Abstract
The Sendai virus C proteins, C′, C, Y1, and Y2, are a nested set of independently initiated carboxy-coterminal proteins translated from a reading frame overlapping the P frame on the P mRNA. The C proteins are extremely versatile and have been shown to counteract the antiviral action of interferons (IFNs), to down-regulate viral RNA synthesis, and to promote virus assembly. Using the stable cell lines expressing the C, Y1, Y2, or truncated C protein, we investigated the region responsible for anti-IFN action and for down-regulating viral RNA synthesis. Truncation from the amino terminus to the middle of the C protein maintained the inhibition of the signal transduction of IFNs, the formation of IFN-stimulated gene factor 3 (ISGF3) complex, the generation of the anti-vesicular stomatitis virus state, and the synthesis of viral RNA, but further truncation resulted in the simultaneous loss of all of these inhibitory activities. A relatively small truncation from the carboxy terminus also abolished all of these inhibitory activities. These data indicated that the activities of the C protein to counteract the antiviral action of IFNs and to down-regulate viral RNA synthesis were not encoded within a region of at least 98 amino acids in its amino-terminal half.
Sendai virus (SeV) is an enveloped virus with a linear, nonsegmented, negative-sense genome RNA of 15,384 nucleotides and belongs to the genus Respirovirus of the family Paramyxoviridae. There are six genes on the genome, the order of which is 3′-(leader)-N-P-M-F-HN-L-(trailer)-5′. The monocistronic mRNAs directing the single translation products are usually transcribed from the genes by viral RNA polymerase (31). However, the P gene expression is exceptional, because it gives rise to multiple protein species by a process known as RNA editing and by the use of an overlapping open reading frame (ORF).
In RNA editing, one nontemplated G residue is cotranscriptionally inserted into a specific position to generate the edited mRNA that encodes the protein termed V. The unedited mRNA that is the exact copy of the P gene encodes the phosphoprotein (P protein), the smaller subunit of RNA polymerase. The P and V proteins are therefore amino coterminous, while a −1 transframe is used to generate the carboxy-terminal half of the V protein (for a review, see reference 31). The V-unique carboxy-terminal region contains seven cysteine residues. These cysteine residues are highly conserved in paramyxovirus V proteins, form zinc finger motifs, and indeed bind Zn2+ (20, 33, 40, 46). Analysis of V knockout SeVs has indicated that the V protein is completely dispensable for viral replication in cultured cells (4, 25) but is required to maintain high viral loads in the lungs and to cause severe pneumonia in mice (20, 24, 25). The contributions of the V protein to pathogenesis were subsequently demonstrated in other paramyxoviruses, such as measles virus (39, 49), human parainfluenza virus type 3 (hPIV3) (7), and Newcastle disease virus (34). Thus, paramyxovirus V proteins are generally nonessential for viral tissue culture replication but have a luxury function required for in vivo pathogenesis (for reviews, see references 35 and 36).
An ORF overlapping the amino-terminal portion of the SeV P ORF in the +1 frame produces a nested set of carboxy-coterminal proteins called C′, C, Y1, and Y2, collectively referred to as the C proteins. Translation of the C′ protein is initiated on a non-AUG codon, ACG, at position 81 on the P mRNA (16), whereas C, Y1, and Y2 start on AUGs at positions 114, 183, and 201, respectively (Fig. 1A) (41). All four C proteins terminate at the same position, 726 (31). Among them, the C protein is the major species expressed in infected cells, at a molar ratio severalfold higher than those of the other three (30, 41). A reading frame overlapping the P ORF is found in viruses belonging to the two genera Respirovirus and Morbillivirus but not in viruses belonging to the genus Rubulavirus. The members of Respirovirus have the potential to encode the nested set of C proteins, but those of Morbillivirus have a single C protein (1, 31).
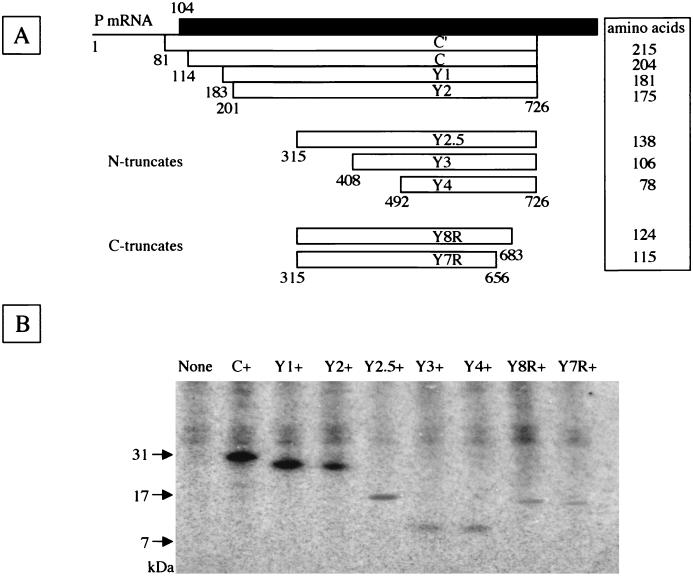
FIG. 1.
Native and truncated C proteins expressed from the reading frame overlapping the P frame on SeV P mRNA. (A) The C proteins (open bars) cloned into the expression vector are shown on the P mRNA. The amino-terminal portion of the P protein is shown as the filled bar. The numbers represent the nucleotide positions from the start site of P mRNA. The amino acid length of each C protein is shown in the box at right. (B) Immunoprecipitates of HeLa cell lines expressing native or respective truncated C proteins are shown. The positions of molecular mass markers (in kilodaltons) are shown at left.
In contrast to the V protein, which is almost completely dispensable for SeV replication in cultured cells, a lack of C proteins could profoundly affect the viral life cycle. Indeed, all-four-C knockout SeV replicated very poorly in cultured cells and embryonated chicken eggs (30). Four-C knockout SeV was found to produce larger amounts of viral mRNAs and translational products but generated smaller amounts of genomic RNAs and viable progeny viruses than did wild-type SeV in infected cells (17). These results indicate that C proteins are required for coordinating the amount of RNA between transcription and replication. The down-regulating activity of RNA synthesis was the first property found for C proteins in vitro (3) and was consistent with the findings obtained with four-C knockout SeV described above. The down-regulating activity is dose dependent and is well correlated with the binding capability of C proteins with the large (L) protein, a catalytic subunit of viral RNA polymerase (15, 19). The down-regulation of leader-primed RNA synthesis is more pronounced than that of trailer-primed RNA synthesis, implying the promoter specificity of inhibition (2, 48).
An additional important role of C proteins is the contribution to virus assembly. The progeny virions from cells infected with four-C knockout SeV have altered morphology and show a lower ratio of infectivity to hemagglutinating activity than does the wild-type virus (17). The SeV matrix (M) protein and envelope glycoproteins, F and HN, appeared to miss the viral assembly in the absence of C proteins (17). The contributions of the C protein to the viral life cycle have been demonstrated for other paramyxoviruses. The measles virus C protein has been shown to down-regulate RNA synthesis in a dose-dependent manner in vitro (43) and to improve the multiplying efficiency in some human primary cells (8, 39).
In addition to the roles described above, SeV C proteins have been shown to counteract the antiviral action of exogenously added and endogenously produced interferons (IFNs) (12, 50). This anti-IFN action was totally lost in cells infected with all-four-C knockout SeV (14, 47). This anti-IFN action of SeV may be required for pathogenesis at an acute phase of infection and also for the establishment of persistent infection in cultured cells or mice. Cell lines stably expressing C, Y1, or Y2 protein equally circumvent the antiviral action of alpha/beta IFN (IFN-α/β) and IFN-γ, indicating that C protein alone is sufficient for the anti-IFN function (22). In the same study, the C-, Y1-, or Y2-expressing cell lines were also shown to inhibit SeV multiplication at the transcriptional level. These data clearly showed that the smallest protein, Y2, was as active as the larger C and Y1 proteins in both counteracting the antiviral action of IFNs and down-regulating viral RNA synthesis (22). At least with respect to these two functions, there appears to be no reason for SeV to produce the C′, C, Y1, and Y2 proteins. Anti-IFN functions have been shown to be present in many other viruses (5, 9, 42, 53). It is the V proteins for simian virus type 5 (6, 52), mumps virus (29), and hPIV2 (37, 38) that counteract the antiviral action of IFNs. These viruses, which belong to the genus Rubulavirus, do not encode the C protein. It is interesting to know the evolutionary pathways common to and distinct between the two genera Rubulavirus and Respirovirus.
In this report, we describe the establishment of cell lines stably expressing the several truncated forms of the SeV C protein and examine which regions of the C protein are responsible for the anti-IFN actions and the down-regulation of viral RNA synthesis. Truncations from the amino terminus up to the middle of the C protein maintained both activities. Further truncation from the amino terminus as well as a relatively small truncation from the carboxy terminus readily resulted in the loss of both activities. The amino-terminal half of the C protein was thus found not to be responsible for either counteracting the antiviral action of IFNs or down-regulating viral RNA synthesis.
MATERIALS AND METHODS
Plasmids.
The DNA fragments encoding the truncated C ORFs were obtained by PCR from the full-length cDNA clone of SeV, pSeV(+) (26). For the amino-terminal truncation of the C protein, three forward primers, Y2.5F (5′-GAATTCHindIII aagcttGCCATG 315AAGGAGAAGTCTCAACAC-3′), Y3F (5′-GAATTCHindIIIaagcttGCC408ATGTTATCGGATTCCTCG-3′), and Y4F (5′-GAATTCHindIIIaagcttGCC492ATGCTGTCCTGTCGAGTG-3′), and one reverse primer, CR (5′-GAATTCBamHIggatccCTA726TTACTCTTGCACTATGTG-3′), were used for PCR amplification. For the carboxy-terminal truncation, one forward primer, Y2.5F, and two reverse primers, Y8R (5′-GAATTCBamHIggatccCTA683TATTACTTGCTCCTCCTT-3′) and Y7R (5′-GAATTCBamHIggatccCTA656CAGGTAGGGATGTACTTC-3′), were used for PCR. The initiation and termination codons are underlined and numbered in accordance with Fig. 1. Each region upstream of the initiation codon (italicized) was modified to optimize for translation according to Kozak's rule (28). These fragments were cut with HindIII and BamHI (superscripts in the primer sequences) and cloned into the same sites of plasmid pKS336 (GenBank accession number AF403737). After verification of the sequences, these plasmids encoding the Y2.5, Y3, Y4, Y8R, and Y7R proteins, named pKS-Y2.5, pKS-Y3, pKS-Y4, pKS-Y8R, and pKS-Y7R, respectively, were used to establish stable transformants.
Transfection.
The stable transformants expressing the variously truncated C protein were established as described previously (22). In brief, HeLa cells maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum were plated at a density of 106 cells/plate in 9-cm-diameter dishes. Ten micrograms of pKS-Y2.5, -Y3, -Y4, -Y8R, or -Y7R plasmid was transfected into cells by using a mammalian transfection kit (Stratagene, La Jolla, Calif.). Two days later, the medium was replaced with DMEM containing blasticidin at 10 μg/ml. Colonies grown in the selection medium were picked up, propagated, and used throughout the study.
35S labeling of HeLa cells.
Parental cells and the native or truncated C-expressing HeLa cells were inoculated to a density of 5 × 105 cells/plate in 35-mm-diameter plates 1 day before the labeling. The next day, the cells were washed once with phosphate-buffered saline (PBS) and replaced in 0.5 ml of DMEM without methionine. After 30 min for starvation, 0.74 MBq (20 μCi) of l-[35S]methionine (43.48 TBq/mmol; ICN Inc., Aurora, Ohio)/plate was added. The cells were incubated at 37°C for 2 h in the 5% CO2 incubator and then washed twice with ice-cold PBS. The harvested cells were then lysed with 0.5 ml of radioimmunoprecipitation assay buffer on ice for 5 min and spun at 15,000 rpm for 5 min at 4°C. Immunoprecipitations were performed as described previously (24) and were analyzed on a 16% acrylamide-Tricine gel (NuPAGE; Invitrogen Corp., Carlsbad, Calif.), which was subsequently treated with Enlightning reagent (Perkin-Elmer Life Sciences NEN, Boston, Mass.) for 30 min, dried, and visualized by fluorography.
Immunoblotting.
Cytoplasmic cell extracts were run through 4 to 12% acrylamide-bis-Tris gel (NuPAGE; Invitrogen Corp.) and electroblotted onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). The membranes were blocked for 1 h in 3% skim milk in TBST (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) and probed with a mixture of anti-STAT1α/β (sc-346; Santa Cruz Biotechnology, Santa Cruz, Calif.) and anti-STAT2 (sc-476; Santa Cruz Biotechnology) antibodies or with anti-PKR (sc-707; Santa Cruz Biotechnology) antibody. Immunoreactivity was visualized by peroxidase color reaction with a Konica immunostain kit (HRP-1000; Seikagaku Corp., Tokyo, Japan) according to the manufacturer's instructions. The visualized bands on the membranes were incorporated digitally by the scanner. The dendrograms of each band were achieved by using NIH-Image software and were used for calibrating the relative intensity.
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed to determine whether or not the IFN-stimulated gene factor 3 (ISGF3) complex was formed. After incubation with or without 1,000 IU of IFN-β/ml for 4 h, HeLa whole-cell extracts were prepared. Cells (2 × 106) were washed three times in ice-cold PBS and lysed in 50 μl of buffer (0.1% NP-40, 20 mM HEPES [pH 7.9], 50 mM NaCl, 10 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin/ml, 1 mM Na3VO4) followed by 10 min of incubation on ice. Nuclei were pelleted by spinning down at 10,000 rpm at 4°C for 5 min in an Eppendorf microcentrifuge. The supernatant was stored as a cytoplasmic extract at −80°C until use. The self-complementary DNA fragment (IFN-stimulated response element [IRE]; 5′-GAGAGGGAAACCGAAACTGAATTAGCTTTCAGTTTCGGTTTCCCTCT-3′) (with the IRE underlined in the sequence) was chemically synthesized and was used as a probe in EMSA (51). IRE was end labeled with T4 polynucleotide kinase (Toyobo Biochemicals, Tokyo, Japan) and [γ-32P]ATP (111 TBq/mmol; Perkin-Elmer Life Sciences NEN) according to the manufacturer's recommendation. Binding reactions were performed by incubating the cytoplasmic extract corresponding to 105 cells with 2 × 104 cpm of labeled IRE (10 fmol), 0.35 μg of salmon sperm DNA, and 3 μg of bovine serum albumin in a binding buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5% glycerol, 1 mM dithiothreitol, 0.5 mM EDTA) for 10 min at 25°C. In some cases, nonlabeled IRE (1 pmol) was added to the binding reaction mixtures. The reacted samples were then loaded onto native 4% acrylamide gels prepared with 1× Tris-borate-EDTA. After the running, the gels were dried and visualized by autoradiography.
Antiviral activity of IFNs.
Parental cells and the native or truncated C-expressing HeLa cells plated at a density of 5 × 104 cells/well in 24-well plates were treated with human IFN-β or IFN-γ at 0, 10, 100, or 1,000 IU/ml for 24 h in serum-free DMEM. The cells were washed once with PBS and infected with vesicular stomatitis virus (VSV) at a multiplicity of infection (MOI) of 2 in 0.1 ml of serum-free DMEM for 1 h at 37°C. After removal of the virus-containing media, cells were further incubated in the same medium without IFN. For viral cytopathic effect assays, cells were fixed and stained with Coomassie brilliant blue when significant cytopathic changes developed.
Minigenome assay.
The RNA genome analog (minigenome), in which the firefly luciferase ORF was substituted for the region beginning at the initiation codon for nucleocapsid (N) ORF and ending at the stop codon of L ORF in the context of the SeV genome, was transcribed from linearized pHVlucRT4(−) with a T7 transcription kit (AmpliScribe; Epicentre Technologies, Madison, Wis.) (26). The minigenome, the pGEM-N, -P, and -L plasmids carrying the SeV N, P, and L genes driven by the T7 promoter (provided by D. Kolakofsky, Geneva, Switzerland), and the pRL-TK plasmid (used as an internal control) were cotransfected into the parental and respective C-expressing HeLa cells as described previously (22). Control plasmid pRL-TK, which contained the herpes simplex virus thymidine kinase promoter region fused to the Renilla luciferase gene, was purchased from Promega (Madison, Wis.). Before transfection, cells were infected with recombinant vaccinia virus strain vTF7-3 expressing T7 polymerase (supplied by B. Moss, National Institutes of Health). Relative luciferase activities were measured at 20 h posttransfection by using a dual-luciferase assay kit (Promega).
Virus infections.
The strain Z of wild-type SeV and its recombinant containing the luciferase gene (rSeV/luci) were inoculated into the parental cells and the native or truncated C-expressing HeLa cells at an MOI of 5 per cell in a six-well plate (18, 23). The infected cells were maintained in serum-free DMEM with or without 2 μg of the MG-132 proteasome inhibitor (Calbiochem, La Jolla, Calif.)/ml. The virus titers in the culture supernatants were measured at several time points postinfection (p.i.). The wild-type SeV titers were measured by cell infection unit (CIU) assay (25). The luciferase activities and the protein quantities in rSeV/luci-infected cells were measured by using a luciferase assay kit (Promega) and a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.), respectively. hPIV1 (Kobe strain) and hPIV3 (Kobe strain) were kindly provided by M. Ito (Osaka Institute of Health, Osaka, Japan) and were inoculated into the parental and Y2-expressing HeLa cells at an MOI of 5 per cell in six-well plates. Measles virus (Edmonston strain) purchased from the American Type Culture Collection and mumps virus (RW strain) kept in our institute were used in this study. The virus titers were measured at several time points by a 50% infective dose of tissue-cultured cells with Vero or LLCMK2 cells.
RESULTS
Establishment of stable cell lines expressing the truncated C proteins.
There are eight in-frame AUG codons in the SeV C ORFs in addition to an ACG initiation codon for the C′ protein. The first three AUG codons, at positions 114, 183, and 201, are used to translate the C, Y1, and Y2 proteins, respectively. These four proteins are collectively called the C proteins, because they form a carboxy-coterminal nested set with heterogeneity at their amino termini (Fig. 1A). We recently found that HeLa cells stably expressing either C, Y1, or Y2 protein blocked the IFN-α/β- and IFN-γ-mediated JAK/STAT signal transduction pathway, resulting in the inhibition of antiviral action of IFNs, and down-regulated viral RNA synthesis (22). In these inhibitory capabilities, the C, Y1, and Y2 proteins were equally active, indicating that even Y2, the smallest of the SeV C proteins, is fully capable of performing these two functions.
To determine the regions responsible for these functions, three DNA fragments encoding the amino-terminally truncated C ORFs, Y2.5, Y3, and Y4 (Fig. 1A), were prepared and transfected into HeLa cells. The reading frames of Y3 and Y4 start from the fourth and fifth AUG codons, at positions 408 and 492 of the SeV P mRNA, respectively, while Y2.5 was prepared by adding an initiation codon at position 315 of the P mRNA following upstream truncation (Fig. 1A). Colonies grown in the selection medium were collected and propagated. Several clones expressing the Y2.5, Y3, or Y4 protein were obtained. As shown in Fig. 1B, the three clones named Y2.5+, Y3+, and Y4+ were found by immunoprecipitation with anti-C serum to express the Y2.5, Y3, and Y4 proteins, which migrated faster in sodium dodecyl sulfate-polyacrylamide gel electrophoresis than did the Y2 protein of the previously established cell line, designated Y2+, and exhibited the respective authentic sizes. In addition to the amino-terminal truncates, HeLa cell lines expressing the carboxy-terminally truncated Y8R or Y7R protein were prepared and designated Y8R+ or Y7R+, respectively (Fig. 1). The reading frames of Y8R and Y7R start at position 315, as does the Y2.5 frame, but stop immediately before the eighth and seventh initiation codons, at positions 684 and 657 of the P mRNA, respectively (Fig. 1A). The Y8R and Y7R proteins exhibited authentic mobility, migrating faster than the Y2.5 protein (Fig. 1B). The immunoprecipitated bands of the truncated proteins were fainter than those of the intact C, Y1, and Y2 proteins in proportion to the length of truncation, according to a gradual loss of methionine residues to be labeled and of antigenic sites to be bound with anti-C serum to be precipitated. No specific coprecipitates with C proteins were found under these conditions (Fig. 1B).
Effect of truncated SeV C proteins on the IFN-β-induced signaling.
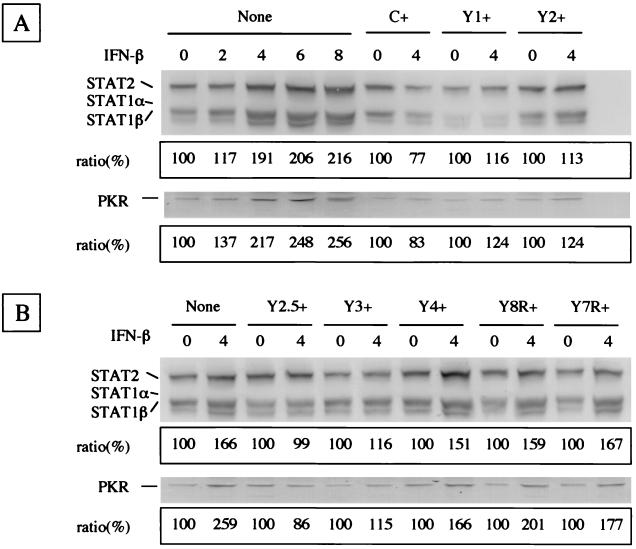
IFN-α and IFN-β are involved in innate responses as major host defense mechanisms, as exemplified by the establishment of the antiviral state through the induction of IFN-stimulated gene (ISG) products via the so-called JAK/STAT pathway (44, 45). To clarify the effect of truncated C proteins on the IFN-β-mediated signaling, the levels of the ISG products, STAT1α/β, STAT2, and PKR, were compared. The intracellular levels of STAT1α/β and STAT2 increased 1.17-, 1.91-, 2.06-, and 2.16-fold in accordance with incubation with IFN-β for 2, 4, 6, and 8 h, respectively (Fig. 2A). Similar IFN-β-mediated stimulation was also found in PKR production. The amount of PKR increased 1.37-, 2.17-, 2.48-, and 2.56-fold during the incubations for 2, 4, 6, and 8 h, respectively. As the synthesis of ISG products thus appeared to be stimulated clearly by 4-h incubation, the stimulation levels of the expressing cells incubated for 4 h with or without IFN-β were compared. No or little appreciable stimulation of STAT1α/β-STAT2 or PKR synthesis by IFN-β was found in C+, Y1+, or Y2+ cells (0.77- to 1.24-fold) (Fig. 2A). Stimulation was found in parental HeLa cells (1.66-fold for STAT1α/β-STAT2 and 2.59-fold for PKR), while just as in these C-expressing cells, the presence or absence of IFN-β did not appreciably change the thickness or intensity of bands for either Y2.5+ or Y3+ cells (Fig. 2B). However, darker and thicker bands of STAT1α/β-STAT2 and PKR were observed in Y4+ cells (1.67- and 1.77-fold, respectively) treated with IFN-β than in those without IFN-β (Fig. 2B). Thus, the amino-terminal truncation of the C protein to position 492 from the 5′ end of P mRNA was suggested to cause the loss of inhibition of IFN signaling. The effect of carboxy-terminal truncations was then studied by using Y8R+ and Y7R+ cells, which express Y2.5 proteins lacking the 3′ region of the reading frame. The stimulations of STAT1α/β and STAT2 in the presence of IFN-β were observed in both Y8R+ (1.59- and 2.01-fold, respectively) and Y7R+ (1.67- and 1.77-fold, respectively) cells (Fig. 2B), suggesting that carboxy-terminal truncation of the C protein to positions 684 and 657 from the 3′ end of P mRNA caused the loss of inhibiting activity. Essentially the same results were obtained when the lysates of cells treated with IFN-α or IFN-γ were immunoblotted (data not shown). These findings confirmed that the C, Y1, and Y2 proteins blocked IFN-α/β- and IFN-γ-mediated signaling equally well without the aid of any other SeV proteins and indicated that the region responsible for this effect resided in the Y2.5 and Y3 proteins but not in the Y4, Y7R, or Y8R protein.
FIG. 2.
Expression of IFN-responsible STAT1, STAT2, and PKR proteins in the parental and C-expressing HeLa cells. The amounts of STAT1α/β, STAT2, and PKR proteins are shown in each immunoblot. The densities of the bands taken in each lane were calibrated as described in Materials and Methods. IFN-β was applied for the indicated number of hours. The stimulation ratios between incubations in the absence and the presence of IFN-β are shown in the boxes. The assays for parental, C+, Y1+, and Y2+ HeLa cells (A) and for cells expressing the respective truncated C proteins (B) are indicated.
Effect of truncated C proteins on the formation of ISGF3 complex.
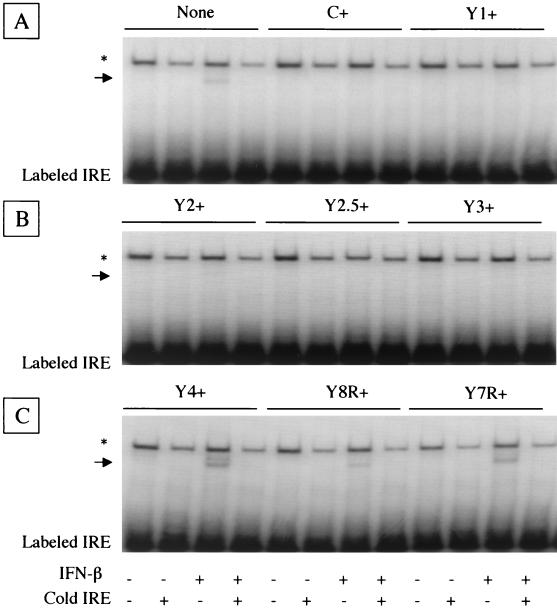
To further clarify the effect of the truncated C proteins on the inhibition of IFN signaling, we studied the formation of ISGF3 complex required for ISG transcription by DNA-binding assay. Cytoplasmic cell extracts prepared from the parental HeLa cells or stable transformants expressing native or truncated C protein were subjected to EMSA with the labeled oligonucleotide probe (IRE). The parental HeLa cell extract pretreated with IFN-β gave a more slowly migrating band than the probe alone in the gel (Fig. 3A). This band was not found when the parental cell extract without IFN-β pretreatment was assayed or when the extract was assayed with a 100-fold excess amount of the same, nonlabeled oligonucleotides, indicating the specific binding of the probe due to the formation of ISGF3 complex in cells. The upper band appeared in all assays in which the probe was incubated with cell extracts (Fig. 3) but disappeared when the probe was electrophoresed alone under the same conditions (data not shown). This common band was thus considered to represent nonspecific binding of some constitutively expressed cellular components with the probe rather than the formation of ISGF3 complex.
FIG. 3.
Formation of ISGF3 complex in the IFN-stimulated cells. The formation of ISGF3 complex was assayed by the mobility of the DNA fragment (IRE). The positions of the shifted IRE fragments bound with the ISGF3 complex are indicated by arrows. The nonspecific bands caused by incubation with cell extracts are indicated by asterisks. The assays for parental, C+, and Y1+ HeLa cells (A), for Y2+, Y2.5+, and Y3+ HeLa cells (B), and for Y4+, Y8R+, and Y7R+ HeLa cells (C) are shown. Labeled IRE, unbound free IRE probe.
The C+, Y1+, and Y2+ cell extracts yielded no specific band under any conditions used (Fig. 3A and B), indicating that ISGF3 complex was not formed in these cells. The Y2.5+ and Y3+ cell extracts did not give the specific band either (Fig. 3B), but Y4+ cell extract pretreated with IFN-β did (Fig. 3C), suggesting the requirement of the amino-terminal region between Y3 and Y4 for inhibiting ISGF3 complex formation. The extracts obtained from Y8R+ and Y7R+ cells expressing the carboxy-terminally truncated Y2.5 proteins gave the specific band when cells were treated with IFN-β (Fig. 3C), suggesting the requirement of the very carboxy-terminal 14 residues of the C protein for the inhibition of ISGF3 complex formation.
Effect of truncated C proteins on the establishment of IFN-β- and IFN-γ-induced antiviral state.
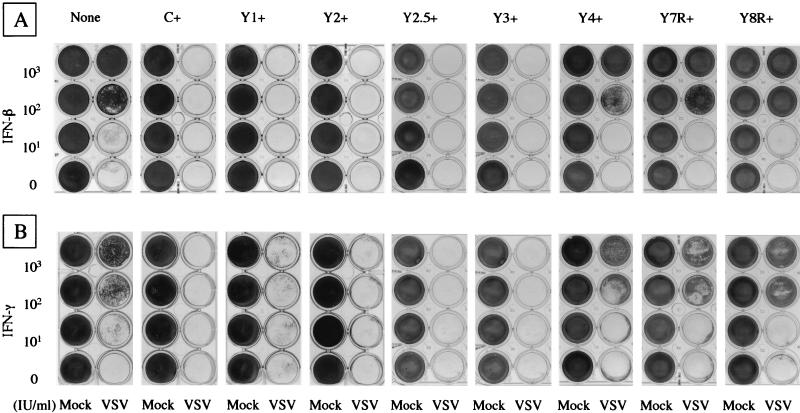
To determine the effects of the truncated C proteins on the establishment of the antiviral state, cells preincubated with IFN-β at 0, 10, 100, or 1,000 IU/ml for 24 h were challenged with VSV. As shown in Fig. 4A, IFN-β alone did not cause any apparent cytopathogenicity in any of the cells tested under these conditions. All of the cells cultured in the absence of IFN-β were detached from the plates by subsequent VSV infection. The parental cells were protected from the VSV-induced cytopathic effect with IFN-β pretreatments at 100 and 1,000 IU/ml. In contrast, C+, Y1+, and Y2+ cells were totally detached by VSV infection even after pretreatment with IFN-β at 1,000 IU/ml. VSV multiplication in the C+, Y1+, and Y2+ cells at the highest concentration of IFN was confirmed by immunoblotting with anti-VSV antibody (data not shown), indicating that the cell detachment was indeed caused by VSV multiplication. Essentially the same results were obtained when IFN-γ was used instead of IFN-β (Fig. 4B).
FIG. 4.
IFN-induced antiviral states of the parental and C-expressing HeLa cells. The cells were incubated for 24 h with various amounts of IFN-β (A) or IFN-γ (B) as indicated. They were then challenged with VSV or mock infected (Mock). Cells that survived the challenge infection and attached to the plates were fixed and stained.
The Y2.5+ and Y3+ cells expressing the amino-terminally truncated C protein were also detached by VSV challenge under all of the conditions tested. In contrast, Y4+ cells expressing the further truncated C protein were protected by IFN-β or IFN-γ pretreatment at 100 or 1,000 IU/ml (Fig. 4), indicating the critical importance of the region between Y3 and Y4 for counteracting the antiviral activity of IFNs. The Y7R+ and Y8R+ cells expressing the carboxy-terminally truncated Y2.5 protein were also protected from VSV challenge by pretreatment with IFN-β or IFN-γ at 100 or 1,000 IU/ml (Fig. 4), indicating the requirement of the very carboxy-terminal residues for generating the anti-IFN action of C proteins. We concluded that the SeV C-protein region responsible for inhibiting the IFN-β- and IFN-γ-mediated signaling and for abolishing the antiviral state resides in the Y2.5 and Y3 proteins.
Effect of truncated C proteins on viral RNA synthesis.
In our previous report, we showed that the Y2 protein, the smallest of the SeV C proteins, was fully active not only in counteracting the antiviral action of IFNs but also in down-regulating viral RNA synthesis (22). The RNA-synthesizing activities were then studied in vitro by using a minigenome with the reporter firefly luciferase gene. The minigenome RNA was cotransfected into cells expressing the respective truncated C protein together with the N, P, and L plasmids. This cotransfection further included the plasmid encoding the Renilla luciferase used as an internal control. The minigenome was replicated and transcribed by the N, P, and L proteins, which were supplied by the respective plasmids driven by T7 polymerase derived from the coinfecting vTF7.3. In the P plasmid, the expression of C proteins had been silenced by a replacement of its translational initiation site with the influenza viral hemagglutinin tag (3). The extent of transcription from the minigenome was monitored by firefly luciferase activities. Potential fluctuations in transfection efficiency and other experimentation were normalized by the expression levels of Renilla luciferase from the control plasmid. The relative luciferase activities were measured in four independent experiments, and the mean value and its deviation were calculated.
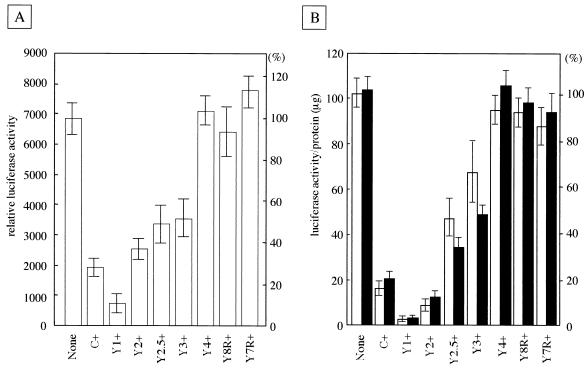
The relative luciferase activities in the C+, Y1+, and Y2+ cells were about 29, 12, and 37% of that in parental cells, respectively (Fig. 5A). As previously shown (22), the degrees of inhibition were strictly dependent upon the amounts of the various C proteins expressed in cells but not on the C protein species. For instance, the obviously stronger inhibition caused by the Y1 protein than by the C or Y2 protein was simply attributable to the fact that the levels of intracellular expression were higher for the Y1 protein than for the C or Y2 protein. The relative luciferase activities in Y2.5+ and Y3+ cells were 50 and 53% of that in parental cells, respectively. Note that the degrees of inhibition caused by the Y2.5 and Y3 proteins were statistically as high as or not much lower than that caused by the Y2 protein. Thus, the Y2.5 and Y3 proteins remained inhibitory for viral RNA synthesis. In contrast, little or no inhibitory capability was found for the Y4, Y8R, or Y7R protein-expressing cells (Fig. 5A). These results indicated that though the Y3 protein retained the ability to down-regulate minigenome RNA synthesis, the further truncation at either the amino or the carboxy terminus caused the loss of inhibitory activity.
FIG. 5.
Reporter gene expression from the SeV minigenome or the rSeV genome in the parental and C-expressing HeLa cells. (A) The firefly and Renilla luciferase activities expressed from the minigenome and the control plasmid, respectively, were calibrated in the various cells. Relative luciferase activities are expressed as mean values with error bars at left, and percentages are given at right. (B) The luciferase activities per microgram of protein, with error bars, are shown at left, and percentages are shown at right. Filled bars and open bars indicate the activities examined under the conditions with and without proteasome inhibitor (MG-132), respectively.
Effect of truncated C proteins on SeV transcription in infected cells.
We further attempted to evaluate the effect of truncated C proteins in the context of viral infection. The recombinant SeV, rSeV/luci, containing the firefly luciferase gene was used in this experiment. The luciferase activity in rSeV/luci-infected cells was well correlated with the amount of luciferase mRNA in the early stage (<12 h) of infection (18, 23). rSeV/luci was inoculated into the parental cells and the native or truncated-C-protein-expressing HeLa cells at an MOI of 5 per cell and incubated for 6 h. Cell lysates were prepared for measuring the luciferase activity and the protein quantity. Potential fluctuations in the cell amounts were normalized by the protein quantity of cell lysate. A high luciferase activity per microgram of protein was found in the parental HeLa cells, whereas the expression in C+, Y1+, and Y2+ cells was 16, 4, and 9% of that in the parental cells, respectively (Fig. 5B). The inhibitions caused by the C proteins were retained at least partially in cells expressing the amino-terminally truncated Y2.5 or Y3 protein, being 46 or 67% of the activity in the parental cells, respectively. In contrast, the Y4 protein generated by further truncation of the Y3 protein did not show significant inhibition (96% of activity). In addition, the Y8R and Y7R proteins generated by the carboxy-terminal truncation of the Y2.5 protein also showed no appreciable inhibition (95 and 86% of activity, respectively). These data were consistent with those obtained with the minigenome (Fig. 5A). However, the inhibitions of RNA synthesis by the Y2.5 and Y3 proteins in infected cells were not as pronounced as those observed in the minigenome assay. This may have been due to the instability of the Y2.5 and Y3 proteins in infected cells, because the addition of a proteasome inhibitor to the culture medium significantly improved the degree of inhibition (35 and 49% of activity, respectively) (Fig. 5B). Note that no such alteration of inhibition was seen for the other C proteins, including Y4, Y8R, and Y7R (Fig. 5B). Taken together, these results strongly suggested that the Y3 protein was the smallest of the proteins tested to down-regulate viral RNA synthesis.
Effect of truncated C proteins on virus growth.
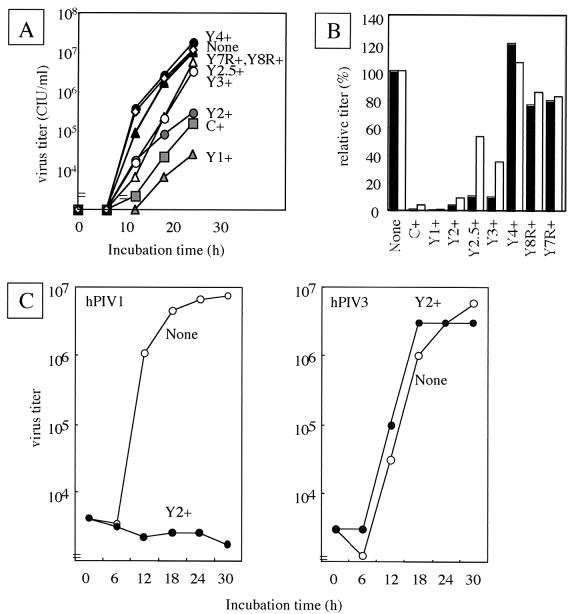
The growth rate and peak titer of all-four-C knockout SeV were much lower than those of the wild-type SeV, even in the IFN-nonproducing Vero cells (17), suggesting that C proteins play an essential role in maximal viral production independently of their anti-IFN action. If they were present in a certain amount in cells prior to infection, the C proteins strongly inhibited the RNA synthesis of infecting SeV and impaired viral growth. This growth impediment caused by the C proteins was reexamined by using the cell lines expressing the truncated C protein. As shown in Fig. 6A, SeV replication not only in C-, Y1-, and Y2-expressing cells but also in Y2.5- and Y3-expressing cells was significantly slower than that in parental cells. Though not by as much as those in Y2-expressing cells, the titers at 24 h p.i. were also lower in Y2.5- and Y3-expressing cells. Little difference in either growth rate or titer was found between parental and Y4-, Y8R-, or Y7R-expressing cells. The growth inhibition by the Y2.5 and Y3 proteins but not by the Y4, Y8R, or Y7R protein was confirmed by the relative virus titers at 18 and 24 h p.i., particularly the former, which were calibrated by standardization, with the titer of the wild-type virus taken as 100% (Fig. 6B). The C, Y1, Y2, Y2.5, and Y3 proteins gave significant inhibition (less than 15%) at 18 h p.i. The inhibition was continued in C+, Y1+, and Y2+ cells, but the virus titers were restored to some extent in Y2.5+ and Y3+ cells at 24 h p.i. This may have been due, at least in part, to the instability of the Y2.5 and Y3 proteins. These results clearly indicated that the trans-expressed 108-amino-acid Y3 protein rendered HeLa cells poorly permissive for SeV replication but that further truncated ones did not.
FIG. 6.
Virus growth in the parental and C-expressing HeLa cells. (A) The SeV titers in the culture supernatants at several incubation times are plotted for the parental and C-expressing HeLa cells. (B) The SeV titers at 18 h (filled bars) and 24 h (open bars) are expressed as percentages of the respective titers of the parent cells. (C) hPIV1 (left) and hPIV3 (right) were inoculated into the parental (open circles) and Y2+ (filled circles) HeLa cells. The virus titers at several incubation times are shown.
Finally, we examined the specificity of SeV C inhibition for some other related viruses. As already shown in Fig. 4, the growth of VSV was not inhibited by the expression of SeV C proteins. Similarly, the growth of measles virus and mumps virus was not impaired in cells expressing the SeV C protein but rather was enhanced, probably because of the inhibition of antiviral action of the endogenously produced IFNs (data not shown). When the growth of hPIV1 and hPIV3, which belong to the genus Respirovirus, was compared between the parental and Y2-expressing HeLa cells, hPIV1 but not hPIV3 showed greatly impaired growth in Y2-expressing cells (Fig. 6C). This hPIV1 inhibition by the SeV Y2 protein was as strong as or even stronger than that for the homologous SeV (Fig. 6A). Note that the homology of the carboxy-terminal half of the C protein required for inhibition is 89.5% between SeV and hPIV1 while it is 48.6% between SeV and hPIV3.
DISCUSSION
The carboxy-terminally nested set of four C proteins (C′, C, Y1, and Y2) is translated from the in-frame initiation codons at positions 81, 114, 183, and 201 of SeV P mRNA (Fig. 1A). Like the wild type, the C′ knockout SeV replicates normally in cultured cells and kills the mouse (32). The C′-C double-knockout SeV and the C′-C-Y1 triple-knockout SeV showed slightly slower replication in cultured cells than the wild-type SeV but replicated poorly in the lungs of mice. The mutant viruses were attenuated for mice (30). Therefore, the roles of C′ appear to be subrogated by the C, Y1, and Y2 proteins, but the roles of C′ and C do not appear to be subrogated by the Y1 and/or Y2 proteins. The growth of all-four-C knockout SeV was hampered greatly even in cultured cells. Because these SeVs are cleared immediately after inoculation, it was totally nonpathogenic for mice (17). The fact that all-four-C knockout SeV was much more attenuated both in vitro and in vivo than others indicated the requirement of all kinds of C proteins for full replication capability of SeV. To date, three functions, the anti-IFN function, the ability to down-regulate viral RNA synthesis, and the activity to promote virus assembly, have been assigned to one or more of the C proteins (3, 12, 17, 22). However, the first two of these functions have been clearly mapped in all of the four C proteins (22), indicating that the C proteins cannot be discriminated one from the other by these functions.
To determine the region responsible for each of these functions in the C proteins, we established HeLa cell lines expressing the truncated C protein and examined whether or not they retained these abilities. The truncated Y3 protein, which consisted of amino acids 99 to 204 of the C protein, retained the abilities to counteract IFNs and to down-regulate viral RNA synthesis, but Y4 protein, which consisted of amino acids 127 to 204 of the C protein, lost both abilities simultaneously. These results indicated that the amino-terminal residues 1 to 98 of the C protein were not essential for blocking the establishment of the antiviral state by IFNs or for down-regulating RNA synthesis. It is noteworthy that the region comprising amino acids 40 to 98, between the Y2 and Y3 proteins, did not essentially contribute to these inhibitory functions but contributed at least to some extent to the stabilization of the protein (Fig. 5B and 6B). Though the Y2.5 protein, which consisted of amino acids 68 to 204 of the C protein, retained the two abilities, two carboxy-terminal truncates consisting of amino acids 68 to 190 (Y8R) and 68 to 181 (Y7R) of the C protein lost both abilities. This result indicated that the carboxy-terminal region (amino acids 191 to 204) of the C protein is also required for generating these two functions. These two functions were thus eventually mapped within a region of 106 amino acids in the carboxy-terminal half of the C protein (99 to 204). However, as the stability of truncated C proteins was not examined in this study, the regions between Y3 and Y4 (99 to 108), Y2.5 and Y8R (191 to 204), and Y2.5 and Y7R (182 to 204) could possibly contribute to the stability of C proteins indirectly in cells rather than contributing directly to the anti-IFN action and to the down-regulation of viral RNA synthesis.
A spontaneous mutation from serine to phenylalanine at amino acid 170 of SeV C protein of the Ohita strain rendered this highly virulent SeV avirulent for mice (13, 21) and less capable of counteracting the IFNs (11). Though the sequence of the C protein differs for the Ohita strain and less virulent Z strain which we used, the sequence of the carboxy-terminal half, including the serine at position 170, is identical for the two strains. This fact is consistent with our conclusion that amino acids 99 to 204 of the C protein were responsible for the inhibition of IFNs from establishing an antiviral state. The carboxy-terminal half region (99 to 204) of SeV C protein showed a striking similarity (89.5%) with the counterpart of hPIV1 but only moderate similarity (48.6%) with that of hPIV3, though the similarity of the entire C protein is 69.1% between SeV and hPIV1 and 44.1% between SeV and hPIV3. Thus, SeV C protein strongly inhibited the growth of hPIV1 but not at all that of hPIV3 (Fig. 6C). These results indicated the importance of the primary sequence of the carboxy-terminal half for the down-regulation of viral RNA synthesis.
The ability of SeV C protein to bind L protein has been shown to correlate with the down-regulating activity of RNA synthesis in vitro, and the charged amino acids important for binding L protein and for down-regulating RNA synthesis have been shown to lie scattered, predominantly within the carboxy-terminal half of the C protein (15). These results were in good agreement with the results of the present truncations. However, the mutant C protein whose amino acids 76-KIID-79 were replaced with 76-AIIA-79 impaired the binding with L protein and the inhibition of RNA synthesis (15), while our Y3 protein without amino acids 1 to 98 retained the inhibiting activity (Fig. 4 and 5). The point mutations may have caused three-dimensional hindrance rather than the direct effects on binding and inhibition.
Garcin et al. reported that all of the C′, C, Y1, and Y2 proteins could equally inhibit the tyrosine phosphorylation at amino acid 701 of STAT1 and hamper the signaling of IFNs (11) but indicated further that the C′ and C proteins could induce the instability of STAT1 and prevent IFNs from establishing an antiviral state, whereas the Y1 and Y2 proteins and the C protein lacking amino acids 10 to 15 could not (10). Based upon these observations, they claimed that the inhibition of signaling was not sufficient to counteract the antiviral activity of IFNs and that the anti-IFN action could be attributed to the amino-terminal region of the C protein. Finally, they concluded that the larger C′ and C and the smaller Y1 and Y2 proteins play different roles (10). These claims are highly contradictory to our previous conclusion that Y2, the smallest of the C proteins, is fully capable of antagonizing both the signaling of IFNs and their antiviral activity (22); these claims also contradict the present results indicating that these functions are primarily encoded by the carboxy-terminal half of the C protein. The discrepancy can be attributed, at least in part, to the different cell lines used, because the strength and duration of IFN signaling vary depending on cell type. More than this, we must emphasize differences in experimental design; we used the cell lines expressing the C protein constitutively, while Garcin et al. used recombinant viruses (rSeVs) with variously mutated C proteins. In the context of wild-type SeV replication, anti-IFN action by C proteins takes place within a relatively short period (2 h) p.i. (14), but for this action to take place, SeV infection must be done at least 30 min prior to IFN treatment (50). Namely, a small but critical amount of intracellular accumulation of C proteins has to be guaranteed prior to IFN treatment in all experiments. This must be even more carefully taken into consideration when mutant rSeVs are used, because they are often attenuated in their replication capability (14, 27). Some of the inoculated rSeVs could be eliminated by the antiviral action of IFNs not because the C proteins had a reduced ability to counteract IFNs but because the amount of C proteins was insufficient to overcome IFNs in cells. It has to be noted that in the study by Garcin et al. (10), when cells were inoculated with rSeVs before the IFN treatment, the results obtained appeared to be similar to ours, but when cells were inoculated with rSeVs and treated with IFN simultaneously or subsequently, the results were in disagreement with ours. The viral life cycle involves amplification of genome replication and mRNA transcription. The quantitative differences in intracellular C proteins that were potentially caused by the differences in replication capability among the rSeVs must become greater as the viral life cycle advances. This may also make precise characterization of rSeV C mutants difficult. These problems, which tend to lead to biased interpretations, can be avoided at least in part through the use of stable transformants expressing relatively invariable amounts of native or mutant C proteins prior to and throughout the experiment. It has to be further noted that the dose-dependent inhibition of RNA synthesis caused by Y2, the smallest native C protein, was essentially the same as those caused by the longer Y1 and C proteins (22).
However, a further enigma remains to be solved. As described above, rSeV lacking the C′ protein replicates as rapidly as the wild-type virus, but rSeVs lacking the C′ and C proteins or the C′, C, and Y1 proteins grow more slowly than the wild type (30, 32). All-four-C knockout SeV is further attenuated (17). It would thus appear that the Y1 and/or Y2 proteins are not representative of all of the roles of C proteins. In addition, all field isolates and laboratory strains of SeV so far sequenced encode or at least have the potential to encode four different C proteins. The enigma to be solved is why SeV should encode four different C proteins. To address this, finely tuned quantitative studies will be needed. On-off expression of each C protein in cells may be used as such to define how much each C protein complements the C′-C-Y1-Y2 knockout SeV replication and whether or not all four C proteins are equally active in all of the relevant functions, including the recruitment of viral structural proteins into the assembly pathway. The generation of additional C knockout viruses and their characterization both in cells and in host animals will also be helpful in answering this question.
Acknowledgments
We thank B. Gotoh, K. Takeuchi, and N. Miyajima for their suggestions.
This work was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and from the Bio-Oriented Technology Research Advancement Institution (BRAIN), Saitama, Japan. Y.O. was the recipient of a BRAIN fellowship.
REFERENCES
- 1.Boeck, R., J. Curran, Y. Matsuoka, R. Compans, and D. Kolakofsky. 1992. The parainfluenza virus type 1 P/C gene uses a very efficient GUG codon to start its C′ protein. J. Virol. 66:1765-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadd, T., D. Garcin, C. Tapparel, M. Itoh, M. Homma, L. Roux, J. Curran, and D. Kolakofsky. 1996. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J. Virol. 70:5067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curran, J., J.-B. Marq, and D. Kolakofsky. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189:647-656. [DOI] [PubMed] [Google Scholar]
- 4.Delenda, C., S. Hausmann, D. Garcin, and D. Kolakofsky. 1997. Normal cellular replication of Sendai virus without the trans-frame, nonstructural V protein. Virology 228:55-62. [DOI] [PubMed] [Google Scholar]
- 5.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbin, A. P., J. M. McAuliffe, P. L. Collins, and B. R. Murphy. 1999. Mutations in the C, D, and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology 261:319-330. [DOI] [PubMed] [Google Scholar]
- 8.Escoffier, C., S. Manie, S. Vincent, C. P. Muller, M. Billeter, and D. Gerlier. 1999. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J. Virol. 73:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 10.Garcin, D., J. Curran, M. Itoh, and D. Kolakofsky. 2001. Longer and shorter forms of Sendai virus C proteins play different roles in modulating the cellular antiviral response. J. Virol. 75:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcin, D., M. Itoh, and D. Kolakofsky. 1997. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology 238:424-431. [DOI] [PubMed] [Google Scholar]
- 14.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-α/β-mediated responses. FEBS Lett. 459:205-210. [DOI] [PubMed] [Google Scholar]
- 15.Grogan, C. C., and S. A. Moyer. 2001. Sendai virus wild-type and mutant C proteins show a direct correlation between L polymerase binding and inhibition of viral RNA synthesis. Virology 288:96-108. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, K. C., and S. Patwardhan. 1988. ACG, the initiator codon for Sendai virus C protein. J. Biol. Chem. 263:8553-8556. [PubMed] [Google Scholar]
- 17.Hasan, M. K., A. Kato, M. Muranaka, R. Yamaguchi, Y. Sakai, I. Hatano, M. Tashiro, and Y. Nagai. 2000. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74:5619-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasan, M. K., A. Kato, T. Shioda, Y. Sakai, D. Yu, and Y. Nagai. 1997. Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3′ proximal first locus. J. Gen. Virol. 78:2813-2820. [DOI] [PubMed] [Google Scholar]
- 19.Horikami, S. M., R. E. Hector, S. Smallwood, and S. A. Moyer. 1997. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology 235:261-270. [DOI] [PubMed] [Google Scholar]
- 20.Huang, C., K. Kiyotani, Y. Fujii, N. Fukuhara, A. Kato, Y. Nagai, T. Yoshida, and T. Sakaguchi. 2000. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J. Virol. 74:7834-7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh, M., Y. Isegawa, H. Hotta, and M. Homma. 1997. Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC-MK2 cells. J. Gen. Virol. 78:3207-3215. [DOI] [PubMed] [Google Scholar]
- 22.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, A., K. Kiyotani, M. K. Hasan, T. Shioda, Y. Sakai, T. Yoshida, and Y. Nagai. 1999. Sendai virus gene start signals are not equivalent in reinitiation capacity: moderation at the fusion protein gene. J. Virol. 73:9237-9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, T. Shioda, and Y. Nagai. 1997. Importance of the cysteine-rich carboxyl terminal half of V protein for Sendai virus pathogenesis. J. Virol. 71:7266-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, A., Y. Sakai, T. Shioda, T. Kondo, M. Nakanishi, and Y. Nagai. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1:569-579. [DOI] [PubMed] [Google Scholar]
- 27.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak, M. 1984. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 12:3873-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 30.Kurotani, A., K. Kiyotani, A. Kato, T. Shioda, Y. Sakai, K. Mizumoto, T. Yoshida, and Y. Nagai. 1998. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells 2:111-124. [DOI] [PubMed] [Google Scholar]
- 31.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins Press, Philadelphia, Pa.
- 32.Latorre, P., T. Cadd, M. Itoh, J. Curran, and D. Kolakofsky. 1998. The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J. Virol. 72:5984-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liston, P., and D. J. Briedis. 1994. Measles virus V protein binds zinc. Virology 198:399-404. [DOI] [PubMed] [Google Scholar]
- 34.Mebatsion, T., S. Verstegen, L. T. De Vaan, A. Romer-Oberdorfer, and C. C. Schrier. 2001. A recombinant Newcastle disease virus with low-level V protein expression is immunogenic and lacks pathogenicity for chicken embryos. J. Virol. 75:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagai, Y. 1999. Paramyxovirus replication and pathogenesis. Reverse genetics transforms understanding. Rev. Med. Virol. 9:83-99. [DOI] [PubMed] [Google Scholar]
- 36.Nagai, Y., and A. Kato. 1999. Paramyxovirus reverse genetics is coming of age. Microbiol. Immunol. 43:613-624. [DOI] [PubMed] [Google Scholar]
- 37.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. M. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 39.Patterson, J. B., D. Thomas, H. Lewicki, M. A. Billeter, and M. B. Oldstone. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80-89. [DOI] [PubMed] [Google Scholar]
- 40.Patterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 41.Patwardhan, S., and K. C. Gupta. 1988. Translation initiation potential of the 5′ proximal AUGs of the polycistronic P/C mRNA of Sendai virus. J. Biol. Chem. 263:4907-4913. [PubMed] [Google Scholar]
- 42.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 43.Reutter, G. L., C. Cortese-Grogan, J. Wilson, and S. A. Moyer. 2001. Mutations in measles virus C protein that up regulate viral RNA synthesis. Virology 285:100-109. [DOI] [PubMed] [Google Scholar]
- 44.Sen, G. C., and R. M. Ransohoff. 1993. Interferon induced antiviral actions and their regulation. Adv. Virus Res. 43:57-102. [DOI] [PubMed] [Google Scholar]
- 45.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 46.Steward, M., A. C. Samson, W. Errington, and P. T. Emmerson. 1995. The Newcastle disease virus V protein binds zinc. Arch. Virol. 140:1321-1328. [DOI] [PubMed] [Google Scholar]
- 47.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6:545-557. [DOI] [PubMed] [Google Scholar]
- 48.Tapparel, C., S. Hausmann, T. Pelet, J. Curran, D. Kolakofsky, and L. Roux. 1997. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J. Virol. 71:9588-9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokoo, J., B. Gotoh, T. Komatsu, K. Takeuchi, and T. Miyadai. 1999. Replication-incompetent Sendai virus can suppress the antiviral action of type I interferon. Arch. Virol. 144:1043-1055. [DOI] [PubMed] [Google Scholar]
- 51.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]