Abstract
Adeno-associated virus type 2 (AAV-2) integrates specifically into a site on human chromosome 19 (chr-19) called AAVS1. To study the kinetics and frequency of chr-19-specific integration after AAV infection, we developed a rapid, sensitive, and quantitative real-time PCR assay for AAV inverted terminal repeat-chr-19-specific junctions. Despite the known variability of junction sites, conditions were established that ensured reliable quantification of integration rates within hours after AAV infection. The overall integration frequency was calculated to peak at between 10 and 20% of AAV-infected, unselected HeLa cells. At least 1 in 1,000 infectious AAV-2 particles was found to integrate site specifically up to day 4 postinfection in the absence of selection. Chromosomal breakpoints within AAVS1 agreed with those found in latently infected clonal cell lines and transgenic animals. Use of this quantitative real-time PCR will greatly facilitate the study of the early steps of wild-type and recombinant AAV vector integration.
Adeno-associated virus (AAV) has evolved a biphasic life cycle to ensure persistence in its primate host. It needs an unrelated helper virus, adenovirus (Ad) or herpesvirus, for productive infection (1). In the absence of a helper, AAV-2 establishes latency by preferential integration into a specific site on human chromosome 19 (chr-19) (19q13.3-qter) called AAVS1 (10). The site specificity of AAV integration is mediated by the AAV Rep78 protein or by its C-terminally spliced variant Rep68 (1, 12). During productive AAV replication, Rep78 or Rep 68 is needed for AAV gene expression and DNA replication. The AAV origins of DNA replication reside in the 145-bp inverted terminal repeats (ITRs) that flank the 4.7-kb single-stranded AAV genome. Rep78 and/or Rep 68 bind to the Rep-binding site (RBS) on the AAV ITR (22) and nick and unwind the ITR at the terminal resolution site (trs) (8). The AAV ITRs also serve as cis elements for efficient chromosomal integration (1, 12). Binding of Rep to the ITR is required for site-specific integration (27), whereas nicking of the trs is not essential (31). DNA sequences homologous to RBS and the trs of the AAV ITR are found in the chr-19 preintegration site (9, 21). In vitro studies demonstrated ternary complex formation of Rep68 with the AAV ITR and chr-19 AAVS1 (27). A 33-bp sequence spanning the chr-19 RBS and the trs homology element is sufficient to mediate site-specific AAV integration in vivo. Either of the elements and proper spacing between them are essential (13, 15).
Mapped chr-19 integration sites have been derived from cloned cell lines generated with and without drug selection. The integration sites are highly variable within a range of a few hundred base pairs from the RBS of AAVS1 (9, 16, 17, 21, 24, 29). Sequence rearrangements are prominent features. It is unclear whether these have evolved during clonal selection. The only system in which integration sequences have been analyzed early after AAV-2 infection is integration into AAVS1 carried by Epstein-Barr virus (EBV) shuttle plasmids that are maintained as nuclear episomes (4). In this system, the integration sites were found to be next to or within the RBS sequence of AAVS1 (3). In this study, we have established a quantitative real-time PCR assay that allows detection and quantification of integration at the authentic chr-19 integration site early after AAV infection. Our analysis shows that integration sites match the ones found in latently infected cell lines.
MATERIALS AND METHODS
Cells.
Cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml).
Plasmids.
Plasmid pTAV2-0 covers the AAV-2 wild-type genome (6), pDG contains AAV-2 rep and cap linked to Ad type 5 (Ad-5) helper genes (5). Plasmid pRVK carries AAVS1 of chr-19 (9). pAAVS1-TR covers an AAV ITR/AAVS1 junction. The AAVS1 sequence spanning nucleotides 1053 to 1967 derived from pRVK (9) is fused to the right AAV-2 ITR, nucleotides 4489 to 4636 derived from psub201(+) (20).
Preparation of AAV stocks.
Virus stocks were prepared by either of the following protocols. HeLa cells were transfected with pTAV2-0 by CaPO4-mediated DNA transfection (2) and infected with Ad-2 (multiplicity of infection [MOI] = 5) 16 h later. Alternatively, 293 cells were cotransfected with pTAV2-0 and pDG at a 1:3 ratio by CaPO4-mediated DNA transfection (2) and 16 h later the medium was replaced without infection. Cells and supernatants were harvested between 48 and 72 h postinfection (p.i.), frozen and thawed three times, and centrifuged for 15 min at 1,500 × g. Ad-infected HeLa cells were heat inactivated for 30 min at 56°C. Cleared lysates were stored in aliquots at −80°C. The AAV titers were determined by end point dilution on Ad-2-infected cells as previously described (26).
AAV infection.
HeLa cells (1.7 × 106) were seeded on three 10-cm-diameter dishes per harvest time point. Cells were infected with AAV-2 at an MOI of 500 for 60 min. AAV was removed, and 10 ml of complete medium was added. Cells were harvested immediately after adsorption (0 h) and at 8, 16, 24, 32, 48, 72, and 96 h and 2 weeks p.i. Total genomic DNA was extracted as previously described (7). The viscosity of the DNA samples was reduced by digestion with BamHI. RNA was digested by treatment with RNase A. DNA samples were then purified by adsorption chromatography with a QIAquick spin kit (Qiagen, Hilden, Germany). Concentrations of the purified DNAs were determined by measurement of optical densities at 260 and 280 nm in a Beckman DU 640 spectrophotometer. High purity and exact determination of DNA content are essential for reliable PCR quantification.
Quantitative real-time PCR for determination of AAVS1-specific integration of AAV-2.
Targeted integration of AAV-2 into AAVS1 of human chr-19 was detected by quantitative real-time PCR of virus-cell junctions. A two-step PCR assay was established as follows. Purified DNA samples (1 μg) were preamplified (13 cycles in a Perkin-Elmer 9600 cycler) in 50 μl of polymerase buffer with 2.5 U of Platinum Taq polymerase (Gibco) and 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 200 nM primer PAAVS1 (5′-TCAGAGGACATCACGTG-3′), and 200 nM primer PITR (5′-TTAACTACAAGGAACCCCTA-3′). Assay conditions were as follows: 94°C for 2 min (hot start); 13 cycles at 94°C for 1 min, 56°C for 1 min, and 72°C for 3 min; and then a final elongation step of 72°C for 10 min. A real-time LightCycler PCR was then performed with 2-μl aliquots from the first PCR diluted to a final volume of 20 μl. The reaction mixture included polymerase (LightCycler Kit Fast Start DNA Master Hybridization Probes; Roche), 4 mM MgCl2, 500 nM primer PAAVS1, 500 nM primer PITR, 200 nM donor probe 3′ end labeled with fluorescein (5′-TGTTGCTGCCCAAGGATGCT-FL; TIB Molbiol, Berlin, Germany), and 200 nM acceptor probe 5′ end labeled with LC Red640 (5′-LC Red640-TTTCCGGAGCACTTCCTTCTCG-p; TIB Molbiol). Amplification conditions were 95°C for 10 min to activate the polymerase, followed by 45 cycles at 95°C for 10 s, 56°C for 8 s, and 72°C for 30 s.
Analysis of PCR products.
PCR products were removed from the assay capillary and directly analyzed by agarose gel electrophoresis. In addition, samples were cloned into pCR4-TOPO (Invitrogen) with the TOPO TA cloning kit for sequencing (Invitrogen). Plasmid DNA was isolated, digested with EcoRI to release the DNA insert, analyzed on a 1.3% agarose gel, and subjected to DNA sequencing (MWG Biotech, Ebersberg, Germany). PCR conditions were optimized by the use of defined copy numbers of standard control plasmid pAAVS1-TR. Concentrations of pAAVS1-TR were determined spectrophotometrically as outlined above. Copy equivalents of pAAVS1-TR/ml were calculated as follows: 1 μg of 1,000 bp = 1.52 pmol, and 1 pmol = 6.23 × 1011 molecules.
RESULTS AND DISCUSSION
Establishment of a sensitive, quantitative real-time PCR assay.
To analyze AAV integration into the authentic chr-19 preintegration site early after AAV-2 infection, we developed a sensitive and quantitative LightCycler-based real-time PCR assay. A variety of primer combinations were tested to ensure specificity for AAV/AAVS1 junctions and reduce nonspecific signals in uninfected cell DNA. The optimized primer combination is outlined in Fig. 1. The AAV ITR-specific primer partially overlaps unique sequences of the right end of AAV-2 so that only one orientation of integration will be detected. To account for the variability of published integration sites on chr-19, the AAVS1-specific primer was positioned outside the sequence region where the majority of published junction breakpoints have been mapped (9, 21, 29). Thus, most integration events should be detected. The internal primer set is needed for specific PCR product detection by fluorescence resonance energy transfer technology (Fig. 1). Real-time PCR conditions were optimized by the aid of a control plasmid, pAAVS1-TR, that carries an artificial AAV ITR/chr-19 junction fragment. Uninfected HeLa cell DNA (1 μg) was spiked with various dilutions of pAAVS1-TR (101, 102, 103, 104, and 105 DNA copies per reaction). As can be seen in Fig. 2A, the standard curve is linear between 102 and 105 junction copies. PCR products of various lengths are expected because of the variability of integration sites within AAVS1. To ensure reliable quantification, assay conditions were controlled for comparable efficiencies of amplification of various fragment lengths. Amplification efficiency is measured as the template reduplication rate, which usually ranges between 1.6 and 1.9. Reduplication efficiencies were measured on a background of 1 μg of HeLa cell DNA spiked with 103 to 106 copies of defined junction fragments derived from cloned junctions as outlined below (Fig. 3). Junction fragments of 265, 622, and 667 bp (i.e., pAAVS1-TR) were analyzed in parallel. The reduplication rates were 1,78, 1,74, and 1,77, respectively. In addition, precautions were taken to ensure efficient amplification of longer PCR fragments. The time for the elongation step was considerably longer than calculated for efficient amplification of control plasmid pAAVS1-TR.
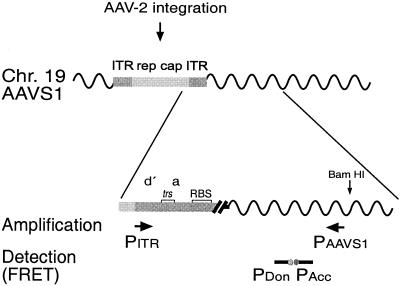
FIG. 1.
Real-time PCR assay for the quantification of site-specific integration of AAV-2 into AAVS1 on chr-19. Sequence elements of integrated AAV-2 are represented as boxes differentially shaded in gray. Amplification, detection, and quantification of site-specific integration were performed with the real-time PCR LightCycler system. The primers used for the amplification of AAV ITR/AAVS1 junctions are indicated by arrows. Primer PITR hybridizes to AAV-2 sequences at positions 4526 to 4545 (23), and primer PAAVS1 hybridizes to AAVS1 on chr-19 19q13.3-qter at positions 1609 to 1593 (9). Sequence elements within the ITR are as outlined in Fig. 3B. Further sequence specificity is guaranteed by the hybridization probe assay format used for detection of the PCR product. Fluorescent dye-labeled probes hybridize to the amplified DNA fragment (donor probe, 1541 to 1560; acceptor probe, 1562 to 1583), thereby bringing the attached dyes into close proximity, thus eliciting fluorescence resonance energy transfer (FRET). Fluorescence emission intensity is directly proportional to the amount of PCR product.
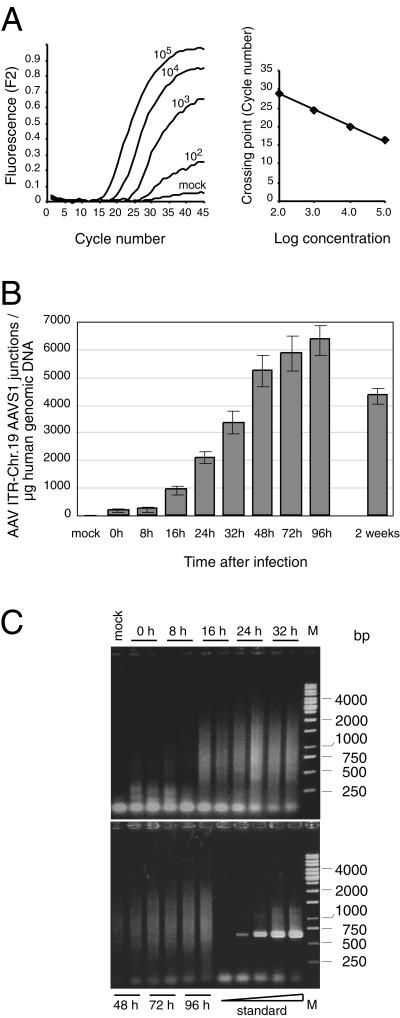
FIG. 2.
Kinetics of AAV site-specific integration. HeLa cells were infected with AAV-2 at an MOI of 500. Total genomic DNA was isolated at different times p.i. Purified DNA (1 μg) was preamplified in 50 μl for 13 cycles by conventional PCR. Samples of 2 μl were subjected to LightCycler PCR as outlined in Materials and Methods. (A) Raw data of the LightCycler analysis of HeLa cell DNA (1 μg) spiked with known copy numbers (102, 103, 104, and 105) of standard plasmid pAAVS1-TR. (B) The copy numbers of AAV ITR/chr-19 integration site junctions per microgram of AAV-infected HeLa DNA were quantified by comparison to values of a standard curve run in parallel as outlined for panel A. Each value represents the mean ± the standard deviation of three independent cultures. (C) LightCycler PCR products were analyzed on agarose gels. Standards were 101, 102, 103, 104, and 105 copies of pAAVS1-TR added to 1 μg of uninfected HeLa cell DNA. The bands in the range below 100 bp present in all lanes represent input primers. M, molecular size marker.
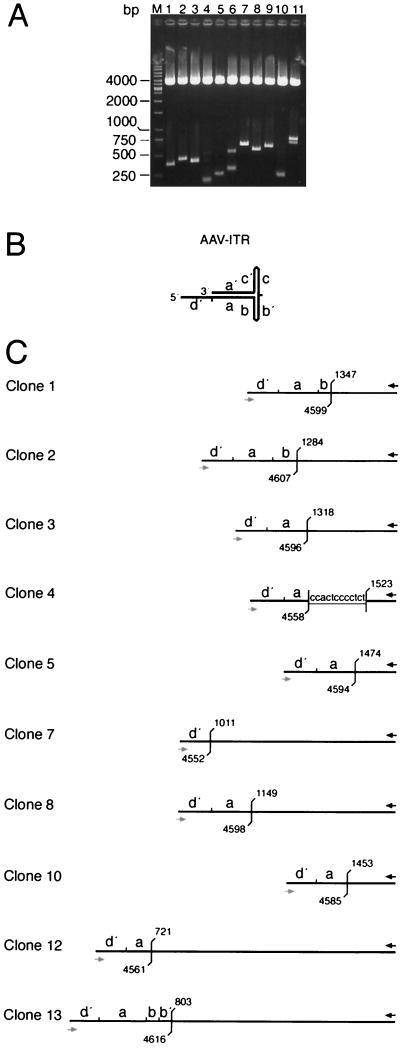
FIG. 3.
Analysis of PCR-amplified AAV ITR/AAVS1 site junctions. PCR fragments of the sample at 96 h p.i. (Fig. 2) were cloned into pCR4-TOPO. Colonies were picked at random. (A) DNAs were digested with EcoRI to release the PCR fragments. Agarose gel electrophoresis visualizes the variability of fragment lengths. (B) The hairpin structure of the AAV ITR is represented in the “flop” orientation. Small letters (d′, a, and b) indicate palindromic sequence elements of the right AAV ITR. (C) Structural maps deduced from DNA sequence analysis of cloned PCR fragments. With the exception of clones 12 and 13, clone numbers refer to the lane numbers in panel A. The black arrow indicates the hybridization site of primer PAAVS1 on chr-19. The gray arrow indicates the binding site of primer PITR on the AAV ITR. Positions of the last unambiguous cellular and/or viral nucleotide are indicated in accordance with the published sequence information (9, 23). Overlapping sequences between the AAV ITR and the chr-19 integration site are underlined.
Kinetics of AAV integration into AAVS1.
To analyze the kinetics of AAV-2 integration into AAVS1, HeLa cells were infected with AAV-2 at an MOI of 500. At the time points indicated, total genomic DNA of three independent plates was extracted. Site-specific integration was analyzed by real-time PCR as outlined in Materials and Methods. Quantification was performed in accordance with a standard curve run in parallel as outlined in Fig. 2A. Site-specific integration became detectable between 8 and 16 h p.i. (Fig. 2B). A plateau was reached at 96 h p.i., when 6,353 ± 524 junctions/μg of genomic DNA were detected. The need to replate the cells in the absence of selection most likely explains the minor reduction at 2 weeks p.i. Several reports have documented that cells harboring latent AAV tend to grow more slowly (25, 28).
Amplified PCR samples were subjected to agarose gel electrophoresis (Fig. 2C). The known variability of junction points resulted in various PCR fragment lengths. The observed DNA smears were therefore expected. The virtual absence of dominant DNA bands in either of the samples further underlines the ability of the assay to coamplify an array of junctions derived from independent integration events without bias or selection. HeLa cell DNA spiked with increasing amounts (101 up to 105 copies) of pAAVS1-TR resulted in unique PCR bands of the expected size (667 bp) (compare Fig. 2C, standard).
The specificity of the assay was documented by a series of negative controls that all scored negative (Table 1); genomic DNAs of mock-infected cells, Ad-infected cells, or plasmids carrying either AAVS1 alone or the entire AAV-2 genome. The specificity for newly generated AAV ITR/AAVS1 junctions was documented by the analysis of AAV-infected cell DNA (72 h p.i.) amplified with either of the external primers alone. No PCR products were detected, whereas the same sample amplified with the combination of both external primers reproduced the expected number of copies (5,865 ± 649) per microgram of genomic DNA (compare Fig. 2A). This excluded the possibility that aberrant, nonjunction PCR products with identical primers at both ends had resulted from potential rearrangements after AAV infection.
TABLE 1.
Specificity of the real-time PCR assay for detection of AAVITR-AAVS1 junctions
| Sample | Primer(s) used | Amt of DNA in PCR (μg) | Mean no. of junctions (SD) |
|---|---|---|---|
| Mock-infected cells | PAAVS1 + PITR | 1 | 0 |
| AAV-infected cells | |||
| 72 h p.i.a | PAAVS1 | 1 | 0 |
| 72 h p.i. a | PITR | 1 | 0 |
| 72 h p.i. | PAAVS1 + PITR | 1 | 5,865 ± 649 |
| Ad-2-infected cells | PAAVS1 + PITR | 1 | 0 |
| Plasmids pRVK + pTAV2-0b | PAAVS1 + PITR | —c | 0 |
The 72-h p.i. sample represented in Fig. 2B was analyzed with only one primer as indicated.
Plasmid pRVK carrying AAVS1 and plasmid pTAV2-0 carrying the wild-type AAV-2 genome were added to the PCR at 105 copies. This equals the number of HeLa genome equivalents per microgram of DNA.
—, 105 copies of each plasmid were used.
Exact quantification of junction copy numbers depends on identical amplification efficiencies of the DNA used as the standard and of the equivalent genomic DNA site in AAV-infected cells. Analyses of serial dilutions of the sample at 96 h p.i. resulted in PCR reduplication efficiencies identical to those of pAAVS1-TR-spiked HeLa DNA. These results are in perfect agreement with the finding that various fragment lengths are efficiently coamplified in parallel (compare Fig. 2C).
Sequence analysis of AAV ITR/chr-19 junctions.
To analyze junction structures, PCR fragments derived from the sample at 96 h p.i. were cloned into pCR4-TOPO as outlined in Materials and Methods. Recombinant colonies were chosen at random. Plasmid DNAs were digested with EcoRI and analyzed by agarose gel electrophoresis (Fig. 3A). The variability of the inserts reflects the variability of integration sites documented by the smears of DNA fragments seen in Fig 2C. The DNA sequences of 10 independent PCR inserts were determined. Each insert had an individual DNA sequence with specific junction points both within the AAV ITR and within the chr-19 integration site. In clones 1, 2, and 13, the amplified AAV sequences extended into the b element, thus representing the “flop” orientation of the AAV ITR. The majority of breakpoints were found within the a sequence (Fig. 3C). As expected from published data (3, 9, 19, 21, 29), the breakpoints within the chr-19 site vary within a range of a few hundred base pairs. None of the 10 PCR inserts analyzed displayed foreign, non-AAV, non-chr-19 integration site sequences.
Comparison of integration sites in unselected cells, in cell lines, and in cells carrying episomal EBV plasmids.
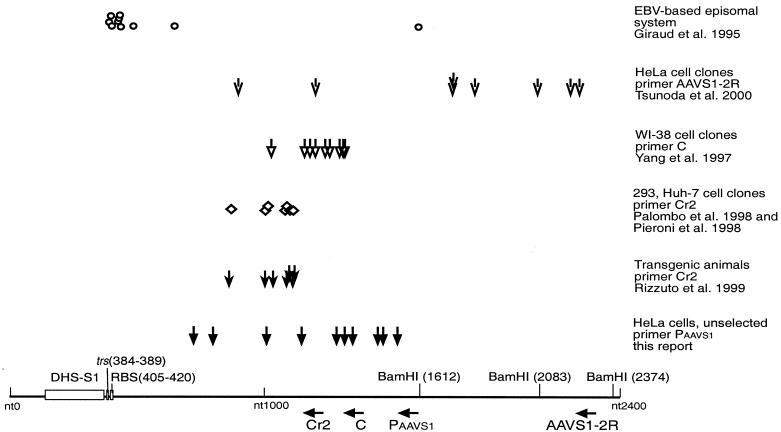
As represented in Fig. 4, crossover points within AAVS1 were compared to those of previous reports. There is good agreement of the AAV integration sites detected with real-time PCR within hours after AAV infection with those described in latently infected cell lines after clonal selection (16-19, 29). Integration sites also agree with those of transgenic mice or rats carrying 3.5 kb of human AAVS1 (19). It is apparent that crossover points near the respective AAVS1 primer are preferentially detected. The reasons for this may be technical in nature, since shorter PCR products are more easily obtained. In the EBV-based episomal system (3), this inherent bias was avoided by detection of integration events through hybridization to an AAV-specific probe. There, integration sites within or next to the RBS of AAVS1 were preferentially detected. Differences between the chromatin structure of the authentic chr-19 region and that on an episomal EBV-based plasmid may explain the divergent findings. The recent description of a DNase I-hypersensitive open chromatin region near the RBS on chr-19 (11) favors the interpretation that chromatin conformation is critical for the spacing between the RBS and a favorable integration site. It will be interesting to see whether integration sites near the RBS can be detected on chr-19 when nearby primers are used for their detection. Our results obtained within hours after AAV infection and in the absence of selection virtually exclude the possibility that early integration sites on chr-19 differ from those detected after clonal selection and extensive chromosomal rearrangements.
FIG. 4.
Compilation of published AAVS1-AAV ITR crossover sequences. Schematic representation of published integration sites within AAVS1 in different cell systems with respect to PCR primers used for their detection. The bottom line represents the nucleotide sequence and numbering as determined by Kotin et al. (9). The trs and RBS homology sequences, as determined by Linden et al. (13), are indicated as boxes, as is the recently described DNase-hypersensitive site (DHS-S1) (11). The primers used for detection of junction fragments are indicated below with the designations used in the respective publications. Nucleotide positions, in the 5′ to 3′ direction, are as follows: Cr2, 1222 to 1201; C, 1345 to 1326; PAAVS1, 1609 to 1593; AAVS1-2R, 2299 to 2280. Integration sites determined in the indicated studies are given as characteristic symbols above. Each symbol represents one integration site. The cell system, the primers used for determination of junction sites, and the respective references are given to the right of the symbols.
Frequency of AAVS1-specific integration per cell.
From our data, the mean site-specific integration frequency can be calculated as follows. Up to 6,500 junction copies are detected per μg of genomic DNA (Fig. 2B). HeLa cells have a hypertriploid (3n+) karyotype (14). Assuming a DNA content of 10 pg per cell, 6 to 7% of HeLa cells are targeted. The EBV-based episomal system is the only system in which site-specific integration frequencies have been quantified without selection (4). Integration frequency per isolated EBV plasmid was found to be in the range of 2%. Since EBV plasmids are maintained at 50 to 100 copies per cell, the frequencies are difficult to compare (4). The most important variables of AAV integration into the authentic chr-19 preintegration site are inherent limitations in the design of the real-time PCR assay. Because of restrictions in primer design, we have used an AAV-specific primer that only detects the right-hand ITR. There is no indication for a preferred orientation of site-specific integration. Preliminary data obtained with a left-hand ITR primer confirm this assumption (D. Hüser and R. Heilbronn, unpublished data). We therefore assume that the true percentage of integration frequencies can be doubled (12 to 14%). In addition, some integration sites have been described beyond the limits of the primers used in this study, as summarized in Fig. 4 (3, 24). Since long PCR products tend to be more difficult to obtain, we deliberately compromised at that point. Some junctions may have DNA sequences that are difficult to amplify for structural reasons. For these reasons, the added missing rate cannot be calculated with certainty. Under the consideration of all variables, we assume that the overall site-specific integration frequency for both orientations at 2 to 4 days p.i. peaks somewhere between 10 and 20% of unselected HeLa cells.
Frequency of AAVS1-specific integration per infectious unit.
To evaluate the probability of site-specific integration per infectious unit, integration frequencies were evaluated with increasing MOIs (Table 2). As expected, the site-specific integration frequency increased with higher MOIs but not proportionally. The 50-fold increase between MOIs of 10 and 500 only resulted in a 13-fold increase in site-specific integration per cell (0.3 to 4.2% for both orientations). By dividing the values in column 3 by the respective AAV MOIs, frequencies of integration per infectious unit were calculated. Up to 0.034% of infectious AAV particles will integrate site specifically at 24 h p.i. Under consideration of the time course data represented in Fig. 2B, the percentage is assumed to reach 0.1% at 96 h p.i. (compare Fig. 2B). Since the above-mentioned confounding variables have been neglected in the calculation, 0.1% represents a low estimate. In summary, at least 1 in 1,000 infectious AAV-2 particles will integrate site specifically within 4 days p.i. in the absence of selective pressure.
TABLE 2.
Site-specific integration frequency at different MOIs
| MOI | Mean no. of right ITR-AAVS1 junctions/μg of HeLa cell DNA (24 h p.i.) ± SDa | Integration frequency (%)/cell (24 h p.i.)c | Integration frequency (%)/infectious unit (24 h p.i.)d | Integration frequency (%)/infectious unit (96 h p.i.)e |
|---|---|---|---|---|
| 10 | 171 ± 96 | 0.34 | 0.034 | 0.103 |
| 30 | 456 ± 191 | 0.91 | 0.030 | 0.091 |
| 100 | 972 ± 446 | 1.94 | 0.019 | 0.058 |
| 500 | 2,097 ± 224b | 4.19 | 0.008 | 0.024 |
HeLa cells were infected with AAV-2 at various MOIs. Total cell DNA was isolated 24 h.p.i. and analyzed by real-time PCR. The results of three experiments are shown.
The MOI of 500 was obtained from the experiment shown in Fig. 2B.
The integration frequency per cell was calculated by assuming a DNA content of 10 pg per HeLa cell and an identical integration frequency of left-and right-hand ITRs as outlined in Results.
The integration frequency per infectious unit was calculated by dividing the frequency in the third column by the respective MOI given in the first column.
The percentages in the fourth column were multiplied by 3.03 to account for the higher peak integration frequencies measured at 96 h p.i. (compare Fig. 2B).
Prospects for the real-time PCR assay for AAV site-specific integration.
Our results show that the real-time PCR assay is not only sensitive but also highly specific for AAV ITR/AAVS1 junctions. In addition, copy numbers can be reliably quantified over a concentration range of several logs. Thus, the natural course of AAV-2 integration into chr-19 19q13.3-qter can be reliably quantified within hours after AAV infection in an unselected cell population. This opens the possibility of rapid monitoring and quantification of the integration efficiencies of AAV ITR and/or rep mutants. In addition, screening for site specificity of recombinant AAV vector integration is possible. Concepts of ways to develop vectors with Rep- and ITR-dependent site-specific integration have been described (16, 18). The surprisingly high frequency of spontaneous, site-specific AAV integration further underlines the attractiveness of these concepts of vector development. The efficient and quantitative short-term assay described in this report opens the possibility of analyzing a series of parallel samples within hours after vector transduction, allowing high-throughput screening for site-specific integration. Last, but not least, screening for AAV/AAVS1 junctions will be useful in the study of the biology of AAV integration in transgenic animals carrying AAVS1 (19, 30), which are the only systems besides primates for studying site-specific AAV integration in a living organism.
Acknowledgments
We thank R. Joncker for expert secretarial assistance and M. Boshart for helpful discussions and critical reading of the manuscript. We are grateful to J. Kleinschmidt, M. Linden, and R. J. Samulski for plasmids.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 506).
REFERENCES
- 1.Berns, K. I. 1996. Parvoviridae: the viruses and their replication, p. 2173-2197. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 2.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giraud, C., E. Winocour, and K. I. Berns. 1995. Recombinant junctions formed by site-specific integration of adeno-associated virus into an episome. J. Virol. 69:6917-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giraud, C., E. Winocour, and K. I. Berns. 1994. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc. Natl. Acad. Sci. USA 91:10039-10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adeno-associated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 6.Heilbronn, R., A. Bürkle, S. Stephan, and H. zur Hausen. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heilbronn, R., and H. zur Hausen. 1989. A subset of herpes simplex virus replication genes induces DNA amplification within the host cell genome. J. Virol. 63:3683-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Im, D.-S., and N. Muzyczka. 1990. The AAV origin-binding protein Rep68 is an ATP-dependent site-specific endonuclease with helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 9.Kotin, R. M., R. M. Linden, and K. I. Berns. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 87:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamartina, S., E. Sporeno, E. Fattori, and C. Toniatti. 2000. Characteristics of the adeno-associated virus preintegration site in human chromosome 19: open chromatin conformation and transcription-competent environment. J. Virol. 74:7671-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. I. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 93:11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linden, R. M., E. Winocour, and K. I. Berns. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA 93:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macville, M., E. Schrock, H. Padilla-Nash, C. Keck, B. M. Ghadimi, D. Zimonjic, N. Popescu, and T. Ried. 1999. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59:141-150. [PubMed] [Google Scholar]
- 15.Meneses, P., K. I. Berns, and E. Winocour. 2000. DNA sequence motifs which direct adeno-associated virus site-specific integration in a model system. J. Virol. 74:6213-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palombo, F., A. Monciotti, A. Recchia, R. Cortese, G. Ciliberto, and N. La Monica. 1998. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus-adeno-associated virus vector. J. Virol. 72:5025-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieroni, L., C. Fipaldini, A. Monciotti, D. Cimini, A. Sgura, E. Fattori, O. Epifano, R. Cortese, F. Palombo, and N. La Monica. 1998. Targeted integration of adeno-associated virus-derived plasmids in transfected human cells. Virology 249:249-259. [DOI] [PubMed] [Google Scholar]
- 18.Recchia, A., R. J. Parks, S. Lamartina, C. Toniatti, L. Pieroni, F. Palombo, G. Ciliberto, F. L. Graham, R. Cortese, N. La Monica, and S. Colloca. 1999. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc. Natl. Acad. Sci. USA 96:2615-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzuto, G., B. Gorgoni, M. Cappelletti, D. Lazzaro, I. Gloaguen, V. Poli, A. Sgura, D. Cimini, G. Ciliberto, R. Cortese, E. Fattori, and N. La Monica. 1999. Development of animal models for adeno-associated virus site-specific integration. J. Virol. 73:2517-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samulski, R. J., L.-S. Chang, and T. Shenk. 1987. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J. Virol. 61:3096-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941-3950. (Erratum, 11:1228, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder, R. O., D.-S. Im, T. Ni, X. Xiao, R. J. Samulski, and N. Muzyczka. 1993. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J. Virol. 67:6096-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsunoda, H., T. Hayakawa, N. Sakuragawa, and H. Koyama. 2000. Site-specific integration of adeno-associated virus-based plasmid vectors in lipofected HeLa cells. Virology 268:391-401. [DOI] [PubMed] [Google Scholar]
- 25.Walz, C., and J. R. Schlehofer. 1992. Modification of some biological properties of HeLa cells containing adeno-associated virus DNA integrated into chromosome 17. J. Virol. 66:2990-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weindler, F. W., and R. Heilbronn. 1991. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J. Virol. 65:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weitzman, M. D., S. R. M. Kyöstiö, R. M. Kotin, and R. A. Owens. 1994. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. USA 91:5808-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winocour, E., M. F. Callaham, and E. Huberman. 1988. Perturbation of the cell cycle by adeno-associated virus. Virology 167:393-399. [PubMed] [Google Scholar]
- 29.Yang, C. C., X. Xiao, X. Zhu, D. C. Ansardi, N. D. Epstein, M. R. Frey, A. G. Matera, and R. J. Samulski. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71:9231-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young, S. M., Jr., D. M. McCarty, N. Degtyareva, and R. J. Samulski. 2000. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 74:3953-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young, S. M., Jr., and R. J. Samulski. 2001. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl. Acad. Sci. USA 98:13525-13530. [DOI] [PMC free article] [PubMed] [Google Scholar]