Abstract
Nucleocytoplasmic trafficking pathways and the status of nuclear pore complex (NPC) components were examined in cells infected with rhinovirus type 14. A variety of shuttling and nonshuttling nuclear proteins, using multiple nuclear import pathways, accumulated in the cytoplasm of cells infected with rhinovirus. An in vitro nuclear import assay with semipermeabilized infected cells confirmed that nuclear import was inhibited and that docking of nuclear import receptor-cargo complexes at the cytoplasmic face of the NPC was prevented in rhinovirus-infected cells. The relocation of cellular proteins and inhibition of nuclear import correlated with the degradation of two NPC components, Nup153 and p62. The degradation of Nup153 and p62 was not due to induction of apoptosis, because p62 was not proteolyzed in apoptotic HeLa cells, and Nup153 was cleaved to produce a 130-kDa cleavage product that was not observed in cells infected with poliovirus or rhinovirus. The finding that both poliovirus and rhinovirus cause inhibition of nuclear import and degradation of NPC components suggests that this may be a common feature of the replicative cycle of picornaviruses. Inhibition of nuclear import is predicted to result in the cytoplasmic accumulation of a large number of nuclear proteins that could have functions in viral translation, RNA synthesis, packaging, or assembly. Additionally, inhibition of nuclear import also presents a novel strategy whereby cytoplasmic RNA viruses can evade host immune defenses by preventing signal transduction into the nucleus.
Picornaviruses are small, nonenveloped viruses that contain RNA genomes of positive polarity. The genomes of all picornaviruses are organized in a similar fashion, with a long 5′ untranslated region (UTR), an open reading frame encoding the viral polyprotein, and a 3′ UTR (reviewed in reference 55). The 5′ UTR contains sequences that are important for replication of the viral genome, as well as an internal ribosomal entry site (IRES), which is required for translation of the viral polyprotein (5, 6, 45). The viral polyprotein is translated from a single large open reading frame and is co- and posttranslationally processed to produce the individual viral gene products (reviewed in reference 55). The 3′ UTR contains a high degree of secondary structure as well as conserved sequences important for viral replication (49, 53, 56, 57).
Numerous interactions between poliovirus and the host cell have been described. For example, during poliovirus infection, the translation initiation factors eIF4GI and -II are cleaved, and translation of capped cellular mRNAs is inhibited (15, 19). Likewise, alterations in cellular transcription rates have been attributed to cleavage of components of the transcriptional apparatus (12, 13, 70-72). In addition, poliovirus infection results in the inhibition of host cell secretion (14) and the induction and subsequent inhibition of apoptosis (3, 65). Recently, we demonstrated that poliovirus infection of HeLa cells results in a dramatic inhibition of nuclear import and the degradation of specific nuclear pore complex (NPC) components (22). Inhibition of nuclear import was shown to result in the cytoplasmic accumulation of a number of nuclear proteins that normally function in RNA biogenesis and nuclear-cytoplasmic transport. Interestingly, some of the relocalized nuclear proteins have been shown to interact with viral gene products or the RNA genome during viral infection, although a direct role for these factors in viral replication in vivo has not been demonstrated (33, 34, 68). Because many antiviral responses involve the transport of cytoplasmic signaling molecules, such as NF-κB and STATs, into the nucleus, we speculated that inhibition of nuclear import may attenuate the antiviral response, leading to a more productive replicative cycle in vivo (22).
In this report, we present data that demonstrate that these events occur in cells infected with rhinovirus type 14. We demonstrate that a number of nuclear proteins that utilize different nuclear import pathways relocalize to the cytoplasm of rhinovirus-infected cells. To demonstrate that nuclear import per se is inhibited, we show that rhinovirus-infected cells are not capable of supporting the nuclear import of cargo in an in vitro import assay. An analysis of NPC components revealed that Nup153 and p62, the same two proteins that were observed to be degraded in poliovirus-infected cells, were also targeted for degradation in rhinovirus-infected cells. Furthermore, we have extended our analysis to show that the degradation of NPC components seen in poliovirus- and rhinovirus-infected cells does not mimic the effects of apoptosis. Cumulatively, these results demonstrate that members of two different genera in the family Picornaviridae target the machinery involved in nucleocytoplasmic trafficking and support the idea that these events are a common feature of infections initiated by this family of viruses.
MATERIALS AND METHODS
Cell lines, viruses, and plasmids.
HeLa cells were maintained as described previously (68). The isolation of cell lines that express enhanced green fluorescent protein (EGFP) and EGFP-nuclear localization signal (NLS) has been described previously (22). Rhinovirus type 14 stocks were prepared by infecting subconfluent HeLa cells with a multiplicity of infection (MOI) of 10. Virus was adsorbed for 30 min at 32°C in CPBS (phosphate-buffered saline [PBS] supplemented with 10 mM MgCl2 and 10 mM CaCl2). Following adsorption, virus was removed, and Dulbecco's modified Eagle's medium containing 10% fetal bovine serum was added. The infected cells were incubated at 32°C for 9 h, at which time the cells were scraped and washed in CPBS. The cell pellets were subjected to three freeze-thaw cycles and centrifuged at 10,000 x g for 5 min, and the supernatant was aliquoted and stored at −20°C. Mahoney type 1 poliovirus stocks were prepared as described previously (68). All infections were initiated at an MOI of 50 and carried out as described above. The construction of pEGFP-M9 and pEGFP-M9G274A has been described previously (22). DNA transfections were performed with the Lipofectin reagent following the manufacturer's recommendations (Gibco/BRL).
Indirect immunofluorescence.
The following antibodies were used for indirect immunofluorescence: MS3 to detect nucleolin (42), SW5 to detect La (62), #SC-333 to detect Sam68 (Santa Cruz Biotechnology), 9H10 to detect heterogeneous nuclear ribonucleoprotein (hnRNP) A1 (a gift from G. Dreyfuss), 12G4 to detect hnRNP K/J (32), 4F4 to detect hnRNP C (11), #S-4045 to detect SC35 (Sigma), 414 to detect nucleoporins (Convance), and #SC-7292 to detect lamins A and C (Santa Cruz Biotechnology). The fixation and permeabilization conditions for the detection of hnRNP A, K, C, SC35, Nups, and lamins A and C have been described previously (22). To detect nucleolin, La and Sam68 cells were fixed and permeabilized by incubation in methanol-acetic acid (3:1) for 10 min at 25°C. Following fixation and permeabilization, cells were incubated overnight with primary antibody at 4°C. Antibody-antigen complexes were detected with fluorescein isothiocyanate (FITC)-conjugated anti-immunoglobulin (Zymed), and nuclei were stained with Hoechst 33258 as previously described (22). Cells were viewed with an Olympus BX-60 fluorescent microscope with a ×60 objective, and images were acquired with a Hamamatsu Orca digital camera and Image Pro Plus software.
In vitro import assay.
Rabbit reticulocyte lysates (RRL; Promega) and glutathione S-transferase (GST)-NLS-EGFP fusion protein (54) were prepared as described previously (22). For import assays, HeLa cells that had been seeded onto 12-mm-diameter glass coverslips 2 days prior were mock infected or infected with rhinovirus for 6 h. The permeabilization and assay conditions were described previously (22). Reactions done in the absence of energy omitted ATP, GTP, creatine kinase, and creatine phosphate.
Immunoblotting.
HeLa cell lysates were prepared by washing cells once in PBS, followed by lysis in Tx buffer (50 mM triethanolamine [pH 7.4]; 500 mM NaCl; 0.5% Triton X-100; 1 mM dithiothreitol; 1 mM phenylmethylsulfonyl fluoride; 10-μg/ml [each] chymostatin, leupeptin, antipain, and pepstatin) (43). Samples were prepared and analyzed as described previously (21). Nup153 and p62 were detected with mouse monoclonal antibody 414 (Convance). Mouse monoclonal antibody MS3 was used to detect nucleolin (42). eIF4GI was detected with rabbit polyclonal sera (4). Poly(ADP-ribose) polymerase (PARP) was detected with #SC-7150 (Santa Cruz Biotechnology).
Induction of apoptosis.
HeLa cells seeded onto 60-mm plates 16 h earlier were induced to undergo apoptosis by adding 5 μg of actinomycin D per ml and 100 μg of cycloheximide per ml and incubating for 6 h at 37°C. Where indicated, the caspase inhibitor z-VAD-fmk was added to a final concentration of 50 μM. Following incubation for the indicated period of time, both floating and adherent cells were collected by scraping and sedimentation at 250 × g for 5 min. The cell pellet was resuspended in 1 ml of PBS and divided into two equal fractions for the isolation of protein or genomic DNA, and cells were sedimented at 250 × g for 5 min. Protein lysates were prepared in Tx buffer as described above. Genomic DNA was prepared by resuspending the cell pellet in 250 μl of lysis buffer (25 mM EDTA, 1% sodium dodecyl sulfate, 0.5 mg of proteinase K per ml) and by incubating this suspension at 50°C for 16 h. Samples were extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform:isoamyl alcohol, precipitated with ethanol, dried, and resuspended in 25 μl of 10 mM Tris-Cl (pH 8). Samples were digested with RNase A (0.8 mg/ml) for 1 h at 37°C and separated on 0.8% agarose gels.
RESULTS
Nucleocytoplasmic relocalization of nucleolin, La, and Sam68 in rhinovirus-infected cells.
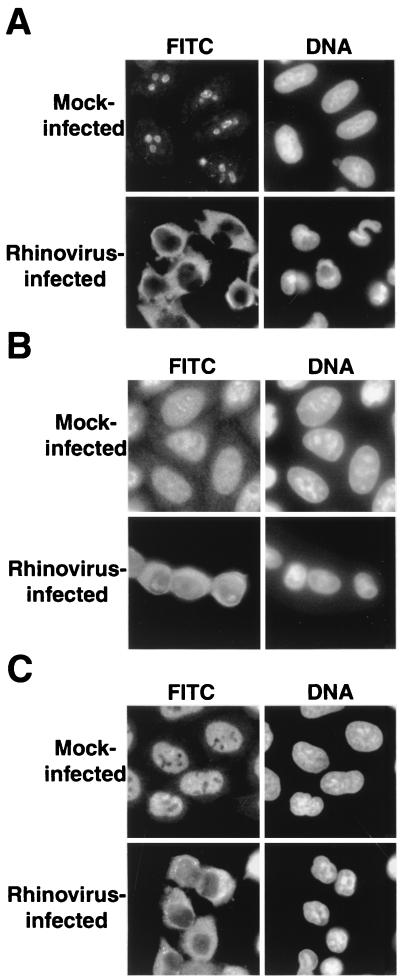
A number of cellular nuclear proteins have been shown to interact with picornaviral RNA genomes or individual viral gene products. These include the La autoantigen, which binds to the picornaviral IRESs and stimulates translation in vitro (34); Sam68, which binds poliovirus RNA-dependent RNA polymerase (33); and nucleolin, which binds to the polioviral 3′ UTR (68). Although roles for these factors in the viral life cycle have yet to be defined, all proteins relocate from the nucleus to the cytoplasm following viral infection. To determine if rhinovirus infection causes the cytoplasmic accumulation of nucleolin, La, and Sam68, the distribution of these proteins was examined in mock- and rhinovirus-infected cells. Figure 1 shows that, as expected, nucleolin, La, and Sam68 all reside in the nucleus in mock-infected cells (17, 62, 69). In contrast, 6 h after infection with rhinovirus, nucleolin, La, and Sam68 have all relocalized from the nucleus to the cytoplasm. These results suggest that rhinovirus causes nucleocytoplasmic relocalization of certain cellular proteins that are normally nuclear residents. The cytoplasmic accumulation of endogenous nuclear proteins was detectable by 4 to 5 h after infection and was complete by 6 to 7 h after infection (data not shown). In cells infected with either poliovirus or rhinovirus, relocalization first became apparent after inhibition of host protein synthesis had occurred, and complete relocalization coincided with the time of maximum viral protein synthesis (data not shown). These results suggest a role for viral proteins in causing the relocalization of cellular proteins from the nucleus to the cytoplasm of infected cells.
FIG. 1.
Intracellular localization of endogenous cellular proteins in uninfected and rhinovirus-infected cells. (A) Uninfected cells (mock infected) or HeLa cells infected with rhinovirus for 6 h were fixed and stained with an antibody directed against nucleolin. In the FITC panels, cells were examined with an FITC filter to detect the indicated antibodies. In the DNA panels, the same field was examined with a UV filter to visualize Hoechst staining of nuclei. (B) Visualization of La was performed as described for panel A. (C) Visualization of Sam68 was performed as described for panel A.
Rhinovirus infection redistributes a reporter protein that contains either a classical or M9 NLS.
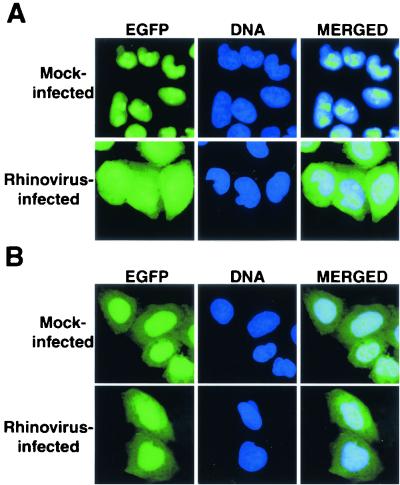
To examine whether proteins that contain NLSs are redistributed to the cytoplasm in cells infected with rhinovirus, we first monitored the distribution of a fusion protein consisting of the EGFP fused in-frame with the NLS from the simian virus 40 (SV40) large T antigen (TAg). The SV40 TAg NLS interacts with a heterodimeric receptor consisting of importin-α, which recognizes the NLS, and importin-β, which mediates docking of the receptor-NLS cargo to the NPC (38). Because the SV40 TAg NLS was the first discovered import sequence, import mediated by this NLS is said to follow the classical import pathway (reviewed in references 25, 31, and 38). When HeLa cells that stably express EGFP-NLS were infected with rhinovirus for 6 h, a significant amount of the EGFP-NLS protein accumulated in the cytoplasm (Fig. 2A). In contrast, the distribution of EGFP molecules that lacked an NLS did not change throughout the course of infection (Fig. 2B). The relocalization of EGFP-NLS was not due to proteolytic removal of the NLS, because immunoblotting revealed no change in the mobility of this protein during infection with rhinovirus (data not shown). These results indicate that either rhinovirus infection inhibits the import of EGFP-NLS or it enhances the export of this protein, although the latter possibility seems unlikely, because the TAg NLS has never been shown to function as a nuclear export signal.
FIG. 2.
Intracellular localization of EGFP and EGFP-NLS molecules in uninfected and rhinovirus-infected cells. (A) HeLa cells stably expressing EGFP-NLS were mock infected or infected with rhinovirus as indicated. Cells were processed and examined by fluorescent microscopy 6 h after infection. EGFP fluorescence was visualized with an FITC filter. In the DNA panels, Hoechst-stained nuclei were examined with a UV filter. The panels on the right show the FITC and Hoechst images merged. (B) HeLa cells stably expressing EGFP fusion proteins were examined as described for panel A.
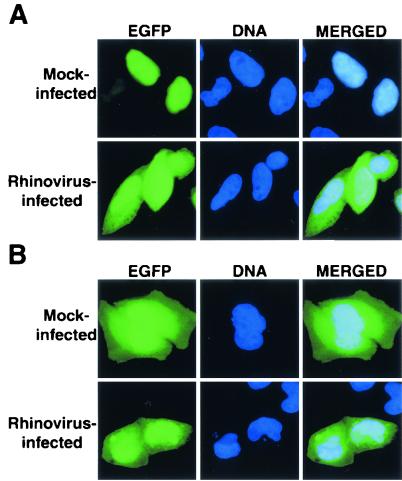
To determine if the effect of rhinovirus infection upon the classical import pathway extends to other nucleocytoplasmic trafficking pathways, we examined the localization of a reporter protein that uses the transportin import pathway. Briefly, transportin is the cellular receptor responsible for the nuclear import of cargos containing an M9 NLS (reviewed in references 25, 31, and 38). The M9 NLS is a glycine-rich, bidirectional shuttling signal originally identified in the hnRNP A1 (51). Cargos that contain the M9 NLS are recognized by transportin, which then targets these cargos to the NPC, analogous to the function of importin-α and -β in the classical import pathway. Nuclear export of these cargos is mediated by an as-yet-unidentified nuclear export receptor. When HeLa cells were transiently transfected with plasmids that express an EGFP-M9 fusion protein, this protein was localized predominantly to the nucleus (Fig. 3A). However, when transfected cells were infected with rhinovirus for 6 h, a significant increase in the amount of cytoplasmic EGFP-M9 was observed. Cytoplasmic accumulation of EGFP-M9 was not due to proteolytic removal of the NLS from EGFP-M9 (data not shown). To confirm that the redistribution of EGFP-M9 was due to a functional NLS, we examined the effect of rhinovirus infection on the distribution of EGFP fused to a mutant form of the M9 NLS (EGFP-M9G274A). This mutant contains a single-amino-acid substitution and does not function as an NLS due to an inability to bind transportin (51). EGFP-M9G274A, like EGFP, resides both in the nucleus and the cytoplasm, because it can diffuse between both of these cellular compartments (Fig. 3B). When cells expressing EGFP-M9G274A were infected with rhinovirus, no change in its distribution was observed (Fig. 3B). These results indicate that the relocalization of EGFP-M9 in rhinovirus-infected cells was due to a disruption of the M9 trafficking pathway. Together with the analysis of the TAg NLS, these results indicate that rhinovirus inhibits at least two different nucleocytoplasmic trafficking pathways.
FIG. 3.
Intracellular localization of EGFP-M9NLS and EGFP-M9G274A molecules in mock-infected and rhinovirus-infected cells. (A) HeLa cells were transiently transfected with plasmid pEGFP-M9NLS (EGFP-M9) and mock infected or infected with rhinovirus at 40 h after transfection and examined 6 h after infection. Labeling of panels is as described in the legend to Fig. 2. (B) Same as panel A, except cells were transfected with plasmid pEGFP-M9G274A (EGFP-M9MT).
Nucleocytoplasmic relocalization of hnRNP A1, K, and C in rhinovirus-infected cells.
During poliovirus infection, several nuclear proteins have been reported to relocalize to the cytoplasm of infected cells (22, 33, 34, 68). To determine whether rhinovirus infection resulted in a similar redistribution of nuclear proteins, we examined the subcellular location of hnRNP A1, K, and C in infected cells.
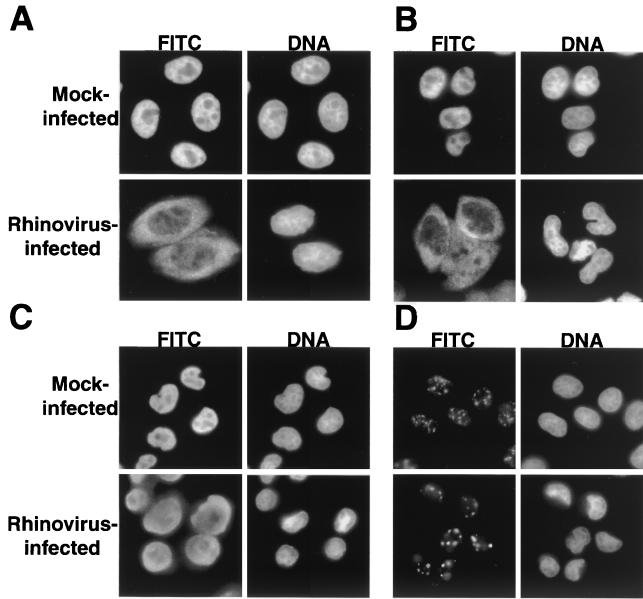
Polypeptide hnRNP A1 has been implicated in pre-mRNA splicing, as well as mRNA export, and shuttles between the nuclear and cytoplasmic compartments due to the presence of the M9 NLS (28, 38). Figure 4A shows that in uninfected cells, hnRNP A1 displays a diffuse nucleoplasmic distribution that is excluded from nucleoli, as previously reported (50). In contrast, when cells that have been infected with rhinovirus for 6 h are examined, a significant amount of hnRNP A1 is found in the cytoplasm (Fig. 4A). Therefore, hnRNP A1, like the M9-containing GFP, relocalizes from the nucleus to the cytoplasm in rhinovirus-infected cells.
FIG. 4.
Intracellular localization of cellular proteins in uninfected and rhinovirus-infected cells. (A) Uninfected cells (mock infected) or HeLa cells infected with rhinovirus for 6 h were fixed and stained with an antibody directed against hnRNP A1. In the FITC panels, cells were examined with an FITC filter to detect indicated antibodies. In the DNA panels, the same field was examined with a UV filter to visualize Hoechst staining of nuclei. (B) Visualization of hnRNP K was performed as described for panel A. (C) Visualization of hnRNP C was performed as described for panel A. (D) Visualization of SC35 was performed as described for panel A.
To determine whether rhinovirus-induced inhibition of nucleocytoplasmic trafficking extended to a third pathway, we examined the distribution of hnRNP K. The shuttling protein hnRNP K has been implicated in transcriptional regulation and translational silencing (reviewed in reference 28). Movement of hnRNP K between the cytoplasmic and nuclear compartments is mediated by the bidirectional K nuclear shuttling (KNS) domain (35). The KNS domain is composed of 40 amino acids and is recognized by import and export receptors that are distinct from those that mediate transport of classical or M9-NLS-containing cargos (35). Indirect immunofluorescence analysis showed that hnRNP K was localized to the nucleus of mock-infected cells (Fig. 4B). Six hours after infection with rhinovirus, however, hnRNP K had relocalized to the cytoplasm of infected cells. This is exactly what was seen for hnRNP A1 and shows that proteins containing classical, M9, and KNS NLSs are relocalized in rhinovirus-infected cells.
Because both hnRNP A1 and K are known to shuttle between the nucleus and cytoplasm, it was possible that relocalization was dependent upon this ability. To determine whether shuttling was required for relocalization during rhinovirus infection, we determined the location of hnRNP C in mock- and rhinovirus-infected cells. The hnRNP C has been implicated in a variety of processes, including packaging, splicing, and stability of mRNA and contains a bipartite NLS, suggesting that it is imported via the classical pathway (28, 60). However, hnRNP C does not shuttle, due to the presence of a 78-amino-acid motif that functions as a nuclear retention signal (37). Indirect immunofluorescence analysis of mock-infected cells confirmed that hnRNP C was confined to the nucleus (Fig. 4C). After 6 h of infection with rhinovirus, however, a significant amount of hnRNP C was found in the cytoplasm. Therefore, the cytoplasmic accumulation of nuclear proteins in rhinovirus-infected cells is not dependent upon the ability to shuttle. As was observed with nucleolin, the cytoplasmic accumulation of these endogenous nuclear proteins was noted by 4 to 5 h after infection and was complete by 6 to 7 h after infection (data not shown).
To determine whether all nuclear proteins redistributed into the cytoplasm during rhinovirus infection, we examined the distribution of the splicing factor SC35 in infected cells. SC35 belongs to a family of essential splicing factors known as SR proteins due to the presence of serine/arginine-rich regions (18). Recently, the import of SR proteins was shown to be dependent upon a novel member of the importin-β family of proteins, known as transportin-SR, that specifically interacts with the SR regions of these proteins (26). Analysis of SC35 by indirect immunofluorescence revealed that it localized to nuclear speckles in uninfected cells, as previously reported (Fig. 4D) (18). Unlike what was seen for the hnRNPs, however, the distribution of SC35 did not change during the course of rhinovirus infection (Fig. 4D). These results could indicate that the transportin-SR import pathway is functional in rhinovirus-infected cells. Alternatively, because SC35 does not possess shuttling activity, it may be sequestered in the nucleus due to interaction with other factors. However, these results demonstrate that not all nuclear proteins relocalize to the cytoplasm of rhinovirus-infected cells.
Inhibition of the classical nuclear import pathway in cells infected with rhinovirus.
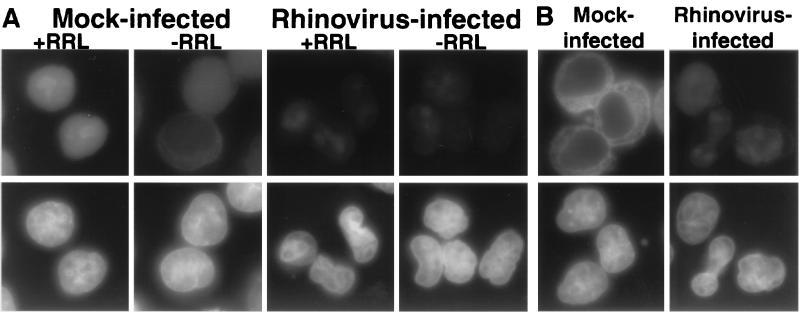
The results presented above are consistent with the hypothesis that rhinovirus infection results in the inhibition of the nuclear import of a number of different proteins. To test the effect of nuclear import directly, we assayed the import of a cargo protein that contained a classical NLS in infected cells that had been permeabilized with digitonin. In this assay, the cells were first treated with digitonin to permeabilize the plasma membrane while leaving the nuclear envelope intact (1). The cells were then washed to remove soluble factors and assayed for nuclear import activity by adding back cytosol, an energy-regenerating system and an import cargo. When mock-infected cells were permeabilized, incubated with the GST-NLS-EGFP cargo, an energy-regenerating system and RRL as a cytosolic source, followed by an extensive wash to remove nontransported cargo, nuclear fluorescence was observed (Fig. 5A). In contrast, only low levels of the GST-NLS-EGFP cargo accumulated in the nucleus when the RRL was omitted (Fig. 5A). The appearance of nuclear fluorescence required the presence of an NLS in the cargo and incubation at temperatures above 0°C (data not shown), as well as the presence of an energy-generating system (Fig. 5B). When permeabilized rhinovirus-infected cells were assayed in the presence or absence of RRL, very little accumulation of the import cargo in the nucleus was observed (Fig. 5A). Therefore, rhinovirus-infected permeabilized cells show a greatly reduced ability to support the nuclear import of classical NLS-containing cargo. Because the cytosol was removed during cell preparation, these data suggest that insoluble components contributed by the permeabilized infected cell are disrupted during infection.
FIG. 5.
Cell-free nuclear import assays. (A) Uninfected cells (mock infected) or cells that had been infected with rhinovirus for 6 h were permeabilized and used in an in vitro nuclear import assay. Assays were carried out in the presence (+RRL) or absence (−RRL) of RRL as a source of cytosolic factors. Top panels show GFP with an FITC filter, and bottom panels show Hoechst staining of DNA with a UV filter. All GFP images were acquired in Photoshop 5.0 (Adobe Systems, Inc.) with identical exposure times and manipulations. (B) Same as panel A, except creatine kinase, creatine phosphate, ATP, and GTP were omitted from the reaction. GFP images were acquired with the same exposure time as panel A.
The first step in nuclear import is recognition of an NLS-containing cargo by an import receptor. The receptor-cargo complex then must dock at the cytoplasmic face of the NPC before it transits the nuclear pore and cargo is released into the nucleoplasm. Previous work has shown that in the absence of energy, or at nonphysiological temperatures, receptor-cargo complexes accumulate at the cytoplasmic face of the NPC, indicative of docking (1). To determine if rhinovirus infection affected docking, we examined the ability of permeabilized cells to accumulate receptor-cargo complexes at the nuclear rim in the absence of energy. Figure 5B shows that, as expected, when permeabilized mock-infected cells were incubated in the absence of ATP, they accumulated import cargo at the nuclear rim, consistent with the notion that docking had occurred. In contrast, when rhinovirus-infected cells were assayed under identical conditions, very little accumulation of the GST-NLS-GFP cargo at the nuclear rim was observed (Fig. 5B). The same results were obtained when the assays were carried out at 0°C (data not shown). These findings demonstrate that one of the earliest steps in the process of nuclear import (i.e., docking of receptor-cargo complexes at the NPC) is disrupted in rhinovirus-infected cells. In these assays, the NPCs are provided by infected cells, while the cargo and cytosolic components are derived from uninfected cells. Therefore, it is likely that rhinovirus infection results in changes to the NPC that prevent docking of receptor-cargo complexes.
Degradation of specific nucleoporins in rhinovirus-infected cells.
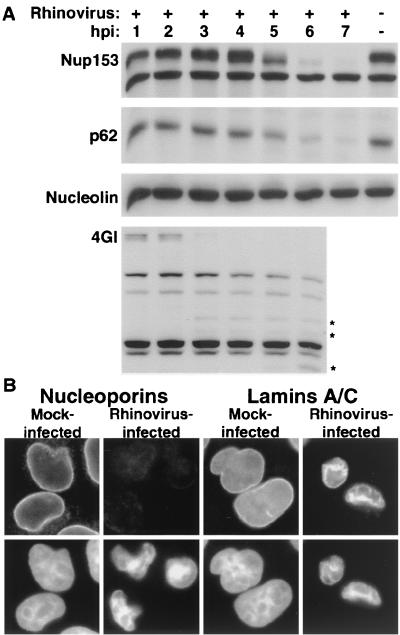
The vertebrate NPC has a mass of approximately 125 MDa and is composed of over 50 different proteins, collectively called nucleoporins (Nups; reviewed in reference 63). Previously, we showed that during poliovirus infection, Nup153 and p62 were degraded, correlating with the relocalization of cellular proteins and the inability of infected cells to mediate import in vitro (22). To determine whether rhinovirus infection also targeted Nup153 and p62 for degradation, we examined the status of these proteins during the course of infection. Immunoblot analysis of whole-cell lysates revealed that the levels of Nup153 and p62 were unaltered through the first 4 h of infection (Fig. 6A), began to decline by 5 h postinfection, and were almost undetectable by 7 h postinfection. These results were not due to differences in loading between lanes or to a general degradation of proteins during rhinovirus infection, because the levels of nucleolin remained unchanged (Fig. 6A). Inhibition of translation by the addition of 100 μg of cycloheximide per ml to infected cells prevented the degradation of Nup153 and p62, indicating that protein synthesis following viral entry was required for degradation (data not shown). Inhibition of cellular transcription by the addition of 5 μg of actinomycin D per ml did not prevent degradation of Nup153 or p62, demonstrating that transcription of a cellular mRNA was not required (data not shown). To determine if the inhibition of nuclear import induced by rhinovirus infection coincides with any other events in the viral life cycle, we examined the cleavage of the translation initiation factor eIF4GI and the onset of inhibition of host protein synthesis. Our results show that very little intact eIF4GI was present at 4 h after infection and that incorporation of radiolabeled methionine into host proteins was inhibited between 3 and 4 h following infection (Fig. 6A) (data not shown). These results are in agreement with those of previous reports (64) and demonstrate that the inhibition of import described here does not occur until after cellular protein synthesis has been inhibited and significant amounts of viral proteins have accumulated.
FIG. 6.
Analysis of nuclear pore complex composition in rhinovirus-infected HeLa cells. (A) Fifty micrograms of whole-cell lysates prepared from mock-infected cells or cells that had been infected with rhinovirus for the indicated length of time was analyzed by immunoblotting with monoclonal antibody 414 to detect Nup153 and p62 or MS3 to detect nucleolin. eIF4GI was detected with rabbit polyclonal sera. An asterisk indicates rhinovirus-specific degradation products of eIF4GI. hpi, hours postinfection. (B) Indirect immunofluorescence with monoclonal antibodies 414 and SC-7292 to detect nucleoporins and lamins, respectively. Cells were either uninfected (mock infected) or infected with rhinovirus for 6 h (rhinovirus infected). The top panels show cells examined with an FITC filter, and the bottom panels show the same fields examined with a UV filter to detect Hoechst staining. FITC images for a given antibody were acquired with identical exposure times and adjustments.
To examine whether the results presented above reflected a loss of Nup153 and p62 from the NPC itself and not just the loss of a more easily extractable fraction, we visualized Nup153 and p62 by indirect immunofluorescence. Analysis of uninfected cells with monoclonal antibody 414, which detects both Nup153 and p62, revealed the punctate nuclear rim staining characteristic of NPC-associated factors recognized by this antibody (Fig. 6B). When rhinovirus-infected cells were examined in this manner, a dramatic decrease in the staining with this antibody was observed (Fig. 6B). To determine whether this reflected a general loss of proteins from the nuclear envelope, we examined the status of the nuclear lamins A and C in uninfected and infected cells. Figure 6B shows that in mock-infected cells, an antibody to lamins A and C stained the nuclear rim. Although the nuclei of cells that have been infected with rhinovirus for 6 h were shrunken and more irregularly shaped than those of mock-infected cells, indirect immunofluorescence with antibody to lamins A and C revealed significant staining that coincided with the Hoechst stain (Fig. 6B). These results demonstrated that the amount of Nup153 and p62 in the NPC of rhinovirus-infected cells is significantly reduced at a time that coincides with the cytoplasmic accumulation of several different nuclear proteins.
Comparison of nucleoporin degradation during apoptosis and picornavirus infection.
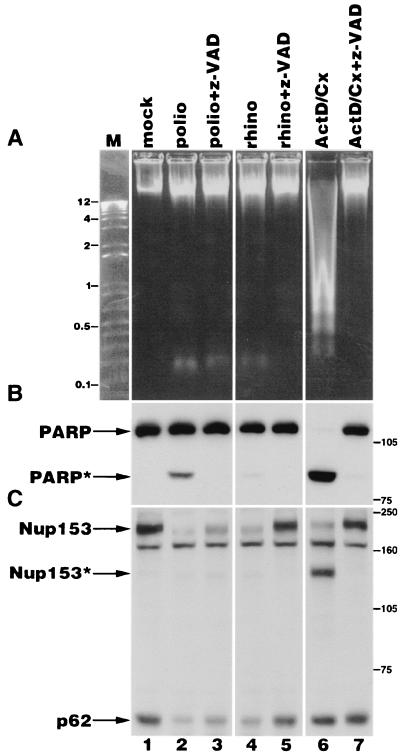
Previous work has shown that in cells undergoing apoptosis, specific components of the NPC are degraded in a caspase 3-dependent manner (10, 16). Poliovirus has been shown to possess a proapoptotic activity that causes the activation of caspases (2). We wished to determine if there were any similarities between the NPC alterations observed during apoptosis and those observed during infection with either poliovirus or rhinovirus. Treatment of cells with the metabolic inhibitors cycloheximide and actinomycin D has been shown to result in the rapid induction of apoptosis in cultured human cells (30). When human HeLa cells were incubated with actinomycin D and cycloheximide, several key features of apoptosis were observed. These included the fragmentation of genomic DNA into low-molecular-mass forms and the appearance of the 89-kDa cleavage product of PARP (Fig. 7A and B, lane 6). Both fragmentation of genomic DNA and degradation of PARP could be prevented by coincubation with the caspase inhibitor z-VAD-fmk, confirming that these events were dependent upon caspase activity (Fig. 7A and B, lane 7). When picornavirus-infected cells were examined in a similar manner, little fragmentation of cellular genomic DNA was observed (Fig. 7A, lanes 2 and 4). The small amount of cleaved PARP seen in poliovirus-infected cells (Fig. 7B, lane 2) is consistent with previous reports showing a small induction of caspase 3 and 7 activity in productively infected cells (3). In agreement with this, cleavage of PARP in poliovirus-infected cells was inhibited by the addition of z-VAD-fmk (Fig. 7B, lane 3). Very little PARP cleavage was observed in rhinovirus-infected cells (Fig. 7B, lane 4).
FIG. 7.
Analysis of Nup153 and p62 in apoptotic and picornavirus-infected cells. (A) Electrophoresis of genomic DNA isolated from HeLa cells following various treatments. mock, lysate from mock-infected cells; polio, lysates from cells that have been infected with poliovirus for 5 h; rhino, lysates from cells that have been infected with rhinovirus for 6 h; ActD/Cx, lysates from cells that have been treated with actinomycin D and cycloheximide for 6 h; z-VAD, z-VAD-fmk added immediately following virus adsorption or coincident with the addition of actinomycin D and cycloheximide; M, molecular size markers (kilobases). (B) Fifty micrograms of whole-cell lysates prepared from cells treated as described in panel A was analyzed by immunoblotting to detect PARP. The positions of the full-length and cleaved forms of PARP are indicated by PARP and PARP∗, respectively. The positions of molecular mass markers (in kilodaltons) are indicated. (C) Fifty micrograms of whole-cell lysates was analyzed by immunoblotting with monoclonal antibody 414 to detect Nup153 and p62. The positions of the full-length and cleaved forms of Nup153 are indicated by Nup153 and Nup153∗, respectively. The positions of molecular mass markers (in kilodaltons) are indicated.
Immunoblot analysis of apoptotic cells confirmed that Nup153 was cleaved to produce a 130-kDa product, and this could be prevented by the addition of z-VAD-fmk (Fig. 7C, lanes 6 and 7) (10, 16). As previously shown, p62 was not a target for degradation in apoptotic cells (Fig. 7C, compare lanes 1 and 6) (10, 16). However, analysis of these NPC components in cells infected with poliovirus or rhinovirus revealed distinct differences when compared to apoptotic cells. Specifically, degradation of Nup153 in both poliovirus- and rhinovirus-infected cells did not reveal the 130-kDa cleavage product, which was readily detected in apoptotic cells (Fig. 7C, compare lanes 2, 4, and 6). Additionally, while p62 was not a target for degradation during apoptosis, infection with either poliovirus or rhinovirus resulted in the loss of this NPC component (Fig. 7C, compare lanes 2, 4, and 6). To test directly whether caspases were involved in degradation of Nup153 and p62, z-VAD-fmk was added to cells following infection with poliovirus or rhinovirus. The addition of z-VAD-fmk had a minor inhibitory effect upon degradation of Nup153 and p62 after 5 h in poliovirus-infected cells, but resulted in very little degradation of these NPC components after 6 h in rhinovirus-infected cells (Fig. 7C, lanes 3 and 5). However, lysates from cells infected with rhinovirus for 8 h in the presence of z-VAD-fmk revealed degraded Nup153 and p62 protein species (data not shown). These results indicate that z-VAD-fmk delays, but does not prevent, the degradation of Nup153 or p62 in rhinovirus-infected cells.
If one of the viral proteases is responsible for degradation of Nup153 and p62, these results would indicate that the rhinovirus and poliovirus proteases have differential sensitivities to z-VAD-fmk. However, analysis of [35S]methionine-labeled lysates prepared from cells infected with rhinovirus or poliovirus in the presence of z-VAD-fmk revealed no differences in the processing patterns of the viral polyprotein, although viral protein synthesis was slightly inhibited (data not shown). Because the activities of picornavirus 2A and 3C proteases were not inhibited by z-VAD-fmk, these results suggest that poliovirus and rhinovirus infections activate either two distinct cellular proteases or a common protease that is more sensitive to z-VAD-fmk at the lower temperature used for rhinovirus infections (32 versus 37°C). While these results do not rule out a possible role for caspases in degradation of NPC components during picornavirus infection, they demonstrate that the targets of degradation and the cleavage products produced during picornavirus infection are different from those seen during apoptosis.
Finally, cells infected with rhinovirus for 6 h in the presence of z-VAD-fmk showed less cytoplasmic accumulation of hnRNP A1 than in the absence of z-VAD-fmk (data not shown). However, if the time of infection was extended in the presence of z-VAD-fmk, relocalization of hnRNP A1 and degradation of Nup153 and p62 were indistinguishable from those seen in the absence of z-VAD-fmk (data not shown). Together, these findings support the idea that the degradation of NPC components correlates with the relocalization of hnRNP A1 to the cytoplasm of picornavirus-infected cells. Cells infected with a low MOI of rhinovirus produced two- to threefold less virus in the presence of z-VAD-fmk than in the absence of z-VAD-fmk, suggesting that treatment with z-VAD-fmk delays relocalization of hnRNP A1 and degradation of nucleoporins, but does not dramatically reduce viral yield.
DISCUSSION
The experiments presented above were designed to determine if rhinovirus causes a disruption in the normal trafficking of proteins between the nucleus and the cytoplasm. Our results demonstrate that a variety of shuttling and nonshuttling proteins that utilize different import pathways accumulate in the cytoplasm of rhinovirus-infected cells. Strikingly, relocalization could be conferred upon a heterologous protein simply by the addition of a functional NLS of either the classical or M9 pathway, suggesting that the nucleocytoplasmic trafficking machinery was impaired during infection. Using an in vitro import assay, we demonstrated directly that infected cells were deficient in their ability to support nuclear import of a cargo containing a classical NLS. Analysis of NPC components in rhinovirus-infected cells revealed that Nup153 and p62 were degraded. These results clearly demonstrate that rhinovirus infection causes an inhibition of the classical nuclear import pathway and suggest that other pathways may also be inhibited.
The finding that viruses belonging to two different genera in the picornavirus family cause an inhibition of nuclear import and target NPC components for degradation suggests that this may be common to most or even all members of this family. In support of this hypothesis, Belov and colleagues have reported that an EGFP-NLS fusion protein similar to the one used here accumulates in the cytoplasm of cells infected with coxsackievirus B3, although the status of Nup153 and p62 was not examined in that study (8).
Other RNA viruses have also been reported to cause the relocalization of nuclear proteins and to inhibit nucleocytoplasmic trafficking. For example, mouse hepatitis virus, a positive-stranded RNA virus belonging to the Coronavirus family, was shown to cause cytoplasmic accumulation of hnRNP A1 (29). The matrix (M) protein of vesicular stomatitis virus, a negative-stranded RNA virus belonging to the family Rhabdovirus, can block multiple transport pathways when expressed in Xenopus oocytes (23, 48, 67). As with the picornaviruses, the ability of the M protein to inhibit transport is conserved across a number of different vesiculoviruses, indicating a selective advantage to maintaining this function (47). The M protein is localized to the NPC, and its ability to inhibit transport is blocked in mutants that have lost the ability to associate with the NPC (47, 48, 67). These findings raise the possibility that disruption or alteration of nuclear transport pathways through targeting of NPC components may be a common occurrence in cells infected by RNA viruses.
Currently, the mechanism responsible for the inhibition of nuclear import in picornavirus-infected cells is not known. Our results demonstrate that degradation of Nup153 and p62 correlated well with the cytoplasmic accumulation of nuclear proteins and the inability of permeabilized infected cells to support import in vitro. Nup153 has been shown to interact with a variety of import and export receptors as well as the GDP-bound form of RAN (9, 36, 39, 58, 59, 73) and has been shown to play a role in the export of mRNA, snRNA, and 5S rRNA (7, 66). Protein p62 is found in the central channel of the NPC (20) and has been shown to interact with importin-β family members (9, 24, 27, 36, 46, 73) and NTF2 (44), a protein critical for the proper recycling of RAN between the nucleus and cytoplasm (52, 61). The loss of either Nup153 or p62 could have effects that extend to multiple transport pathways and result in the effects described here. While the proteolysis of Nup153 and p62 in rhinovirus-infected cells suggests a relatively simple mechanism for the inhibition of import reported here, it is not clear if the loss of these two nucleoporins is necessary or sufficient to cause these effects. The NPC is composed of over 50 different proteins, and our analysis has examined the status of only 2 of them. One possibility, therefore, is that rhinovirus and poliovirus target as-yet-unidentified nucleoporins, which directly inhibit import or trigger the degradation of Nup153 and p62.
Several studies suggest that, in cells, stress-responsive signaling pathways exist that lead to the redistribution of cellular proteins and, in some cases, alterations in NPC composition. For example, in the arrest of secretion response (ASR), yeast cells that harbor temperature-sensitive mutations in genes required for the proper function of the protein secretory apparatus, redistribute nuclear proteins and a number of NPC components to the cytoplasm at the nonpermissive temperature (40, 41). These results suggest the presence of a signaling mechanism that monitors the proper functioning of the secretory system. Poliovirus infection is known to inhibit protein secretion (14), raising the possibility that poliovirus activates a mammalian counterpart to the yeast ASR, which may be responsible for some or all of the effects described here.
As described above, a variety of cellular signaling pathways result in alterations to NPC structure and nucleocytoplasmic trafficking pathways. The effect of poliovirus and rhinovirus infection on nuclear import may therefore be part of the response of the host cell to the stress of infection. Regardless of whether these effects are caused by viral or cellular activities, it will be interesting to elucidate the mechanisms underlying the inhibition of nuclear import and the degradation of NPC components in picornavirus-infected cells.
Acknowledgments
We are very grateful to Karla Kirkegaard for critical reading of the manuscript. We also thank Harris Busch, Gideon Dreyfuss, and Ger Prujin for antibodies.
This work was supported by a grant from the National Institutes of Health (AI25105). K.E.G. is a recipient of a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
REFERENCES
- 1.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agol, V. I., G. A. Belov, K. Bienz, D. Egger, M. S. Kolesnikova, N. T. Raikhlin, L. I. Romanova, E. A. Smirnova, and E. A. Tolskaya. 1998. Two types of death of poliovirus-infected cells: caspase involvement in the apoptosis but not cytopathic effect. Virology 252:343-353. [DOI] [PubMed] [Google Scholar]
- 3.Agol, V. I., G. A. Belov, K. Bienz, D. Egger, M. S. Kolesnikova, L. I. Romanova, L. V. Sladkova, and E. A. Tolskaya. 2000. Competing death programs in poliovirus-infected cells: commitment switch in the middle of the infectious cycle. J. Virol. 74:5534-5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldabe, R., E. Feduchi, I. Novoa, and L. Carrasco. 1995. Efficient cleavage of p220 by poliovirus 2Apro expression in mammalian cells: effects on vaccinia virus. Biochem. Biophys. Res. Commun. 215:928-936. [DOI] [PubMed] [Google Scholar]
- 5.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastos, R., A. Lin, M. Enarson, and B. Burke. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belov, G. A., A. G. Evstafieva, Y. P. Rubtsov, O. V. Mikitas, A. B. Vartapetian, and V. I. Agol. 2000. Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology 275:244-248. [DOI] [PubMed] [Google Scholar]
- 9.Bonifaci, N., J. Moroianu, A. Radu, and G. Blobel. 1997. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc. Natl. Acad. Sci. USA 94:5055-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buendia, B., A. Santa-Maria, and J. C. Courvalin. 1999. Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J. Cell Sci. 112:1743-1753. [DOI] [PubMed] [Google Scholar]
- 11.Choi, Y. D., and G. Dreyfuss. 1984. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J. Cell Biol. 99:1997-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, M. E., T. Hammerle, E. Wimmer, and A. Dasgupta. 1991. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 10:2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark, M. E., P. M. Lieberman, A. J. Berk, and A. Dasgupta. 1993. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 13:1232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. B. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000 dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 16.Ferrando-May, E., V. Cordes, I. Biller-Ckovric, J. Mirkovic, D. Gorlich, and P. Nicotera. 2001. Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ. 8:495-505. [DOI] [PubMed] [Google Scholar]
- 17.Freeman, J. W., A. Chatterjee, B. E. Ross, and H. Busch. 1985. Epitope distribution and immunochemical characterization of nucleolar phosphoprotein C23 using ten monoclonal antibodies. Mol. Cell Biochem. 68:87-96. [DOI] [PubMed] [Google Scholar]
- 18.Fu, X. D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 19.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan, T., S. Muller, G. Klier, N. Pante, J. M. Blevitt, M. Haner, B. Paschal, U. Aebi, and L. Gerace. 1995. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell 6:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustin, K. E., and M. J. Imperiale. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Her, L. S., E. Lund, and J. E. Dahlberg. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science 276:1845-1848. [DOI] [PubMed] [Google Scholar]
- 24.Hu, T., T. Guan, and L. Gerace. 1996. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 134:589-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izaurralde, E., and S. Adam. 1998. Transport of macromolecules between the nucleus and the cytoplasm. RNA 4:351-364. [PMC free article] [PubMed] [Google Scholar]
- 26.Kataoka, N., J. L. Bachorik, and G. Dreyfuss. 1999. Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 145:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kehlenbach, R. H., A. Dickmanns, A. Kehlenbach, T. Guan, and L. Gerace. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 29.Li, H. P., X. Zhang, R. Duncan, L. Comai, and M. M. Lai. 1997. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci.USA 94:9544-9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, S. J., S. V. Lennon, A. M. Bonham, and T. G. Cotter. 1990. Induction of apoptosis (programmed cell death) in human leukemic HL-60 cells by inhibition of RNA or protein synthesis. J. Immunol. 145:1859-1867. [PubMed] [Google Scholar]
- 31.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 32.Matunis, M. J., W. M. Michael, and G. Dreyfuss. 1992. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 12:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride, A. E., A. Schlegel, and K. Kirkegaard. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 93:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moroianu, J., M. Hijikata, G. Blobel, and A. Radu. 1995. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA 92:6532-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakielny, S., and G. Dreyfuss. 1996. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 134:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 39.Nakielny, S., S. Shaikh, B. Burke, and G. Dreyfuss. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18:1982-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nanduri, J., S. Mitra, C. Andrei, Y. Liu, Y. Yu, M. Hitomi, and A. M. Tartakoff. 1999. An unexpected link between the secretory path and the organization of the nucleus. J. Biol. Chem. 274:33785-33789. [DOI] [PubMed] [Google Scholar]
- 41.Nanduri, J., and A. M. Tartakoff. 2001. The arrest of secretion response in yeast. Signaling from the secretory path to the nucleus via Wsc proteins and Pkc1p. Mol. Cell 8:281-289. [DOI] [PubMed] [Google Scholar]
- 42.Olson, M. O., K. Guetzow, and H. Busch. 1981. Localization of phosphoprotein C23 in nucleoli by immunological methods. Exp. Cell Res. 135:259-265. [DOI] [PubMed] [Google Scholar]
- 43.Pante, N., R. Bastos, I. McMorrow, B. Burke, and U. Aebi. 1994. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 126:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paschal, B. M., and L. Gerace. 1995. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell Biol. 129:925-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 46.Percipalle, P., W. D. Clarkson, H. M. Kent, D. Rhodes, and M. Stewart. 1997. Molecular interactions between the importin alpha/beta heterodimer and proteins involved in vertebrate nuclear protein import. J. Mol. Biol. 266:722-732. [DOI] [PubMed] [Google Scholar]
- 47.Petersen, J. M., L. S. Her, and J. E. Dahlberg. 2001. Multiple vesiculoviral matrix proteins inhibit both nuclear export and import. Proc. Natl. Acad. Sci. USA 98:8590-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petersen, J. M., L.-S. Her, V. Varvel, E. Lund, and J. E. Dahlberg. 2000. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol. Cell. Biol. 20:8590-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pilipenko, E. V., K. V. Poperechny, S. V. Maslova, W. J. Melchers, H. J. Slot, and V. I. Agol. 1996. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary ('kissing') interactions. EMBO J. 15:5428-5436. [PMC free article] [PubMed] [Google Scholar]
- 50.Pinol-Roma, S., and G. Dreyfuss. 1991. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science 253:312-314. [DOI] [PubMed] [Google Scholar]
- 51.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 52.Ribbeck, K., G. Lipowsky, H. M. Kent, M. Stewart, and D. Gorlich. 1998. NTF2 mediates nuclear import of Ran. EMBO J. 17:6587-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohll, J. B., D. H. Moon, D. J. Evans, and J. W. Almond. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69:7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosorius, O., P. Heger, G. Stelz, N. Hirschmann, J. Hauber, and R. H. Stauber. 1999. Direct observation of nucleocytoplasmic transport by microinjection of GFP-tagged proteins in living cells. BioTechniques 27:350-355. [DOI] [PubMed] [Google Scholar]
- 55.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philidelphia, Pa.
- 56.Sarnow, P. 1989. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol. 63:467-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarnow, P., H. D. Bernstein, and D. Baltimore. 1986. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc. Natl. Acad. Sci. USA 83:571-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shah, S., and D. J. Forbes. 1998. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 8:1376-1386. [DOI] [PubMed] [Google Scholar]
- 59.Shah, S., S. Tugendreich, and D. Forbes. 1998. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 141:31-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, A., A. Brownawell, and I. G. Macara. 1998. Nuclear import of Ran is mediated by the transport factor NTF2. Curr. Biol. 8:1403-1406. [DOI] [PubMed] [Google Scholar]
- 62.Smith, P. R., D. G. Williams, P. J. Venables, and R. N. Maini. 1985. Monoclonal antibodies to the Sjogren's syndrome associated antigen SS-B (La). J. Immunol. Methods 77:63-76. [DOI] [PubMed] [Google Scholar]
- 63.Stoffler, D., B. Fahrenkrog, and U. Aebi. 1999. The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 11:391-401. [DOI] [PubMed] [Google Scholar]
- 64.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, T. A. Ivannikova, E. A. Smirnova, N. T. Raikhlin, and V. I. Agol. 1995. Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J. Virol. 69:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ullman, K. S., S. Shah, M. A. Powers, and D. J. Forbes. 1999. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell 10:649-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Kobbe, C., J. M. A. van Deursen, J. P. Rodrigues, D. Sitterlin, A. Bachi, X. Wu, M. Wilm, M. Carno-Fonseca, and E. Izaurralde. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell 6:1243-1252. [DOI] [PubMed] [Google Scholar]
- 68.Waggoner, S., and P. Sarnow. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong, G., O. Muller, R. Clark, L. Conroy, M. F. Moran, P. Polakis, and F. McCormick. 1992. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell 69:551-558. [DOI] [PubMed] [Google Scholar]
- 70.Yalamanchili, P., R. Banerjee, and A. Dasgupta. 1997. Poliovirus-encoded protease 2APro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. J. Virol. 71:6881-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yalamanchili, P., U. Datta, and A. Dasgupta. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yalamanchili, P., K. Weidman, and A. Dasgupta. 1997. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology 239:176-185. [DOI] [PubMed] [Google Scholar]
- 73.Yaseen, N. R., and G. Blobel. 1997. Cloning and characterization of human karyopherin β3. Proc. Natl. Acad. Sci. USA 94:4451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]