Abstract
Hepatitis C virus (HCV) sets up a persistent infection in patients that likely involves a complex virus-host interaction. We previously found that the HCV nonstructural 5A (NS5A) protein interacts with growth factor receptor-binding protein 2 (Grb2) adaptor protein and inhibits the activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) by epidermal growth factor (EGF). In the present study, we extended this analysis and investigated the specificity of the Grb2-NS5A interaction and whether the subversion of mitogenic signaling involves additional pathways. NS5A containing mutations within the C-terminal proline-rich motif neither bound Grb2 nor inhibited ERK1/2 activation by EGF, demonstrating that NS5A-Grb2 binding and downstream effects were due to direct interactions. Interestingly, NS5A could also form a complex with the Grb2-associated binder 1 (Gab1) protein in an EGF treatment-dependent manner. However, the NS5A-Gab1 association, which appeared indirect, was not mediated by direct NS5A-Grb2 interaction but was likely dependent on direct NS5A interaction with the p85 subunit of phosphatidylinositol 3-kinase (PI3K). The in vivo association of NS5A with p85 PI3K required the N-terminal, but not the C-terminal, region of NS5A. The downstream effects of the NS5A-p85 PI3K interaction included increased tyrosine phosphorylation of p85 PI3K in response to EGF. Consistent with this observation and the antiapoptotic properties of NS5A, we also detected enhanced tyrosine phosphorylation of the downstream AKT protein kinase and increased serine phosphorylation of BAD, a proapoptotic factor and an AKT substrate, in the presence of NS5A. These results collectively suggest a model in which NS5A interacts with Grb2 to inhibit mitogenic signaling while simultaneously promoting the PI3K-AKT cell survival pathway by interaction with p85 PI3K, which may represent a crucial step in HCV persistence and pathogenesis.
Hepatitis C virus (HCV), a Flaviviridae family member, contains a positive-sense, single-stranded RNA genome that encodes about 10 mature viral structural and nonstructural (NS) proteins (41). Infecting approximately 2% of the world population, HCV is the global leading cause of chronic liver disease and has become a major public health problem in the United States (10, 11). In the majority of cases, acute infection with HCV results in persistent viral replication and establishment of a chronic infection. Chronic hepatitis C frequently leads to progressive liver disease, including liver fibrosis and cirrhosis, and is strongly associated with the onset of hepatocellular carcinoma. HCV research has been hampered by the lack of an efficient tissue culture system or an adequate animal model of HCV infection (18). As a result, the mechanisms of HCV replication, persistence, and pathogenesis remain poorly understood. Consequently, our general understanding of the impact of HCV infection on cellular signaling is far from complete or clear.
HCV-host interactions have been intensely investigated despite the lack of a robust virus infection system. The literature has primarily focused on the interactions among the HCV core, the viral capsid structural protein, and the cellular signaling machinery (34). The HCV NS5A protein itself became the subject of intense investigation following the observation that amino acid substitutions within a region of NS5A, termed the interferon (IFN) sensitivity-determining region, were correlated with the IFN response of patients infected with HCV genotype 1b (15, 16). Although the exact molecular mechanism of IFN resistance mediated by the NS5A protein remains to be elucidated, our previous studies showed that the NS5A protein from IFN-resistant HCV strains can act as a potent inhibitor of IFN-induced, double-stranded RNA (dsRNA)-dependent protein kinase (PKR), a key mediator of the host IFN antiviral and antiproliferative response (17, 20, 21). It is noteworthy that the E2 envelope protein also interacts with and inhibits PKR (54), indicating that HCV may employ multiple strategies to perturb a major host cell antiviral function. NS5A can also confer IFN resistance on encephalomyocarditis virus and vesicular stomatitis virus, viruses normally sensitive to the antiviral actions of IFN (1, 19, 38, 40, 49), and it reverses the IFN-sensitive phenotype of a vaccinia virus (VV) lacking the E3L gene (25). NS5A provides resistance to apoptosis induced by PKR agonists, such as dsRNA, and can cause cell transformation and solid-tumor growth in vivo through both PKR-dependent and -independent mechanisms (19). In addition, NS5A has also been reported to modulate cell cycle regulatory genes and protect against tumor necrosis factor alpha-mediated apoptotic cell death (22, 23). However, the exact molecular mechanisms by which NS5A regulates cell survival and apoptosis await further characterization.
Recently, we found that HCV may utilize the viral NS5A protein to perturb host intracellular signaling pathways. Specifically, we demonstrated that NS5A directly interacts with the cellular adaptor protein growth factor receptor-binding protein-2 (Grb2) and inhibits activation of the extracellular signal-regulated kinase 1 and 2 (ERK1/2) mitogen-activated protein kinases (MAPK) by epidermal growth factor (EGF) (25, 53). However, the exact mechanism(s) by which NS5A interaction with Grb2 subverts the downstream MAPK pathway remains unclear. In the present report, we describe the nature of the NS5A-Grb2 interaction and provide evidence that NS5A associates with the insulin receptor substrate family signaling complex Grb2-associated binder 1 (Gab1) and directly interacts with the p85 subunit of phosphatidylinositol 3-kinase (PI3K). Finally, we describe the downstream effects of the NS5A-p85 PI3K interaction and present a model depicting the multifaceted molecular mechanism of NS5A action upon the cellular signaling machinery.
MATERIALS AND METHODS
Antibodies and immunoblot analysis.
NS5A-specific monoclonal antibody was purchased from ID Labs. Monoclonal antibody specific to HCV NS4A was purchased from Maine Biotechnology Services. Grb2-specific monoclonal antibody was purchased from Transduction Laboratories. Antibodies specific to Gab1, phospholipase C-γ (PLC-γ), and glutathione S-transferase (GST) were purchased from Santa Cruz Biotechnology. Antibodies specific to the p85 subunit of PI3K and phosphotyrosine (4G10) were purchased from Upstate Biotechnology. Antibodies specific to AKT, phospho-AKT (Ser473), the phosphorylated forms of Erk1/2 (Thr202/Tyr 204), total Erk1/2, phospho-Bad (Ser136), and Bad were purchased from Cell Signaling Technology. Human antiserum from HCV-positive patients for the detection of NS5A was described previously (39). Secondary antibodies conjugated to horseradish peroxidase were purchased from Jackson ImmunoResearch. The immunoblot analysis was performed as previously described (25, 53). The relative levels of protein phosphorylation shown in Fig. 1 and 6 were determined by quantifying the immunoblots with ImageQuant (version 5.1). The signals from the phosphospecific immunoblots were normalized against their individual control signals, and the ratio of phosphospecific signal to control signal was determined as described previously (25).
FIG. 1.
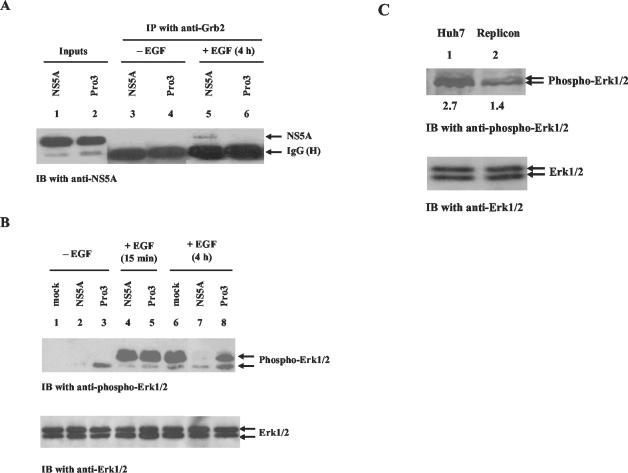
The C-terminal proline-rich, SH3 domain-binding motif of NS5A is required for NS5A-Grb2 interaction and inhibition of ERK1/2 MAPK activation. (A) Tet-Off HeLa cells were transiently transfected with either pTRE-NS5A-wild type (NS5A) or pTRE-NS5A-Pro3 (Pro3). At 24 h posttransfection, the cells were either treated with EGF (lanes 5 and 6) or left untreated (lanes 3 and 4). At 4 h post-EGF treatment, cell lysates were collected and coimmunoprecipitation assays were performed with anti-Grb2 antibody. The immunocomplexes were resolved by SDS-12% PAGE and transferred to a nitrocellulose membrane. IgG (H) denotes the heavy chain of mouse immunoglobulin G. Lanes 1 and 2 show total lysates from cells transfected with either pTRE-NS5A-wild type (NS5A) or pTRE-NS5A-Pro3 (Pro3), respectively, probed with anti-NS5A antibody to show NS5A expression levels. (B) Cell lysates from the experiment described above were resolved by SDS-PAGE and subjected to immunoblot (IB) analysis with antibody specific to the dually phosphorylated, activated forms of Erk1/2 (top). The same blot was stripped and probed with anti-Erk1/2 antibody to show total Erk1/2 protein levels (bottom). (C) Equal amounts (30 μg) of protein from actively proliferating (∼70% confluent) Huh7 and HCV replicon cells were resolved by SDS-PAGE, transferred to nitrocellulose membrane, and probed with antibody specific to phosphorylated Erk1/2 MAPKs (top). The same membrane was stripped and reprobed with antibody specific to Erk1/2 MAPKs (bottom). The values under the antiphospho-Erk1/2 blot are the relative levels of Erk1/2 phosphorylation. IP, immunoprecipitation.
FIG. 6.
NS5A enhances activation of the PI3K-AKT pathway and modulates the downstream apoptosis pathway. (A) Tet-Off HeLa cells stably expressing wild-type NS5A (lanes 1 to 4) or not expressing NS5A (lanes 5 to 8) were either treated with EGF (lanes 2 to 4 and 6 to 8) or left untreated (lanes 1 and 5). Cell lysates were collected at different time points post-EGF treatment, as indicated. Coimmunoprecipitation assays were performed with antibody specific to p85 PI3K. The immunocomplexes were resolved by SDS-12%c PAGE; this was followed by immunoblot (IB) analysis with phosphotyrosine-specific antibody (top). The same membrane was stripped and probed with p85-specific antibody to show total p85 protein levels (bottom). The values under the antiphosphotyrosine blot are the relative levels of p85 PI3K phosphorylation. (B) Tet-Off HeLa cells stably expressing wild-type NS5A (lanes 1 and 2) or not expressing NS5A (lanes 3 and 4) were either treated with EGF (lanes 2 to 4) or left untreated (lanes 1 and 3), and cell lysates were collected at 30 min after EGF treatment. The cell lysates were resolved by SDS-12% PAGE; this was followed by immunoblot analysis with antibody specific to the phosphorylated, activated form of AKT (top). The same blot was stripped and probed with anti-AKT antibody to show AKT levels (bottom). The values under the antiphospho-AKT blot are the relative levels of AKT phosphorylation. (C) HeLa S3 cells were infected with either a recombinant VV carrying the NS5A-encoding gene (vpNS5A) (lanes 4 to 6) or the control VV (vp1080) (lanes 1 to 3). Cell lysates were collected at 2, 4, and 6 h postinfection (p.i.) and resolved by SDS-12% PAGE (12%); this was followed by immunoblot analysis with antibody specific to the phosphorylated form of AKT (top). The same blot was stripped and probed with anti-AKT antibody to show total AKT protein levels (bottom). The values under the anti-phospho-AKT blot are the relative levels of AKT phosphorylation. (D) Stable Tet-Off HeLa cell lines carrying either full-length NS5A-NR (lanes 1 to 4) or N-terminal deletion mutant NS5A (lanes 5 to 8) were either induced to express NS5A (lanes 3, 4, 7, and 8) or not induced (lanes 1, 2, 5, and 6). At 24 h after induction, the cells were either treated with EGF for 30 min (lanes 2, 4, 6, and 8) or left untreated (lanes 1, 3, 5, and 7). Cell lysates were collected and subjected to immunoblot analysis with anti-phospho-AKT antibody (top). The same blot was stripped and probed with anti-AKT antibody to show AKT levels (bottom). The values under the antiphospho-AKT blot are the relative levels of AKT phosphorylation. (E) Tet-Off HeLa cells stably expressing wild-type NS5A (lanes 3 and 4) or not expressing NS5A (lanes 1 and 2) were either treated with EGF (lanes 2 to 4) or left untreated (lanes 1 and 3). Cell lysates were collected 30 min after EGF treatment and resolved by SDS-12% PAGE; this was followed by immunoblot analysis with antibody specific to the phosphorylated form of BAD (top). The same blot was stripped and reprobed with anti-BAD antibody to show BAD levels (bottom). The values under the antiphospho-BAD blot are the relative levels of BAD phosphorylation. (F) HeLa S3 cells were infected with either a recombinant VV carrying the NS5A-encoding gene (vpNS5A) (lanes 4 to 6) or the control VV (vp1080) (lanes 1 to 3). Cell lysates were collected at 2, 4, and 6 h postinfection and resolved by SDS-12% PAGE (12%); this was followed by immunoblot analysis with antibody specific to the phosphorylated form of BAD (top). The same blot was stripped and probed with anti-BAD antibody to show total BAD protein levels (bottom). The values under the antiphospho-BAD blot are the relative levels of BAD phosphorylation. IP, immunoprecipitation; ND, not detectable.
Plasmids and GST fusion proteins.
All molecular cloning techniques were performed as previously described (44). To generate the pTRE-NS5A-1b-wild type, pTRE-NS5A-Pro3, and pTRE-NS5A-1b5 constructs, the entire NS5A coding sequences, fused with the FLAG epitope tag at the N terminus from pFlag-NS5A-1b-wild type, pFlag-NS5A-Pro3, and pFlag-NS5A-1b5 (21), were placed into the XbaI site of plasmid pTRE (Clontech) (53). pTRE-FL-NS5A-NR, pTRE-NS5A-ΔN110, and pTRE-NS5A-ΔC117 were previously described (39). GST, GST-NS5A, and GST-Grb2 were previously described (21, 53). The GST-Gab1 construct was kindly provided by A. J. Wong (Thomas Jefferson University). The GST-p85α construct was kindly provided by B. Vanhaesebroeck (Ludwig Institute for Cancer Research). GST-p85 C-SH2 domain and GST-p85 N-SH2 domain were purchased from Upstate Biotechnology. GST-PLC-γ1 was purchased from Santa Cruz Biotechnology.
Purification of recombinant NS5A protein.
Isolation of recombinant baculovirus DNA containing the full-length NS5A-encoding gene under the transcriptional control of the polyhedrin promoter, transfection of Sf9 cells (Invitrogen), and infection with recombinant baculovirus were performed as previously described (53). After 4 days of infection, cells were collected and protein was purified by conventional chromatographic methods as previously described (53).
Cell lines and tissue culture.
The Tet-Off Gene Expression System and Tet-Off HeLa cells (Clontech) were used to establish the tetracycline-regulated stable NS5A-1b-wild-type-expressing cell lines (53). The stable Tet-Off HeLa cell lines expressing full-length NS5A-NR and NS5A-ΔN222 were previously described (39). Maintenance of stable Tet-Off HeLa cells, induction of NS5A expression, and EGF treatment (serum starvation overnight, followed by EGF treatment at 20 ng/ml for 3 min) were done as previously described (25, 53). Cell lysates were prepared before and at different time points after EGF stimulation as previously described (25, 53). Transient transfection of pTet-Off HeLa cells was performed with SuperFect (Qiagen) in accordance with the manufacturer's instructions. Specifically, 106 cells in 60-mm-diameter tissue culture dishes were transfected with 3 μg of plasmid DNA. At 24 h posttransfection, the cells were treated with EGF and cell lysates were prepared as described above. Huh7 human hepatoma cells were cultured in DMEM supplemented with 2 mM L-glutamine, nonessential amino acids, penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% fetal calf serum at 37°C in the presence of 5% CO2. The HCV replicon cell lines were constructed and maintained as previously described (2, 33).
Construction of NS5A-expressing recombinant VV and virus infection.
In vivo recombination and selection of recombinant VV were performed as previously described (25, 53). HeLa S3 cells (ATCC CCL-2.2) were maintained in DMEM supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM L-glutamine at 37°C in 5% CO2. Cell monolayers (70% confluent) were infected with control recombinant VV (vp1080) or recombinant VV expressing NS5A-1b-wild type (vpNS5A) as previously described (25, 53). At 2, 4, and 6 h postinfection, cell lysates were prepared as previously described (25, 53).
GST pulldown analysis.
GST fusion proteins were purified and coprecipitation assays were performed as previously described (53). Purified GST-Grb2 proteins were cleaved with bovine thrombin (20 cU/ml; Amersham Pharmacia Biotech) at room temperature for 16 h, and 0.1 μg of the cleaved Grb2 was used for GST pulldown analysis. Purified recombinant NS5A (0.1 μg) or lysates (500 μg) from NS5A-expressing cells were incubated with 1.0 μg of each GST fusion protein bound to glutathione-agarose beads for 1 h at 4°C on a rotator. The glutathione-agarose resin was pelleted at 14,000 × g for 2 min and washed three times with ice-cold 1× phosphate-buffered saline. Precipitated proteins were suspended in 2× sodium dodecyl sulfate (SDS) protein-loading buffer (44), boiled for 5 min, and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (12% gel); this was followed by electrotransfer and immunoblot analysis done as previously described (25, 53).
Coimmunoprecipitation analysis.
Cell lysates (500 μg) used for coimmunoprecipitation analysis were first diluted in low-salt lysis buffer (25, 53), and the lysate mixtures (500 μl) were precleared by incubation with 50 μl of 50% protein A agarose or protein G agarose (Boehringer Mannheim) for 2 h at 4°C. After centrifugation of the mixture at 14,000 × g for 5 min, the supernatant was recovered and incubated with 2 μg of each primary antibody at 4°C for 1 h. Protein A agarose or protein G agarose (50 μl of a 50% suspension) was then added, and the mixture was incubated for an additional 2 h at 4°C. All incubations were performed on a rotator to ensure proper mixing. The immune complexes were then pelleted at 14,000 × g for 2 min, washed three times with ice-cold 1× phosphate-buffered saline, diluted in 2× SDS protein loading buffer, and boiled for 5 min. Immunoprecipitation products were subjected to SDS-PAGE (12% gel); this was followed by electrotransfer and immunoblot analysis as described above.
RESULTS
Because of the lack of an HCV infection system, we utilized four different surrogate systems to gain insights into the interaction between NS5A and cell signaling pathways: stable or transiently transfected Tet-Off cell lines expressing HCV NS5A, a recombinant, E3L-deficient VV expressing NS5A, and the recently developed HCV replicon cells expressing HCV NS proteins (2, 33). The Tet-Off cell line system allows the characterization of NS5A function in a tightly controlled, inducible manner. The recombinant VV system provides the context of viral infection, at least activation of the dsRNA-dependent antiviral pathway (25), which is likely important for studying the function of a viral protein. Finally, the HCV replicon system, consisting of human liver cells replicating an HCV subgenomic RNA construct and expressing several HCV NS proteins (NS3, NS4A, NS4B, NS5A, and NS5B), is considered the “gold standard” of HCV model systems and enables the examination of NS5A behavior in the presence of viral genome replication and other HCV proteins.
A mutant form of NS5A, NS5A-Pro3, is unable to interact with Grb2 and inhibit EGF-induced ERK MAPK activation.
Grb2 is an adaptor protein that mediates signaling by nucleating the formation of signal transduction complexes in response to growth factor stimulation (14, 46). We previously showed that NS5A binds Grb2 in an Src homology 3 (SH3) domain/ligand-dependent manner and inhibits ERK1/2 MAPK activation by EGF (53). As an extension of these observations, we determined whether mutations within the C-terminal proline-rich SH3 domain-binding motif affects the ability of NS5A to inhibit EGF-dependent ERK1/2 activation. Consistent with previous findings (53), transient expression of NS5A in Tet-Off HeLa cells and in vivo coimmunoprecipitation analysis revealed that NS5A and Grb2 interaction was dependent on EGF stimulation of the cells (Fig. 1A). NS5A-Pro3, which carries three proline→alanine point mutations within the proline-rich motif, did not interact with Grb2, indicating that the proline-rich motif of NS5A is essential for its interaction with Grb2 (Fig. 1A). This finding was further supported by GST pulldown experiments with a GST-Grb2 fusion protein and lysates from HeLa cells transfected with either wild-type NS5A or NS5A-Pro3 constructs (data not shown). Importantly, the ability of NS5A-Pro3 to inhibit EGF-dependent ERK activation was severely abrogated while wild-type NS5A inhibited Erk1/2 phosphorylation in a transient manner, as shown in our previous studies (25, 53) (Fig. 1B). These results support the notion that NS5A binds Grb2 via an SH3 domain/ligand-dependent manner and demonstrate the requirement and specificity of the interaction for mediating downstream effects on cellular signaling. NS5A-Pro3 also provides an effective approach by which to test whether any effect of NS5A is dependent on Grb2 interaction.
In order to examine the physiological relevance of these results, we compared the level of ERK1/2 MAPK activation in Huh7 human liver cells with its level of activation in HCV replicon cells, which are Huh7-derived human liver cells that carry an HCV subgenomic construct and express several HCV NS proteins, including NS5A (2, 33). In this analysis, we found that activation of the Erk1/2 MAPKs was inhibited by approximately twofold in actively proliferating HCV replicon cells, compared with that in the parental Huh7 cells (Fig. 1C). This finding suggests that NS5A is able to perturb the MAPK pathway in the context of other HCV NS proteins and in the presence of viral genome replication. However, these factors may also exert their own effects on cell signaling pathways and may regulate or counteract the actions of NS5A. This may partially explain the fact that inhibition of the MAPK pathway in the replicon cells was not as dramatic as that observed in Tet-Off cells expressing NS5A only. In addition, the Tet-Off and replicon cells are of different types and may express different levels of NS5A protein.
NS5A indirectly associates with the Gab1 signaling complex in a Grb2-independent manner.
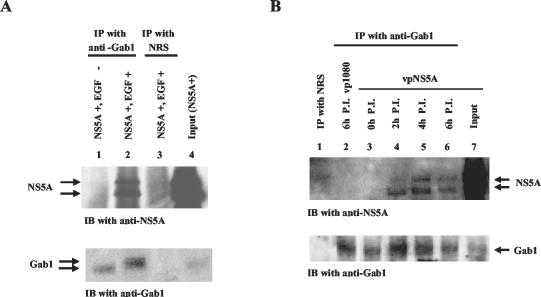
Members of the insulin receptor substrate family of proteins, of which Gab1 is one, play pivotal roles in cell signaling by associating with different growth factor, cytokine, and antigen receptors and by providing docking sites for components of multiple downstream pathways (26, 56). Since Gab1 also associates with the EGF receptor, either via the adaptor protein Grb2 or directly, and since it contributes to activation of the MAPK pathway by providing binding sites for the SH2 domain of Grb2 (32, 45), we tested whether NS5A could associate with the Gab1 signaling complex. Coimmunoprecipitation experiments were performed with anti-Gab1 antibody on lysates from untreated or EGF-treated, wild-type NS5A-expressing HeLa cells. NS5A was detected in the same immunocomplex as Gab1 only after EGF treatment, indicating that this association is regulated by cell signaling events (Fig. 2A), similar to the association between NS5A and Grb2 (53). An NS5A-Gab1 complex was not detectable when a parallel coimmunoprecipitation was performed with a normal-serum control (Fig. 2A). To characterize the function of NS5A in the setting of a virus infection, we examined the association of NS5A with the Gab1 signaling complex in HeLa S3 cells infected with recombinant VV expressing NS5A protein (Fig. 2B). The VV system offers an additional advantage in that VV encodes an EGF homologue that can mimic EGF-induced tyrosine protein phosphorylation (4, 42). Coimmunoprecipitation analyses were carried out with lysates from HeLa S3 cells infected with a recombinant VV expressing NS5A (vpNS5A) or a recombinant VV control (vp1080) (Fig. 2B). Infection of HeLa S3 cells with vpNS5A, but not infection with vp1080, produced an NS5A-Gab1 complex, as revealed by coprecipitation of NS5A by anti-Gab1 antibody. When a parallel experiment was performed with a normal-serum control, neither NS5A nor Gab1 was detectable. Collectively, these results suggest that NS5A associates with the Gab1 multiprotein signaling complex in the context of both viral infection and EGF-induced cellular signaling.
FIG. 2.
NS5A associates with the Gab1 signaling complex. (A) Lysates from wild-type NS5A-expressing Tet-Off HeLa cells treated with EGF (lanes 2 and 3) or left untreated (lane 1) were immunoprecipitated with either anti-Gab1 antibody (lanes 1 and 2) or a normal rabbit serum (NRS) control (lane 3). The immunocomplexes were resolved by SDS-12% PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot (IB) analysis with anti-NS5A antibody (top). Lane 4 shows NS5A expression in total cell lysates. The same membrane was stripped and probed with anti-Gab1 antibody (bottom) to show Gab1 protein levels. (B) HeLa S3 cells were infected with either a recombinant VV carrying the NS5A gene (vpNS5A) or the control VV (vp1080). Cell lysates were collected at 2, 4, and 6 h postinfection (P.I.) and used for coimmunoprecipitation assays with either anti-Gab1 antibody (lanes 2 to 6) or NRS (lane 1); this was followed by immunoblot analysis as described above. Lane 7 shows expression of NS5A (top) and Gab1 (bottom) in total lysates from vpNS5A-infected cells at 6 h postinfection. IP, immunoprecipitation.
To test whether NS5A associates with Gab1 directly or through a bridging molecule, such as Grb2, we used recombinant GST-Gab1 as an affinity matrix to examine whether NS5A and Gab1 could form a complex in cell lysates prepared from wild-type NS5A-expressing Tet-Off HeLa cells. Indeed, we were able to demonstrate the retention of NS5A on glutathione-agarose beads with GST-Gab1, but not with GST or buffer alone (Fig. 3A). Also, the interaction of NS5A with GST-Gab1 was dependent on EGF treatment, further indicating that the interaction is regulated by cellular signaling events. We next examined whether purified NS5A could bind to purified GST-Gab1 in the absence of other proteins (Fig. 3B). While NS5A was efficiently coprecipitated with GST-Grb2, no direct interaction between NS5A and GST-Gab1 was detected. However, when cell lysates prepared from actively proliferating HeLa cells were added to the reaction mixture, NS5A was coprecipitated with GST-Gab1. These results suggest that the association between Gab1 and NS5A is mediated by an additional cellular factor(s) and is thus indirect.
FIG. 3.
The NS5A-Gab1 association is indirect and independent of NS5A-Grb2 interaction. (A) Tet-Off HeLa cells stably expressing wild-type NS5A (lanes 1, 2, 4, and 5) or not expressing NS5A (lane 3) were either treated with EGF (lanes 1, 3, and 5) or left untreated (lanes 2 and 4), and cell lysates were collected at 4 h post-EGF treatment. GST pulldown assays were performed with either GST-Gab1 (lanes 1, 2, and 3) or a GST control (lanes 4 and 5). The pulldown products were resolved by SDS-12% PAGE and transferred to nitrocellulose membrane; this was followed by immunoblot (IB) analysis with NS5A-specific antibody. Lane 6 shows NS5A expression in total lysates from NS5A-expressing cells. (B) Recombinant NS5A protein purified from insect cells was used for GST pulldown assays with either GST-Gab1 (lanes 3 to 6) or GST-Grb2 (lane 2) as described above. Lysates from actively proliferating HeLa S3 cells were added in increasing amounts (10, 30, and 50 μg) to reconstitute the NS5A-Gab1 association (lanes 4 to 6). Lane 1 shows purified NS5A protein immunoblotted with NS5A-specific antibody. (C) Recombinant NS5A protein purified from insect cells was used for GST pulldown assays with either GST-Gab1 (lanes 3 and 4) or GST-Grb2 (lane 2) as described above. Recombinant Grb2 protein purified from Escherichia coli was added to the reaction mixture in lane 4. Lane 1 shows purified NS5A protein immunoblotted with NS5A-specific antibody. (D) Tet-Off HeLa cells were transiently transfected with the pTRE vector (vector lanes), pTRE-NS5A-wild type (NS5A lanes), or pTRE-NS5A-Pro3 (Pro3 lanes). At 24 h after transfection, the cells were either treated with EGF for an additional 4 h or left untreated. Cell lysates were then used for coimmunoprecipitation analysis with anti-Gab1 antibody or a normal rabbit serum (NRS) control. The immunocomplexes were then resolved by SDS-12% PAGE; this was followed by Western transfer and immunoblot analysis with anti-NS5A antibody. Lanes 1 to 3 show total lysates of cells transfected with pTRE, pTRE-NS5A-wild type, or pTRE-NS5A-Pro3 immunoblotted with anti-NS5A antibody. IP, immunoprecipitation.
We specifically tested if Grb2 could mediate the Gab1-NS5A association by using the in vitro GST pulldown system (Fig. 3C). As described above, GST-Gab1 was unable to interact with purified NS5A directly whereas GST-Grb2 efficiently interacted with NS5A in the same assay. Even when recombinant Grb2, purified from the cleavage product of GST-Grb2 fusion protein, was added to the system, no GST-Gab1-NS5A association was detected. The purity and amount of the recombinant Grb2 protein were confirmed by immunoblot analysis (data not shown). This indicates that Grb2 is unable to mediate Gab1-NS5A association, at least under the conditions used in this system. In order to further address the question of whether NS5A-Gab1 association is bridged by the Grb2 adaptor molecule, we tested the ability of wild-type NS5A and NS5A-Pro3, the mutant that does not interact with Grb2, to associate with the Gab1 signaling complex. The wild-type and mutant NS5A proteins were expressed in Tet-Off HeLa cells by transient transfection. Coimmunoprecipitation experiments were performed with lysates from HeLa cells expressing either wild-type NS5A or NS5A-Pro3 with a Gab1-specific antibody. Both the wild-type and mutant NS5A proteins coimmunoprecipitated with Gab1 in an EGF treatment-dependent manner (Fig. 3D). The same cell lysates were also used for GST pulldown assays with GST-Gab1 or the GST control, and similar results were obtained (data not shown). Combined, these results suggest that NS5A-Gab1 association is not dependent on direct interaction of NS5A with Grb2. Rather, the association between NS5A and Gab1 is likely bridged by additional signaling molecules in the Gab1 signaling complex, such as PI3K and PLC-γ, which possess SH3 domains (Fig. 4A) that can potentially interact with the other proline-rich motifs of NS5A (53).
FIG. 4.
NS5A interacts with the p85 subunit of PI3K. (A) Organization of the functional domains of p85 PI3K and PLC-γ. (B) Recombinant NS5A protein purified from insect cells was used for GST pulldown assays with GST-Grb2 (lane 2), GST-Gab1 (lane 3), GST-p85 PI3K (lane 4), GST-C-terminal SH2 domain of p85 (lane 5), GST-N-terminal SH2 domain of p85 (lane 6), or GST-PLC-γ1 (lane 7). The precipitates were resolved by SDS-12% PAGE; this was followed by Western transfer and immunoblot (IB) analysis with anti-NS5A antibody. Lane 1 shows purified NS5A protein immunoblotted with NS5A-specific antibody. The same blot was stripped and probed with anti-GST antibody to show GST fusion protein levels (bottom). The values to the left are molecular masses in kilodaltons. (C) Tet-Off HeLa cells either expressing (lanes 3 to 6) or not expressing wild-type NS5A (lanes 1 and 2) were either treated with EGF (lanes 1 to 3, 5, and 6) or left untreated (lane 4), and cell lysates were collected at different time points post-EGF treatment as indicated. Coimmunoprecipitation assays were performed with p85 PI3K-specific antibody (lanes 1, 2, and 4 to 6) or a normal rabbit serum (NRS) control (lane 3); this was followed by SDS-12% PAGE and immunoblot analysis with NS5A-specific antibody (top). The same blot was stripped and probed with anti-p85 antibody to show p85 protein levels (bottom). (D) HeLa S3 cells were infected with either vpNS5A (lanes 3 to 7) or vp1080 (lanes 1 and 2). Cell lysates were collected at different time points postinfection (P.I.), as indicated, and used for coimmunoprecipitation assays with p85-specific antibody (lanes 1 to 6) or an NRS control (lane 7); this was followed by SDS-12% PAGE and immunoblot analysis with anti-NS5A antibody (top). The same blot was stripped and probed with anti-p85 antibody (bottom). Lane 8 shows NS5A (top) and p85 PI3K (bottom) expression in total lysates from vpNS5A-infected cells. (E and F) HCV replicon cells were treated with EGF for 1 h (lanes 1, 2, and 4) or left untreated (lane 3), and cell lysates were subjected to coimmunoprecipitation analysis with p85 PI3K-specific antibody (lanes 3 and 4) or an NRS control (lane 2). The immunocomplexes were resolved by SDS-12% PAGE; this was followed by immunoblot analysis with anti-NS5A (E) or anti-NS4 (F) antibody. Lane 1 shows NS5A expression in total lysates of HCV replicon cells. IP, immunoprecipitation.
NS5A directly interacts with the p85 subunit of PI3K, which, in turn, mediates the NS5A-Gab1 association.
Primed by the observation that the NS5A-Gab1 association is not mediated by the Grb2 adaptor molecule, we tested whether NS5A is able to interact with other signaling molecules involved in the Gab1 complex, specifically, the SH3 domain-containing p85 subunit of PI3K and PLC-γ. A GST pulldown analysis was performed with purified NS5A protein and a panel of different purified GST fusion proteins (Fig. 4B). GST-Grb2 did, and GST-Gab1 did not, precipitate NS5A as previously shown. Significantly, the full-length GST-p85 protein, but not the two different SH2 domain fragments of p85 (p85 C-SH2 and p85 N-SH2) (Fig. 4A), interacted with NS5A. In addition, GST-PLC-γ, another candidate NS5A-interacting protein, did not interact with NS5A in the same experiments. These results demonstrated that NS5A is able to directly and specifically interact with the p85 subunit of PI3K. This interaction did not seem to be mediated by the SH2 domains of p85 and thus likely involved other regions of p85, especially the SH3 domain.
In order to test the in vivo interaction of NS5A with p85 PI3K, lysates from untreated or EGF-treated wild-type NS5A-expressing Tet-Off HeLa cells were used for coimmunoprecipitation assays with an antibody specific for the p85 subunit of PI3K, followed by immunoblot assays to detect the presence of NS5A and p85 in the immunocomplex. NS5A associated with p85 PI3K in EGF-treated cells but not in untreated cells (Fig. 4C). The association of NS5A with p85 PI3K was also demonstrated in the recombinant VV infection system by coimmunoprecipitation assays. NS5A was found to associate with p85 PI3K in vpNS5A-infected HeLa cells but not in cells infected with the control virus (vp1080) (Fig. 4D). These results showed that NS5A could interact with the p85 subunit of PI3K in vivo during both EGF-induced cell signaling and viral infection, alluding to a mechanism by which NS5A associates with the Gab1 signaling complex. In addition, coimmunoprecipitation analysis showed that both wild-type NS5A and NS5A-Pro3 interacted with p85 PI3K in HeLa cells treated with EGF (data not shown), suggesting that NS5A interacts with p85 PI3K in a Grb2 interaction-independent manner. Thus, NS5A interacts with p85 PI3K and Grb2 through different mechanisms.
Next, we tested the NS5A-p85 PI3K interaction in the HCV replicon system in order to better determine the physiological relevance of this observation. Lysates of untreated or EGF-treated HCV replicon cells were used for a coimmunoprecipitation analysis with antibody specific to the p85 subunit of PI3K (Fig. 4E). Significantly, the NS5A-p85 interaction was also demonstrated in the HCV replicon cells, in an EGF treatment-inducible manner. These results strongly support the physiological relevance of the NS5A-p85 interaction. Under the same conditions, p85 was not found to associate with NS4A, another HCV NS protein present in the replicon cells (Fig. 4F), further demonstrating the specificity of the NS5A-p85 PI3K interaction.
In order to determine the NS5A regions involved in the interaction with p85 PI3K, the p85 PI3K-interacting ability of wild-type NS5A and different N- or C-terminal deletion mutant forms of NS5A was examined next (Fig. 5A). A genotype 1b, full-length NS5A isolate, wild-type NS5A-NR, and NS5A-NR-derived deletion mutants lacking either the N-terminal 110 amino acids (ΔN110) or the C-terminal 117 amino acids (ΔC117) (39) were separately expressed in Tet-Off HeLa cells by transient transfection. After EGF treatment, lysates of these cells expressing the different NS5A proteins were subjected to coimmunoprecipitation analysis with anti-p85 PI3K antibody as described above. Interestingly, both full-length NS5A and the C-terminal deletion mutant form of NS5A were able to interact with p85 PI3K. On the other hand, the mutant NS5A lacking the N-terminal region was unable to associate with p85. The expression of these different forms of NS5A was confirmed by immunoblot analysis, and similar expression levels were observed (data not shown). These results suggest that the NS5A-PI3K interaction involves the N-terminal, but not the C-terminal, region of NS5A. It is likely that the NS5A-PI3K interaction is mediated by the other highly conserved, proline-rich, SH3-binding motif within the N-terminal region of NS5A (53). In addition, the association of these wild-type and mutant forms of NS5A with Gab1 was also tested by performing coimmunoprecipitation analysis with anti-Gab1 antibody as described above (Fig. 5B). Consistent with the above results and our hypothesis that NS5A associates with the Gab1 complex through direct NS5A-p85 PI3K interaction, both the full-length and the C-terminal deletion forms, but not the N-terminal deletion form, of NS5A associated with Gab1 in EGF-treated cells. These results strongly suggest that the NS5A-Gab1 association requires direct NS5A-p85 PI3K interaction, although the possibility that other, unknown, proteins mediate the NS5A-Gab1 association cannot be completely excluded.
FIG. 5.
The NS5A-Gab1 association requires NS5A-p85 PI3K interaction. Tet-Off HeLa cells were transiently transfected with pTRE-NS5A-NR (full length) (lane 1), pTRE-NS5A-ΔN110 (lane 2), or pTRE-NS5A-ΔC117 (lane 3), and at 24 h posttransfection, the cells were treated with EGF for 1 h. Cell lysates were then collected and used for coimmunoprecipitation analysis with either anti-p85 PI3K antibody (A) or anti-Gab1 antibody (B); this was followed by immunoblot (IB) analysis with human serum from HCV patients to detect the presence of NS5A proteins. IgG(H) denotes the heavy chain of human immunoglobulin G. wt, wild type; IP, immunoprecipitation.
NS5A enhances activation of the PI3K-Akt pathway and regulates the downstream apoptosis pathway.
Upon stimulation with growth factors such as EGF, the multisite docking protein Gab1 is phosphorylated at multiple tyrosine residues, which could serve as docking sites for SH2 domain-containing signaling molecules, including previously identified Gab1-binding proteins p85 PI3K, PLC-γ, Shc, and Shp2 (26). Knowing that NS5A directly interacts with p85 PI3K and associates with the Gab1 complex, we next investigated whether NS5A expression affects the activation of any Gab1-mediated signaling pathway, in particular, the PI3K pathway, which plays a pivotal role in regulating cell growth and survival (5, 29) and is a popular target of viral modulation of host cell signaling (9, 13). Lysates from the stable wild-type NS5A-expressing Tet-Off HeLa cell line were subjected to coimmunoprecipitation analysis by using anti-p85 PI3K antibody and immunoblot analysis with antiphosphotyrosine antibody. Following EGF stimulation, cells expressing NS5A displayed enhanced tyrosine phosphorylation of p85 PI3K (Fig. 6A) but not PLC-γ (data not shown), indicating sustained activation of PI3K by NS5A. On average, NS5A expression caused an approximately twofold increase in PI3K tyrosine phosphorylation at early time points after EGF treatment (Fig. 6A). No significant effect of NS5A on Gab1 tyrosine phosphorylation was observed (data not shown), consistent with the model in which NS5A interacts directly with p85 PI3K and associates with the Gab1 complex indirectly.
The AKT serine/threonine protein kinase is activated by phosphatidylinositol 3,4-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate, which are lipid products of PI3K, and AKT acts as an important signal mediator downstream of PI3K, regulating cell survival and proliferation (6, 27). We therefore tested the effect of NS5A expression on AKT pathway activation. For these studies, Tet-Off HeLa cells expressing NS5A were treated with EGF and immunoblot assays with antibody specific to the phosphorylated/activated forms of AKT kinase (Fig. 6B) were performed as described previously (30, 48, 52). Consistent with the observation that NS5A enhances p85 PI3K phosphorylation, NS5A expression also enhanced AKT phosphorylation by about threefold in HeLa cells treated with EGF for 30 min. In addition, NS5A expression enhanced AKT phosphorylation/activation in HeLa S3 cells infected with recombinant VV expressing NS5A, compared with cells infected with the control VV (Fig. 6C). On average, in recombinant VV-infected cells, NS5A expression resulted in an approximately 2.5-fold increase in the AKT phosphorylation level. More importantly, NS5A expression did not cause a significant change in the level of p85 PI3K or AKT protein. Collectively, these results show that NS5A expression can enhance the PI3K-AKT pathway in both cellular and viral systems.
In order to test whether the effect of NS5A on the PI3K-AKT pathway requires NS5A-p85 PI3K interaction, AKT phosphorylation levels were compared in two stable Tet-Off HeLa cell lines expressing either full-length, wild-type NS5A-NR or an N-terminal deletion mutant form of NS5A (ΔN222) that does not interact with p85 PI3K. We found that expression of full-length NS5A-NR, which is capable of interacting with p85 PI3K, also enhanced AKT phosphorylation, showing that the effect of NS5A on the PI3K-AKT pathway is not limited to a particular NS5A isolate (Fig. 6D, lanes 3 and 4). On the other hand, in cells expressing the N-terminal deletion form of NS5A, which lost its ability to interact with p85 PI3K, the ability to enhance the PI3K-AKT pathway was also lost (Fig. 6D, lanes 7 and 8). These results suggest that the enhancement of the PI3K-AKT pathway by NS5A requires its interaction with p85 PI3K. In addition, by comparing HeLa cells infected with recombinant VV expressing either wild-type NS5A or NS5A-Pro3, we found that both wild-type NS5A and NS5A-Pro3 enhanced AKT phosphorylation to similar levels (data not shown). These results further support the notion that the ability of NS5A to associate with p85 PI3K and enhance the PI3K-AKT pathway is independent of Grb2 interaction.
The PI3K-AKT pathway plays a pivotal role in the regulation of cell survival and apoptosis. For example, AKT is known to cause serine phosphorylation of BAD, a proapoptotic Bcl-2 family member, and to inhibit BAD function, thus contributing to a blockade of apoptosis (8). In order to examine the effect of NS5A upregulation of the PI3K-AKT pathway on the downstream apoptosis pathway, serine phosphorylation levels of BAD were compared in Tet-Off cells expressing wild-type NS5A either in the presence or in the absence of EGF treatment (Fig. 6E). As revealed by immunoblot analysis with antibody specific to serine-phosphorylated BAD, NS5A expression also enhanced AKT-mediated serine 136 phosphorylation of BAD, consistent with the effect of NS5A on the PI3K-AKT pathway. Similarly, NS5A expression also enhanced the serine phosphorylation levels of BAD in recombinant VV-infected cells (Fig. 6F), demonstrating the ability of NS5A to modulate the apoptosis machinery through the PI3K-AKT pathway during both cellular signaling and viral infection. Upregulation of the PI3K-AKT-BAD pathway may contribute to the antiapoptotic and oncogenic abilities of NS5A (19, 22).
DISCUSSION
A model of NS5A action: a multifaceted attack.
Our laboratory has focused extensively on the HCV NS5A protein and its role in conferring IFN resistance and disrupting signal transduction pathways. In this report, we show that NS5A expression enhanced the activation of the PI3K-AKT-BAD pathway in both cellular and viral systems, suggesting a mechanism by which NS5A regulates apoptosis. Indeed, NS5A expression was previously observed to reduce cell death in both recombinant VV-infected HeLa cells (25) and Tet-Off cells treated with tumor necrosis factor alpha and poly(riboinosine-ribocytosine) (19, 22). More interestingly, Tet-Off cells expressing full-length NS5A were found to be more resistant to serum starvation-induced apoptosis than those expressing the N-terminal deletion mutant NS5A, suggesting that the interaction between NS5A and the PI3K-AKT pathway plays a role in regulating apoptosis (D. Grandgirard et al., unpublished results). However, the inhibition of apoptosis by NS5A is unlikely limited to the PI3K-AKT pathway-dependent mechanism, since NS5A may also regulate cell death through PKR-dependent pathways (19). These findings, combined with our previous results (53), have led us to hypothesize that NS5A, by targeting Grb2, acts to downregulate the Ras-Raf-MEK-MAPK pathway, which may influence viral persistence and the IFN response (24, 25, 53), while promoting the cell survival and antiapoptotic activities of the PI3K-AKT-BAD pathway (Fig. 7). We suggest that the association of NS5A with p85 PI3K enhances the activity of the PI3K-AKT pathway. This results in inhibition of apoptosis to help HCV persist in host cells and might contribute to the development of liver cancer associated with HCV infection, among other possible consequences (Fig. 7).
FIG. 7.
Model of the modulation of cellular signaling pathways by NS5A. NS5A, through its C-terminal proline-rich SH3-binding motif, directly interacts with Grb2 to perturb the downstream MAPK pathway, which may contribute to regulation of translation (25) and perturbation of IFN signaling (24). On the other hand, the N-terminal region of NS5A directly interacts with the p85 subunit of PI3K, which enhances the PI3K-AKT pathway and regulates the downstream apoptosis machinery. The modulation of the PI3K-AKT pathway by NS5A may contribute to cell survival and inhibition of apoptosis in virus-infected cells, resulting in viral persistence and contributing to pathogenesis.
We initially hypothesized that NS5A forms a complex with Gab1 through a Grb2-dependent bridging mechanism, possibly with NS5A and Gab1 associating with the N-terminal SH3 domain and the C-terminal SH3 domain of Grb2, respectively. Indeed, we found an indirect association between NS5A and Gab1. However, the association of NS5A with Gab1 was mediated not by Grb2 but very likely through the p85 subunit of PI3K, which we showed to directly interact with NS5A in vitro and to associate with NS5A in vivo. Similar to other viral proteins (12, 57), by interacting with PI3K, NS5A upregulates activation of the downstream PI3K-AKT pathway, which may provide an important mechanism for regulation of the viral life cycle and viral pathogenesis. Although the exact mechanism by which NS5A enhances p85 PI3K activation remains unclear, it is likely that NS5A somehow facilitates p85 PI3K activation by upstream pathways, considering the fact that NS5A enhances p85 phosphorylation, which is carried out by upstream kinases. However, it remains possible that NS5A directly upregulates p85 PI3K phosphorylation and activation through a direct interaction, even though NS5A is not known to possess any enzymatic activity. It is also possible that NS5A upregulates the downstream AKT-BAD pathway through a p85 PI3K-independent mechanism, for example, by directly interacting with AKT (C. Coito et al., unpublished data).
Viruses and cell signaling: targeting of the PI3K-AKT pathway.
Viruses have developed different strategies to interfere with host cell signaling, and such interference can be an important component of the viral life cycle and pathogenesis (28). After growth factor or cytokine signaling, p85 PI3K becomes tyrosine phosphorylated and binds to the p110 catalytic subunit of PI3K. Activated PI3K generates intracellular secondary messengers by phosphorylating phosphoinositides and regulates diverse cellular responses, including cell growth, inhibition of apoptosis, insulin-dependent metabolism of glucose, cytoskeletal reorganization, and tumorigenesis (29, 31). Thus, the perturbation of the PI3K pathway by NS5A may influence various important processes in host cells. Although our studies have used EGF as a model cell system, it is likely that NS5A also perturbs these cellular pathways during stimulation by other growth factors and cytokines, such as IFN-α/β (24) and hepatocyte growth factor (Coito et al., unpublished), which likely play important roles in liver metabolism and HCV pathogenesis. AKT is a serine/threonine kinase and has been recognized as a major target of PI3K (6, 27). AKT phosphorylates cellular targets involved in multiple biological processes, such as apoptosis, glycogen metabolism, and gene transcription. AKT phosphorylation of Bad results in inhibition of apoptosis (8). In fact, modification of the BAD pathway by the US3 protein kinase of herpes simplex virus type 1 prevents programmed cell death in the absence of other viral proteins, highlighting the pivotal role of this pathway (37).
Because of its central role in the regulation of host cell survival, the PI3K-AKT pathway has become the target of numerous viruses. Some viral proteins, such as the hepatitis B virus X protein (30, 48), Epstein-Barr virus (EBV) LMP2A (latent membrane protein 2A) (47, 52), and the polyomavirus middle T antigen (7, 35, 51, 55), can activate the PI3K-AKT pathway, which contributes to suppression of apoptosis and causes cell transformation. Interestingly, two human immunodeficiency virus type 1 (HIV-1) proteins, Tat (3) and Nef (57), both activate the PI3K-AKT pathway, thus providing antiapoptotic signals. It is thought that activation of the PI3K-AKT pathway by these HIV-1 proteins may regulate viral replication and contribute to AIDS pathogenesis. Among the viral proteins known to modulate the PI3K-AKT pathway, both HIV-1 Nef (57) and the polyomavirus middle T antigen (12) interact directly with and activate PI3K, similar to the mechanism utilized by NS5A. In addition, some retroviruses, such as the AKT8 retrovirus, encode viral versions of the PI3K-AKT pathway components, further suggesting the importance of this pathway in viral pathogenesis (50).
In summary, our study illustrates an interesting example of a single viral protein that differentially modulates two pivotal cellular pathways that regulate cell growth/differentiation and survival, the MAPK and the PI3K-AKT pathways. In fact, this strategy is not limited to HCV, since another viral protein, EBV LMP2A, which transforms epithelial cells and inhibits cell differentiation, also activates the PI3K-AKT pathway, whereas activation of the MAPK pathway is not observed (47). More interestingly, a naturally occurring mutant EGF receptor, EGFRvIII, which causes cell transformation, constitutively activates the PI3K pathway while downregulating the MAPK pathway (36), resembling the manner of action employed by NS5A and EBV LMP2A. It is noteworthy that AKT cross talks with and inhibits the Raf-MEK-MAPK pathway, which regulates cell differentiation and proliferation in different biological systems (43, 58). It therefore appears that differential regulation of the MAPK and PI3K pathways constitutes a normal mechanism of cell life control and that this mechanism has been hijacked by viruses to facilitate the viral life cycle instead.
Acknowledgments
Y.H. and H.N. contributed equally to this study.
We thank T. Imagawa for providing the NS5A-specific antibody. We thank Albert Wong for providing the GST-Gab1 construct and Bart Vanhaesebroeck for providing the GST-p85α construct. We are grateful to Marcus J. Korth for editorial assistance; Marlene Wambach, Semih Tareen, and Rick Ferguson for technical support; and Christina A. Heid for administrative support.
S.J.P. is partially supported by a Liver Scholar Award from the American Liver Foundation. This work was supported by grants from the National Institutes of Health (AI-22646, RR-00166, AI-41629, AI-47304, and AI-48214) to M.G.K.
REFERENCES
- 1.Aizaki, H., S. Saito, T. Ogino, N. Miyajima, T. Harada, Y. Matsuura, T. Miyamura, and M. Kohase. 2000. Suppression of interferon-induced antiviral activity in cells expressing hepatitis C virus proteins. J. Interferon Cytokine Res. 20:1111-1120. [DOI] [PubMed]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Borgatti, P., G. Zauli, M. L. Colamussi, D. Gibellini, M. Previati, L. L. Cantley, and S. Capitani. 1997. Extracellular HIV-1 Tat protein activates phosphatidylinositol 3- and Akt/PKB kinases in CD4+ T lymphoblastoid Jurkat cells. Eur. J. Immunol. 27:2805-2811. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. P., D. R. Twardzik, H. Marquardt, and G. J. Todaro. 1985. Vaccinia virus encodes a polypeptide homologous to epidermal growth factor and transforming growth factor. Nature 313:491-492. [DOI] [PubMed] [Google Scholar]
- 5.Cantrell, D. A. 2001. Phosphoinositide 3-kinase signalling pathways. J. Cell Sci. 114:1439-1445. [DOI] [PubMed] [Google Scholar]
- 6.Coffer, P. J., J. Jin, and J. R. Woodgett. 1998. Protein kinase B (c-Akt): a multifunctional mediator of phosphatidylinositol 3-kinase activation. Biochem. J. 335:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl, J., A. Jurczak, L. A. Cheng, D. C. Baker, and T. L. Benjamin. 1998. Evidence of a role for phosphatidylinositol 3-kinase activation in the blocking of apoptosis by polyomavirus middle T antigen. J. Virol. 72:3221-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta, S. R., H. Dudek, X. Tao, S. Masters, H. Fu, Y. Gotoh, and M. E. Greenberg. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91:231-241. [DOI] [PubMed] [Google Scholar]
- 9.Diao, J., R. Garces, and C. D. Richardson. 2001. X protein of hepatitis B virus modulates cytokine and growth factor related signal transduction pathways during the course of viral infections and hepatocarcinogenesis. Cytokine Growth Factor Rev. 12:189-205. [DOI] [PubMed]
- 10.Di Bisceglie, A. M. 1998. Hepatitis C. Lancet 351:351-355. [DOI] [PubMed] [Google Scholar]
- 11.Di Bisceglie, A. M., R. L. Carithers, and G. J. Gores. 1998. Hepatocellular carcinoma. Hepatology 28:1161-1165. [DOI] [PubMed] [Google Scholar]
- 12.Dilworth, S. M., C. E. Brewster, M. D. Jones, L. Lanfrancone, G. Pelicci, and P. G. Pelicci. 1994. Transformation by polyoma virus middle T-antigen involves the binding and tyrosine phosphorylation of Shc. Nature 367:87-90. [DOI] [PubMed] [Google Scholar]
- 13.DiMaio, D., C. C. Lai, and O. Klein. 1998. Virocrine transformation: the intersection between viral transforming proteins and cellular signal transduction pathways. Annu. Rev. Microbiol. 52:397-421. [DOI] [PubMed] [Google Scholar]
- 14.Downward, J. 1994. The GRB2/Sem-5 adaptor protein. FEBS Lett. 338:113-117. [DOI] [PubMed] [Google Scholar]
- 15.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. [DOI] [PubMed] [Google Scholar]
- 17.Gale, M., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale, M., Jr., and M. R. Beard. 2001. Molecular clones of hepatitis C virus: applications to animal models. ILAR J. 42:139-151. [DOI] [PubMed] [Google Scholar]
- 19.Gale, M., B. Kwieciszewski, M. Dossett, H. Nakao, and M. G. Katze. 1999. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J. Virol. 73:6506-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gale, M. J., M. J. Korth, and M. G. Katze. 1998. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin. Diagn. Virol. 10:157-162. [DOI] [PubMed] [Google Scholar]
- 21.Gale, M. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh, A. K., M. Majumder, R. Steele, K. Meyer, R. Ray, and R. B. Ray. 2000. Hepatitis C virus NS5A protein protects against TNF-α mediated apoptotic cell death. Virus Res. 67:173-178. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh, A. K., R. Steele, K. Meyer, R. Ray, and R. B. Ray. 1999. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J. Gen. Virol. 80:1179-1183. [DOI] [PubMed] [Google Scholar]
- 24.He, Y., and M. Katze. 2002. To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol. 15:95-119. [DOI] [PubMed] [Google Scholar]
- 25.He, Y., S. L. Tan, S. U. Tareen, S. Vijaysri, J. O. Langland, B. L. Jacobs, and M. G. Katze. 2001. Regulation of mRNA translation and cellular signaling by hepatitis C virus nonstructural protein NS5A. J. Virol. 75:5090-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hibi, M., and T. Hirano. 2000. Gab-family adapter molecules in signal transduction of cytokine and growth factor receptors, and T and B cell antigen receptors. Leuk. Lymphoma 37:299-307. [DOI] [PubMed] [Google Scholar]
- 27.Kandel, E. S., and N. Hay. 1999. The regulation and activities of the multifunctional serine/threonine kinase Akt/PKB. Exp. Cell Res. 253:210-229. [DOI] [PubMed] [Google Scholar]
- 28.Krajcsi, P., and W. S. Wold. 1998. Viral proteins that regulate cellular signalling. J. Gen. Virol. 79:1323-1335. [DOI] [PubMed] [Google Scholar]
- 29.Krasilnikov, M. A. 2000. Phosphatidylinositol-3 kinase dependent pathways: the role in control of cell growth, survival, and malignant transformation. Biochemistry (Moscow) 65:59-67. [PubMed] [Google Scholar]
- 30.Lee, Y. I., S. Kang-Park, and S. I. Do. 2001. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 276:16969-16977. [DOI] [PubMed] [Google Scholar]
- 31.Leevers, S. J., B. Vanhaesebroeck, and M. D. Waterfield. 1999. Signalling through phosphoinositide 3-kinases: the lipids take center stage. Curr. Opin. Cell Biol. 11:219-225. [DOI] [PubMed] [Google Scholar]
- 32.Lock, L. S., I. Royal, M. A. Naujokas, and M. Park. 2000. Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J. Biol. Chem. 275:31536-31545. [DOI] [PubMed] [Google Scholar]
- 33.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 34.McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepatitis 7:2-14. [DOI] [PubMed] [Google Scholar]
- 35.Meili, R., P. Cron, B. A. Hemmings, and K. Ballmer-Hofer. 1998. Protein kinase B/Akt is activated by polyomavirus middle-T antigen via a phosphatidylinositol 3-kinase-dependent mechanism. Oncogene 16:903-907. [DOI] [PubMed] [Google Scholar]
- 36.Moscatello, D. K., M. Holgado-Madruga, D. R. Emlet, R. B. Montgomery, and A. J. Wong. 1998. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J. Biol. Chem. 273:200-206. [DOI] [PubMed] [Google Scholar]
- 37.Munger, J., and B. Roizman. 2001. The US3 protein kinase of herpes simplex virus 1 mediates the posttranslational modification of BAD and prevents BAD-induced programmed cell death in the absence of other viral proteins. Proc. Natl. Acad. Sci. USA 98:10410-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paterson, M., C. D. Laxton, H. C. Thomas, A. M. Ackrill, and G. R. Foster. 1999. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology 117:1187-1197. [DOI] [PubMed] [Google Scholar]
- 39.Polyak, S. J., K. S. Khabar, D. M. Paschal, H. J. Ezelle, G. Duverlie, G. N. Barber, D. E. Levy, N. Mukaida, and D. R. Gretch. 2001. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J. Virol. 75:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polyak, S. J., D. M. Paschal, S. McArdle, M. J. Gale, Jr., D. Moradpour, and D. R. Gretch. 1999. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology 29:1262-1271. [DOI] [PubMed] [Google Scholar]
- 41.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 42.Reisner, A. H. 1985. Similarity between the vaccinia virus 19K early protein and epidermal growth factor. Nature 313:801-803. [DOI] [PubMed] [Google Scholar]
- 43.Rommel, C., B. A. Clarke, S. Zimmermann, L. Nunez, R. Rossman, K. Reid, K. Moelling, G. D. Yancopoulos, and D. J. Glass. 1999. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science 286:1738-1741. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Schaeper, U., N. H. Gehring, K. P. Fuchs, M. Sachs, B. Kempkes, and W. Birchmeier. 2000. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J. Cell Biol. 149:1419-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlessinger, J. 1994. SH2/SH3 signaling proteins. Curr. Opin. Genet. Dev. 4:25-30. [DOI] [PubMed] [Google Scholar]
- 47.Scholle, F., K. M. Bendt, and N. Raab-Traub. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shih, W. L., M. L. Kuo, S. E. Chuang, A. L. Cheng, and S. L. Doong. 2000. Hepatitis B virus X protein inhibits transforming growth factor-beta-induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 275:25858-25864. [DOI] [PubMed] [Google Scholar]
- 49.Song, J., M. Fujii, F. Wang, M. Itoh, and H. Hotta. 1999. The NS5A protein of hepatitis C virus partially inhibits the antiviral activity of interferon. J. Gen. Virol. 80:879-886. [DOI] [PubMed] [Google Scholar]
- 50.Staal, S. P. 1987. Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proc. Natl. Acad. Sci. USA 84:5034-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Summers, S. A., L. Lipfert, and M. J. Birnbaum. 1998. Polyoma middle T antigen activates the Ser/Thr kinase Akt in a PI3-kinase-dependent manner. Biochem. Biophys. Res. Commun. 246:76-81. [DOI] [PubMed] [Google Scholar]
- 52.Swart, R., I. K. Ruf, J. Sample, and R. Longnecker. 2000. Latent membrane protein 2A-mediated effects on the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 74:10838-10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan, S. L., H. Nakao, Y. He, S. Vijaysri, P. Neddermann, B. L. Jacobs, B. J. Mayer, and M. G. Katze. 1999. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. USA 96:5533-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 55.Webster, M. A., J. N. Hutchinson, M. J. Rauh, S. K. Muthuswamy, M. Anton, C. G. Tortorice, R. D. Cardiff, F. L. Graham, J. A. Hassell, and W. J. Muller. 1998. Requirement for both Shc and phosphatidylinositol 3′ kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol. Cell. Biol. 18:2344-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White, M. F., and L. Yenush. 1998. The IRS-signaling system: a network of docking proteins that mediate insulin and cytokine action. Curr. Top. Microbiol. Immunol. 228:179-208. [DOI] [PubMed] [Google Scholar]
- 57.Wolf, D., V. Witte, B. Laffert, K. Blume, E. Stromer, S. Trapp, P. d'Aloja, A. Schurmann, and A. S. Baur. 2001. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat. Med. 7:1217-1224. [DOI] [PubMed] [Google Scholar]
- 58.Zimmermann, S., and K. Moelling. 1999. Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 286:1741-1744. [DOI] [PubMed] [Google Scholar]