Abstract
The envelope glycoprotein complex (gp120-gp41) of human immunodeficiency virus type 1 (HIV-1) promotes the fusion of viral and cellular membranes through formation of the fusion-active six-helix bundle in the gp41 ectodomain. This gp41 core structure consists of three C-terminal helices packed in an antiparallel manner into hydrophobic grooves on the surface of the N-terminal trimeric coiled coil. Alanine mutations that destabilize the N- and C-terminal interhelical packing interactions also reduce viral infectivity. Here we show that viruses bearing these mutations exhibit a marked potentiation of inhibition by peptides that make up the gp41 core. By contrast, these viruses are unchanged in their sensitivities to soluble CD4, the CXCR4 coreceptor ligand SDF-1α, and human anti-HIV immunoglobulin, reagents that impact the initial, receptor-induced conformational changes in the envelope glycoprotein. Our results support the notion that these alanine mutations specifically affect the conformational transition to the fusion-active gp41 structure. The mutations also increase viral sensitivity to the gp41-directed monoclonal antibody 2F5, suggesting that this broadly neutralizing antibody may also interfere with this transition. The conformational activation of the HIV-1 envelope glycoprotein likely represents a viable target for vaccine and antiviral drug development.
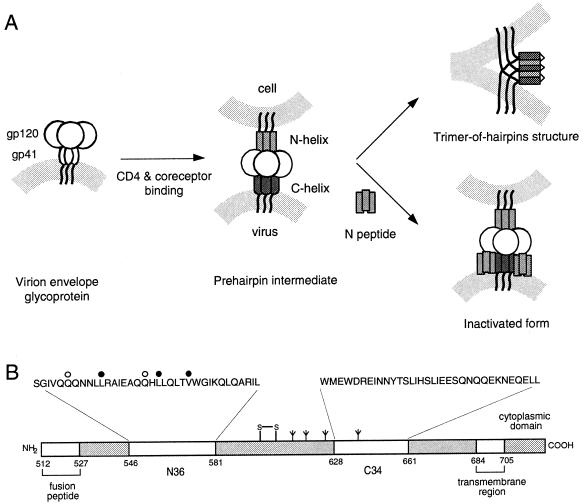
The envelope glycoprotein of human immunodeficiency virus type 1 (HIV-1) mediates viral entry by promoting the fusion of viral and cellular membranes. On the virion surface, the envelope glycoprotein complex exists as an oligomeric spike comprising the receptor-binding subunit gp120, which is anchored to the viral membrane through a noncovalent association with the transmembrane subunit gp41 (for a review, see reference 32). Considerable evidence indicates that HIV-1 membrane fusion requires a series of conformational changes in gp41. Numerous studies lead to the following working model for envelope glycoprotein-mediated membrane fusion (for a review, see reference 13 and references therein) (Fig. 1A). The native form of gp41 on the surface of the virus is stabilized by interactions with gp120, which are altered upon the binding of gp120 to CD4 and a coreceptor. The prehairpin intermediate of gp41 is subsequently formed by folding of the N-terminal trimeric coiled coil, leading to the insertion of the N-terminal hydrophobic fusion-peptide region into the target membrane. This prehairpin intermediate then collapses to form the six-helix bundle structure in which the C-terminal regions pack into the hydrophobic grooves of the N-terminal coiled-coil trimer in an antiparallel manner. The formation of this trimer-of-hairpins structure brings the viral and cellular membranes into close apposition. The free energy made available by the formation of this highly stable gp41 core is thought to contribute to overcoming the energy barrier to membrane fusion (e.g., 20).
FIG. 1.
A working model for gp41-mediated membrane fusion and its inhibition by peptides. (A) The native envelope glycoprotein complex on the virion surface undergoes conformational changes upon binding to CD4 and the coreceptor, and the gp41 subunit is released to form a prehairpin intermediate in which the hydrophobic N-terminal fusion peptide of gp41 is inserted into the cellular membrane and the N-terminal heptad-repeat regions (N helix) form a trimeric coiled coil. Productive infection is mediated by the collapse of the prehairpin intermediate to form the fusion-active gp41 core in which the C-terminal heptad-repeat regions (C helix) pack into the hydrophobic grooves of the N-terminal coiled coil. The formation of the trimer-of-hairpins brings the viral and cellular membranes into apposition to promote fusion. Inhibition of infection by N and C peptides is mediated by peptide binding to the cognate gp41 helix. The N peptide is shown binding to the C helix as a trimeric coiled coil. Peptide binding inactivates the envelope glycoprotein complex and blocks viral infectivity. (B) The amino acid sequences of the N36 and C34 peptides are shown above the N- and C-terminal heptad-repeat regions in this linear representation of gp41. The positions of the interhelical alanine mutations that result in fusion-intact (open circles) and fusion-deficient (closed circles) envelope glycoproteins are indicated. The residues are numbered according to their positions in HXB2 gp160.
The crucial role of interhelical packing interactions between the N- and C-terminal regions of the gp41 ectodomain provides an opportunity for antiviral intervention (13, 41). For example, peptides corresponding to the C-terminal heptad-repeat region of gp41, termed C peptides, are capable of inhibiting entry of HIV-1 at nanomolar concentrations (24, 28, 49). One such peptide, T20, has been shown to have antiviral activity in humans (25). Biochemical and structural studies strongly suggest that C peptides act in a dominant-negative manner by binding to the N-terminal coiled-coil region of gp41 in its prehairpin intermediate state, thereby interfering with its transition to the fusion-active six-helix bundle structure (6, 8, 17, 28, 45, 49). Similarly, peptides derived from the N-terminal heptad-repeat region of gp41 (called N peptides) are thought to block HIV-1 fusion by binding to the C-helix region of the gp41 intermediate (Fig. 1A) (7, 28, 48). N peptides may also act by intercalating into the N-helix coiled-coil structure (3). Because N peptides aggregate in solution, they are less potent inhibitors than the C peptides (28). However, soluble forms of the trimeric N peptide inhibit with nanomolar potency similar to that of the C peptides (12, 38).
Mutations in gp41 that alter the interhelical packing interactions can destabilize the fusion-active gp41 core structure and reduce the ability of the envelope glycoprotein to mediate membrane fusion (4, 21, 22, 31, 40, 44, 46, 47). By using alanine-scanning mutagenesis, we have recently identified several amino acid residues in the interhelical e and g positions of the N-peptide region that are critical for membrane fusion activity (31). Alanine substitutions at these positions decrease the thermal stability of the six-helix bundle and markedly reduce the ability of the mutant envelope glycoproteins to mediate cell-cell fusion (31). These results suggest that the destabilizing alanine mutations render the formation of the trimer-of-hairpins structure less favorable and perhaps increase exposure of the transient prehairpin intermediate (31).
According to this idea, we surmised that these alanine mutations might render the virus more sensitive to inhibition by N and C peptides that target the prehairpin intermediate. In order to directly test this hypothesis, we constructed recombinant viruses bearing single alanine mutations at the interhelical Leu-556, Leu-565, and Val-570 positions. We also generated recombinant viruses bearing mutations at Gln-551 and Gln-563, mutations that have little or no effect on the stability of the gp41 core or on the fusion activity of the envelope glycoprotein (Fig. 1B and Table 1).
TABLE 1.
Summary of mutant envelope glycoproteins and viruses
| Envelope glycoprotein | Cell-cell fusion activitya | Tm of gp41 six-helix bundle (°C)a,b | Viral infectivityc | gp120 per viriond (arbitrary units) |
|---|---|---|---|---|
| Wild type | +++ | 70 | 4,500 | 0.15 |
| L556A | ± | 64 | 330 | 0.13 |
| L565A | ± | 50 | 120 | 0.20 |
| V570A | ± | 56 | 15 | 0.16 |
| Q551A | +++ | 74 | 2,500 | 0.16 |
| Q563A | +++ | 68 | 21,000 | 0.27 |
The results are from reference 31.
The melting temperature (Tm) of the gp41 six-helix bundle was determined in the N34(L6)C28 model (29).
p24 was determined by antigen capture enzyme-linked immunosorbent assay (Beckman Coulter, Inc.). Nominal viral infectivity (number of foci per nanogram of p24) was calculated by using a dilution of virus that produced 100 foci in a 96-well U87-CD4-CXCR4 cell microculture assay.
The relative amount of gp120 per virion was estimated by Western blot analysis of centrifugally purified virions (51). gp120 was determined with the deglycosylated 55-kDa polypeptide of gp120 (31) and the anti-gp120 MAb Chessie 12 (1). Virion core protein p17 was assessed using immunoglobulin from HIV-infected persons (HIVIG). Antibody binding was visualized by ECL-Plus (Amersham Pharmacia Biotech), and fluorescence was quantitated using a Fuji FLA-3000G analyzer. The ratio of fluorescence intensities is shown.
Characterization of mutant viruses.
Infectious HIV-1 NL4-3 (2) proviruses bearing the alanine mutations in gp41 were constructed for this study. These mutant envelope glycoproteins included fusion-deficient phenotypes (viz., L556A, L565A, and V570A) as well as fusion-intact controls (viz., Q551A and Q563A) (31). The alanine mutations were transferred from the HXB2 envelope glycoprotein expression plasmid (31) to the NL4-3 provirus by using a 1.6-kb NheI-XhoI restriction endonuclease fragment which encodes the envelope glycoprotein from the C3 region of gp120 through the entire gp41 protein. The wild-type HXB2 fragment was also transferred to generate the wild-type provirus bearing a chimeric NL4-3/HXB2 env gene. The molecularly cloned NL4-3 and HXB2 env genes were derived from the same human T-cell leukemia virus strain III/lymphadenopathy-associated virus/HIV-1 strain IIIb population and are >97% identical (2, 36). In the case of Q551A, the provirus was constructed with the use of a BamHI site within the sequence encoding the cytoplasmic domain of gp41 rather than the XhoI site.
The wild-type and mutant provirus plasmids were used to generate infectious virus stocks by FuGene-6-mediated transfection of COS-7 cells (51). Western blot analysis of centrifugally purified virions demonstrated comparable levels of envelope glycoprotein in the wild-type and mutant virion particles (data not shown). These results confirmed that the synthesis, transport, proteolytic cleavage, and virion incorporation of these fusion-deficient and fusion-intact glycoproteins were unaffected by the alanine mutations (Table 1).
Infectivity of the mutant virions was determined by serial dilution in a U87-CD4-CXCR4 cell infectivity assay (19, 26) (Table 1). Among the viruses bearing a fusion-deficient envelope glycoprotein, L556A was 14-fold-less infectious than the wild-type virus, whereas L565A and V570A were approximately 40-fold- and 300-fold-less infectious, respectively. This ordering of viral infectivity is identical to that observed for the envelope glycoproteins in cell-cell fusion assays (31) and generally corresponds to the measured reduction in the thermal stability of the gp41 core. The least destabilizing mutation (L556A) resulted in the most infectious virus whereas the more highly destabilized mutants (L565A and V570A) were severely debilitated in infectivity. The infectivity of virions containing the stable and fusion-intact envelope glycoproteins (Q551A and Q563A) was similar to that of the wild-type virus.
Destabilizing mutations do not affect early steps in binding and entry.
The working model for envelope glycoprotein-mediated fusion (Fig. 1A) defines two sequential steps comprising (i) the initial interactions between the gp120 moiety of the envelope glycoprotein complex and its cellular receptor and coreceptor, and (ii) the triggering of the membrane-fusion activity of gp41. Based on this model, we anticipated that mutations that destabilize the interhelical packing interactions within the gp41 fusion-active core should not perturb the early processes of CD4 and coreceptor binding.
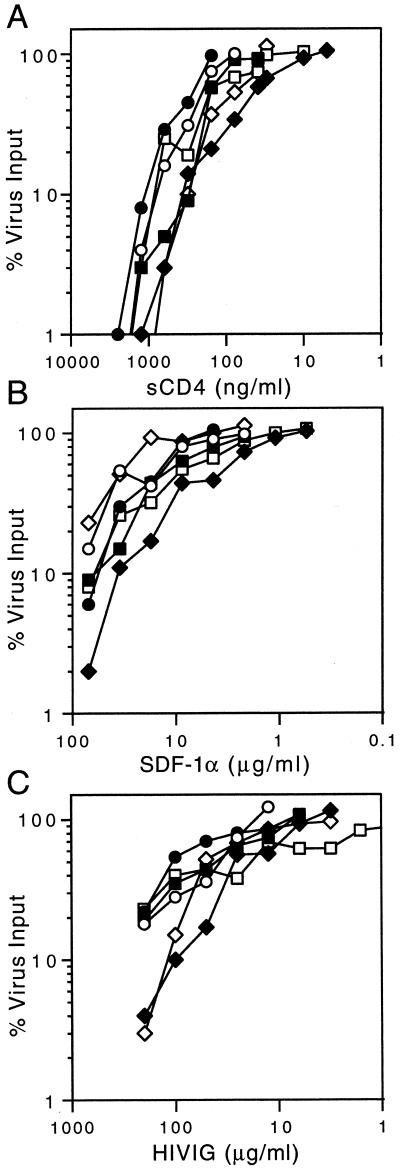
To directly test this hypothesis, we examined the sensitivity of the viruses to two inhibitors that target early, gp120-mediated steps in binding and entry: soluble CD4 (sCD4) (10) and the CXCR4 chemokine SDF-1α (34). We reasoned that potential alterations in the process of CD4 and coreceptor binding might be manifested in differential sensitivities to these specific inhibitors. In these studies, wild-type and mutant viruses were exposed to serial dilutions of sCD4 or SDF-1α for 1 h at 37°C, and the mixture was used to infect U87-CD4-CXCR4 cells. The virus input into the assay was predetermined to yield approximately 100 to 200 infected-cell foci in the 96-well microculture assay. Two days after infection, cells were immunochemically stained and infected cells were enumerated microscopically (26). As demonstrated by the results shown in Fig. 2A and 2B, inhibition of infection by sCD4 or by blockade of the CXCR4 coreceptor was entirely unaffected in the fusion-deficient (L556A, L565A, and V570A) and fusion-intact (Q551A and Q563A) viruses.
FIG. 2.
Sensitivity to soluble CD4, SDF-1α and HIVIG. The U87-CD4-CXCR4 cell microculture virus inhibition assay is described in the text (26). Inhibition is indicated as the percent reduction in the number of infected-cell foci relative to the control virus input. (A) The sCD4 ectodomain was obtained from Progenics Pharmaceuticals, and (B) the CXCR4 chemokine SDF-1α was kindly provided by Ian Clark-Lewis (University of British Columbia). (C) HIVIG, an immunoglobulin preparation from HIV-infected persons, was provided by Fred Prince (New York City Blood Center). In each case, the results of a representative study performed in duplicate are shown; replicate experiments yielded concordant results. Wild type (circle) and fusion-intact mutants Q551A (square) and Q563A (diamond) are shown as open symbols. Fusion-deficient mutants L556A (square), L565A (circle), and V570A (diamond) are shown as closed symbols.
We extended these findings to examine the sensitivity of the fusion-deficient and fusion-intact viruses to neutralization by HIVIG, an immunoglobulin preparation from HIV-infected persons (35). The bulk of HIVIG's neutralization activity against laboratory-adapted strains such as those studied here is directed to the third variable (V3) loop of gp120 (43). Antibody binding to the V3 loop may affect neutralization by perturbing CD4-induced changes that contribute to the formation of the coreceptor binding site in gp120 (37). As shown in Fig. 2C, sensitivity to neutralization by HIVIG was largely comparable in the fusion-deficient and fusion-intact viruses. Thus, neutralization by HIVIG does not appear to interact with the gp41-destabilizing mutations and in this regard is analogous to the inhibition seen by the other gp120-directed reagents, sCD4 and SDF-1α.
From these results, we conclude that the destabilizing mutations within the N-terminal helix do not significantly affect the structures and conformational changes that mediate CD4 and coreceptor binding. Nor do the destabilizing mutations impart global or pleiotropic effects upon the envelope glycoprotein complex that render the debilitated viruses fragile in a nonspecific manner.
Destabilizing mutations affect conformational changes in gp41.
We reasoned that mutations that destabilize the interhelical packing interactions within the fusion-active gp41 trimer-of-hairpins might correspondingly favor the preceding prehairpin intermediate. Therefore, we examined the sensitivities of the mutant viruses to peptides that specifically target the N- and C-terminal helices within the prehairpin intermediate. N36 and C34 peptides (Fig. 1B) were produced as described previously (30). We initially focused on the N36 peptide because this peptide binds to wild-type C-terminal helix sequences that are identical in all the mutant viruses.
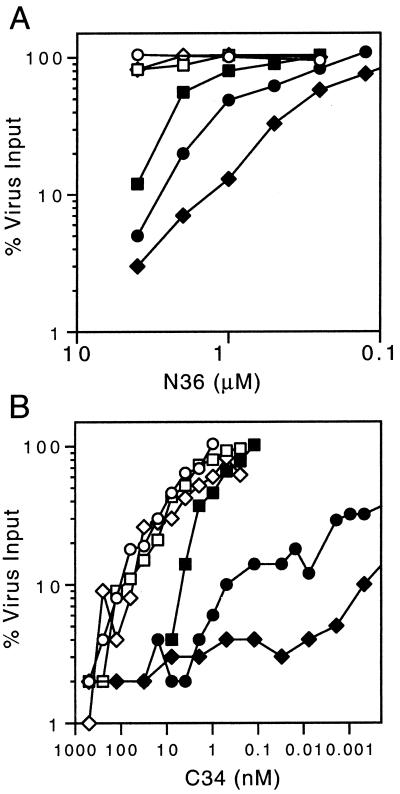
Wild-type and mutant viruses were incubated with serial dilutions of the N36 peptide, and inhibition was assessed in U87-CD4-CXCR4 cell microcultures. N36 was soluble to 4 μM, and higher concentrations were precluded by peptide insolubility in the cell culture medium. Among wild-type and fusion-intact (Q551A and Q563A) viruses (Fig. 3A), little or no inhibition was observed in the presence of a 4-μM concentration of N36, consistent with previous findings that N peptides exhibit relatively low potency (27, 28, 48).
FIG. 3.
Inhibition by N36 and C34 peptides. The U87-CD4-CXCR4 cell inhibition assay and the mutant viruses are described in the legend to Fig. 2. Inhibition by the N36 peptide (A) and the C34 peptide (B) is shown. The measurement of C36 inhibition was extended to 1 fM of peptide (data not presented).
By contrast, all three fusion-deficient alanine mutations that destabilize the interhelical packing interaction in the six-helix bundle structure dramatically increased the sensitivity of the corresponding mutant viruses to inhibition by N36 (Fig. 3A). Ninety percent inhibition (IC90) of the fusion-deficient viruses L556A, L565A, and V570A was seen at 4, 2.8, and 1.2 μM concentrations of N36, respectively. The rank ordering of the susceptibilities to inhibition by N36 corresponds to the relative reduction in viral infectivity. Because peptide binding is thought to be with wild-type C-terminal helix sequence in all cases, these findings suggest that the N-helix mutations that destabilize the trimer-of-hairpin structure increase exposure of the vulnerable prehairpin target.
These data suggest that the destabilizing alanine mutations in the L556A, L565A, and V570A viruses exert a specific and circumscribed effect on the latter steps of gp41-mediated membrane fusion and in particular on the formation of the fusion-active gp41 core. These mutations that destabilize the interhelical packing interactions within the six-helix bundle in vitro appear not to favor the conformational transition from the prehairpin intermediate in vivo. Interference in this transition within the dynamic envelope glycoprotein strongly diminishes viral infectivity and specifically increases viral sensitivity to N peptides that target this critical process.
In contrast to that of the N36 peptide, the stability of binding of the C34 peptide to the different alanine-substituted N-terminal helices is expected to vary in accordance with the thermal stabilities determined in the N34(L6)C28 model (Table 1). Thus, the analysis of inhibition by C34 offers the opportunity to probe the balance between two complex and counteracting influences: (i) the reduced stability of peptide binding to the mutated N-terminal helix and (ii) the increased exposure of the helical regions in the prehairpin intermediate target. In these studies, the fusion-deficient viruses L556A, L565A, and V570A again showed a marked increase in sensitivity to inhibition by C34 (IC90 values of 5, 0.5, and 0.0004 nM, respectively [Fig. 3B]). The wild-type and fusion-intact mutant (Q551A and Q563A) viruses with an IC90 of approximately 100 nM were inhibited. Thus, the predominant effect is one of increased sensitivity to peptide inhibition. It appears that in a dynamic system, the increased exposure of the prehairpin target contributes to more inhibition than does the counteracting influence of decreased peptide binding stability. The mechanistic basis for this outcome is unclear but likely reflects the complexity of the competing processes within the intact envelope glycoprotein.
This complexity is also reflected in the broad range of inhibition of L565A and V570A, the most destabilized and debilitated viruses. For instance, L565A virus infectivity was inhibited by 50% at 0.008 pM C34, a concentration approximately 105-fold below the IC90. This finding suggests a second route of inhibition in these mutants, although the mechanism is unknown.
Neutralization by MAb 2F5.
Our data support the simple model that alanine mutations that destabilize interhelical packing interactions act specifically by rendering the conformational transition to the fusion-active trimer-of-hairpins structure less favorable. We therefore wanted to utilize these specific effects to study the mechanism of virus inhibition by the unique and broadly neutralizing gp41-directed human monoclonal antibody (MAb) 2F5 (9, 11, 42). This MAb targets a predominantly linear epitope (ELDKWA) that lies distal to the C-terminal heptad-repeat sequence (33) in the membrane-proximal region of gp41, a region that has been independently identified as an important determinant of membrane fusion (39, 50).
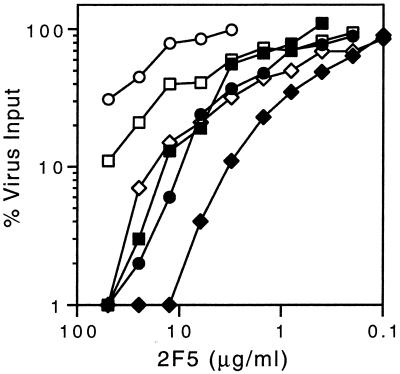
To assess whether neutralization by MAb 2F5 was affected by the destabilizing mutations, we determined the neutralization sensitivity of the fusion-intact and fusion-deficient viruses. Ninety-percent inhibition of the wild-type virus and the Q551A fusion-intact mutant required more than 50 μg of MAb 2F5/ml (Fig. 4). By contrast, the fusion-deficient viruses L556A and L565A were inhibited by 90% at 10 μg/ml, and the highly debilitated V570A virus displayed an IC90 of 3 μg/ml. Thus, neutralization by MAb 2F5 was strikingly and specifically potentiated by the destabilizing mutations.
FIG. 4.
Neutralization by MAb 2F5. The assay and the mutant viruses are as described in the legend to Fig. 2. MAb 2F5, a gp41-directed and broadly neutralizing MAb, from Hermann Katinger (Institute for Applied Microbiology) was provided by Anthony Conley (Merck Research Laboratories).
Interestingly, the Q563A fusion-intact virus displays a sensitivity phenotype similar to that of the less destabilized mutants L556A and L565A (IC90 of Q563A, ∼20 μg/ml). This mutant virus was also somewhat sensitive to neutralization by HIVIG (Fig. 2C) and may suffer a more generalized vulnerability to antibody-mediated neutralization.
Taken together, our results suggest that MAb 2F5 may act by inhibiting the conformational transition to the fusion-active structure. Indeed, Gorny and Zolla-Pazner (18) have previously shown that MAb 2F5 binding to a synthetic C43 peptide is occluded by prior formation of the C43-N51 peptide complex. Our findings with mutant viruses provide biological evidence that the neutralization activity of MAb 2F5 targets the transition to the fusion-active gp41 structure.
Among other gp41-directed MAbs tested (MAbs F240 [5], NC-1 [23], 50-69D, 1281, 98-6D, and 246-D [18]), none neutralized the parental virus and none was able to evoke neutralization of the fusion-deficient mutants (data not shown). These examples may reflect insufficient potency or simply the lack of antibody binding.
Discussion.
Our study provides a novel demonstration in support of the current model for gp41-mediated membrane fusion and its inhibition by N and C peptides. Based on extensive genetic and biophysical evidence (for a review, see reference 13 and references therein), the model proposes a metastable prehairpin gp41 intermediate that folds upon itself to form the stable trimer-of-hairpins structure in which C-terminal helices pack into hydrophobic grooves on a central coiled-coil trimer of N-terminal helices. The energy made available by formation of the stable gp41 core is thought to contribute to membrane fusion. Destabilization of the N- and C-interhelical packing interactions by mutation renders the transition to the fusion-active trimer-of-hairpins less favorable and reduces the ability of the mutant virus to complete membrane fusion and initiate infection.
Our study provides direct evidence for the existence of a discrete and relatively long-lived prehairpin intermediate that is targeted by N and C peptides (6, 17, 40, 45). We suggest that mutations that destabilize the interhelical packing interactions in the trimer-of-hairpins structure correspondingly increase the lifetime of the prehairpin intermediate. In this model, the relative antiviral activity of gp41 peptides is determined by a competition between formation of the trimer-of-hairpins structure and the binding of peptide to the prehairpin intermediate. As the rate of trimer-of-hairpins formation is decreased by mutation, there is more opportunity for peptide binding and thus more of the prehairpin intermediate is diverted into an inactivated form.
Inhibitors that target the conformational transition of gp41 to the fusion-active state represent a new and promising class of antiviral drugs (14, 16). Such antiviral agents might be expected to interact synergistically to enhance the antiviral potency of C peptides that are currently in clinical study, by increasing exposure of the vulnerable prehairpin intermediate.
We have analyzed the interactions between the effects of specific envelope glycoprotein mutations and those of defined antiviral agents in order to obtain additional insight into the respective molecular targets (15). In particular, our finding that neutralization by MAb 2F5 is potentiated by the destabilizing mutations suggests that this broadly neutralizing MAb may itself bind to a prefusogenic structure and may act by interfering with the formation of the fusogenic trimer-of-hairpins. Recently, other neutralizing human MAbs directed to the same membrane-proximal region of gp41 have been identified (52) and these may act similarly to MAb 2F5.
Further characterization of the conformational changes in the HIV-1 envelope glycoprotein will facilitate efforts to develop novel therapeutics and vaccine immunogens. In particular, mutations in the envelope glycoprotein that specifically destabilize the trimer-of-hairpins structure may enhance the presentation of intermediate conformations for vaccine development. Continued analysis of this critical and vulnerable transition in gp41 may point to unique targets for antiviral intervention.
Acknowledgments
This work was supported by National Institutes of Health grants AI42382 (M.L.) and AI44669 (J.H.N.).
We thank Lisa Cavacini, Ian Clark-Lewis, Anthony Conley, Hermann Katinger, George Lewis, Fred Prince, Norbert Schuelke, and Susan Zolla-Pazner for providing reagents used in these studies. We are grateful to Meg Trahey and Richard J. Field for helpful discussions during preparation of the manuscript. The infectious NL4-3 provirus plasmid, from Malcolm Martin, was obtained through the AIDS Research and Reference Reagent Program (NIH), and additional DNA sequence information was kindly provided by Charles Buckler.
REFERENCES
- 1.Abacioglu, Y. H., T. R. Fouts, J. D. Laman, E. Claassen, S. H. Pincus, J. P. Moore, C. A. Roby, R. Kamin-Lewis, and G. K. Lewis. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retroviruses 10:371-381. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bewley, C. A., J. M. Louis, R. Ghirlando, and G. M. Clore. 2002. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J. Biol. Chem. 277:14238-14245. [DOI] [PubMed] [Google Scholar]
- 4.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67:2747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavacini, L. A., C. L. Emes, A. V. Wisnewski, J. Power, G. Lewis, D. Montefiori, and M. R. Posner. 1998. Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res. Hum. Retroviruses 14:1271-1280. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. H., T. J. Matthews, C. B. McDanal, D. P. Bolognesi, and M. L. Greenberg. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 69:3771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S. S.-L., C.-N. Lee, W.-R. Lee, K. McIntosh, and T.-H. Lee. 1993. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane protein. J. Virol. 67:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conley, A. J., J. A. Kessler, I. I., L. J. Boots, J. S. Tung, B. A. Arnold, P. M. Keller, A. R. Shaw, and E. A. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proceedings of the National Academy of Sciences USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deen, K. C., J. S. McDougal, R. Inacker, G. Folena-Wasserman, J. Arthos, J. Rosenberg, P. J. Maddon, R. Axel, and R. W. Sweet. 1988. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature 331:82-84. [DOI] [PubMed] [Google Scholar]
- 11.D'Souza, M. P., D. Livnat, J. A. Bradac, S. H. Bridges, and AIDS Clinical Trials Group Antibody Selection Working Group 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 12.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. USA 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 14.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 15.Elion, G. B., S. Singer, and G. H. Hitchings. 1954. Antagonists of nucleic acid derivatives. VIII. Synergism in combinations of biochemically related antimetabolites. J. Biol. Chem. 208:477-488. [PubMed] [Google Scholar]
- 16.Ferrer, M., T. M. Kapoor, T. Strassmaier, W. Weissenhorn, J. J. Skehel, D. Oprian, S. L. Schreiber, D. C. Wiley, and S. C. Harrison. 1999. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat. Struct. Biol. 6:953-960. [DOI] [PubMed] [Google Scholar]
- 17.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 18.Gorny, M. K., and S. Zolla-Pazner. 2000. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 74:6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, C. M., H. Deng, D. Unutmaz, V. N. Kewalramani, L. Bastiani, M. K. Gorny, S. Zolla-Pazner, and D. R. Littman. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71:6296-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jelesarov, I., and M. Lu. 2001. Thermodynamics of trimer-of-hairpins formation by the SIV gp41 envelope protein. J. Mol. Biol. 307:637-656. [DOI] [PubMed] [Google Scholar]
- 21.Ji, H., C. Bracken, and M. Lu. 2000. Buried polar interactions and conformational stability in the simian immunodeficiency virus (SIV) gp41 core. Biochemistry 39:676-685. [DOI] [PubMed] [Google Scholar]
- 22.Ji, H., W. Shu, F. T. Burling, S. Jiang, and M. Lu. 1999. Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion. J. Virol. 73:8578-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 25.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 26.LaCasse, R. A., K. E. Follis, T. Moudgil, M. Trahey, J. M. Binley, V. Planelles, S. Zolla-Pazner, and J. H. Nunberg. 1998. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J. Virol. 72:2491-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawless, M. K., S. Barney, K. I. Guthrie, T. B. Bucy, S. R. Petteway Jr, and G. Merutka. 1996. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry 35:13697-13708. [DOI] [PubMed] [Google Scholar]
- 28.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 29.Lu, M., H. Ji, and S. Shen. 1999. Subdomain folding and biological activity of the core structure from human immunodeficiency virus type 1 gp41: implications for viral membrane fusion. J. Virol. 73:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, M., and P. S. Kim. 1997. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 15:465-471. [DOI] [PubMed] [Google Scholar]
- 31.Lu, M., M. O. Stoller, S. Wang, J. Liu, M. B. Fagan, and J. H. Nunberg. 2001. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 75:11146-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luciw, P. A. 1996. Human immunodeficiency viruses and their replication, p. 1881-1952. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melinick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields Virology. Lippincott-Raven, Philadelphia, Pa.
- 33.Muster, T., R. Guinea, A. Trkola, M. Purtscher, A. Klima, F. Steindl, P. Palese, and H. Katinger. 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68:4031-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J.-L. Virelizier, F. Arenzana-Seisdedos, O. Schwartz, J.-M. Heard, I. Clark-Lewis, D. F. Legler, M. Loetscher, M. Baggiolini, and B. Moser. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382:833-835. [DOI] [PubMed] [Google Scholar]
- 35.Prince, A. M., H. Reesink, D. Pascual, B. Horowitz, I. Hewlett, K. K. Murthy, K. E. Cobb, and J. W. Eichberg. 1991. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res. Hum. Retroviruses 7:971-973. [DOI] [PubMed] [Google Scholar]
- 36.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starich, S. J. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, L. Ivanoff, S. R. J. Petteway, M. L. Pearson, J. A. Lautenberger, P. T. S., J. Ghrayeb, N. T. Chang, R. C. Gallo, and F. Wong-Staal. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 37.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 38.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 39.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu, W., J. Liu, H. Ji, L. Radigen, S. Jiang, and M. Lu. 2000. Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides. Biochemistry 39:1634-1642. [DOI] [PubMed] [Google Scholar]
- 41.Sodroski, J. G. 1999. HIV-1 entry inhibitors in the side pocket. Cell 99:243-246. [DOI] [PubMed] [Google Scholar]
- 42.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas, I. I. I., D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-immunoglobulin G. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanCott, T. C., V. R. Polonis, L. D. Loomis, N. L. Michael, P. L. Nara, and D. L. Birx. 1995. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res. Hum. Retroviruses 11:1379-1390. [DOI] [PubMed] [Google Scholar]
- 44.Wang, S., J. York, W. Shu, M. O. Stoller, J. H. Nunberg, and M. Lu. Helical packing interactions in the HIV-1 gp41 core: implications for the activation of membrane fusion. Biochemistry, in press. [DOI] [PubMed]
- 45.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 46.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng, Y., Z. Yang, and C. D. Weiss. 2000. Structure-function studies of the self-assembly domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 74:5368-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, K. A., A. L. Maerz, and P. Poumbourios. 2001. Evidence that the transmembrane domain proximal region of the human T-cell leukemia virus type 1 fusion glycoprotein gp21 has distinct roles in the prefusion and fusion-activated states. J. Biol. Chem. 276:49466-49475. [DOI] [PubMed] [Google Scholar]
- 51.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. H. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J. Virol. 75:2741-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]