Abstract
To elucidate cytolytic mechanisms in the channel catfish, lysates from catfish lymphoid and fibroblast cell lines were screened by Western blot analysis using a panel of antibodies reactive with components of the mammalian apoptotic pathway. Strong reactivity with three proteins (approximate Mr 70,000, 37,000, and 15,000) was seen using an antibody targeted to mammalian Fas ligand (FasL). The sizes of the two smaller proteins are consistent with their tentative designation as membrane-bound (37,000 Mr) and soluble (15,000 Mr) FasL. Treatments known to induce FasL in mammalian systems (e.g., PMA/calcium ionophore, UV-irradiation) induced expression of the 37,000-Mr protein in catfish T-cell lines. Moreover, expression of the 37,000-Mr protein in clonal T cells was up-regulated by increasing cell density. At the nucleotide level, homologues of Fas receptor (FasR), FADD, and caspase 8 were identified and characterized. These gene products likely constitute the teleost equivalent of the death-inducing signaling complex (DISC). FADD was constitutively expressed in all (T, B, macrophage, and fibroblast) cell lines examined as well as in peripheral blood lymphocytes (PBL), whereas FasR and caspase 8 were expressed in all cell lines except CCO, a FasL-positive fibroblast line. In contrast to FasL, expression of FasR and caspase 8 was inversely proportional to cell density. Collectively these studies identified four membrane-proximal proteins involved in the initiation of apoptosis in channel catfish and suggest that mechanisms of cell-mediated cytotoxicity in teleosts are similar to those used by mammals.
Keywords: Catfish, Apoptosis, FasL, FasR, Caspase 8
Introduction
Channel catfish (Ictalurus punctatus) possess a diverse repertoire of cytotoxic cells, including alloantigen-dependent cytotoxic T cells, nonspecific cytotoxic cells (NCC), and NK-like cells (Evans et al. 1984; Graves et al. 1985; Yoshida et al. 1995; Stuge et al. 2000; Zhou et al. 2001; Shen et al. 2002, 2004). Although PBL-derived NK-like effectors and alloantigen-specific cytotoxic cells kill via apoptosis, the molecular mechanisms underlying this process have not yet been fully elucidated. Inhibitor studies suggest that PBL effectors kill via a calcium-dependent process that is likely mediated by perforin/granzyme (Hogan et al. 1999), whereas alloantigen-dependent cytotoxic cells may utilize both perforin/granzyme and FasR/FasL systems (Zhou et al. 2001). Similarly, both perforin/granzyme and FasR/FasL systems may play a role in NCC-mediated cell death (Carlson et al. 1985; Evans et al. 2001; Bishop et al. 2002). To further elucidate teleost cytotoxic mechanisms, apoptosis-related proteins/genes were identified in channel catfish using molecular and serological approaches. Our results indicate that catfish express four key membrane-proximal genes critical for cell-mediated cytotoxicity: FasL, FasR, FADD, and caspase 8.
In mammals, FasR, a member of the tumor necrosis factor receptor/nerve growth factor receptor (TNFR/NGFR) superfamily, is a “death receptor” that, when bound to its ligand, FasL, transduces an apoptotic signal. The TNFR/NGFR family is comprised of type I transmembrane proteins whose extracellular regions consist of one to five cysteine-rich domains (CRD) and whose cytoplasmic tail contains a single death domain (DD). The FasR DD, essential for transducing the apoptotic signal, consists of an ∼60-amino-acid bundle of six alpha helices that interacts with a second DD-containing adaptor protein designated FasR-associated DD (FADD; Bodmer et al. 2002; Jones et al. 1989; Locksley et al. 2001; McDonald et al. 2001). FasL–FasR interaction results in the trimerization of the FasR and the subsequent binding of FADD via the homologous DD of FADD and FasR. The association of FADD with FasR results in the rapid recruitment and activation of caspase 8 via the interaction of homologous death effector domains (DEDs) contained within FADD and the prodomain of caspase 8. Subsequently, active caspase 8 cleaves downstream caspases, which execute the apoptotic cascade (reviewed by Wallach et al. 1999).
This study confirms and extends previous reports of FasL in catfish (Evans et al. 2001; Bishop et al. 2002) and is the first molecular description of caspase 8 in catfish and the first report of a full-length gene for FasR in teleosts. Collectively, these findings strengthen the suggestion that ectothermic vertebrates, like their mammalian counterparts, possess the molecular machinery required to trigger apoptosis by the FasR/FasL pathway.
Materials and methods
Cell lines
The following long-term channel catfish lymphoid cell lines (LCL) were used in this study: G14D and G14C, T-cell lines of unknown specificity (Hogan et al. 1999); TS32.15, TS32.17, and TS32.43, alloantigen-dependent cytotoxic T-cell lines (Stuge et al. 2000); 3B11 and 1G8, B-cell lines (Miller et al. 1994a,b); and 42TA, a macrophage-like cell line (Miller et al. 1994a). In addition, two adherent cell lines, CCO (channel catfish ovary, Bowser and Plumb 1980) and BB (brown bullhead, ATCC no. CCL 59), were also used. Catfish LCL were cultured in AL-5 medium, a medium composed of AIM-V media, L-15 media, and de-ionized water (in the ratio 45:45:10) containing 50 mm 2-mercaptoethanol and 5% heatinactivated catfish serum (Miller et al. 1994a). CCO and BB were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS). Cell cultures were incubated at 26°C in a humidified incubator in 5% CO2/95% air. PBL were obtained from catfish by venipuncture of the caudal sinus and isolated following centrifugation onto a cushion of lymphoprep (Miller et al. 1994a).
Characterization of channel catfish FasL (CF FasL) by Western blot analysis
Catfish cells (5×106) were lysed in 50 μl 2× direct sample buffer (2× DSB: 125 mm Tris pH 6.8, 4% SDS, 10% glycerol, 0.006% bromophenol blue, 2% β-mercaptoethanol), analyzed by electrophoresis on 10% SDS-polyacrylamide gels, and cellular proteins transferred to nitrocellulose (Laemmli 1970; Sambrook and Russel 2001). Filters, blocked overnight at 4°C with 5% nonfat dry milk in TBST (137 mm NaCl, 20 mm Tris–HCl pH 8.0, 0.05% Tween-20), were washed and incubated for 1 h at room temperature with mouse anti-human FasL mAb (clone 33, Transduction Laboratories) diluted 1:1,000 in TBSTM (TBST containing 5% nonfat dry milk). After removal of the primary antibody, the filters were washed, incubated for an additional hour at room temperature with a 1:5,000 dilution of HRP-conjugated goat anti-mouse IgG (Southern Biotechnology, Birmingham, Ala.), washed, and the reactive proteins identified using an enhanced chemiluminescence system (ECL, Amersham).
Induction of putative CF FasL
G14D T cells were exposed to conditions known to induce expression of FasL in mammalian systems and the levels of putative CF FasL were monitored by Western blot analysis as described above. G14D cells were either treated with phorbol 12-myristate-13-acetate (PMA, 0.05 μg/ml) and calcium ionophore (A23187, 0.5 μg/ml) or exposed to varying doses of UV light (Algeciras-Schimnich and Paya 2000; Caricchio et al. 1998). In the former, 5×106 cells in 1 ml AL5 were treated with PMA/A23187 and cell lysates prepared at 0.5, 1, 2, 6, and 24 h post-treatment. In the latter, a BioRad GS GeneLinker was used to deliver 1, 5, or 10 mJ UV light to 5×106 G14D cells in 1 ml AL5, and lysates were prepared for electrophoresis 4 h following UV irradiation. Finally, to determine if FasL expression varied with changes in cell density, lysates were prepared from logarithmically growing, stationary and declining G14D cultures.
Sequencing of catfish FasR and catfish caspase 8 cDNAs
Plasmids encoding putative catfish FasR (CF FasR) and caspase 8, were identified following single-pass sequencing and BLASTX (Altschul et al. 1997) analysis of cDNA libraries (http://morag.umsmed.edu) constructed from B (3B11) cells and macrophages (42TA), respectively.
Plasmids containing FasR and caspase 8 cDNAs were initially sequenced on both strands using the universal sequencing primers M13F and M13R. Contigs were assembled using the SEQMAN program within DNASTAR (Madison, Wis.). Based on the overlapping contigs, additional primers were designed and used to determine the complete sequences. Because the plasmid bearing the FasR gene did not contain full-length cDNA, a 5′ rapid amplification of cDNA ends (5′RACE) was used to determine the authentic 5′ end of the transcript using a commercially available kit (GibcoBRL). Briefly, 1 μg total RNA from 1G8 B cells was reverse transcribed using SuperScript II RT and the FasR-specific primer 01-20R. Nested primers used in the final reactions were 01-23R and 01-18R. The sequence, gene specificity, and nucleotide position of all primers used in this study are shown in Table 1.
Table 1.
Primers used for sequencing and expression analysis of CF FasR, CF FADD and CF caspase 8
| Primer namea | Positionb | Sequence (5′ to 3′) |
|---|---|---|
| 01-15F (CF FasR) | 262–283 | GGATCACGCGAACGCTCTGTCA |
| 01-18R (CF FasR) | 461–482 | ACTGTGCGTCGCTGGTGAGGTT |
| 01-19F (CF FasR) | 784–802 | TCCTCATGACGCGAGCGAA |
| 01-20R (CF FasR) | 784–802 | TTCGCTCGCGTCATGAGGA |
| 01-23R (CF FasR) | 612–630 | TTCAGTGGTTCCGGTGGTT |
| 01-13F (CF FADD) | 295–315 | ATCAACTGTGCAATAGAAGTG |
| 01-17R (CF FADD) | 526–549 | GAGATTCATCTGCACAACATCAGC |
| 01-21F (CF caspase 8) | 731–748 | ATGGAGTACACGGCACAG |
| 01-22R (CF caspase 8) | 731–748 | CTGTGCCGTGTACTCCAT |
| 01-24F (CF caspase 8) | 351–369 | TGTGGATCTCTTGCAGCGA |
| 01-10F (CF actin) | 611–630 | ACTACCTGATGAAGACCCTG |
| 01-10R (CF actin) | 1,108–1,126 | GGCTGATCCACATCTGTTG |
| 03-16F (CF 18S rRNA) | 840–863 | CCCGCCCAACTCGCCTGAATACCT |
| 03-17R (CF 18S rRNA) | 1,325–1,348 | GGCCATGCACCACCACCCACAGAA |
The name within the parentheses indicates the gene for which the primer is specific.
Position indicates the nucelotide position within the gene of interest.
Multiple alignments of the deduced amino acid sequences of CF FasR and CF caspase 8 with their representative homologues were generated using the CLUSTAL V program within MEGALIGN (DNASTAR, Madison, Wis.) and the percent similarities between CF FasR and caspase 8 and their homologous counterparts determined. Phylogenetic trees based on these multiple alignments were generated using the neighbor-joining (NJ) algorithm within MEGA version 2.1 (http://www.megasoftware.net, Kumar et al. 2001). Pairwise protein sequence divergence was corrected for multiple hits by using the Poisson correction. The degree of confidence for each branch point was validated by the bootstrap method (1,000 replications). Signal peptide prediction was performed using the SignalP program, version 2.0.b2 (Nielsen et al. 1997). Transmembrane spanning sequences were identified by the TMPRED program (Hofmann and Stoffel 1993). The Prosite database was searched to identify motifs within the deduced amino acid sequences (Falquet et al. 2002).
Catfish FasR, FADD, and caspase 8 expression studies: RT-PCR
RT-PCR analysis was conducted to determine which cell lines expressed CF FasR, FADD, and caspase 8 as well as to determine if expression of these genes was regulated by cell density. In the former, RNA was isolated from various catfish LCLs, PBLs, and adherent CCO and BB cells using RNAwiz (Ambion, Austin, Tex.), treated with RNase-free DNase (Invitrogen), and used as template in RT-PCR reactions. Total RNA (2.5 μg) was reverse transcribed with an oligo(dT) primer and Superscript II RNase H-minus reverse transcriptase (Invitrogen) according to the manufacturer's instructions. One microliter of the resulting cDNA was used as template in PCR reactions. PCR reactions (50 μl total volume) contained 50 mm KCl, 10 mm Tris–HCl pH 8.3, 1.5 mm MgCl2, 200 μm each dATP, dCTP, dGTP, dTTP, 500 ng forward and reverse primers, and 2.5 units AmpliTaq Gold DNA polymerase (Perkin–Elmer). Primer pairs 01-15F and 01-20R were used to amplify CF FasR; 01-13F and 01-17R were used to amplify CF FADD; 01-24F and 01-22R were used to amplify CF caspase 8. Cycling conditions were: one cycle of 94°C for 3 min; 30 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min; 1 cycle of 72°C for 3 min. Products were visualized by electrophoresis on 1.2% agarose gels containing 150 ng/ml ethidium bromide. As a control for RNA integrity, RT-PCR was also performed using primers specific for catfish actin (01-10F and 01-10R) and catfish 18S rRNA (03-16F and 03-17R). To determine if cell density influenced gene expression, RNA was isolated from G14D cultures during logarithmic growth as well as from stationary and declining cultures.
GenBank accession numbers
CF FasR (AY553585), CF FADD (AY555194), CF caspase 8 (AY555576). The sequence of FADD was reported previously by Hawke and co-workers (1999).
Results
Characterization of catfish FasL
Previous work demonstrated that channel catfish cytotoxic cells kill their targets by both necrotic and apoptotic mechanisms (Hogan et al. 1999; Zhou et al. 2001). To determine whether the induction of apoptosis in catfish utilizes genes homologous to those found in mammals, we attempted to identify catfish apoptotic genes at the serological and nucleotide levels. Initially catfish homologues were sought by their cross-reactivity with monoclonal antibodies targeted to mammalian apoptotic proteins. This approach has proven successful with other catfish proteins, e.g., NFAT (Park et al. 2002) and STAT 6 (Rycyzyn et al. 1998). Lysates from catfish cell lines were screened by immunoblotting against a panel of 12 antibodies (Apoptosis Sampler Kit S82080, Transduction Laboratories) directed against various components of the mammalian apoptotic pathway. Cross-reactivity was observed only with a mouse anti-human FasL mAb (clone 33, Transduction Laboratories) that had previously reacted with an ∼40,000-Mr protein from tilapia NCC (Jaso-Friedmann et al. 2000). As shown in Fig. 1, reactivity was seen with three proteins (approximate Mr 70,000, 37,000, and 15,000). The molecular weights of the two smaller proteins are consistent with the reported sizes of the transmembrane (37,000 Mr) and soluble (16,000–26,000 Mr) forms of mammalian FasL. The 70,000-Mr protein is considerably larger than mammalian FasL and likely represents adventitious cross-reactivity. As depicted in Fig. 1, Western blot analysis showed that autonomous catfish T (G14D and G14C) and fibroblast (BB and CCO) cells expressed the 37,000-Mr protein. The 37,000-Mr isoform was absent in macrophages (42TA), non-autonomous T cells (TS32.15, TS32.17, and TS32.43), and B cells (3B11). Furthermore expression of the 37,000-Mr protein in G14D T cells appeared to be regulated in a cell density-dependent manner (see below). An ∼15,000-Mr protein was observed in the TS32.17 and TS32.43 cell lines and may be similar to an alternatively spliced 16,000-Mr soluble FasL protein present in resting mouse lymphocytes (Ayroldi et al. 1999). Interestingly, TS32.15, a cytotoxic T-cell clone distinct from TS32.17 and TS32.43 (Zhou et al. 2001), possessed neither the 37,000-Mr nor the 15,000-Mr form of FasL. These results suggest that catfish cells possess both membrane bound and soluble FasL. Although FACS analysis detected intracellular FasL, attempts at identifying membrane-bound FasL (mFasL) were unsuccessful (data not shown). This latter result most likely reflects the reported inability of this specific mAb to react with native cell-surface associated FasL on human cells (Kiener et al. 1997).
Fig. 1.
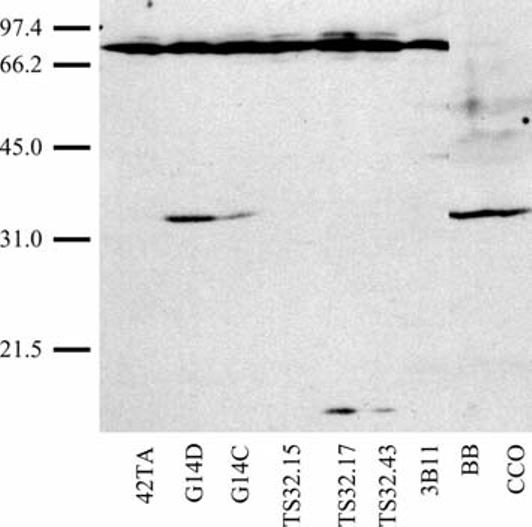
Western blot analysis: reactivity with human anti-FasL mAb. Lysates from 5×106 macrophage (42TA), T (G14D, G14C), cytotoxic T (TS32.15, TS32.17, TS32.43), B (3B11), and fibroblast (BB, CCO) cells were subjected to 10% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated with a mouse anti-human FasL mAb (Transduction Laboratories, Lexington, Ky.) and reactive proteins detected by chemiluminescence
In order to determine if the 37,000-Mr protein is authentic FasL, G14D T cells were exposed to conditions known to induce FasL expression in mammalian systems, and levels of putative FasL were determined by Western blot analysis. Cell lysates were obtained from G14D cells treated with PMA and calcium ionophore, or exposed to varying doses of UV light. Consistent with its designation as FasL, both treatments induced expression of the 37,000-Mr protein. With PMA/calcium ionophore, putative FasL was seen as early as 30 min following induction and was not detectable after 6 h, indicating that, as in mammals, induction was rapid and transient (Fig. 2a). Likewise, CF FasL was not detected in mock-irradiated cells, but was induced by low doses of UV-irradiation (Fig. 2b). Since expression of the 37,000-Mr FasL protein in nonstimulated G14D cells varied among experiments (Figs. 1, 2a,b), we speculated that FasL levels might be regulated by cell density. As a result, the correlation of FasL expression and cell density was examined. As depicted in Fig. 2c, FasL expression rose with increasing cell density and reached maximum levels in stationary and declining cultures. Taken together, these findings indicate that catfish cells express a protein which cross-reacts with a mAb directed against human FasL and is regulated in a similar manner.
Fig. 2a-c.
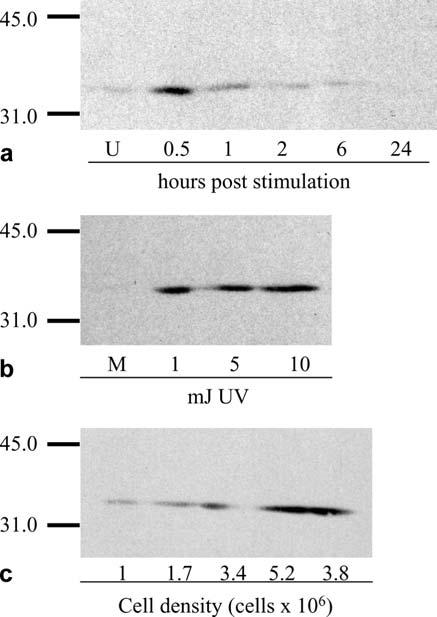
Induction and expression of a 37,000-Mr protein cross-reactive with anti-FasL. G14D T cells were exposed to conditions known to induce FasL expression in mammalian systems. Western blots using anti-FasL were performed on cell lysates (5×106 cell equivalents) obtained from G14D cells a treated with 0.05 μg/ml PMA and 0.5 μg/ml calcium ionophore, or b exposed to varying doses of UV-irradiation expressed in mJoules (mJ). Mitogen-treated cells were harvested at the times indicated in the figure. UV-irradiated cells were harvested 4 h post-irradiation. c Western blots using anti-FasL were performed on G14D cells harvested from logarithmic (1×106, 1.7×106, and 3.4×106 cells/ml), stationary (5.2×106 cells/ml), or declining (3.8×106 cells/ml) cultures. Each lane contains the equivalent of 5×106 cells (U unstimulated, M mock)
In order to facilitate the identification of a cDNA encoding the catfish homologue of FasL, a multiple alignment of known mammalian (i.e., human, mouse, and rat), avian (i.e, chicken) and teleost (fugu and zebrafish) FasL sequences was made. Degenerate primers were constructed based on conserved regions and used to screen cDNAs prepared from UV-irradiated G14D cells by RTPCR. Using this approach, no clones were obtained with sequence homology to FasL. In another attempt to identify CF FasL cDNA, zebrafish FasL (GenBank accession no. CK030329) was blasted against the catfish EST database maintained at http://www.tigr.org. This approach generated a hit to an EST made from an intestine cDNA library (GenBank accession no. CF261600), which was annotated as a protein of unknown identity. BLASTX analysis of this EST revealed a match to mouse TNF-related apoptosis inducing ligand (TRAIL; bit score 46; E value 5×10−4). Among mammalian TNF family members, TRAIL shows the highest similarity to FasL (Wiley et al. 1995) and, if this relationship holds true for teleosts, may facilitate the future identification of CF FasL cDNA. Nonetheless, our present efforts to identify the cDNA encoding the catfish homologue of FasL have not been successful.
Identification of catfish FasR: sequence analysis
BLASTX analysis of an EST library constructed from the autonomous B-cell line 3B11 identified a cDNA with sequence homology to human and rabbit FasR. The catfish sequence showed 40% identity/52% similarity to human FasR (over a region of 108 amino acids) and 38% identity/50% similarity to rabbit FasR (over a region of 119 amino acids). Because sequence analysis revealed that the original cDNA clone did not contain the 5′ end of the message, 5′RACE was performed. The complete FasR sequence (Fig. 3) contained a 19-bp 5′-untranslated region (UTR), a 939-bp open reading frame encoding a 312-amino-acid protein (inferred Mr of 34,900), and a 627-bp 3′UTR. Within the 3′UTR, two mRNA instability motifs (ATTTA) were identified (Taniguichi 1988), and a polyadenylation signal matching the consensus (AATAAA) was located 21 nucleotides upstream from the start of the poly(A) tail (Darnell et al. 1990). Analysis with the SignalP program detected a classical signal peptide and predicted a cleavage site between amino acids 22 and 23 (Nielsen et al. 1997). A 20-amino-acid isoleucine-rich membrane spanning region (IIIPILIIAFIIIIIIIAAV) was identified by the TMPRED program at amino acid position 167–186 (Hofmann and Stoffel 1993). These results predict a transmembrane protein with a 144-amino-acid extracellular region, a 20-amino-acid membrane spanning region, and a 126-amino-acid cytoplasmic tail.
Fig. 3.
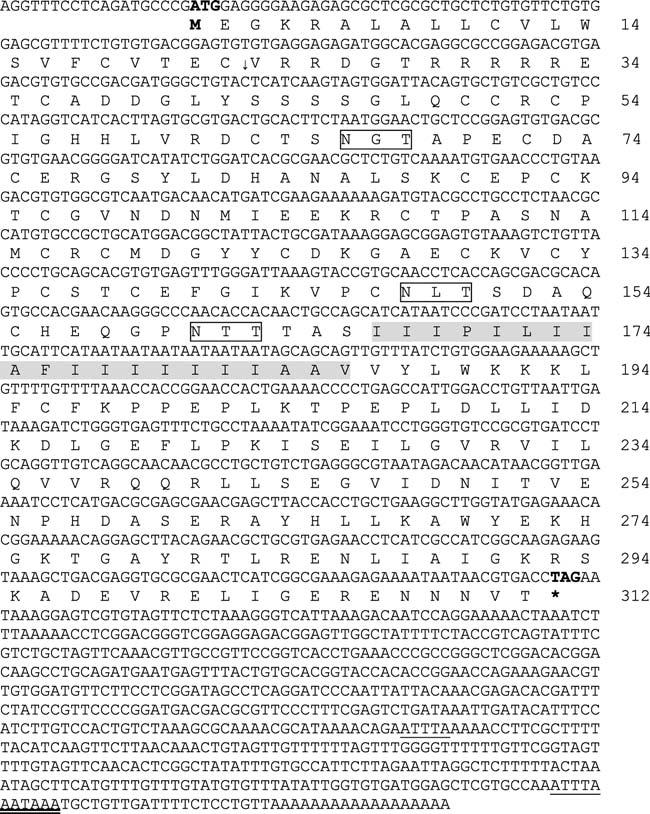
Catfish FasR cDNA and deduced amino acid sequences. The translation start and stop codons are indicated in bold type. Two mRNA instability motifs (ATTTA) within the 3′ UTR are underlined, a polyadenylation signal (AATAAA) is double-underlined, and three potential glycosylation sites are boxed. The downward-pointing arrow between 22C and 23V indicates the probable cleavage site for removing the predicted signal peptide. The transmembrane spanning region is shaded
The Prosite database identified a cysteine-rich region from amino acid positions 50 to 139, typical of the members of the TNFR/NGFR superfamily to which FasR belongs (Falquet et al. 2002; Nagata and Golstein 1995). Moreover within the predicted extracellular domain, the Prosite database identified two TNFR/NGFR family cysteine-rich region domains [C-X4,6-(FHY)-X5,10-CX0,2-C-X2,3-C-X7,11-C-X4,6-(DNEQSKP)-X2-C] at positions 75–116 and 118–155. Proteins that contain this domain include FasR, TNFR type I and II, lymphotoxin beta receptor, CD40, CD27, and CD30 (Mallet and Barclay 1991; Sprang 1990; Krammer and Debatin 1992; Bazan 1993). It has been shown in mammals that the six cysteines present within this domain participate in intrachain disulfide bond formation such that an intrachain disulfide bond is formed between the first two cysteines, the third and fifth cysteine, and the fourth and sixth cysteine of the domain (Banner et al. 1993). Both domains found in the catfish match the consensus in all places except for the number of residues between Y80-C90 and Y123-C133, where nine amino acids rather than ten separate the two. From this, it is predicted that intrachain disulfide bonds are located between residues 75/90, 93/108, 96/116, 118/133, 136/147, and 139/155. Also consistent with its designation as FasR, the Prosite database detected a death domain, necessary for transducing the apoptotic signal, within the cytoplasmic tail between amino acid positions 243 and 300. Finally, three potential N-linked glycosylation sites were located at positions 66–68, 148–150, and 161–163.
To determine the relatedness of CF FasR to other known FasR sequences, a multiple alignment was performed (Fig. 4). Although the percent similarities of CF FasR to its mammalian homologues were low (∼23%), the cysteines that form the three CRDs corresponding to CF FasR amino acids 36–73, 74–117, and 118–155 were highly conserved (Smith et al. 1994; Bajorath 1999). Within the CRD, all 20 cysteines found within the mature FasR of cows, pigs, sheep, rats, and catfish are conserved, whereas 18/20 are conserved in humans, and 19/20 in rabbits. Moreover, within the CRD, CF FasR is approximately 37% identical to its mammalian counterparts. In mammals, FasL/FasR interactions occur mainly in the second and third CRD (Locksley et al. 2001). The high level of conservation in the second and third CRD of CF FasR implies that it may also bind FasL within this region. Taken together, these results suggest that despite considerable overall sequence diversity, the cysteine backbone of CF FasR is maintained, as well as key amino acids.
Fig. 4.
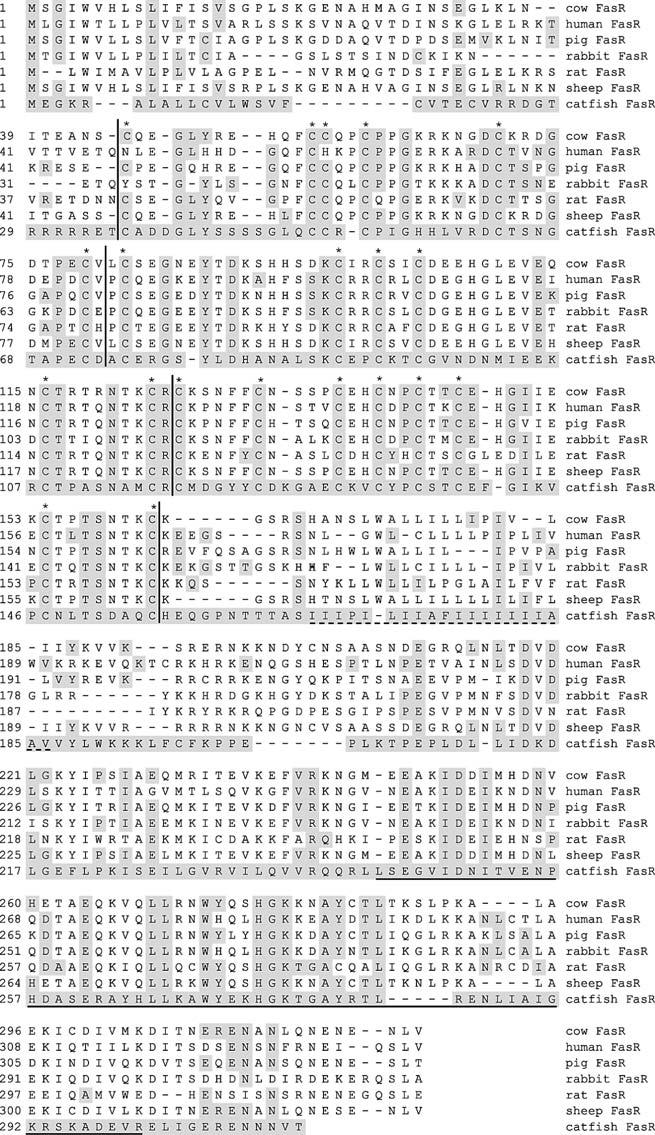
Multiple alignment of CF FasR with select mammalian homologues. The inferred amino acid sequence of CF FasR was aligned with the indicated FasR sequences using the CLUSTAL program within MEGALIGN (DNASTAR). Residues that match the CF FasR sequence are shaded. Vertical lines flank the three CRDs and asterisks denote conserved cysteines within those domains. The dashed line indicates the transmembrane spanning region, whereas the solid horizontal line denotes the death domain. GenBank accession numbers: cow FasR (NP_777087), human FasR (NP_000034), pig FasR (077736), rabbit FasR (BAA78428), rat FasR (NP_631933), sheep FasR (BAA37093), and catfish FasR (AY553585)
Identification of catfish caspase 8: sequence analysis
Analysis of an EST library generated from the macrophage cell line 42TA identified a cDNA with homology to mammalian caspases. Sequencing identified a 144-bp 5′ UTR, a 1,101-bp open reading frame encoding a 367-amino-acid protein (inferred Mr of 41,400), and a 164-bp 3′UTR (Fig. 5). BLASTP analysis of the complete transcript showed highest homology to chicken and human caspase 8. Within the 3′UTR, a single mRNA instability motif was identified (Taniguichi 1988). No consensus polyadenylation signal was identified, suggesting that the cDNA may have been generated from oligo(dT) priming of an A-rich region within the 3′UTR. Consistent with its intracellular location, analysis with the SignalP and the TMPRED programs failed to detect a classical signal peptide or a transmembrane spanning domain (Nielsen et al. 1997; Hofmann and Stoffel 1993). Two potential N-linked glycosylation sites and an integrin binding site (KGD) were identified at amino-acid positions 41–43, 162–164, and 61–63, respectively (Ruoslahti 1996). The active catalytic site (QACXG) at amino-acid position 233–237 and the caspase family signature HX2–4(S,C)X4(L,I,V,M,F)2(S,T)HG at amino-acid position 180–193 were conserved (Laing et al. 2001). Amino acids (Cys-235 and His-192) involved in catalysis were also maintained. The Prosite database identified the caspase p10 (position 271–366) and p20 (117–239) domains as well as a CARD (caspase recruitment domain) at position 1–74 suggesting that this protein is produced as an inactive proenzyme that must be cleaved to be functional. A conserved aspartic acid residue (among caspase 8 homologues) at amino acid position 269 is the likely cleavage site for releasing the p10 subunit.
Fig. 5.
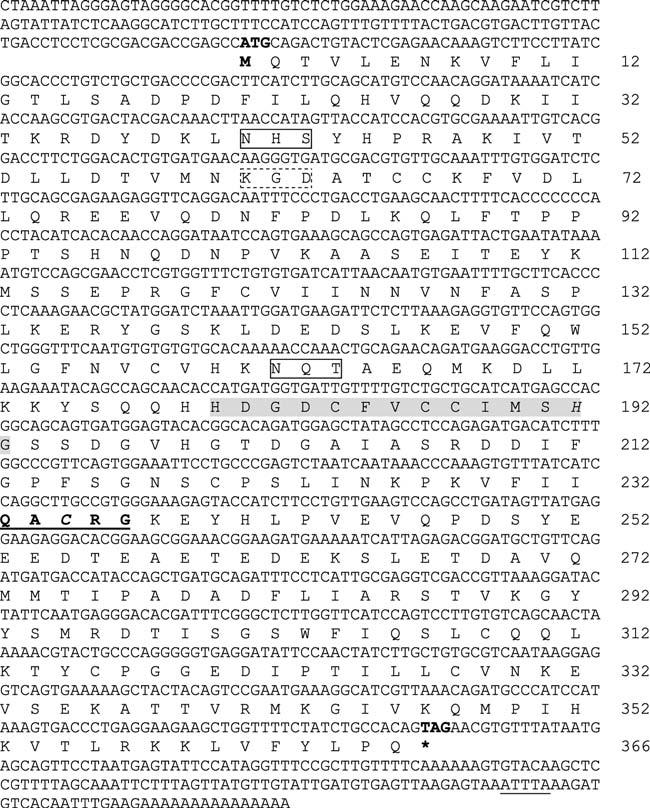
Catfish caspase 8 cDNA and deduced amino-acid sequences. The translation start and stop codons are indicated in bold type. Two potential glycosylation sites are boxed (solid lines), a single integrin binding site is boxed (dashed line), and a single mRNA instability motif (ATTTA) is underlined. The putative catalytic site (QACRG) is indicated in bold type and underlined and the caspase family signature is shaded. Amino acids involved in catalysis (Cys-235 and His 192) are italicized
To better determine the class to which this caspase belongs, a phylogenetic tree was constructed based on a multiple alignment of the deduced amino acid sequences of the CF caspase with representative mammalian and fish caspase 8 sequences, as well as with representative CARD-containing caspases (Fig. 6). Our initial designation of the gene product as caspase 8 was supported by the observation that the highest similarities following pairwise comparison were with other caspase 8 proteins. For example, CF caspase 8 was 30.5% and 30% similar to human and chicken caspase 8, respectively, but only 24.8% and 23.2% similar to the CARD-containing human and chicken caspase 9, respectively (Table 2). When CF caspase 8 was compared with other CARD-containing caspases (i.e., caspases 1, 2, 4, 5, and 12), percent similarity fell below 20%. The phylogenetic tree depicted in Fig. 6 shows that CF caspase 8 clusters apart from representative CARD-containing caspases and, more importantly that it forms a distinct, well-supported cluster with mammalian and zebrafish caspase 8 proteins. Interestingly, however, catfish caspase 8 appears basal and not sister to zebrafish caspase 8. This was surprising in that it was anticipated that the catfish and zebrafish sequences would have shared a higher degree of similarity. We speculate that the low level of similarity (i.e., 25.6%) reflects the long divergence time between the separation of catfish and zebrafish. Additionally, we note that similarity among mammalian and avian caspase 8 homologues is not very high. For example, human caspase 8 is 66, 64, and 55% similar to mouse, rat, and chicken caspase 8, respectively (data not shown).
Fig. 6.
Phylogenetic tree of CF caspase 8. An unrooted phylogenetic tree was constructed using the NJ algorithm within MEGA version 2.1 (Kumar et al. 2001). The tree was constructed using the Poisson correction and was validated by 1,000 bootstrap repetitions (shown as a percentage). Branch lengths are drawn to scale and a scale bar representing the number of amino acid changes is shown. GenBank accession numbers: chicken caspase 1 (AAC69917), mouse caspase 1 (NP_033937), human caspase 2 (NP_116764), human caspase 4 (NP_150649), human caspase 5 (NP_004338), mouse caspase 12 (NP_033938), chicken caspase 9 (AAL23701), human caspase 9 (P55211), mouse caspase 9 (NP_056548), rat caspase 9 (NP_113820), Xenopus caspase 9 (BAA94750), chicken caspase 8 (AAL23700), human caspase 8 (AAL87628), mouse caspase 8 (NP_033942), rat caspase 8 (NP_071613), zebrafish caspase 8 (NP_571585), catfish caspase 8 (AY555576)
Table 2.
Percent similarities between catfish caspase 8 and its homologues. Percent similarities were determined by the MegAlign program within DNASTAR. Percent similarity = 100 × (matches/matches + mismatches + gaps). Protein alignment scores are affected by partial matches, derived from the PAM250 residue weight table
| Species/gene | Percent similarity |
|---|---|
| Chicken caspase 1 | 18.7 |
| Mouse caspase 1 | 19.9 |
| Human caspase 2 | 20.4 |
| Human caspase 4 | 19.9 |
| Human caspase 5 | 19.6 |
| Mouse caspase 12 | 15.8 |
| Human caspase 9 | 24.8 |
| Mouse caspase 9 | 22.3 |
| Rat caspase 9 | 24.3 |
| Xenopus caspase 9 | 23.2 |
| Chicken caspase 8 | 30.0 |
| Human caspase 8 | 30.5 |
| Zebrafish caspase 8 | 25.6 |
Expression of catfish FasR, FADD, and caspase 8
To analyze the expression of the various apoptotic genes, specific primers were designed for the two genes identified here (FasR and caspase 8) as well as the previously identified CF FADD (Hawke et al. 1999). Total RNA was isolated from T (G14D), cytotoxic T (TS 32.17), B (1G8 and 3B11), macrophage (42TA) and fibroblast (CCO) cell lines as well as from PBLs, and reverse transcribed using an oligo(dT) primer. The resulting cDNA was analyzed by RT-PCR. PCR using FasR gene-specific primers (01-15F and 01-20R) gave rise to a single product of 538 bp, whereas FADD (01-13F and 01-17R) and caspase 8 (01-24F and 01-22R) gene-specific primers yielded products of 250 bp and 398 bp, respectively. As shown in Fig. 7, FasR, FADD, and caspase 8 were expressed in all lymphoid cells and cell lines tested. However, little if any FasR and caspase 8 appeared to be expressed in FasL positive CCO cells.
Fig. 7.
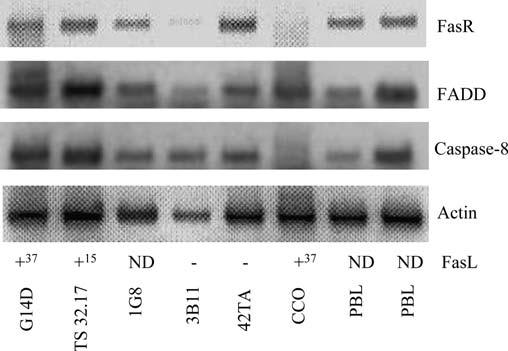
Expression of FasR, FADD, and caspase 8 in PBL and various catfish LCL. Total RNA was isolated from T (G14D), cytotoxic T (TS 32.17), B (1G8, 3B11), macrophage (42TA), and fibroblast (CCO) cell lines as well as PBLs from two fish and reverse transcribed using an oligo[dT] primer. The resulting cDNA was used in PCR along with gene-specific primers. Actin was included as a positive control. The presence or absence of FasL cross-reactive proteins as determined previously by Western blotting is indicated: +37 denotes the 37,000-Mr membrane form of FasL, whereas +15 denotes the 15,000-Mr soluble form. ND denotes not detected
To determine if expression of these genes was influenced by cell density, RNA was extracted from cultures during various stages of the growth cycle, and the levels of FasR, FADD, and caspase 8 expression determined by RT-PCR. Figure 8 shows that FADD was consistently expressed at all stages of the growth cycle. In contrast, FasR and caspase 8 were expressed at the highest level in cultures undergoing logarithmic growth (2.7×106 cells/ml and 4.8×106 cells/ml) but were lowest in stationary (5.5×106 cells/ml) and declining (3.4×106 cells/ml and 2.4×106 cells/ml) cultures.
Fig. 8.
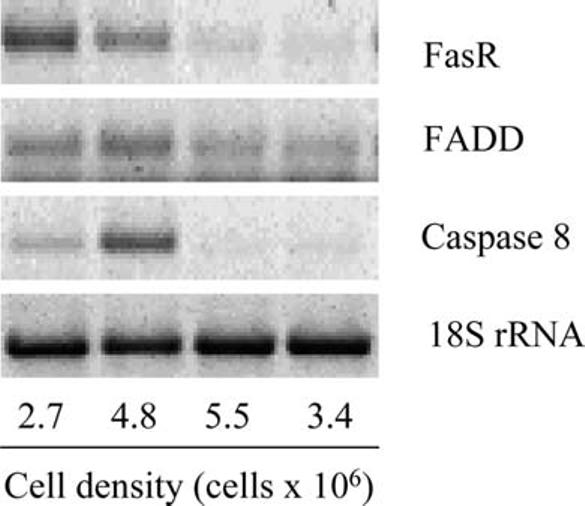
CF FasR, CF FADD, and CF caspase 8 are regulated by cell density. Expression levels of FasR, FADD, and caspase 8 at various times in the growth curve were determined by RT-PCR. Total RNA was isolated from G14D cells at the indicated cell densities and reverse transcribed using an oligo(dT) primer. The resulting cDNA was used in PCR along with primers specific for FasR, FADD, and caspase 8; 18S rRNA was included as a positive control. The last lane represents cells harvested from declining cultures
Discussion
Although catfish possess a variety of cytotoxic cells, the precise mechanisms by which these effectors mediate cytotoxicity remain unclear. Since inhibitor studies implicated both FasR-based and perforin/granzyme-based mechanisms in programmed cell death in catfish, attempts to identify the genes and/or proteins responsible for apoptosis were made. Here we report the identification of channel catfish cDNAs homologous to mammalian FasR and caspase 8. Although various caspases have been identified in other fish species, this is the first report identifying FasR in teleosts. These data also confirm and extend the earlier observations of Evans and co-workers (2000) and Bishop and co-workers (2002) that teleosts express proteins cross-reactive with antibodies to mammalian FasL. Coupled with the earlier identification of a cDNA encoding CF FADD (Hawke et al. 1999), these results suggest that, as in mammals, teleosts possess the molecular machinery required to trigger apoptosis.
FasL
An ∼40,000-Mr protein was initially identified in tilapia by Western blot analysis and subsequently detected in both tilapia and channel catfish by FACS analysis of permeabilized NCCs using an anti-human FasL mAb (Transduction Laboratories; Evans et al. 2000; Bishop et al. 2002). Analysis showed that apoptosis of FasR-positive HL-60 tumor cells followed interaction with either tilapia or catfish NCC. In contrast to systems which utilized membrane-bound FasL, NCC-mediated killing involved the release of an ∼40,000-Mr isoform of soluble FasL (sFasL) from NCC effectors. In addition, killing was blocked by pre-incubating HL-60 cells with anti-FasR monoclonal antibody (Bishop et al. 2002). Herein we extend these findings and demonstrate anti-FasL reactivity with the previously identified ∼40,000-Mr isoform as well as with two additional proteins (Mr ∼70,000 and ∼15,000). The ∼37,000-Mr protein identified in our studies was expressed in autonomous T-cell lines (G14C and G14D) and in two fibroblast lines, one from brown bullheads (BB), the second from channel catfish (CCO). A smaller protein (∼15 Mr) was detected in lysates of two previously identified cytotoxic T-cell clones (TS32.17 and TS32.43), but not in another cytotoxic T-cell clone TS32.15 or in other catfish lymphoid and non-LCL. The 15,000-Mr protein is similar in size to a soluble FasL protein present in mouse lymphocytes as well as to a 19,000-Mr cytosolic FasL protein identified in gilthead seabream using an anti-FasL mAb different from the one used in this study (clone Kay-10, Pharmingen; Ayroldi et al. 1999; Cuesta et al. 2003). The presence of the 15,000-Mr protein in TS32.17 and TS32.43 (Group II CTLs), coupled with its absence in TS32.15 (Group I CTLs), supports the suggestion of Zhou and co-workers (2001) that Group II CTLs may kill via a FasL mediated pathway, whereas the Group I cell lines predominately kill via perforin/granzyme mechanisms. To strengthen the suggestion that the 37,000-Mr protein was authentic FasL, induction experiments were performed under conditions shown to induce FasL expression in mammalian cells. Following treatment with PMA/calcium ionophore (Fig. 2a) or exposure to UV-irradiation (Fig. 2b) induction of the 37,000-Mr cross-reactive protein was clearly detected.
Variation in the expression of the 37,000-Mr protein in G14D cells suggested that CF FasL might be regulated in a cell density-dependent manner. Catfish lymphoid cell cultures, including G14D, experience rapid exponential growth and reach densities of 5×106 cells/ml or greater before declining. It is not known if the observed cell senescence and death is a response to a reduction in a key growth factor, the synthesis of a pro-apoptotic molecule (i.e., FasL), or the “stress” of increasing cell density. To determine if cell density influenced expression of FasL, G14D T cells were sampled over the course of a typical growth cycle. Freshly subdivided cultures expressed little FasL, whereas those at higher densities, or declining cultures, expressed high levels of the 37,000-Mr FasL protein (Fig. 2c). This result is reminiscent of events in mammalian cell cultures where density-dependent death has been attributed to apoptosis. For example, in the HL-60 system, apoptosis occurs when the culture density exceeds ∼106 cell/ml (Saeki et al. 1997). In another system, expression of FasL and FasR in human articular chondrocytes was shown to be regulated by cell density (Kuhn et al. 1999). Given these findings, it would be important to determine if density-dependent death in catfish LCL is mediated by FasL and is due to apoptosis. Nonetheless, these results suggest that the general features of CF FasL metabolism resemble those of mammals. However, despite the presence of at least one conserved epitope among mammalian and catfish FasL-like homologues, our attempts at identifying the gene encoding CF FasL by degenerative PCR were unsuccessful. While the reason for this is not known, the marked sequence diversity seen among other catfish apoptosis-related genes is likely responsible.
FasR
Although the amino-acid sequence similarity of CF FasR was low compared with six representative mammalian FasR proteins (∼23%), the sequence designated CF FasR is likely authentic for the following reasons: (1) as in mammalian FasR, three CRD (located between amino-acid positions 36 and 155) were identified, (2) two TNFR/NGFR family signatures were detected, and (3) a death domain within the predicted cytoplasmic tail was identified (Falquet et al. 2002; Nagata and Golstein 1995; Mallet and Barclay 1991; Sprang 1990). Within the three CRD, the percent identity of CF FasR to representative mammalian FasRs increased to 37%. Moreover, the cysteine backbone of the molecule was remarkably conserved. For example, 20/20 cysteines are conserved among cows, pigs, rats, sheep, and catfish.
Caspase 8
Consistent with its designation as a caspase, CF caspase 8 contains the caspase family signature, the active catalytic site, and the p10 and p20 subunits (Cohen 1997; Nicholson and Thornberry 1997). Although tentatively identified as caspase 8 based on similarity with mammalian homologues, the presence of a CARD motif in the prodomain challenges this classification. Mammalian caspase 8 contains two DEDs in its prodomain that interact with the DED of the adaptor molecule FADD (Cohen 1997). In contrast, caspases that contain CARD motifs typically interact with other CARD-containing proteins such as the adaptor protein RAIDD. While CARDs are functionally similar to DEDs, they have not been found in caspase 8 homologues. Instead CARD motifs have been found in mammalian caspases 1, 2, 4, 5, 9 and 12 (Duan and Dixit 1997; Hofmann et al. 1997; Chinnaiyan et al. 1997; Irmler et al. 1997). Despite the presence of a CARD motif, the tentative designation of this molecule as CF caspase 8 is supported by phylogenetic analysis, which clusters putative CF caspase 8 with zebrafish caspase 8 and several mammalian caspase 8 proteins (Fig. 6).
The presence within CF caspase 8 of a CARD motif coupled to mammalian caspase 8-like catalytic domains is intriguing. Given this unique arrangement, at least two scenarios for caspase 8 activation can be envisioned: (1) CF caspase 8 may interact with putative CF FasR via a CARD-containing adapter molecule such as RAIDD, or (2) CF caspase 8 may interact with CF FADD via CARD–DED binding. While homotypic (i.e., DED–DED), rather than heterotypic (i.e., DED–CARD) interactions are the rule for proteins containing “death folds” (i.e., DD, DED, CARD, PYRIN), there is some evidence supporting heterotypic (e.g., PYRIN–CARD) interaction (Weber and Vincenz 2001). Moreover, the homologous nature of DED, DD, CARD and PYRIN motifs, all of which form a death domain fold composed of six or seven anti-parallel α-helices, and the observation that only CARD domains are found in lower organisms (e.g., C. elegans) suggest that DED and DD may have evolved from an ancestral prototype that gave rise to the CARD family of proteins (Bouchier-Hayes and Martin 2002; Tibbetts et al. 2003). Perhaps phylogenetically primitive organisms such as fish contain death folds that are more promiscuous in their choice of binding partners than their more phylogenetically advanced mammalian counterparts. In view of this, we note that two recently identified zerbrafish caspases that are homologous to human caspases 1 and 5 within their catalytic domains, possess a PYRIN motif, rather than a CARD motif, within their prodomain (Masumoto et al. 2003). As sequences of additional fish proteins within the death domain superfamily become available, their relationship to other members within the superfamily (i.e., DD, DED, CARD, or PYRIN) will be elucidated.
The above studies indicate that catfish, and perhaps other teleosts, possess the three membrane-proximal components of the death-inducing signaling complex (DISC): FasR, FADD, and caspase-8. In mammals, the DISC transduces the apoptotic signal triggered by FasL. Formation of the DISC is initiated by trimerization of FasR following its interaction with FasL. Subsequently, FADD associates with trimerized FasR via its own death domain and recruits procaspase 8 to the nascent complex through interaction between its DED and the DED of procaspase 8. High-localized concentrations of procaspase 8 lead to cleavage of the proenzyme. Following release of native caspase 8 from the complex, caspase 8 cleaves downstream amplifying caspases, which, in turn, cleave the effector caspases that target proteins maintaining cellular homeostatis and DNA integrity, e.g., lamin, PARP, and ICAD (Enari 1998; Wallach et al. 1999). The molecular identification of CF FasR, FADD, and caspase 8, coupled with the serological identification of FasL, provide compelling evidence that catfish contain the machinery required to mediate apoptotic death following FasR–FasL interaction.
Expression studies
Expression studies indicate that FADD, FasR, and caspase 8 were widely expressed, and that transcription of the latter two was down-regulated in response to increasing cell density. This was especially noticeable in CCO cells in which FasL expression was readily detected in confluent cultures, but which showed little if any expression of FasR and caspase 8 (Fig. 7). This result is similar to that seen in several mammalian cell lines (e.g., THP-1, HL-60, and HCW-2) that constitutively express FasL and, in response to increasing cell density, down-regulate FasR to prevent FasL-induced apoptosis (Bremner et al. 1999). Consistent with this observation, we noted that FasL and FasR/caspase 8 were differentially expressed during growth of G14D cells. FasR and caspase 8 were detected during log growth, but were down-regulated in stationary and declining cultures (Fig. 8), whereas FasL was expressed in stationary and declining cultures (Fig. 2c). Perhaps, the reciprocal regulation of FasL versus FasR and caspase 8 during various phases of the G14D growth cycle represents an attempt by these cells to forestall apoptosis.
The above work demonstrates that four of the membrane-proximal proteins (FasL, FasR, FADD, and caspase 8) involved in cytotoxicity and apoptosis in mammals are also present in channel catfish. Coupled with the identification of catfish granzyme B and perforin (Evenhuis and Wilson, unpublished observations), these results indicate that catfish possess death pathways similar to those of mammals. Interestingly, despite the apparently similar pathways, the proteins charged with mediating those events show considerable sequence diversity. Perhaps the need to deal with different pathogens or simply evolution over ∼450 million years has resulted in considerable genetic diversification among these proteins (Kumar and Hedges 1998).
Acknowledgements
We wish to thank Noel Hawke (Lineberger Cancer Center, University of North Carolina Medical School) for contributing the catfish FADD sequence. This work was supported by grants from the US Department of Agriculture (NRI/CGP 99-35204-7944 and 2002-35204-12211) and the National Institutes of Health (ROI-AI-19530). Nucleotide sequencing was performed in collaboration with Dr. Greg Warr and Darlene Middleton (Medical University of South Carolina).
References
- Algeciras-Schimnich A, Paya C. Transcriptional regulation of the FasL gene. In: Sitkovsky M, Henkart P, editors. Cytotoxic cells: basic mechanisms and medical applications. Lippincott, Williams & Wilkins; Philadelphia: 2000. pp. 239–247. [Google Scholar]
- Altschul S, Maden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroldi E, D'Adamio F, Zollo O, Agostini M, Moraca R, Cannarile L, Migliorati G, Delfino D, Riccardi C. Cloning and expression of a short Fas ligand: a new alternatively spliced product of the mouse Fas ligand. Blood. 1999;94(10):3456–3467. [PubMed] [Google Scholar]
- Bajorath J. Identification of the ligand binding site in Fas (CD95) and analysis of Fas–ligand interactions. Proteins: Struct Funct Genet. 1999;35:475–482. [PubMed] [Google Scholar]
- Banner D, D'Acry A, Janes W, Gentz R, Schoenfeld H-J, Broger C, Loetscher H, Lesslauer W. Crystal structure of the soluble human 55 kD TNF receptor-human TNFβ complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- Bazan J. Emerging families of cytokines and receptors. Curr Biol. 1993;3:603–606. doi: 10.1016/0960-9822(93)90009-d. [DOI] [PubMed] [Google Scholar]
- Bishop G, Taylor S, Jaso-Friedmann L, Evans D. Mechanisms of nonspecific cytotoxic regulation of apoptosis: cytokine-like activity of Fas ligand. Fish Shellfish Immunol. 2002;13:47–67. doi: 10.1006/fsim.2001.0380. [DOI] [PubMed] [Google Scholar]
- Bodmer J-L, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;271:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- Bouchier-Hayes L, Martin SJ. CARD games in apoptosis and immunity. EMBO Rep. 2002;3:616–621. doi: 10.1093/embo-reports/kvf139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser P, Plumb J. Growth rates of a new cell line from the channel catfish ovary and channel catfish virus replication at different temperatures. Can J Fish Aquat Sci. 1980;37:871–873. [Google Scholar]
- Bremner T, Chatterjee D, Han Z, Tsan M, Wyche J. THP-1 monocytic leukemia cells express Fas ligand constitutively and kill Fas-positive Jurkat cells. Leuk Res. 1999;23:865–870. doi: 10.1016/s0145-2126(99)00101-0. [DOI] [PubMed] [Google Scholar]
- Caricchio R, Reap E, Cohen P. Fas/Fas ligand interactions are involved in ultraviolet-B-induced human lymphocyte apoptosis. J Immunol. 1998;161:241–251. [PubMed] [Google Scholar]
- Carlson RL, Evans DL, Graves SS. Nonspecific cytotoxic cells in fish (Ictalurus punctatus) V. metabolic requirements of lysis. Dev Comp Immunol. 1985;9:271–280. doi: 10.1016/0145-305x(85)90118-1. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan A, Chaudhary D, O'Rourke K, Koonin E, Dixit V. Role of CED-4 in the activation of CED-3. Nature. 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- Cohen G. Caspases: the executioners of apoptosis. J Biochem. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta A, Esteban A, Meseguer J. Identification of a FasL-like molecule in leucocytes of the teleost fish gilthead seabream (Sparus aurata L.) Dev Comp Immunol. 2003;27:21–27. doi: 10.1016/s0145-305x(02)00041-1. [DOI] [PubMed] [Google Scholar]
- Darnell J, Lodish H, Baltimore D. Molecular cell biology. Scientific American/Freeman; New York: 1990. [Google Scholar]
- Duan H, Dixit V. RAIDD is a new ‘death’ adaptor protein. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- Enari M. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Evans D, Graves S, Cobb D, Dawe D. Nonspecific cytotoxic cells in fish (Ictalurus punctatus). II. Parameters of target cell lysis and specificity. Dev Comp Immunol. 1984;8:303–312. doi: 10.1016/0145-305x(84)90037-5. [DOI] [PubMed] [Google Scholar]
- Evans D, Taylor S, Leary J, Bishop G, Eldar A, Jaso-Friedmann L. In vivo activation of tilapia nonspecific cytotoxic cells by Streptococcus iniae and amplification with apoptosis regulatory factor(s) Fish Shellfish Immunol. 2000;10:419–434. doi: 10.1006/fsim.1999.0250. [DOI] [PubMed] [Google Scholar]
- Evans D, Leary H, Jaso-Friedmann L. Nonspecific cytotoxic cells and innate immunity: regulation by programmed cell death. Dev Comp Immunol. 2001;25:791–805. doi: 10.1016/s0145-305x(01)00036-2. [DOI] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;2002:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves S, Evans D, Dawe D. Antiprotozoan activity of nonspecific cytotoxic cells (NCC) from the channel catfish (Ictalurus punctatus) J Immunol. 1985;134:78–85. [PubMed] [Google Scholar]
- Hawke N, Yoder J, Litman G. Expanding our understanding of immunoglobulin, T-cell antigen receptor, and novel immunetype receptor genes: a subset of the immunoglobulin gene superfamily. Immunogenetics. 1999;50:124–133. doi: 10.1007/s002510050588. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. Tmbase: a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hofmann K, Bucher P, Tschopp J. The CARD domain is a new apoptotic signaling motif. Trends Biochem Sci. 1997;22:15–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- Hogan R, Taylor W, Cuchens M, Naftel J, Clem LW, Miller NW, Chinchar VG. Induction of target cell apoptosis by channel catfish cytotoxic cells. Cell Immunol. 1999;195:110–118. doi: 10.1006/cimm.1999.1523. [DOI] [PubMed] [Google Scholar]
- Irmler M, Hofmann K, Vaux D, Tschopp J. Direct physical interactions between the Caenorhabditis elegans ‘death proteins’ CED-3 and CED-4. FEBS Lett. 1997;406:189–190. doi: 10.1016/s0014-5793(97)00271-8. [DOI] [PubMed] [Google Scholar]
- Jaso-Friedmann L, Leary J, Evans D. Role of nonspecific cytotoxic cells in the induction of programmed cell death of pathogenic protozoans: participation of the Fas ligand–Fas receptor system. Exp Parasitol. 2000;96:75–88. doi: 10.1006/expr.2000.4561. [DOI] [PubMed] [Google Scholar]
- Jones E, Stuart D, Walker N. Structure of tumor necrosis factor. Nature. 1989;338:225–228. doi: 10.1038/338225a0. [DOI] [PubMed] [Google Scholar]
- Kiener P, Davis P, Rankin B, Klebanoff S, Ledbetter J, Starling G, Liles WC. Human monocytic cells contain high levels of intracellular Fas ligand. J Immunol. 1997;159:1594–1598. [PubMed] [Google Scholar]
- Krammer P, Debatin K-M. A gene for a cell surface molecule known as Fas or APO-1 that can trigger apoptosis has been identified as the target of mutations that cause lymphoproliferation and autoimmunity in mice. Curr Biol. 1992;2:383–385. [Google Scholar]
- Kuhn K, Hashimoto S, Lotz M. Cell density modulates apoptosis in human articular chondrocytes. J Cell Physiol. 1999;180:439–447. doi: 10.1002/(SICI)1097-4652(199909)180:3<439::AID-JCP15>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges S. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen I, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during assembly of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laing K, Holland J, Bonilla S, Cunningham C, Secombes C. Cloning and sequencing of caspase 6 in rainbow trout (Oncorhynchus mykiss), and its expression under conditions known to induce apoptosis. Dev Comp Immunol. 2001;25:303–312. doi: 10.1016/s0145-305x(00)00061-6. [DOI] [PubMed] [Google Scholar]
- Locksley R, Kileen N, Lenardo M. The TNF and TNFR superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Mallet S, Barclay A. A new superfamily of cell surface proteins related to the nerve growth factor receptor. Immunol Today. 1991;12:220–223. doi: 10.1016/0167-5699(91)90033-P. [DOI] [PubMed] [Google Scholar]
- Masumoto J, Zhou W, Chen FF, Su F, Kuwada Y, Hidaka E, Katsuyama T, Sagara J, Taniguchi S, Ngo-Hazelett P, Postlethwait JH, Nunez G, Inohara N. Caspy, a zebrafish caspase, activated by ASC oligomerization is required for pharyngeal arch development. J Biol Chem. 2003;278:4268–4276. doi: 10.1074/jbc.M203944200. [DOI] [PubMed] [Google Scholar]
- McDonald E, III, Chui P, Martelli P, Dicker D, El-Deiry W. Death domain mutagenesis of KILLER/DR5 reveals residues critical for apoptotic signaling. J Biol Chem. 2001;276:14939–14945. doi: 10.1074/jbc.M100399200. [DOI] [PubMed] [Google Scholar]
- Miller NW, Chinchar VG, Clem LW. Development of leukocyte cell lines from the channel catfish (Ictalurus punctatus) J Tissue Cult methods. 1994a;16:1–7. [Google Scholar]
- Miller NW, Rycyzyn MA, Wilson MR, Warr GW, Naftel JP, Clem LW. Development and characterization of channel catfish long term B cell lines. J Immunol. 1994b;152:2180–2189. [PubMed] [Google Scholar]
- Nagata S, Golstein P. The Fas death factor. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- Nicholson D, Thornberry N. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Park H, Zhou H, Bengten E, Wilson M, Chinchar VG, Clem LW, Miller NW. Activation of channel catfish (Ictalurus punctatus) T cells involves NFAT-like transcription factors. Dev Comp Immunol. 2002;26:775–784. doi: 10.1016/s0145-305x(02)00037-x. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E. RGD and other recognition sites for integrins. Dev Biol. 1996;12:696–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- Rycyzyn M, Wilson M, Bengten E, Warr G, Clem LW, Miller NW. Mitogen and growth factor-induced activation of a STAT-like molecule in channel catfish lymphoid cells. Mol Immunol. 1998;35(2):127–136. doi: 10.1016/s0161-5890(98)00006-6. [DOI] [PubMed] [Google Scholar]
- Saeki K, You A, Kato M, Miyazonon K, Yazaki Y, Takaku F. Cell density-dependent apoptosis in HL-60 cells, which is mediated by an unknown soluble factor, is inhibited by transforming growth factor β1 and overexpression of Bcl-2. J Biol Chem. 1997;272:20003–20010. doi: 10.1074/jbc.272.32.20003. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. 3rd edn. Cold Spring Harbor Laboratory; New York (Plainview): 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- Shen L, Stuge TB, Zhou H, Khayat M, Barker KS, Quiniou S, Wilson M, Bengten E, Chinchar VG, Clem LW, Miller NW. Channel catfish cytotoxic cells: a mini-review. Dev Comp Immunol. 2002;26:141–149. doi: 10.1016/s0145-305x(01)00056-8. [DOI] [PubMed] [Google Scholar]
- Shen L, Stuge T, Bengten E, Wilson M, Chinchar VG, Naftel J, Bernanke J, Clem LW, Miller NW. Identification and characterization of clonal NK-like cells from channel catfish (Ictalurus punctatus) Dev Comp Immunol. 2004;28:139–152. doi: 10.1016/s0145-305x(03)00119-8. [DOI] [PubMed] [Google Scholar]
- Smith C, Farrah T, Goodwin R. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- Sprang S. The divergent receptors for TNF. Trends Biochem Sci. 1990;15:366–368. doi: 10.1016/0968-0004(90)90228-4. [DOI] [PubMed] [Google Scholar]
- Stuge TB, Wilson MR, Zhou H, Barker KS, Bengten E, Chinchar G, Miller NW, Clem LW. Development and analysis of various clonal alloantigen-dependent cytotoxic cell lines from channel catfish. J Immunol. 2000;164:2971–2977. doi: 10.4049/jimmunol.164.6.2971. [DOI] [PubMed] [Google Scholar]
- Taniguichi T. Regulation of cytokine gene expression. Annu Rev Immunol. 1988;6:439–464. doi: 10.1146/annurev.iy.06.040188.002255. [DOI] [PubMed] [Google Scholar]
- Tibbetts MD, Zheng L, Lenardo MJ. The death effector domain protein family: regulators of cellular homeostasis. Nat Immunol. 2003;4:404–409. doi: 10.1038/ni0503-404. [DOI] [PubMed] [Google Scholar]
- Wallach D, Varfolomeev E, Malinin N, Goltsev Y, Kovalenko A, Boldin M. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu Rev Immunol. 1999;17:331–367. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- Weber CH, Vincenz C. The death domain superfamily: a tale of two interfaces? Trends Biochem Sci. 2001;26:475–481. doi: 10.1016/s0968-0004(01)01905-3. [DOI] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Stuge T, Miller NW, Clem LW. Phylogeny of lymphocyte heterogeneity: cytotoxic activity of channel catfish peripheral blood leukocytes directed against allogeneic targets. Dev Comp Immunol. 1995;19:71–77. doi: 10.1016/0145-305x(94)00053-i. [DOI] [PubMed] [Google Scholar]
- Zhou H, Stuge T, Miller NW, Bengten E, Naftel J, Bernarke J, Chinchar VG, Clem LW, Wilson M. Heterogeneity of channel catfish CTL with respect to target recognition and cytotoxic mechanisms employed. J Immunol. 2001;167:1325–1332. doi: 10.4049/jimmunol.167.3.1325. [DOI] [PubMed] [Google Scholar]



