Abstract
The temperature dependence of the unimolecular kinetics for dissociation of the heme group from holo-myoglobin (Mb) and holo-hemoglobin α-chain (Hb-α) was investigated with blackbody infrared radiative dissociation (BIRD). The rate constant for dissociation of the 9 + charge state of Mb formed by electrospray ionization from a “pseudo-native” solution is 60% lower than that of Hb-α at each of the temperatures investigated. In solutions of pH 5.5–8.0, the thermal dissociation rate for Mb is also lower than that of HB-α (Hargrove, M. S. et al. J. Biol. Chem. 1994, 269, 4207–4214). Thus, Mb is thermally more stable with respect to heme loss than Hb-α both in the gas phase and in solution. The Arrhenius activation parameters for both dissociation processes are indistinguishable within the current experimental error (activation energy 0.9 eV and pre-exponential factor of 108–10 s−1). The 9+ to 12+ charge states of Mb have similar Arrhenius parameters when these ions are formed from pseudo-native solutions. In contrast, the activation energies and pre-exponential factors decrease from 0.8 to 0.3 eV and 107 to 102 s−1, respectively, for the 9 + to 12 + charge states formed from acidified solutions in which at least 50% of the secondary structure is lost. These results demonstrate that gas-phase Mb ions retain clear memory of the composition of the solution from which they are formed and that these differences can be probed by BIRD.
Noncovalent interactions play a critical role in the function of biomolecules in solution [1]. Under gentle conditions, both electrospray ionization [2–16] and matrix-assisted laser desorption [17–20] can produce intact gas-phase ions of a variety of noncovalent biomolecule complexes. Recent studies indicate that information about the relative binding affinities of noncovalent complexes in solution can be obtained from the relative abundances of the corresponding ions observed in a mass spectrum [12–15, 21]. Careful control experiments are critical [22], because other factors, such as ion source conditions and gas-phase chemistry, can influence these abundances. An important question is what structural changes occur when noncovalent complexes enter the gas phase, that is, are specific interactions retained in the absence of solvent, and if so, do their bindings strengths reflect those in solution? Recent studies, which probe the gas-phase conformation of ions by using H/D exchange [23, 24], collisional cross section [5, 25] and ion mobility [26, 27], proton transfer reactivity [28, 29], and high-energy surface-impact collisions [6], show that many proteins can have a highly compact conformation in the gas phase, although the exact nature of these conformations is not known. These results suggest the possibility that ions can retain some conformational attributes of their solution structure.
Noncovalent heme-binding proteins such as myoglobin (Mb) and hemoglobin (Hb) have been extensively studied in solution [30–42] as well as in the gas phase [3–8, 10, 11, 43]. In solution, the binding of ligands such as O2 and CO [30, 38, 39], and the affinity of the protein for the heme group, have been studied [30–35, 40–42]. The relative solution-phase binding affinities of a wide variety of heme-binding proteins have been determined by heme transfer experiments [30, 32] in which the transfer of the heme group from a holo-protein to the apo form of a different heme-binding protein is measured. These experiments are typically monitored by changes in the optical absorption upon heme binding in the proteins and can be measured as a function of solution conditions such as pH, temperature, and so forth. Binding affinities can be obtained from the temperature dependence of the rate constants for heme transfer [32].
In gas-phase studies, McLuckey and Ramsey [7] reported that holo-Mb ions could be trapped for 1 s in a quadrupole ion trap mass spectrometer. Dissociation of the 9 + charge state resulted in the production of singly charged heme and the 8 + charge state of the protein [7]. Ganem and co-workers [43] reported dissociation of the 9 + charge state of both holo-Mb and Hb α-chain by collisional activation in a triple quadrupole mass spectrometer. They observed both loss of neutral and singly charged heme and the complementary 9 + and 8 + protein products. Under identical collision conditions, the extent of dissociation was greater for the more weakly bound Hb complex. Konishi and Feng [8] showed that fewer collisions were required to dissociate heme from the 8 + ion of reduced Hb α-chain than the 9 + ion of reduced Mb.
Blackbody infrared radiative dissociation (BIRD) [44–47], in which ions trapped in a cell of a Fourier-transform mass spectrometer are activated by absorption of photons emitted from the heated walls of the vacuum chamber, has been used to dissociate large biomolecule ions [44–46]. From the temperature dependence of the unimolecular dissociation rate constants, Arrhenius activation energies and pre-exponential factors can be determined. For large ions, the rate of energy exchange with the vacuum chamber walls can greatly exceed the rate of dissociation. In this rapid energy exchange limit, ions have a Boltzmann distribution of internal energy. The rates of dissociation at a given temperature thus give direct information about the relative thermal stability of the ions, and the activation energies directly reflect dissociation threshold energies [46].
Here it is demonstrated that the relative gas-phase thermal dissociation rates of heme from the two proteins, Mb and hemoglobin α-chain (Hb-α), which have “strikingly similar” tertiary structures [48], are indicative of their relative thermal stabilities in solution. In addition, the dissociation activation parameters depend on the composition of the solution from which these ions are formed, that is, these ions retain clear memory of their solution-phase structure and these differences can be probed by BIRD.
Experimental Methods
Mass Spectrometry
Ions were generated by electrospray ionization in an external ion source Fourier-transform mass spectrometer (FTMS), which has been described previously [49]. The ions were desolvated in a heated stainless steel capillary (~ 170°C). A mechanical shutter was opened to allow ions to accumulate in the ion cell for 10 s (Vtrap = 4.0 V) during which time a pulse of N2 was introduced to the main vacuum chamber to a pressure of 2 × 10 −6 torr to enhance ion trapping and thermalization. Ions above m/z 800 were ejected by single-frequency resonant excitations and those below were ejected by a broadband frequency sweep excitation (50 Hz/μs). The trapping voltage was reduced to 1.0 V for detection.
Dissociation Rate Constants
The vacuum chamber containing the ion cell is heated with a resistive heating blanket and heating tapes [44]. The intensities of the isolated holo-protein and the apo-protein one charge state lower were measured as a function of delay time. Normalized intensities were obtained by correcting these values for the different charge state of the ions. First-order rate constants were obtained from the slope of a plot of the natural log of the normalized intensity versus the delay time.
Sample Preparation
Mb (equine skeletal muscle) and Hb (human) were obtained from Sigma Chemical Company (St. Louis, MO) and were used without further publication. Mb was electrosprayed from two different solutions: ~ 3 × 10 −5 M in water/methanol (80/20%) and water/methanol/acetic acid (50/50/0.1%), referred to as “pseudo-native” and “denaturing,” respectively. The pH of these solutions was ~ 5.5 and ~ 3.4, respectively, measured with a semi-micro combination pH electrode (Corning Instruments, Corning, NY). Hb was eletrosprayed from a water/methanol (80/20%) solution at a concentration of ~ 2 × 10 −5 M (each chain). New solutions were made prior to acquiring kinetics at each temperature. Sodium adduction, which was prevalent in the water/methanol (80/20%) solutions of both proteins when made in glass vials, was minimized by making the solution in plastic vials.
Results and Discussion
Electrospray Mass Spectrometry of Myoglobin and Hemoglobin
Figure 1 shows typical electrospray mass spectra of Mb from two different solutions referred to as denaturing and pseudo-native. A significant difference in the amount of holo-Mb is evident in these two spectra. In addition, higher charge states of the apo-Mb are observed from the denaturing solution. These spectra have significant amounts of the apo-protein indicating that heme is dissociated from the protein either in solution or in the electrospray interface. In aqueous solution, Mb is in its native conformation in the pH range 4.5–7.0. The pseudo-native solution (pH ~ 5.5) contains 20% methanol, which was added to maintain stable and reproducible ion signal over the long duration of these experiments. The presence of methanol is likely to have an effect on the protein's solution conformation. However, two pieces of evidence suggest that a significant amount of the tertiary structure is retained even in the presence of 20% methanol. First, the abundance of the holo-protein from the pseudo-native solution is significantly higher than from the denaturing solution. Second, the rates of heme loss from Mb electrosprayed from the pseudo-native solution were compared to those from a 100% water solution at two temperatures. These rates differed by ≤ 5%, whereas the rates from the pseudo-native and denaturing solutions differed by ~ 15%, averaged over all the charge states.
Figure 1.
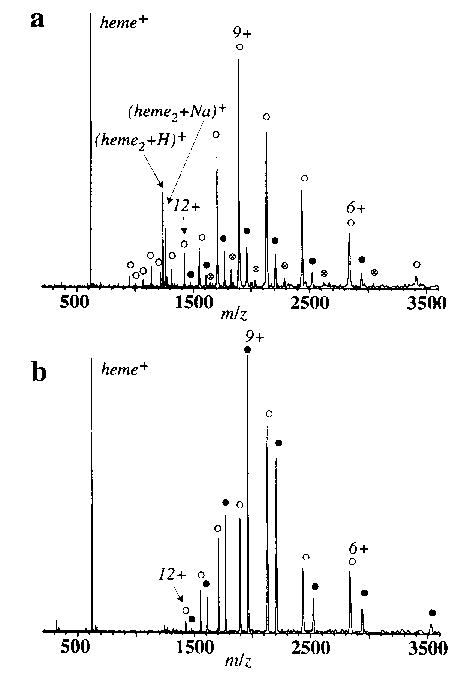
Electrospray ionization Fourier-transform mass spectra of myoglobin from (a) denaturing (50/50/0.1% water/methanol/acetic acid) and (b) pseudo-native (80/20% water/methanol) solutions, showing both holo (filled circle) and apo (open circle) myoglobin ions. A low-intensity distribution of a complex of myoglobin with two hemes (circled multi) is also observed from the denaturing solution.
Holo-Mb ions are observed from the denaturing solution if gentle source conditions are used; the intensity of the signal due to the heme attached protein is often as much as ~ 10% of that of the apo-protein (Figure 1a). The existence of holo-protein from the denaturing solution could be due to nonspecific complex formation. For example, the 2/1 heme/protein complex (Figure 1a) indicates that some nonspecific complexation does occur. Resonance Raman measurements of aqueous ferric-Mb solution in the pH range 2.6–4.0 indicate that ~ 50% of the secondary structure is lost, but the heme remains bound [37]. Under these acidic conditions, the proximal histidine becomes protonated and the bond between it and the heme iron is broken. However, the structure of the heme binding pocket is largely unchanged. Thus, some tertiary structure may remain in the denatured solution, but significantly less than that in the pseudo-native solution.
For the tetrameric protein hemoglobin electrosprayed from the pseudo-native solution, both the apo and the holo forms of each individual chain (α and β) are observed with the α-chain in significantly greater abundance than the β-chain in both cases. No dimeric or tetrameric forms of this protein were seen, consistent with the fact that these species are unstable in acidic solutions [50]. The charge state distribution extends from 11 + to 6 + for both proteins. No holo-hemoglobin was detected from more acidic solutions and no nonspecific 2/1 heme/protein complex was observed.
Dissociation of holo-Myoglobin and holo-Hemoglobin from Pseudo-Native Solutions
The BIRD spectra of both Mb and Hb-α show exclusive loss of a singly charged heme containing iron in the 3 + oxidation state. No loss of neutral heme is observed. The selective dissociation observed here is likely the result of the long time frame of these experiments and is consistent with ion trap results [7]. The isolated holo ions of Mb and Hb-α are stable for greater than 180 s at cell temperatures up to ~ 110°C. This demonstrates that the ions have insufficient energy to dissociate on the time scale of this experiment unless they are heated in the FTMS cell. An example of the kinetic data from which unimolecular dissociation rate constants are obtained is shown in Figure 2 for the 9 + ion of Mb formed from the pseudo-native solution, with the cell temperature at 119–179°C. Kinetic data were measured for times up to 200 s; experiments at longer times were precluded by our inability to store these high mass-to-charge ratio ions at a magnetic field strength of 2.7 T. From the temperature dependence of these rate constants. Arrhenius activation parameters are obtained. These data for the 9 + ion clearly show that Mb dissociates more slowly than Hb-α; the rate constant is ~ 60% lower than that of Hb-α at each of the temperatures investigated (Figure 3). This is consistent with the results of Konishi and Feng [8], who showed that the reduced form of Hb-α 8 + required fewer collisions to undergo 50% dissociation than did the reduced form of Mb 9 +. From BIRD kinetics of the 9 + ions of Mb and Hb-α, Arrhenius activation energies (Ea) of 0.9 eV and pre-exponential factors (A) of 108 ± 101.1 and 1010 ± 101.1 s−1 are obtained, respectively. Thus, these structurally similar proteins have indistinguishable gas-phase heme-dissociation parameters despite their clearly different dissociation kinetics. To determine whether the difference in the kinetics is due to differences in either the Ea or A (or both) would require improvements to the precision of these measurements. We are currently investigating modifications to the experiment that will make this possible. It should be noted that if the A factor is the same for these dissociation processes, then a ΔEa = 0.04 eV would account for the different kinetics. This illustrates that the BIRD kinetics are a very sensitive probe of small differences in the Arrhenius parameters.
Figure 2.
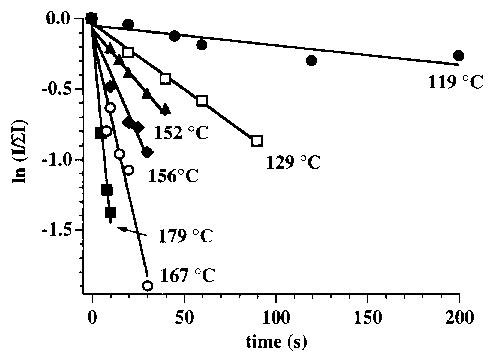
Semilog plots showing normalized holo-myoglobin abundance as a function of time for the 9 + ion electrosprayed from the pseudo-native solution. The six temperatures shown correspond to the highest, lowest, and four intermediate values at which these blackbody infrared radiative dissociation experiments were performed.
Figure 3.
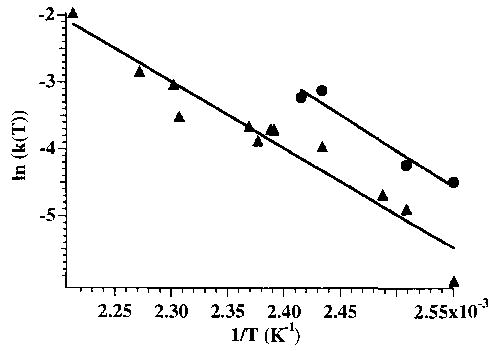
Arrhenius plots for the 9 + charge states of myoglobin (filled triangle) and hemoglobin α-chain (filled circle), from which an activation energy for loss of heme + ~ 0.9 eV is obtained for both ions.
Solution-phase dissociation rate constants for these processes have not been reported for the same form of the proteins studied here. However, some information can be obtained by comparing values for proteins that have been measured. In solution at pH 5.5–8.0, the dissociation rate constants for heme loss from sperm whale Mb are significantly lower than those for human Hb-α in the Hb tetramer; for example, at pH 5.5 (37°C) these values are 0.1 and 1.0 h−1, respectively [32]. At pH 5, the rate constant for heme loss from horse Mb is 2.5 times that from sperm whale Mb [35]. AT pH 9, isolated Hb-α loses heme 100 times faster than Hb-α in the α β subunit pair, which loses heme at a rate similar to that of the tetramer [51]. These results suggest that equine Mb is thermally more stable than human Hb-α in solution and thus the relative thermal stability of Mb and Hb-α is the same in both the gas phase and solution.
In solution at pH 5.0, Hargrove et al. [32] report that the activation energies of Mb and human Hb-α are comparable (0.4–0.6 eV; no activation energies were reported at higher pH). The activation energies in the gas phase are ~ 80% larger than those in solution. This difference likely reflects both differences in solution and gas-phase structure, as well as the shielding effect of solvent on the electrostatic interactions. In the gas phase, the charged heme group can be significantly solvated by polarizable groups in the folded protein. In the transition state, the departing charged heme group would not be significantly solvated in the gas phase but would be solvated in solution. The increased stabilization of the transition state in solution relative to that in the gas phase would result in a lower dissociation activation energy in solution.
The activation energies we measure (0.9 eV) are comparable to the value for the dissociation of ribonuclease S into S-peptide and S-protein measured by Smith and co-workers [52] by using a heated capillary electrospray interface. As noted by the authors in that study, heating in the early part of the capillary presumably influences solution behavior of the complexes in the electrospray droplet, and in the latter stages effects the gas-phase properties. In addition, the dissociation kinetics can be influenced by interface conditions and by the effects of ion desolvation. BIRD has the advantage that the kinetics do not depend on either interface conditions or ion desolvation. In addition, the ion temperature in BIRD experiments is better characterized [44–46]. Additional measurements should provide both a better indication of the range of gas-phase dissociation energies possible for large non-covalently bound complexes and an improved understanding of how these values compare to those in solution.
Effects of Solution Composition
The Arrhenius activation parameters for the loss of heme from the 9 + to 12 + charge states of myoglobin electrosprayed from the pseudo-native and denaturing solutions are given in Table 1. The Arrhenius plots for all charge states are shown in Figure 4. The Arrhenius activation parameters for all the charge states from the pseudo-native solution are similar. In contrast, the activation parameters from the denaturing solution show a clear trend with increasing charge state. As the charge state increases from 9 + to 12 +, the activation energy decreases from 0.8 to 0.3 eV, and the A-factor decreases from 107 to 102 s−1. Under the slow reaction conditions used in these experiments, all of these large ions have likely equilibrated with the blackbody radiation field inside the vacuum chamber [46]. Thus, the low A factors indicate unusually “tight” (entropically unfavored) transition states [53] consistent with complicated dissociation processes involving multiple steps or multiple coordination sites. These results demonstate that these gas-phase ions retain clear memory of their solution-phase conformations and that differences in these conformations influence the gas-phase dissociation kinetics and energetics.
Table 1.
Arrhenius activation energy (Ea) and pre-exponential ( A) for heme dissociation from myoglobin electrosprayed from pseudo-native and denaturing solutionsa
| Charge state | A(s−1) (± ≤ factor of 101.1) | Ea (eV) (± < 0.1 eV) |
|---|---|---|
| Pseudo-native solution | ||
| 12+ | 108 | 0.8 |
| 11+ | 109 | 0.9 |
| 10+ | 109 | 0.9 |
| 9+ | 108 | 0.9 |
| Denaturing solution | ||
| 12+ | 102 | 0.3 |
| 11+ | 104 | 0.5 |
| 10+ | 104 | 0.6 |
| 9+ | 107 | 0.8 |
Errors reported are from the linear least square fit of the data in the Arrhenius plots.
Figure 4.
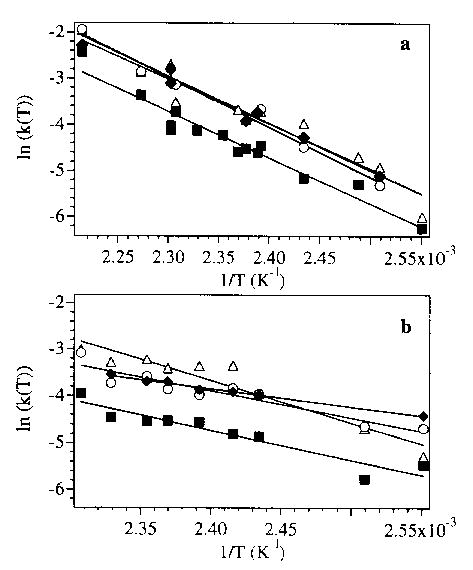
Arrhenius plots for the heme dissociation from the 9+ to 12+ charge states of myoglobin (open triangle, filled square, open circle, and filled diamond, respectively) electrosprayed from (a) pseudo-native and (b) denaturing solutions.
Also evident from these plots is that the rate of heme loss from the 10 + Mb ion from both solutions is consistently approximately half that of the other charge states over the temperature range investigated. The reason for the extra stability of this charge state is not known. Similar results were found for Na + adducted ions of both Mb and Hb-α, which had dissociation rates that decreased with increasing numbers of adducts.
Conclusions
Mb is thermally more stable than Hb-α with respect to heme loss both in the gas phase and in solution (solution pH > 5.5). In addition, the dissociation kinetics and energetics depend on the solution conformation of the ions. These results indicate that BIRD could be a useful method for obtaining information about the gas-phase stabilities and activation energies for dissociation of noncovalent complexes. The relative thermal stabilities of Mb and Hb-α in solution are a sensitive function of the pH [32]. We are investigating methods to improve the electrospray signal from pure-aqueous or buffered solutions over the long time frame required for these experiments to evaluate gas-phase stabilities of ions formed from a wide range of solution pH. Obviously, additional studies of other noncovalent complexes with known solution-phase binding energies are necessary to determine if there is a general relationship between gas-phase and solution values. To the extent that the dissociation activation energies in the gas phase parallel those in solution, BIRD could be used to evaluate relative thermal stabilities and would be particularly useful for the rapid screening of complicated mixtures of noncovalent complexes formed from combinatorial libraries [54, 55].
Acknowledgments
The authors thank Professor J. S. Olson for generously providing preprints regarding heme–globin binding in solution and acknowledge generous financial support from NSF (grant no. CHE-9258178), NIH (grant no. 1R29GM50336-01A2), and Finnigan MAT through sponsorship of the 1994 ASMS Research Award (E.R.W.).
References
- 1.Creighton, T. E. Proteins, 2nd ed.; Freeman: New York, 1993.
- 2.Gao J, Cheng X, Chen R, Sigal GB, Bruce JB, Schwartz BL, Hofstadler SA, Anderson GA, Smith RD, White-sides GA. J Med Chem. 1996;39:1949–1955. doi: 10.1021/jm960013g. [DOI] [PubMed] [Google Scholar]
- 3.Loo JA. Bioconjugate Chem. 1995;6:644–665. doi: 10.1021/bc00036a600. [DOI] [PubMed] [Google Scholar]
- 4.Smith DL, Zhang ZQ. Mass Spectrom Rev. 1994;13:411–429. [Google Scholar]
- 5.Collings BA, Douglas DJ. J Am Chem Soc. 1996;118:4488–4489. [Google Scholar]
- 6.Sullivan PA, Axelsson J, Altmann S, Quist AP, Sunqvist BUR, Reimann CT. J Am Soc Mass Spectrom. 1996;7:329–341. doi: 10.1016/1044-0305(95)00702-4. [DOI] [PubMed] [Google Scholar]
- 7.McLuckey SA, Ramsey RS. J Am Soc Mass Spectrom. 1994;5:324–327. doi: 10.1016/1044-0305(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 8.Konishi Y, Feng R. Biochemistry. 1994;33:9706–9711. doi: 10.1021/bi00198a041. [DOI] [PubMed] [Google Scholar]
- 9.Li YT, Hsieh YL, Henion JD, Senko MW, McLafferty FW, Ganem B. J Am Chem Soc. 1993;115:8409–8413. [Google Scholar]
- 10.Jacquinod M, Lieze E, Potier N, Albrecht AM, Shanzer A, Van Dorsselear A. Tetrahedron Lett. 1993;34:2771–2774. [Google Scholar]
- 11.Katta V, Chait BT. J Am Chem Soc. 1991;213:8534–8535. [Google Scholar]
- 12.Gale DC, Smith RD. J Am Soc Mass Spectrom. 1995;6:1154–1164. doi: 10.1016/1044-0305(95)00530-7. [DOI] [PubMed] [Google Scholar]
- 13.Feng R, Castelhano AL, Billedeau R, Yuan ZY. J Am Soc Mass Spectrom. 1995;6:1105–1111. doi: 10.1016/1044-0305(95)00548-X. [DOI] [PubMed] [Google Scholar]
- 14.Lim HK, Hsieh YL, Ganem B, Henion JD. J Mass Spectrom. 1995;30:708–714. [Google Scholar]
- 15.Doktycz MJ, Habibigoudarzi S, McLuckey SA. Anal Chem. 1994;66:3416–3422. doi: 10.1021/ac00092a019. [DOI] [PubMed] [Google Scholar]
- 16.Ganem B, Li YT, Henion JD. J Am Chem Soc. 1991;113:6294–6296. [Google Scholar]
- 17.Tang X, Callahan JH, Zhou P, Vertes A. Anal Chem. 1995;67:4542–4548. doi: 10.1021/ac00120a018. [DOI] [PubMed] [Google Scholar]
- 18.Rosinke B, Strupat K, Hillenkamp F, Rosenbuch J, Dercher N, Kruger U, Galla HJ. J Mass Spectrom. 1995;30:1462–1468. [Google Scholar]
- 19.Woods AS, Buchsbaum JC, Worrall TA, Berg JM, Cotter RJ. Anal Chem. 1995;67:4462–4465. [Google Scholar]
- 20.Juhasz P, Biemann K. Proc Natl Acad Sci USA. 1994;91:4333–4337. doi: 10.1073/pnas.91.10.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganem B, Li YT, Henion JD. J Am Chem Soc. 1991;123:7817–7819. [Google Scholar]
- 22.Smith RD, Light-Wahl KJ. Biol Mass Spectrom. 1993;22:493–501. doi: 10.1002/bms.1200220203. [DOI] [PubMed] [Google Scholar]
- 23.Wood TD, Chorush RA, Wampler FM, Little DP, O’Connor PB, McLafferty FW. Proc Natl Acad Sci USA. 1995;92:2451–2454. doi: 10.1073/pnas.92.7.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winger BE, Light-Wahl KJ, Rockwood AL, Smith RD. J Am Chem Soc. 1992;134:5897–5898. [Google Scholar]
- 25.Covey T, Douglas DJ. J Am Soc Mass Spectrom. 1993;4:616–623. doi: 10.1016/1044-0305(93)85025-S. [DOI] [PubMed] [Google Scholar]
- 26.von Helden G, Wyttenbach T, Bowers MT. Science. 1995;267:1483–1485. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- 27.Clemmer DE, Hudgins RR, Jarrold MF. J Am Chem Soc. 1995;117:10141–10142. [Google Scholar]
- 28.Gross DS, Schnier PD, Rodriguez-Cruz SE, Fagerquist CK, Williams ER. Proc Natl Acad Sci USA. 1996;93:3143–3148. doi: 10.1073/pnas.93.7.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loo RRO, Smith RD. J Am Soc Mass Spectrom. 1994;5:207–220. doi: 10.1016/1044-0305(94)85011-9. [DOI] [PubMed] [Google Scholar]
- 30.Antonini, E.; Brunori, M. Hemoglobin and Myoglobin in Their Reactions With Ligands, Vol. 21; North-Holland: Amsterdam, 1971.
- 31.Hargrove MS, Krzywda S, Wilkinson AJ, Dou Y, Ikeda-Saito I, Olson JS. Biochemistry. 1994;133:11767–11775. doi: 10.1021/bi00205a012. [DOI] [PubMed] [Google Scholar]
- 32.Hargrove MS, Singleton EW, Quillen ML, Ortiz LA, Phillips JGN, Olson JS, Mathews AJ. J Biol Chem. 1994;269:4207–4214. doi: 10.2210/pdb1mgn/pdb. [DOI] [PubMed] [Google Scholar]
- 33.Hargrove MS, Barrick D, Olson JS. Biochemistry. 1996;35:11293–11299. doi: 10.1021/bi960371l. [DOI] [PubMed] [Google Scholar]
- 34.Hargrove MS, Wilkinson AJ, Olson JS. Biochemistry. 1996;35:11300–11309. doi: 10.1021/bi960372d. [DOI] [PubMed] [Google Scholar]
- 35.Hargrove MS, Olson JS. Biochemistry. 1996;35:11310–11318. doi: 10.1021/bi9603736. [DOI] [PubMed] [Google Scholar]
- 36.Palaniappan V, Bocian DE. Biochemistry. 1994;33:14264–14274. doi: 10.1021/bi00251a039. [DOI] [PubMed] [Google Scholar]
- 37.Sage JT, Morikis D, Champion PM. Biochemistry. 1991;30:1227–1237. doi: 10.1021/bi00219a010. [DOI] [PubMed] [Google Scholar]
- 38.Nienhaus GU, Mourant JR, Chu K, Frauenfelder H. Biochemistry. 1994;33:13413–13430. doi: 10.1021/bi00249a030. [DOI] [PubMed] [Google Scholar]
- 39.Frauenfelder H, Wolynes PG. Science. 1985;229:337–345. doi: 10.1126/science.4012322. [DOI] [PubMed] [Google Scholar]
- 40.Whitaker TL, Berry MB, Ho EL, Hargrove MS, Phillips GN, Komiyama NH, Nagai K, Olson JS. Biochemistry. 1995;34:8221–8226. doi: 10.1021/bi00026a002. [DOI] [PubMed] [Google Scholar]
- 41.Gryczynski Z, Lubkowski J, Bucci E. J Biol Chem. 1995;270:19232–19237. doi: 10.1074/jbc.270.33.19232. [DOI] [PubMed] [Google Scholar]
- 42.Smith ML, Paul J, Ohlsson PL, Hjortsberg K, Paul KG. Proc Natl Acad Sci USA. 1991;88:882–886. doi: 10.1073/pnas.88.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YT, Hsieh YL, Henion JD, Ganem B. J Am Soc Mass Spectrom. 1993;4:631–637. doi: 10.1016/1044-0305(93)85027-U. [DOI] [PubMed] [Google Scholar]
- 44.Price WD, Schnier PD, Williams ER. Anal Chem. 1996;68:859–866. doi: 10.1021/ac951038a. [DOI] [PubMed] [Google Scholar]
- 45.Schnier PD, Price WD, Jockusch RA, Williams ER. J Am Chem Soc. 1996;118:7178–7189. doi: 10.1021/ja9609157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price WD, Schnier PD, Jockusch RA, Strittmatter EF, Williams ER. J Am Chem Soc. 1996;118:10640–10644. doi: 10.1021/ja961812r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunbar RC, McMahon TB, Tholmann D, Tonner DS, Salahub DR, Wei D. J Am Chem Soc. 1996;117:12819–12825. [Google Scholar]
- 48.Stryer, L. Biochemistry, 3rd ed.; Freeman: New York, 1988.
- 49.Gross DS, Williams ER. J Am Chem Soc. 1995;117:883–890. [Google Scholar]
- 50.Chiancone E, Gilbert GA. J Biol Chem. 1965;240:3866–3867. [PubMed] [Google Scholar]
- 51.Gattoni M, Boffi A, Sarti P, Chiancone E. J Biol Chem. 1996;271:10130–10139. doi: 10.1074/jbc.271.17.10130. [DOI] [PubMed] [Google Scholar]
- 52.Goodlett DR, Ogorzalek Loo RR, Loo JA, Wahl JH, Udseth HR, Smith RD. J Am Soc Mass Spectrom. 1994;5:614–622. doi: 10.1016/1044-0305(94)85002-X. [DOI] [PubMed] [Google Scholar]
- 53.Gilbert, R. C.; Smith, S. C. Theory of Unimolecular and Recombination Reactions; Blackwell Scientific: London, 1990.
- 54.Gallop MA, Barrett RW, Dower WJ, Fodor SPA, Gordon EM. J Med Chem. 1994;37:1233–1251. doi: 10.1021/jm00035a001. [DOI] [PubMed] [Google Scholar]
- 55.Ellmann JA. Acc Chem Res. 1996;29:132–143. [Google Scholar]


