacson. 1999. Gene transfer to the nigrostriatal system by hybrid herpes simplex virus/adeno-associated virus amplicon vectors. Hum. Gene Ther. 10:2481-2494. [DOI] [PubMed] [Google Scholar]
- 6.Fraefel, C., D. R. Jacoby, C. Lage, H. Hilderbrand, J. Y. Chou, F. W. Alt, X. O. Breakefield, and J. A. Majzoub. 1997. Gene transfer into hepatocytes mediated by helper virus-free HSV/AAV hybrid vectors. Mol. Med. 3:813-825. [PMC free article] [PubMed] [Google Scholar]
- 7.Fraefel, C., S. Song, F. Lim, P. Lang, L. Yu, Y. Wang, P. Wild, and A. I. Geller. 1996. Helper virus-free transfer of herpes simplex virus type 1 plasmid vectors into neural cells. J. Virol. 70:7190-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geller, A. I., and X. O. Breakefield. 1988. A defective HSV-1 vector expresses Escherichia coli beta-galactosidase in cultured peripheral neurons. Science 241:1667-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraud, C., E. Winocour, and K. I. Berns. 1995. Recombinant junctions formed by site-specific integration of adeno- associated virus into an episome. J. Virol. 69:6917-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraud, C., E. Winocour, and K. I. Berns. 1994. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc. Natl. Acad. Sci. USA 91:10039-10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermonat, P. L. 1992. Inhibition of bovine papillomavirus plasmid DNA replication by adeno-associated virus. Virology 189:329-333. [DOI] [PubMed] [Google Scholar]
- 12.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 14.Horsburgh, B. C., M. M. Hubinette, D. Qiang, M. L. MacDonald, and F. Tufaro. 1999. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 6:922-930. [DOI] [PubMed] [Google Scholar]
- 15.Jin, B. K., M. Belloni, B. Conti, H. J. Federoff, R. Starr, J. H. Son, H. Baker, and T. H. Joh. 1996. Prolonged in vivo gene expression driven by a tyrosine hydroxylase promoter in a defective herpes simplex virus amplicon vector. Hum. Gene Ther. 7:2015-2024. [DOI] [PubMed] [Google Scholar]
- 16.Johnston, K. M., D. Jacoby, P. A. Pechan, C. Fraefel, P. Borghesani, D. Schuback, R. J. Dunn, F. I. Smith, and X. O. Breakefield. 1997. HSV/AAV hybrid amplicon vectors extend transgene expression in human glioma cells. Hum. Gene Ther. 8:359-370. [DOI] [PubMed] [Google Scholar]
- 17.Kotin, R. M., R. M. Linden, and K. I. Berns. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 87:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laughlin, C. A., J. D. Tratschin, H. Coon, and B. J. Carter. 1983. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 23:65-73. [DOI] [PubMed] [Google Scholar]
- 20.Lim, F., D. Hartley, P. Starr, P. Lang, S. Song, L. Yu, Y. Wang, and A. I. Geller. 1996. Generation of high-titer defective HSV-1 vectors using an IE 2 deletion mutant and quantitative study of expression in cultured cortical cells. BioTechniques 20:460-469. [DOI] [PubMed] [Google Scholar]
- 21.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. I. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 93:11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linden, R. M., E. Winocour, and K. I. Berns. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA 93:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lusby, E., K. H. Fife, and K. I. Berns. 1980. Nucleotide sequence of the inverted terminal repetition in adeno- associated virus DNA. J. Virol. 34:402-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palombo, F., A. Monciotti, A. Recchia, R. Cortese, G. Ciliberto, and N. La Monica. 1998. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus-adeno-associated virus vector. J. Virol. 72:5025-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persichetti, F., F. Trettel, C. C. Huang, C. Fraefel, H. T. Timmers, J. F. Gusella, and M. E. MacDonald. 1999. Mutant huntingtin forms in vivo complexes with distinct context- dependent conformations of the polyglutamine segment. Neurobiol. Dis. 6:364-375. [DOI] [PubMed] [Google Scholar]
- 26.Pieroni, L., C. Fipaldini, A. Monciotti, D. Cimini, A. Sgura, E. Fattori, O. Epifano, R. Cortese, F. Palombo, and N. La Monica. 1998. Targeted integration of adeno-associated virus-derived plasmids in transfected human cells. Virology 249:249-259. [DOI] [PubMed] [Google Scholar]
- 27.Recchia, A., R. J. Parks, S. Lamartina, C. Toniatti, L. Pieroni, F. Palombo, G. Ciliberto, F. L. Graham, R. Cortese, N. La Monica, and S. Colloca. 1999. Site-specific integration mediated by a hybrid adenovirus/adeno- associated virus vector. Proc. Natl. Acad. Sci. USA 96:2615-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizzuto, G., B. Gorgoni, M. Cappelletti, D. Lazzaro, I. Gloaguen, V. Poli, A. Sgura, D. Cimini, G. Ciliberto, R. Cortese, E. Fattori, and N. La Monica. 1999. Development of animal models for adeno-associated virus site-specific integration. J. Virol. 73:2517-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeki, Y., C. Fraefel, T. Ichikawa, X. O. Breakefield, and E. A. Chiocca. 2001. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Mol. Ther. 3:591-601. [DOI] [PubMed] [Google Scholar]
- 30.Saeki, Y., T. Ichikawa, A. Saeki, E. A. Chiocca, K. Tobler, M. Ackermann, X. O. Breakefield, and C. Fraefel. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum. Gene Ther. 9:2787-2794. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, I. L., M. A. Hardwicke, and R. M. Sandri-Goldin. 1992. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 186:74-86. [DOI] [PubMed] [Google Scholar]
- 34.Spaete, R. R., and N. Frenkel. 1982. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell 30:295-304. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stavropoulos, T. A., and C. A. Strathdee. 1998. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J. Virol. 72:7137-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun, T. Q., E. Livanos, and J. M. Vos. 1996. Engineering a mini-herpesvirus as a general strategy to transduce up to 180 kb of functional self-replicating human mini-chromosomes. Gene Ther. 3:1081-1088. [PubMed] [Google Scholar]
- 38.Surosky, R. T., M. Urabe, S. G. Godwin, S. A. McQuiston, G. J. Kurtzman, K. Ozawa, and G. Natsoulis. 1997. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 71:7951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsunoda, H., T. Hayakawa, N. Sakuragawa, and H. Koyama. 2000. Site-specific integration of adeno-associated virus-based plasmid vectors in lipofected HeLa cells. Virology 268:391-401. [DOI] [PubMed] [Google Scholar]
- 40.Wade-Martins, R., E. R. Smith, E. Tyminski, E. A. Chiocca, and Y. Saeki. 2001. An infectious transfer and expression system for genomic DNA loci in human and mouse cells. Nat. Biotechnol. 19:1067-1070. [DOI] [PubMed] [Google Scholar]
- 41.Wang, S., and J. M. Vos. 1996. A hybrid herpesvirus infectious vector based on Epstein-Barr virus and herpes simplex virus type 1 for gene transfer into human cells in vitro and in vivo. J. Virol. 70:8422-8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, Y., C. Fraefel, F. Protasi, R. A. Moore, J. D. Fessenden, I. N. Pessah, A. DiFrancesco, X. Breakefield, and P. D. Allen. 2000. HSV-1 amplicon vectors are a highly efficient gene delivery system for skeletal muscle myoblasts and myotubes. Am. J. Physiol. Cell. Physiol. 278:C619-C626. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Y., S. M. Camp, M. Niwano, X. Shen, J. C. Bakowska, X. O. Breakefield, and P. D. Allen. 2002. Herpes simplex virus type 1/adeno-associated virus rep+ hybrid amplicon vector improves the stability of transgene expression in human cells by site-specific integration. J. Virol. 76:7150-7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weitzman, M. D., K. J. Fisher, and J. M. Wilson. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willis, R. A., W. J. Bowers, M. J. Turner, T. L. Fisher, C. S. Abdul-Alim, D. F. Howard, H. J. Federoff, E. M. Lord, and J. G. Frelinger. 2001. Dendritic cells transduced with hsv-1 amplicons expressing prostate-specific antigen generate antitumor immunity in mice. Hum. Gene Ther. 12:1867-1879. [DOI] [PubMed] [Google Scholar]
- 46.Yang, Q., F. Chen, and J. P. Trempe. 1994. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J. Virol. 68:4847-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]