Abstract
The γ134.5 protein of herpes simplex virus type 1 (HSV-1) is required for viral neurovirulence in vivo. In infected cells, this viral protein prevents the shutoff of protein synthesis mediated by double-stranded-RNA-dependent protein kinase PKR. This is accomplished by recruiting protein phosphatase 1 to dephosphorylate the α subunit of translation initiation factor eIF-2 (eIF-2α). Moreover, the γ134.5 protein is implicated in viral egress and interacts with proliferating cell nuclear antigen. In this report, we show that the γ134.5 protein encoded by HSV-1(F) is distributed in the nucleus, nucleolus, and cytoplasm in transfected or superinfected cells. Deletion analysis revealed that the Arg-rich cluster from amino acids 1 to 16 in the γ134.5 protein functions as a nucleolar localization signal. The region from amino acids 208 to 236, containing a bipartite basic amino acid cluster, is able to mediate nuclear localization. R215A and R216A substitutions in the bipartite motif disrupt this activity. Intriguingly, leptomycin B, an inhibitor of nuclear export, blocks the cytoplasmic accumulation of the γ134.5 protein. L134A and L136A substitutions in the leucine-rich motif completely excluded the γ134.5 protein from the cytoplasm. These results suggest that the γ134.5 protein continuously shuttles between the nucleus, nucleolus, and cytoplasm, which may be a requirement for the different activities of the γ134.5 protein in virus-infected cells.
The γ134.5 gene of herpes simplex viruses (HSVs) is located in the inverted repeats of the viral genome flanking the unique long sequence and is present in two copies per genome (1, 13, 14). In HSV type 1 (HSV-1) strain F, the γ134.5 gene encodes a protein of 263 amino acids consisting of an amino-terminal domain, a linker region of three amino acid repeats (Ala-Thr-Pro), and a carboxyl-terminal domain (13). The triplet repeats are a constant feature of the γ134.5 protein in HSV-1, but the number of repeats varies from strain to strain (3, 13, 16). In HSV-2, these triplet repeats are not present in the γ134.5 protein, as determined by nucleotide sequence analysis (32). Interestingly, the carboxyl-terminal domain of the γ134.5 protein is partially homologous to the corresponding domains of the murine myeloid differentiation primary-response protein MyD116 (28; D. J. McGeoch and B. C. Barnett, Letter, Nature 353:609, 1991), the human and hamster growth arrest and DNA damage response protein GADD34 (46), and virulence factor NL/I14L of African swine fever virus (18, 47).
The γ134.5 protein is essential for HSV to display neurovirulence in experimental animal models (10, 29, 44). Deletion or nonsense mutation in the γ134.5 gene abates the ability of HSV to replicate in the central nervous system neurons, and therefore, the mutant is incapable of causing encephalitis (10, 37). In human neuroblastoma cell lines infected with HSV-1, the γ134.5 protein is expressed to prevent the shutoff of protein synthesis mediated by the double-stranded-RNA-dependent protein kinase (PKR) (9, 11). This function requires the carboxyl terminus of the γ134.5 protein to recruit cellular protein phosphatase 1 (PP1), forming a high-molecular-weight complex that dephosphorylates the α subunit of the translation initiation factor 2 (eIF-2α) (8, 12, 21, 22). In virus-infected cells, the γ134.5 protein-mediated eIF-2α dephosphorylation contributes to HSV resistance to the antiviral effect of alpha/beta interferon (7).
The carboxyl-terminal domains of the γ134.5 protein and GADD34/MyD116 are functionally interchangeable in the context of the HSV genome (20). GADD34/MyD116 belongs to a family of proteins induced under conditions of genotoxic stress, growth arrest, differentiation, and apoptosis (23, 28, 46). GADD34 promotes apoptosis induced by ionizing radiation or methyl methanesulfate, and this activity is negatively regulated by Src kinase Lyn (19, 23). It is also involved in the negative regulation of a stress-inducible gene, CHOP (36). Like the γ134.5 protein, GADD34 complexes with proliferating cell nuclear antigen (PCNA) through its carboxyl-terminal domain (6). It has been proposed that the interaction of GADD34 or the γ134.5 protein with PCNA may release cells from growth arrest and facilitate viral replication in HSV-infected cells (6). Studies show that the γ134.5 protein is required for viral glycoprotein processing and maturation from infected cells (3, 5). In mouse 3T6 cells, the γ134.5 deletion mutant is defective in viral egress and the growth of the mutant is severely affected in resting but not in actively dividing cells (4, 5).
Early studies indicate that the γ134.5 protein of HSV-1(F) is a soluble protein that accumulates in both the nucleus and cytoplasm late in infection (1). However, studies with the γ134.5 protein of HSV-1(17+) suggest that it is a cytoplasmic protein (33, 34). In analyzing the domain function of the γ134.5 protein, we found that although it is predominantly found in the cytoplasmic fraction, the γ134.5 protein is also present in the nuclear fraction (20). Interestingly, deletion of the carboxyl terminus or substitution of it with the corresponding region of MyD116 leads to an increased accumulation of the γ134.5 protein in the nuclear fraction (20). These observations raise the possibility that the γ134.5 protein may contain cis elements that determine its subcellular localization.
In the present study, we further examined the cellular localization of the γ134.5 protein derived from strain HSV-1(F). We show that in transfected cells the γ134.5 protein is distributed in the nucleus, nucleolus, and cytoplasm. We demonstrate, by the use of deletions and site-specific mutations, that the γ134.5 protein contains nuclear import and export signals that control its nuclear, nucleolar, and cytoplasmic accumulation. We also find that leptomycin B inhibits the cytoplasmic localization of the γ134.5 protein. These findings suggest that the γ134.5 protein continuously shuttles between the nucleus, nucleolus, and cytoplasm. While this work was in progress, it was reported that the cellular localization of the γ134.5 protein is affected by the number of ATP repeats and the arginine-rich cluster in the amino terminus (31).
MATERIALS AND METHODS
Cells and reagents.
HeLa, Vero, and human neuroblastoma SK-N-SH cell lines were obtained from the American Type Culture Collection and propagated in Dulbecco's modified Eagle's medium supplemented with 5% (HeLa and Vero cells) or 10% (SK-N-SH cells) fetal bovine serum. Recombinant virus R3616, which has a 1-kb deletion in the γ134.5 gene, was generously provided by Bernard Roizman (10). Leptomycin B and 4′,6′-diamidino-2-phenylindole (DAPI) were purchased from Sigma and Vector Laboratories, respectively.
Plasmids.
Mammalian expression vectors pEYFP-C1 and pEYFP-N1 were purchased from Clontech. pRB143 has the BamHI S fragment of HSV-1(F) encoding the γ134.5 protein (21). pGF9907 contains a BamHI-StuI fragment that encodes the entire coding region of the γ134.5 protein (8). To construct pGF9912, a BamHI-BspEI fragment from pRB143 was blunt-ended with Klenow fragment and cloned into the EcoRV site of pTet-Splice (GIBCO BRL). In this plasmid, the expression of the γ134.5 gene is driven by a cytomegalovirus promoter. To construct pGF2101, a NcoI/Klenow-HindIII fragment from plasmid pGF9907 was cloned into the BspEI/Klenow and HindIII sites of pEYFP-C1. In this plasmid, the full-length γ134.5 protein was fused in frame to the carboxyl terminus of the green fluorescent protein (GFP). Deletions in the γ134.5 gene were generated by PCR with Platinum high-fidelity Taq polymerase (GIBCO BRL) and pRB143 as a template (8). To construct plasmid pGF2110(ΔN16), oligo17-N (ATGCGCTAGCCATGGGGCCCACGGGCGCCGTCCCAACCGCACAG) and oligoR34.5-Hind (ATCGAAGCTTTATATGCGCGGCTCCTGCCATCGTCTCTCC) were used. The PCR fragment was digested with NcoI/Klenow and HindIII and then ligated into the BspEI/Klenow and HindIII sites of pEYEP-C1. In this plasmid, the DNA fragment encoding amino acids 17 to 263 of the γ134.5 protein was fused in frame to the carboxyl terminus of GFP. In a similar way, the following deletion mutants were generated: pGF2109, with a deletion of amino acids 1 to 28 (ΔN28); pGF2104, with a deletion of amino acids 1 to 52 (ΔN52); pGF2105, with a deletion of amino acids 1 to 83(ΔN83); pGF2106, with a deletion of amino acids 1 to 116 (ΔN116); pGF2107, with a deletion of amino acids 1 to 146 (ΔN146); pGF2117, with a deletion of amino acids 1 to 187 (ΔN187); and pGF2118, with a deletion of amino acids 1 to 216 (ΔN216). In the PCR amplification, oligoR34.5-Hind was used as a downstream primer. The upstream primers were oligo29-N (ATGCGCTAGCCATGGTAACCTCCACGCCCAACTCGGAACCCGCG) for pGF2109(Δ28), oligo53-N (ATGCGCTAGCCATGGCCAGTGGGCCCCCGCCTTCTTGTTCGC) for pGF2104(Δ52), oligo84-N (ATGCGCTAGCCATGGACAGCCCCCCGCCCGAGCCGGCGCCAG) for pGF2105(Δ83), oligo117-N (ATGCGCTAGCCATGGCTAACCCCTCCCACCCCCCCTCACGCCCCTTCCG) for pGF2106(Δ116), oligo147-N (ATGCGCTAGCCATGGTGCGCCTGCGACGCGCGGGCGGGGAGGGGGCG) for pGF2107(Δ146), oligo188-N (ATGCGCTAGCCATGGCGACCCCCGCGCGGGTGCGCTTCTCGC) for pGF2117(Δ187), and oligo217-N (ATGCGCTAGCCATGGGCTCGTGGGCCCGCGAGCGGGCCGACC) for pGF2118(Δ216). To construct pGF2121(N16), oligoN16-Nhe (CTAGCCATGGCCCGCCGCCGCCGCCATCGCGGCCCCCGCCGCCCCCGGCCGCCCC) and its complement, oligoN16-Xho (TCGAGGGGCGGCCGGGGGCGGCGGGGGCCGCGATGGCGGCGGCGGCGGGCCATGG), were cloned into the NheI and XhoI sites of pEYFP-N1. In this plasmid, amino acids 1 to 16 of the γ134.5 protein were fused to the amino terminus of GFP. To construct pGF2142(188-236), PCR amplification was carried out with oligo188-N and oligo236-C (ATGCCTCGAGGGCCTCCGCCACCCGGCGCCGGAACCGAGC) and the PCR fragment digested with NheI and XhoI was ligated into the NheI and XhoI sites of pEYFP-N1. In this plasmid, amino acids 188 to 236 of the γ134.5 protein were fused to the amino terminus of GFP. Similarly, to construct pGF2143(208-236), oligoN208-Nhe (CTAGCCATGGCCTCGGCCGCCCGCCTGGCGCGCCGCGGCTCGTGGGCCCGCGAGCGGGCCGACCGGGCTCGGTTCCGGCGCCGGGTGGCGGAGGCCC) and its complement, oligoN236-Xho (TCGAGGGCCTCCGCCACCCGGCGCCGGAACCGAGCCCGGTCGGCCCGCTCGCGGGCCCACGAGCCGCGGCGCGCCAGGCGGGCGGCCGAGGCCATGG), were cloned into the NheI and XhoI sites of pEYFP-N1. In this plasmid, amino acids 208 to 236 of the γ134.5 protein were fused to the amino terminus of GFP. To construct pGF2231, oligoN208/2R-Nhe (CTAGCCATGGCCTCGGCCGCCCGCCTGGCGGCCGCCGGCTCGTGGGCCCGCGAGCGGGCCGACCGGGCTCGGTTCCGGCGCCGGGTGGCGGAGGCCC) and its complement, oligoN236/2R-Xho (TCGAGGGCCTCCGCCACCCGGCGCCGGAACCGAGCCCGGTCGGCCCGCTCGCGGGCCCACGAGCCGGCGGCCGCCAGGCGGGCGGCCGAGGCCATGG), were cloned into the NheI and XhoI sites of pEYFP-N1. In this plasmid, amino acids 208 to 236 of the γ134.5 protein were fused to the amino terminus of GFP, with Arg215 and Arg216 replaced by Ala. To construct plasmid pGF2144, a two-step PCR was performed by using plasmid pRB143 as the template. In the first round of PCR, two fragments were amplified. One fragment was amplified with oligoR34.5-Nco (ATGCGCTAGCCATGGCCCGCCGCCGCCGCCATCGCGGC) and oligoNES-C (GCTCTGCGGTGACGCGCGCGCGGGCGGCGAGGCGCGGCG), and the other was amplified with oligoNES-N (CGCCGCGCCTCGCCGCCCGCGCGCGCGTCACCGCAGAGC) and oligoR34.5-Hind. Using a mixture of two PCR fragments as a template, a second round of PCR was carried out with oligoR34.5-Nco and oligoR34.5-Hind, and the PCR fragment digested with NcoI/Klenow and HindIII was cloned into the BspEI/Klenow and HindIII sites of pEYFP-C1. In this plasmid, the full-length γ134.5 protein, with both Leu134 and Leu136 mutated to Ala, was fused in frame to the carboxyl terminus of GFP.
Antibodies.
Plasmid pRB4893 encoding a glutathione S-transferase-γ134.5 fusion protein (amino acids 146 to 263) was transformed into Escherichia coli BL21. Expression of the fusion protein was induced by addition of isopropyl-β-d-thiogalactopyranoside to the bacterial culture, followed by affinity purification of the fusion protein from lysates on agarose beads conjugated with glutathionine (22). The fusion protein was used for immunization of rabbits for production of polyclonal anti-γ134.5 antibody. Mouse monoclonal anti-C23 antibody was purchased from Santa Cruz Biotechnology.
Transfection and leptomycin B treatment.
Cells were grown on glass coverslips to 60% confluency. An aliquot of plasmid DNA (0.5 to 1.0 μg) was mixed with 3 μl of Lipofectamine reagent (GIBCO BRL) and added to cells as suggested by the manufacturer. Five hours after incubation, the cells were placed in serum-containing medium and further incubated for 36 h at 37°C. Leptomycin B (Sigma) was added to the medium at a final concentration of 5 nM at 36 h after transfection. The incubation was continued with 5% CO2 at 37°C for 3 h.
Fluorescence confocal microscopy.
After transfection, the medium was aspirated and cells were washed twice with phosphate-buffered saline, fixed with 4% paraformaldehyde for 20 min, and examined directly by a fluorescence microscope. For indirect-immunofluorescence analysis, cells plated on glass coverslips were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.2% Triton X-100 for 30 min, blocked with 1% bovine serum albumin, and then incubated with the rabbit polyclonal anti-γ134.5 antibody (1:250 dilution) or mouse monoclonal anti-C23 antibody (1:100 dilution) overnight at 4°C. After extensive washing with phosphate-buffered saline, cells were incubated with goat anti-rabbit or mouse secondary antibody conjugated with Rhodamine (Santa Cruz Biotechnology) at a 1:100 dilution. Cells were then visualized with a fluorescence microscope. The procedure for DAPI staining was carried out as described previously (27). Briefly, after direct- or indirect-fluorescence analysis, cells were counterstained with DAPI (1.5 μg/ml) in the VECTASHIELD mounting medium (Vector Laboratories). The slides were then examined by an Olympus IX70 inverted digital confocal microscope. For each sample, approximately 100 cells from different fields were examined. All images, representing a middle focal plane of cells, were captured with a Cooke Sensican camera, analyzed with Slidebook3 software (Intelligent Imaging Innovation, Inc.), and presented by use of Adobe Photoshop 5.0.
RESULTS
The γ134.5 protein encoded by HSV-1(F) is localized in the cytoplasm and a distinct region in the nucleus.
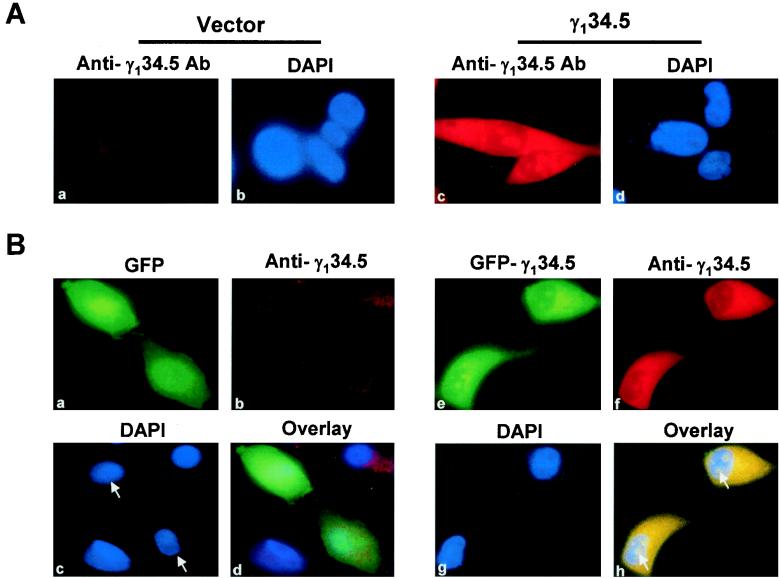
In order to examine the distribution pattern of the γ134.5 protein in intact cells, a DNA fragment encoding the full-length γ134.5 protein of HSV-1(F) was cloned into the mammalian expression vector pTet-Splice. This plasmid construct or a vector alone was transfected into HeLa cells. Thirty-six hours after transfection, the cells were probed with the anti-γ134.5 antibody and visualized by indirect immunostaining by using a Rhodamine-conjugated secondary antibody. As shown in Fig. 1A, a very weak fluorescent signal (panel a), which may have resulted from background staining, was observed in cells transfected with a vector. In cells transfected with plasmid expressing the γ134.5 protein, an intense fluorescent signal was seen both in the cytoplasm and nucleus (Fig. 1A, panel c). It is obvious that besides being concentrated in the cytoplasm, the γ134.5 protein was localized to distinct regions of the nucleus, most likely the nucleoli (Fig. 1A, panels c and d), which are recognized by their exclusion from counterstaining by DAPI, a dye that specifically reacts with double-stranded DNA. Moreover, the γ134.5 protein was also present in a much lower level in the region between the cytoplasm and discrete areas in the nucleus.
FIG. 1.
(A) Cellular localization of the γ134.5 protein. HeLa cells grown on glass coverslips were transfected with either vector (panels a and b) or pGF9912 expressing the γ134.5 protein of HSV-1(F) (panels c and d). Thirty-six hours after transfection, cells were fixed with 4% paraformaldehyde and the samples were processed as described in Materials and Methods. Cells were visualized by indirect immunostaining with anti-γ134.5 serum, followed by incubation with Rhodamine-conjugated goat anti-rabbit secondary antibody (Santa Cruz) (panels a and c). DNA in the nucleus was stained with DAPI (1.5 μg/ml) (panels b and d). (B) Localization of GFP-γ134.5 fusion protein. HeLa cells were transfected with a GFP vector (panels a to d) or GFP-γ134.5 construct (panels e to h), and samples were processed as described in Materials and Methods. Cells were visualized by direct-fluorescence microscopy analysis (panels a and e) or indirect immunostaining with anti-γ134.5 serum, followed by incubation with Rhodamine-conjugated goat anti-rabbit secondary antibody (panels b and f). DNA in the nucleus was stained with DAPI (1.5 μg/ml) (panels c and g). Overlaid images indicate the superimposed signals of GFP and anti-γ134.5 antibody, which are shown in yellow (panels d and h). Arrows denote the corresponding cells transfected with vector or GFP-γ134.5 construct.
To facilitate the assay, the full-length γ134.5 protein was fused to the GFP in vector pEYFP. When expressed in HeLa cells, this fusion protein, like the wild-type γ134.5 protein, is able to mediate dephosphorylation of eIF-2α (data not shown). After transfection of HeLa cells with a vector or plasmid expressing the GFP-γ134.5 fusion protein, the intracellular localization of these proteins was examined by fluorescence confocal microscopy. The results shown in Fig. 1B indicate that the GFP control diffused throughout the cells (panels a and c). Although the GFP-γ134.5 fusion protein was also seen in both the cytoplasm and nucleus, its distribution within the nucleus was quite different, with the fusion protein mainly targeted to the subnuclear structures (Fig. 1B, panels e and g). Among the 100 cells from different fields that were examined, over 90% displayed this localization pattern. To confirm that the observed green fluorescence represents the intracellular localization of the γ134.5 protein, the same cells were probed with the anti-γ134.5 antibody and visualized by indirect immunostaining using a Rhodamine-conjugated secondary antibody. As shown in Fig. 1B, panels b and f, the anti-γ134.5 antibody reacted strongly with the GFP-γ134.5 fusion protein, with the γ134.5 protein distributed similarly in the cytoplasm and distinct regions of the nucleus. The faint fluorescence in the GFP-transfected cells is likely due to the background staining. The overlaid images (Fig. 1B, panels d and h) clearly indicate that the bulk of the protein is present in the cytoplasm and the distinct regions of the nucleus. These results indicate that localization of the GFP-γ134.5 fusion protein does not differ from that of the wild-type γ134.5 protein. When Vero cells or human neuroblastoma SK-N-SH cell lines were transfected with the GFP-γ134.5 construct, a similar distribution pattern was observed (data not shown).
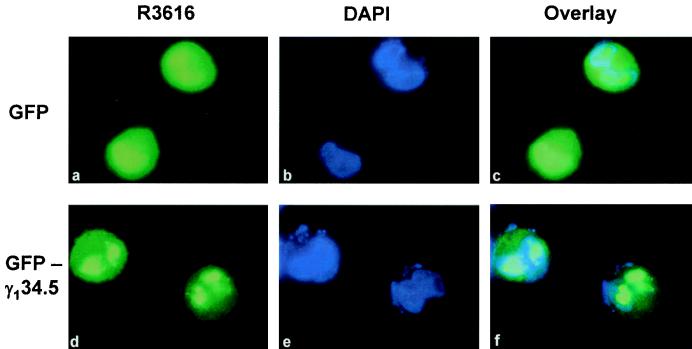
To examine whether virus infection affects the localization of the γ134.5 protein, HeLa cells were transfected with either the GFP-γ134.5 construct or vector plasmid. Sixteen hours after transfection, cells were infected with the HSV-1 mutant R3616 (5 PFU/cell), from which the γ134.5 gene is deleted (10). At 9 h postinfection, cells were examined for the cellular localization of the γ134.5 protein by fluorescence microscopy. The confocal microscopy images shown in Fig. 2 indicate that while GFP alone spread throughout cells (panels a, b, and c), the GFP-γ134.5 fusion protein was localized primarily to the cytoplasm and the discrete areas of the nucleus (panels d, e, and f). The distorted nucleus structures in the round cells were the result of virus infection. Essentially, the localization of the GFP-γ134.5 fusion protein is similar to that seen in cells transfected but not infected with virus. We conclude from these collective experiments that the γ134.5 protein of HSV-1(F) is a cytoplasmic and nuclear protein.
FIG. 2.
Cellular localization of GFP-γ134.5 protein in HSV-1-infected cells. HeLa cells were transfected with a GFP vector (panels a to c) or GFP-γ134.5 construct (panels d to f) as described in Materials and Methods. Sixteen hours after transfection, cells were infected with recombinant virus R3616, from which the γ134.5 gene has been deleted (5 PFU/cell). Nine hours after infection, cells were fixed with 4% paraformaldehyde and processed for direct-fluorescence analysis. DNA in the nucleus was stained with DAPI (1.5 μg/ml) (panels b and e). Overlaid images indicate the superimposed signals of GFP and DAPI staining (panels c and f).
Deletions in the γ134.5 protein affect intracellular localization.
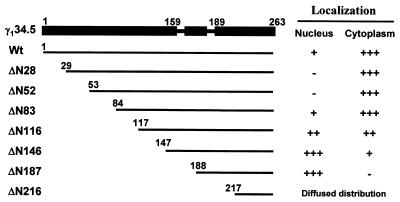
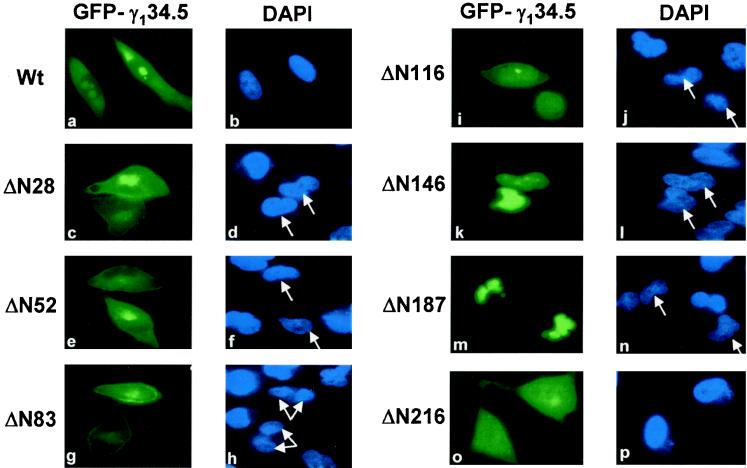
The γ134.5 protein of HSV-1(F) has 159 amino acids in the amino-terminal domain, 30 amino acids in the central domain arranged as triplet repeats (ATP), and 74 amino acids in the carboxyl-terminal domain (Fig. 3). Given that it is distributed in the cytoplasm and nucleus, we examined whether deletions in the γ134.5 protein had any effect on its intracellular distribution. For this purpose, we constructed a series of γ134.5 deletion mutants that were fused to the carboxyl terminus of GFP (Fig. 3). These mutants, with deletions spanning amino acids 1 to 216, include ΔN28 (a deletion of amino acids 1 to 28), ΔN52 (a deletion of amino acids 1 to 52), ΔN83 (a deletion of amino acids 1 to 83), ΔN116 (a deletion of amino acids 1 to 116), ΔN146 (a deletion of amino acids 1 to 146), ΔN187 (a deletion of amino acids 1 to 187), and ΔN216 (a deletion of amino acids 1 to 216). When transfected into HeLa cells, these mutants were expressed, as detected by Western blot analysis, with anti-GFP and anti-γ134.5 antibodies (data not shown). HeLa cells transfected with these mutants were then examined for intracellular distribution by confocal microscopy. The fluorescence confocal microscopy images shown in Fig. 4, panels a to p, represent a middle focal plane of cells transfected with the various GFP-γ134.5 constructs and stained with DAPI. Unlike the full-length GFP-γ134.5 fusion protein, ΔN28 and ΔN52 were primarily found in the cytoplasm, and no obvious nuclear accumulation was observed in over 90% of transfected cells. This suggests that the region containing amino acids 1 to 28 is required for the γ134.5 protein to be localized in the distinct region within the nucleus.
FIG. 3.
Schematic diagrams of the full-length γ134.5 protein of HSV-1(F) and deletion mutants. The designations for the wild-type (Wt) γ134.5 protein and each mutant are indicated on the left. The filled bar at the top represents the domain structure, and the numbers indicate amino acid positions. The amino-terminal domain contains amino acids 1 to 159, the ATP repeat region contains amino acids 160 to 188, and the carboxyl-terminal domain contains amino acids 189 to 263. Thin lines indicate the coding regions retained in the wild-type γ134.5 protein or deletion mutants. The number at the left of each line denotes the starting amino acid in each construct. All constructs were fused in frame to the carboxyl terminus of GFP. A summary of the cellular localization of wild-type γ134.5 or mutants is presented on the right. −, negative signal; +, positive signal, with each additional + indicating a stronger signal.
FIG. 4.
Fluorescence microscopy images of the full-length γ134.5 protein and deletion mutants. HeLa cells, transfected with plasmid constructs as described in the legend to Fig. 3, were fixed with 4% paraformaldehyde at 36 h after transfection and visualized by direct-fluorescence microscopy as described in the legend to Fig. 1. The intracellular distribution of each mutant is shown along with DAPI staining for nuclear DNA. Arrows denote the corresponding cells transfected with various GFP-γ134.5 constructs. Wt, wild type.
Further analysis indicated that mutants ΔN83 and ΔN116 exhibited different patterns (Fig. 4, panels g to j). These mutants were found in both the cytoplasm and nucleus. The mutant ΔN83 appeared to be localized to regions resembling the plasma membrane. Moreover, this mutant was distributed either in the cytoplasm or throughout the cells. The frequency of cytoplasmic localization was approximately 80% in cells expressing the ΔN83 mutant. While present throughout the cells, the mutant ΔN116 displayed a more nuclear localization pattern. Intriguingly, the next-shorter truncation mutant, ΔN146, was almost completely restricted to the nucleus, displaying a uniform distribution pattern (Fig. 4, panels k and l). These results may be interpreted as indicating that the region containing amino acids 83 to 146 is likely involved in either nuclear export or cytoplasmic anchoring. As shown in Fig. 4, panels m and n, mutant ΔN187 was also present only in the nucleus. Thus, the lack of the amino-terminal domain and the central domain containing the triplet repeats had no effect on the nuclear localization. However, further deletion of amino acids to amino acid 216, as represented by ΔN216, resulted in a diffused distribution pattern throughout the cells (Fig. 4, panels o and p). Notably, an additional deletion of amino acids 188 to 216 from the mutant ΔN187 completely abolished its exclusive nuclear distribution. The diffused distribution pattern is similar to that seen for GFP alone (Fig. 1B, panels a and d), which is likely due to passive diffusion. These results indicate that the region spanning amino acids 187 to 263 is required for nuclear localization. Taken together, these data strongly suggest that both the amino-terminal and the carboxyl-terminal domains have cis elements that determine the intracellular localization of the γ134.5 protein.
A stretch of basic amino acids from amino acids 1 to 16 in the γ134.5 protein directs nucleolar localization.
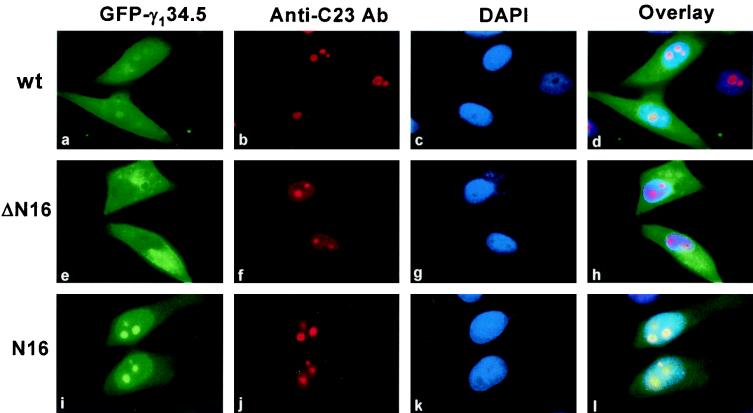
The results of the deletion analysis shown in Fig. 4, panels a to f, indicate that the first 28 amino acids are involved in the translocation of the γ134.5 protein into discrete regions within the nucleus. This region, from amino acids 1 to 16, consists of a cluster of arginine residues (MARRRRHRGPRRPRPP). This cluster is rich in basic amino acids that are reminiscent of nucleolar localization signals found in the Rev protein of human immunodeficiency virus (HIV) (30), the Rex protein of human T-cell leukemia virus type 1 (HTLV-1) (38), the MEQ protein of Marek's disease virus (27), and the NL/I14L protein of African swine fever virus (18). To precisely define the function of this motif, two mutants were constructed. In the mutant ΔN16, the γ134.5 protein with a deletion of amino acids 1 to 16 was fused to the carboxyl terminus of GFP. In the mutant N16, only amino acids 1 to 16 from the γ134.5 protein were fused to the amino terminus of GFP. When examined for cellular localization, the two mutants displayed different staining patterns. As shown in Fig. 5, panels a to l, a deletion of amino acids 1 to 16 from the γ134.5 protein resulted in a cytoplasmic localization. Essentially, the mutant ΔN16 was excluded from the nucleus. In sharp contrast, the mutant N16 was accumulated in the discrete regions within the nucleus resembling the nucleoli or nucleolus-organizing regions. To verify that these distinct regions or structures within the nucleus represent the nucleoli, the same cells were stained for the major nucleolar protein C23 or nucleolin with the anti-C23 antibody (26). The punctate structures within the nucleus, as shown in Fig. 5, panels b, f, and j, were recognized specifically by anti-C23 antibody. Remarkably, localization of the full-length GFP-γ134.5 protein and the mutant N16 coincided well with these intensely stained spots within the nucleus (Fig. 5, panels a, b, i, and j). This is clearly indicated in the superimposed images (Fig. 5, panels d and l). These data demonstrate that the motif containing amino acids 1 to 16 functions independently as a nucleolar localization signal.
FIG. 5.
Characterization of nucleolar localization signal in the γ134.5 protein. HeLa cells transiently expressing the full-length GFP-γ134.5 fusion protein (wt), mutant ΔN16, or mutant N16 were fixed with 4% paraformaldehyde and visualized by direct-fluorescence microscopy. Mutant ΔN16 has a deletion of amino acids 1 to 16 (MARRRRHRGPRRPRPP) from the γ134.5 protein which is fused to the carboxyl terminus of GFP. Mutant N16 expresses only amino acids 1 to 16 from the γ134.5 protein which is fused to the amino terminus of GFP. The C23 protein in the nucleolus was detected by indirect-immunofluorescence staining (panels b, f, and j). Cells were incubated with mouse anti-C23 monoclonal antibody, followed by reaction with Rhodamine-conjugated goat anti-mouse immunoglobulin G. DNA in the nucleus was stained with DAPI (panels c, g, and k). Overlaid images represent the superimposed signals of GFP, anti-C23 antibody, and DAPI (panels d, h, and l).
The region from amino acids 208 to 236 in the carboxyl terminus of the γ134.5 protein is able to mediate nuclear localization.
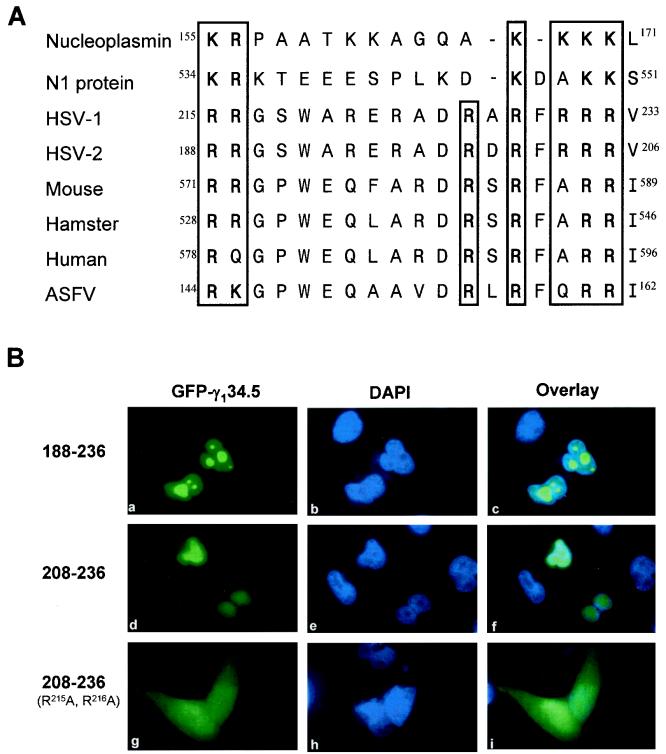
Figure 4, panels m to p, shows that the mutant ΔN187, which expresses amino acids 188 to 263, was capable of localizing to the nucleus, whereas the mutant ΔN216, which expresses amino acids 217 to 263, diffused throughout the cells. One interpretation of this phenotype is that amino acids 188 to 217 in the carboxyl terminus of the γ134.5 protein are crucial for nuclear localization. To examine the role of this domain, two additional deletion mutants were constructed. In the mutant 188-236, the region from amino acids 188 to 236 in the γ134.5 protein was fused to GFP. Similarly, in the mutant 208-236, the region from amino acids 208 to 236 was fused to GFP. Fluorescence confocal microscopy analysis showed that both mutants were predominantly localized to the nucleus in over 90% of transfected cells (Fig. 6B, panels a to f). Interestingly, the mutant 188-236 also concentrated to the regions resembling nucleoli (Fig. 6B, panels a to c). These results demonstrate that the region containing amino acids 208 to 236 is sufficient to direct nuclear localization. Inspection of the amino acid sequence of this region revealed two clusters of basic amino acids separated by 9 to 11 spacer amino acids (Fig. 6A), indicating that it bears the signature motif of bipartite nuclear localization signals identified in other proteins, such as nucleoplasmin and N1 from Xenopus laevis (15).
FIG. 6.
Analysis of the bipartite nuclear localization signal in the γ134.5 protein. (A) Amino acid sequence alignment of bipartite nuclear localization signals of nucleoplasmin and N1 from Xenopus laevis (15); the γ134.5 protein from HSV-1 and HSV-2 (13, 32); GADD34 from human (23), mouse (28), and hamster (46); and NL/I14L from African swine fever virus (47). (B) Immunofluorescence confocal microscopy images of the γ134.5 deletion mutants. Mutant 188-236 contains amino acids 188 to 236 of the γ134.5 protein fused to GFP. Mutant 208-236 contains amino acids 208 to 236 of the γ134.5 protein fused to GFP. Mutant 208-236sb was derived from mutant 208-236 by R215A and R216A substitutions. These mutants were transfected into HeLa cells, and samples were processed as described in Materials and Methods. Cells were visualized by direct-fluorescence microscopy (panels a, d, and g). DAPI was used to stain DNA in the nucleus (panels b, e, and h). The superimposed signals of GFP and DAPI are indicated by overlaid images (panels c, f, and i).
To further analyze the bipartite motif in the γ134.5 protein, site-specific mutations were introduced into the mutant 208-236. In the 18-amino-acid sequences of the γ134.5 protein shown in Fig. 6A, two conserved basic amino acids (R215 and R216) in the first cluster were replaced by alanine. The images shown in Fig. 6B, panels g to i, indicate that the mutant 208-236sb, with the R215A and R216A substitutions, diffused throughout the cells. Compared to mutant 208-236, there is a significant increase in the cytoplasmic accumulation and a concomitant decrease in nuclear localization for mutant 208-236sb (Fig. 6B, panels f and i). This result indicates that the two conserved arginines are required for efficient nuclear localization, which is consistent with the feature of bipartite nuclear localization signals (15). Combined with the diffused distribution of the mutant ΔN216 (Fig. 4, panels o and p), in which the amino-terminal deletion extended to both R215 and R216 in the first cluster, these data strongly suggest that the bipartite motif in the γ134.5 protein is critical for nuclear localization. It is interesting that this motif is present not only in the γ134.5 protein from HSV-1 and HSV-2 but also in the NL/I14L protein from African swine fever virus and GADD34 from mouse, hamster, and human (Fig. 6A).
Leptomycin B inhibits the cytoplasmic accumulation of the γ134.5 protein.
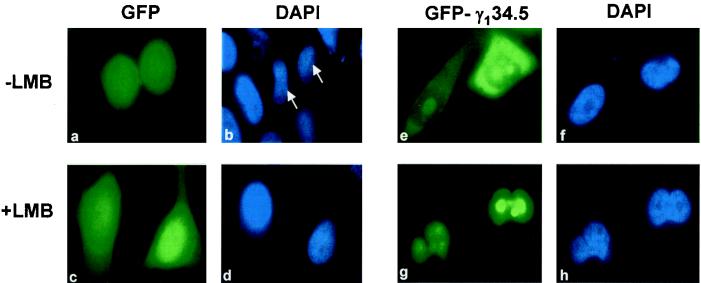
Since the γ134.5 protein is found in the cytoplasm, we examined whether nuclear export or cytoplasmic anchoring is required for its cellular localization. Specifically, HeLa cells were transfected with the GFP or GFP-γ134.5 construct. Thirty-six hours after transfection, cells were left untreated or treated with leptomycin B, a specific inhibitor of CRM1-mediated active nuclear export (17, 45). Subcellular localization of GFP or GFP-γ134.5 was determined by fluorescence microscopy. As shown in Fig. 7, panels a to h, prior to treatment with leptomycin B, the GFP control alone diffused throughout the cells. GFP-γ134.5 predominantly localized in the cytoplasm and in the nucleolus. After leptomycin B treatment was carried out as reported previously (17), the GFP control still showed a diffused staining pattern whereas all GFP-γ134.5 became localized in the nucleus and nucleolus. The strong staining in the nucleolus is attributed to the nucleolar signal present in the amino terminus of the γ134.5 protein. These results suggest that active nuclear export via the CRM1 pathway contributes to the cytoplasmic localization of the γ134.5 protein.
FIG. 7.
Effect of leptomycin B (LMB) on cellular localization of the γ134.5 protein. HeLa cells transfected with a GFP vector (panels a to d) or GFP-γ134.5 construct (panels e to h) were either left untreated or treated with leptomycin B (5 nM) for 3 h, fixed with 4% paraformaldehyde, and visualized by direct-fluorescence microscopy. DAPI was used to stain DNA in the nucleus (panels b, d, f, and h). Arrows denote the corresponding cells transfected with GFP vector.
L134A and L136A substitutions in the leucine-rich motif result in nuclear accumulation of the γ134.5 protein.
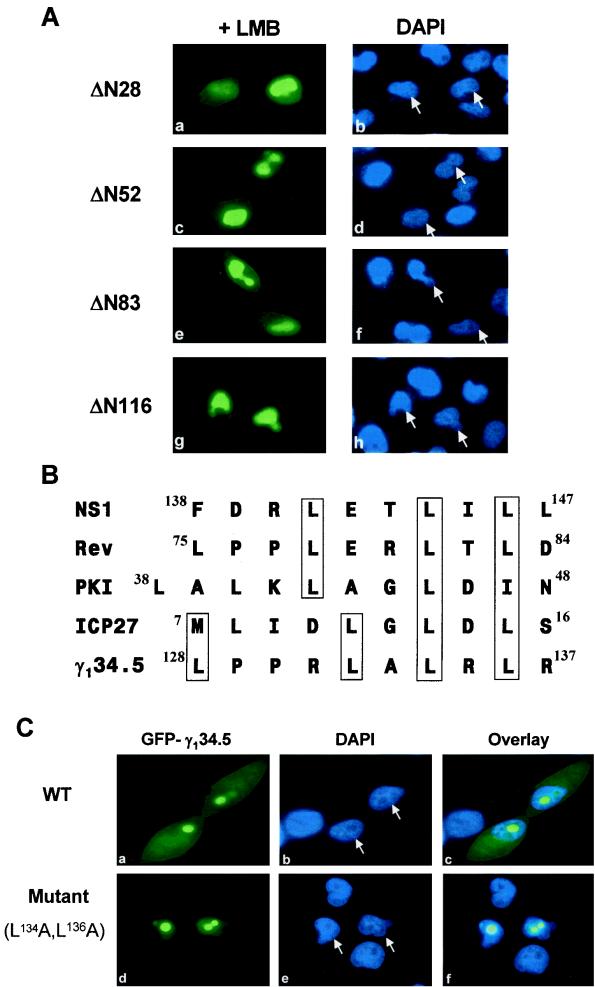
To map the cis element required for nuclear export, HeLa cells were transfected with the deletion mutants ΔN28, ΔN52, ΔN83, and ΔN116, and the effect of leptomycin B on intracellular distribution of these mutants was examined. As shown in Fig. 8A, all of these mutants were primarily localized in the nucleus, which is different from the phenotype seen in the cells not treated with leptomycin B (Fig. 4). The data indicate that all of these mutants are still capable of moving in and out of the nucleus. Comparison of the cellular distribution of all of these mutants with that of ΔN146 (Fig. 4, panels k and l), which is restricted to the nucleus, suggests that the sequence consisting of amino acids 116 to 146 is required for nuclear export.
FIG. 8.
Characterization of the nuclear export signal of the γ134.5 protein. (A) HeLa cells transfected with the indicated GFP-γ134.5 deletion mutants were treated with 5 nM leptomycin B (LMB) for 3 h, fixed with 4% paraformaldehyde, and visualized by direct-fluorescence microscopy (panels a, c, e, and g). DAPI was used to stain DNA in the nucleus (panels b, d, f, and h). (B) Alignment of the nuclear export signals of protein kinase A inhibitor (PKI) (43), NS1 from influenza virus (25), REV from HIV (42), ICP27 (39) and γ134.5 (this work) from HSV-1. (C) Cellular localization of the L134A L136A mutant. HeLa cells expressing wild-type (WT) γ134.5 (panels a, b, and c) or the mutant, in which both L134 and L136 have been replaced by A (panels d, e, and f), were analyzed by direct-fluorescence microscopy. The superimposed signals of GFP and DAPI are indicated by overlaid images (panels c and f). Arrows denote the corresponding cells transfected with the GFP-γ134.5 constructs.
Based on the results described above, we analyzed the amino acid sequence between amino acids 116 to 146 and identified a leucine-rich motif containing LPPRLALRLR from amino acids 128 to 137. This cluster bears a signature sequence of nuclear export signals present in protein kinase A inhibitor (43), REV from HIV (42), NS1 from influenza A virus (25), and ICP27 from HSV-1 (39) (Fig. 8B). To further demonstrate the function of this leucine-rich motif in nuclear export, we introduced the L134A and L136A substitutions in the full-length γ134.5 protein. This mutant was examined for intracellular localization in transfected HeLa cells. As shown in Fig. 8C, while the wild-type γ134.5 protein was found in the nucleus, nucleolus, and cytoplasm, the mutant was exclusively present in the nucleus and nucleolus (panels a to f). This localization pattern was observed in 90% of the cells examined. These results strongly suggest that the leucine-rich motif is required for the cytoplasmic localization of the γ134.5 protein through active nuclear export.
DISCUSSION
We have demonstrated that the γ134.5 protein of HSV-1(F) is present in the nucleus, nucleolus, and cytoplasm in transfected or superinfected cells. These results are consistent with previous findings that the γ134.5 protein of HSV-1 accumulates in both the nucleus and cytoplasm late in infection (1). Nuclear and cytoplasmic localization of the γ134.5 protein was also demonstrated by a recent study of different HSV-1 isolates (31). Furthermore, the data presented here indicate that the amino-terminal domain of the γ134.5 protein contains a nucleolar localization signal and nuclear export signals, whereas the carboxyl-terminal domain contains a nuclear localization signal. An important observation emerging from these studies is that the γ134.5 protein of HSV-1(F) continuously shuttles between the nucleus, nucleolus, and cytoplasm.
Our studies indicate that the region containing amino acids 1 to 16 in the γ134.5 protein is a nucleolar localization signal. This region is rich in arginine residues and resembles the nucleolar localization signals found in other viral proteins such as Rev of HIV type 1 (30), Rex of HTLV-1 (38), MEQ of Marek's disease virus (27), and NL/I14L of African swine fever virus (18). A common feature of the nucleolar localization signals of these virus proteins is an array of a various number of basic amino acids (R/K), which are required to direct nucleolar localization. As shown in Fig. 5, deletion of this region led to the complete cytoplasmic localization of the γ134.5 protein. Further, addition of this sequence to GFP is sufficient to mediate nucleolar localization. It was recently reported that the Arg-rich motif of the γ134.5 protein derived from different HSV-1 strains directs proteins to both the cytoplasm and discrete regions of the nucleus (31). However, as shown in Fig. 5, when amino acids 1 to 16 were fused to GFP, the protein was highly concentrated in the nucleolus and very little of it was seen in the cytoplasm. These different phenotypes could result from differences in the strains or in the numbers of amino acids in the γ134.5 protein fused to GFP. Nevertheless, the data presented in this study strongly suggest that amino acids 1 to 16 of the γ134.5 protein of HSV-1(F) primarily target the protein to the nucleolus.
Deletion mutagenesis suggests that a bipartite nuclear localization signal is located in the carboxyl terminus of the γ134.5 protein (Fig. 6A and B). While both the mutant 188-236 and the mutant 208-236 are capable of directing nuclear accumulation of GFP, the mutant 188-236 tends to concentrate in areas resembling the nucleolus. It is likely that the presence of a region containing amino acids 188 to 208 may affect the activity of the bipartite motif. Importantly, these mutants contain a sequence with two clusters of basic amino acids that is similar to the pattern of bipartite nuclear localization signals defined in nucleoplasmin and N1 from Xenopus laevis (15). These signals typically consist of two clusters of basic amino acids separated by a short spacer of 10 to 12 amino acids. Notably, substitution of alanine for arginine215 and arginine216 in the first cluster of the bipartite motif significantly increased the cytoplasmic localization (Fig. 6B, panels g to i). In agreement with this observation, ΔN216, in which the amino-terminal deletion extended into arginine215 and arginine216, lost this exclusive nuclear localization pattern (Fig. 4, panels m to p). These results suggest that the first two arginines in the bipartite motif are critical for nuclear import. The nuclear localization sequence in the γ134.5 protein of HSV-1 seems to fit the bipartite pattern with 9 to 11 spacer amino acids. This sequence is also conserved in the γ134.5 protein from HSV-2, the NL/I14L protein from African swine fever virus, and GADD34 from human, hamster, and mouse (Fig. 6A). It is likely that this element may play a role that is common to these proteins.
Although harboring a bipartite basic motif, the full-length γ134.5 protein is predominantly found in the nucleolus and the cytoplasm (Fig. 1A and B). One possibility is that the bipartite motif normally functions to direct nuclear import, but it works less efficiently than the nucleolar localization signal or nuclear export signal in the γ134.5 protein. The net outcome is the predominantly nucleolar and cytoplasmic distribution. The notion that the bipartite basic motif of the γ134.5 protein is a functional element is supported by the fact that the mutant ΔN28, which normally localizes to the cytoplasm, accumulates in the nucleus after leptomycin B treatment (Fig. 4, panel c, and 8A, panel a). Since the nucleolar localization signal from amino acids l to 16 was deleted in ΔN28, this phenotype was due to a block in nuclear export and continued nuclear import by the bipartite motif. Another possibility is that the bipartite nuclear localization signal is an element that functions only under certain conditions such as posttranslational modifications or protein-protein interactions. Interpreted within the framework of this model, it is notable that the γ134.5 protein is found in a high-molecular-weight complex containing PP1 in virus-infected cells (8, 21). Interestingly, the γ134.5 protein encoded by the KOS strain of HSV-1 colocalizes with PP1 in the nucleus (31). In addition, replacement of the carboxyl terminus of the γ134.5 protein with the corresponding region from MyD116/GADD34 leads to an increased nuclear accumulation of the γ134.5 protein (20).
The amino-terminal domain of the γ134.5 protein has a nuclear export signal. This signal sequence contains a leucine-rich cluster found in the nuclear export signals of many proteins such as the NS1 protein of influenza A virus (25), the Rev protein of HIV (42), protein kinase A inhibitor (43), and p53 (40). Two lines of evidence support this conclusion. First, leptomycin B, an inhibitor of the nuclear export receptor CMR1 (17), blocked the cytoplasmic accumulation of the γ134.5 protein (Fig. 7). Second, as shown in Fig. 8C, amino acid substitutions (L134A and L136A) in the leucine-rich cluster completely abolished the cytoplasmic localization of the γ134.5 protein. This result indicates that the leucine-rich-cluster-mediated nuclear export is critical for maintaining the cytoplasmic pool of the γ134.5 protein. Because the leucine-rich cluster is present in the γ134.5 protein of HSV-1 and HSV-2 but not in the NL/I14L protein from African swine fever virus or GADD34/MyD116 (Fig. 8B), we speculate that this nuclear export signal might be related to the functions that are unique to the γ134.5 protein.
Our analysis is consistent with the model that the γ134.5 protein shuttles continuously between different cellular compartments. Both the amino-terminal and carboxyl-terminal domains are involved in this process. Presently, it is not clear whether the cellular localization of the γ134.5 protein derived from HSV-1(F) is a regulated process. However, it should be noted that the amino-terminal and the carboxyl-terminal domains of the γ134.5 protein are relatively constant, but the number of triplet repeats (ATP) in the central region varies among different HSV-1 strains (3, 13, 16). The results of recent experiments have indicated that variations in the number of triplet repeats in the γ134.5 protein affect its intracellular distribution (31). The γ134.5 protein with a long ATP repeat tends to be present in the cytoplasm (31). In light of these observations, it is conceivable that the triplet repeat may indirectly affect the cellular localization of the γ134.5 protein by masking or unmasking the nuclear import or export signals within the γ134.5 protein.
Finally, the question arises as to why the γ134.5 protein shuttles dynamically between the nucleus and the cytoplasm. A number of studies have indicated that the γ134.5 protein of HSV is critical for promoting virulence in experimental animal models (2, 10, 24, 37, 41). More than one function associated with this viral factor has been suggested to contribute to the observed phenotype in vivo (4-6, 10, 20, 35). In cell culture, this viral protein is required to prevent the shutoff of protein synthesis mediated by the double-stranded-RNA-dependent protein kinase PKR (9, 11). In doing so, the γ134.5 protein binds to and redirects PP1 to dephosphorylate the α subunit of eIF-2α (8, 21, 22). Consistent with previous findings, a recent study demonstrated that the γ134.5 protein and PP1 colocalize in the cytosol (31). Therefore, the cytoplasmic γ134.5 protein is more likely involved in mediating eIF-2α dephosphorylation and therefore blocks the cellular antiviral response mediated by interferon. This activity does not exclude the additional functions it performs in the cytoplasm. The γ134.5 protein is also implicated in viral egress and glycoprotein processing (3, 5). An additional function of the γ134.5 protein is suggested by the observation that it associates with PCNA involved in DNA replication and cell cycle regulation (6). Thus, in principle, the nuclear γ134.5 protein may be required to perform these last two functions. These processes require the γ134.5 protein to be rapidly shuttled between the different cellular sites. It remains an open question whether the nuclear and cytoplasmic shuttling of the γ134.5 protein has as-yet-unidentified functions. Further experiments are needed to explore these possibilities.
Acknowledgments
We thank David McDonald for helpful discussion and suggestions and Melissa Cerveny for critical reading of the manuscript.
This work was supported by grant AI 46665 (B.H.) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Ackermann, M., J. Chou, M. Sarmiento, R. A. Lerner, and B. Roizman. 1986. Identification by antibody to a synthetic peptide of a protein specified by a diploid gene located in the terminal repeats of the L component of herpes simplex virus genome. J. Virol. 58:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolovan, C. A., N. M. Sawtell, and R. L. Thompson. 1994. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J. Virol. 68:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bower, J. R., H. Mao, C. Durishin, E. Rozenbom, M. Detwiler, D. Rempinski, T. L. Karban, and K. S. Rosenthal. 1999. Intrastrain variants of herpes simplex virus type 1 isolated from a neonate with fatal disseminated infection differ in the ICP34.5 gene, glycoprotein processing, and neuroinvasiveness. J. Virol. 73:3843-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, S. M., J. Harland, A. R. MacLean, J. Podlech, and J. B. Clements. 1994. Cell type and cell state determine differential in vitro growth of non-neurovirulent ICP34.5-negative herpes simplex virus types 1 and 2. J. Gen. Virol. 75:2367-2377. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. M., A. R. MacLean, J. D. Aitken, and J. Harland. 1994. ICP34.5 influences herpes simplex virus type 1 maturation and egress from infected cells in vitro. J. Gen. Virol. 75:3679-3686. [DOI] [PubMed] [Google Scholar]
- 6.Brown, S. M., A. R. MacLean, E. A. McKie, and J. Harland. 1997. The herpes simplex virus virulence factor ICP34.5 and the cellular protein MyD116 complex with proliferating cell nuclear antigen through the 63-amino-acid domain conserved in ICP34.5, MyD116, and GADD34. J. Virol. 71:9442-9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, G., M. E. Brett, and B. He. 2001. Val193 and Phe195 of the γ134.5 protein of herpes simplex virus 1 are required for viral resistance to interferon-α/β. Virology 290:115-120. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, G., M. Gross, M. E. Brett, and B. He. 2001. AlaArg motif in the carboxyl terminus of the γ134.5 protein of herpes simplex virus type 1 is required for the formation of a high-molecular-weight complex that dephosphorylates eIF-2α. J. Virol. 75:3666-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 11.Chou, J., and B. Roizman. 1992. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, J., and B. Roizman. 1994. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc. Natl. Acad. Sci. USA 91:5247-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou, J., and B. Roizman. 1990. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J. Virol. 64:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou, J., and B. Roizman. 1986. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J. Virol. 57:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 16.Dolan, A., E. McKie, A. R. MacLean, and D. J. McGeoch. 1992. Status of the ICP34.5 gene in herpes simplex virus type 1 strain 17. J. Gen. Virol. 73:971-973. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 18.Goatley, L. C., M. B. Marron, S. C. Jacobs, J. M. Hammond, J. E. Miskin, C. C. Abrams, G. L. Smith, and L. K. Dixon. 1999. Nuclear and nucleolar localization of an African swine fever virus protein, I14L, that is similar to the herpes simplex virus-encoded virulence factor ICP34.5. J. Gen. Virol. 80:525-535. [DOI] [PubMed] [Google Scholar]
- 19.Grishin, A. V., O. Azhipa, I. Semenov, and S. J. Corey. 2001. Interaction between growth arrest-DNA damage protein 34 and Src kinase Lyn negatively regulates genotoxic apoptosis. Proc. Natl. Acad. Sci. USA 98:10172-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, B., J. Chou, D. A. Liebermann, B. Hoffman, and B. Roizman. 1996. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the γ134.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J. Virol. 70:84-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, B., M. Gross, and B. Roizman. 1998. The γ134.5 protein of herpes simplex virus 1 has the structural and functional attributes of a protein phosphatase 1 regulatory subunit and is present in a high molecular weight complex with the enzyme in infected cells. J. Biol. Chem. 273:20737-20743. [DOI] [PubMed] [Google Scholar]
- 22.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollander, M. C., Q. Zhan, I. Bae, and A. J. Fornace, Jr. 1997. Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J. Biol. Chem. 272:13731-13737. [DOI] [PubMed] [Google Scholar]
- 24.Leib, D. A., M. A. Machalek, B. R. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y., Y. Yamakita, and R. M. Krug. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. USA 95:4864-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y. P., R. K. Busch, B. C. Valdez, and H. Busch. 1996. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur. J. Biochem. 237:153-158. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J. L., L. F. Lee, Y. Ye, Z. Qian, and H. J. Kung. 1997. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J. Virol. 71:3188-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lord, K. A., B. Hoffman-Liebermann, and D. A. Liebermann. 1990. Sequence of MyD116 cDNA: a novel myeloid differentiation primary response gene induced by IL6. Nucleic Acids Res. 18:2823.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean, A., L. Robertson, E. McKay, and S. M. Brown. 1991. The RL neurovirulence locus in herpes simplex virus type 2 strain HG52 plays no role in latency. J. Gen. Virol. 72:2305-2310. [DOI] [PubMed] [Google Scholar]
- 30.Malim, M. H., S. Bohnlein, J. Hauber, and B. R. Cullen. 1989. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell 58:205-214. [DOI] [PubMed] [Google Scholar]
- 31.Mao, H., and K. S. Rosenthal. 2002. An N-terminal arginine rich cluster and a proline-alanine-threonine repeat region determines the cellular localization of the herpes simplex virus type-1 ICP34.5 protein and its ligand, protein phosphatase 1. J. Biol. Chem. 277:11423-11431. [DOI] [PubMed] [Google Scholar]
- 32.McGeoch, D. J., C. Cunningham, G. McIntyre, and A. Dolan. 1991. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J. Gen. Virol. 72:3057-3075. [DOI] [PubMed] [Google Scholar]
- 33.McKay, E. M., B. McVey, H. S. Marsden, S. M. Brown, and A. R. MacLean. 1993. The herpes simplex virus type 1 strain 17 open reading frame RL1 encodes a polypeptide of apparent M(r) 37K equivalent to ICP34.5 of herpes simplex virus type 1 strain F. J. Gen. Virol. 74:2493-2497. [DOI] [PubMed] [Google Scholar]
- 34.McKie, E. A., R. G. Hope, S. M. Brown, and A. R. MacLean. 1994. Characterization of the herpes simplex virus type 1 strain 17+ neurovirulence gene RL1 and its expression in a bacterial system. J. Gen. Virol. 75:733-741. [DOI] [PubMed] [Google Scholar]
- 35.Mohr, I., D. Sternberg, S. Ward, D. Leib, M. Mulvey, and Y. Gluzman. 2001. A herpes simplex virus type 1 γ34.5 second-site suppressor mutant that exhibits enhanced growth in cultured glioblastoma cells is severely attenuated in animals. J. Virol. 75:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novoa, I., H. Zeng, H. P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravi, V., P. G. Kennedy, and A. R. MacLean. 1998. Functional analysis of the herpes simplex virus type 2 strain HG52 RL1 gene: the intron plays no role in virulence. J. Gen. Virol. 79:1613-1617. [DOI] [PubMed] [Google Scholar]
- 38.Siomi, H., H. Shida, S. H. Nam, T. Nosaka, M. Maki, and M. Hatanaka. 1988. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell 55:197-209. [DOI] [PubMed] [Google Scholar]
- 39.Soliman, T. M., and S. J. Silverstein. 2000. Identification of an export control sequence and a requirement for the KH domains in ICP27 from herpes simplex virus type 1. J. Virol. 74:7600-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valyi-Nagy, T., M. U. Fareed, J. S. O'Keefe, R. M. Gesser, A. R. MacLean, S. M. Brown, J. G. Spivack, and N. W. Fraser. 1994. The herpes simplex virus type 1 strain 17+ γ34.5 deletion mutant 1716 is avirulent in SCID mice. J. Gen. Virol. 75:2059-2063. [DOI] [PubMed] [Google Scholar]
- 42.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 43.Wen, W., S. S. Taylor, and J. L. Meinkoth. 1995. The expression and intracellular distribution of the heat-stable protein kinase inhibitor is cell cycle regulated. J. Biol. Chem. 270:2041-2046. [DOI] [PubMed] [Google Scholar]
- 44.Whitley, R. J., E. R. Kern, S. Chatterjee, J. Chou, and B. Roizman. 1993. Replication, establishment of latency, and induced reactivation of herpes simplex virus γ134.5 deletion mutants in rodent models. J. Clin. Investig. 91:2837-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 46.Zhan, Q., K. A. Lord, I. Alamo, Jr., M. C. Hollander, F. Carrier, D. Ron, K. W. Kohn, B. Hoffman, D. A. Liebermann, and A. J. Fornace, Jr. 1994. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol. Cell. Biol. 14:2361-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zsak, L., Z. Lu, G. F. Kutish, J. G. Neilan, and D. L. Rock. 1996. An African swine fever virus virulence-associated gene NL-S with similarity to the herpes simplex virus ICP34.5 gene. J. Virol. 70:8865-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]