Abstract
During the early phase of infection, the E1B-55K protein of adenovirus type 5 (Ad5) counters the E1A-induced stabilization of p53, whereas in the late phase, E1B-55K modulates the preferential nucleocytoplasmic transport and translation of the late viral mRNAs. The mechanism(s) by which E1B-55K performs these functions has not yet been clearly elucidated. In this study, we have taken a proteomics-based approach to identify and characterize novel E1B-55K-associated proteins. A multiprotein E1B-55K-containing complex was immunopurified from Ad5-infected HeLa cells and found to contain E4-orf6, as well as several cellular factors previously implicated in the ubiquitin-proteasome-mediated destruction of proteins, including Cullin-5, Rbx1/ROC1/Hrt1, and Elongins B and C. We further demonstrate that a complex containing these as well as other proteins is capable of directing the polyubiquitination of p53 in vitro. These ubiquitin ligase components were found in a high-molecular-mass complex of 800 to 900 kDa. We propose that these newly identified binding partners (Cullin-5, Elongins B and C, and Rbx1) complex with E1B-55K and E4-orf6 during Ad infection to form part of an E3 ubiquitin ligase that targets specific protein substrates for degradation. We further suggest that E1B-55K functions as the principal substrate recognition component of this SCF-type ubiquitin ligase, whereas E4-orf6 may serve to nucleate the assembly of the complex. Lastly, we describe the identification and characterization of two novel E1B-55K interacting factors, importin-α1 and pp32, that may also participate in the functions previously ascribed to E1B-55K and E4-orf6.
The adenovirus type 5 (Ad5) E1B-55K protein performs several functions critical to the virus's life cycle. In the early phase of infection, E1B-55K counteracts the E1A-induced stabilization of p53 that may adversely affect viral replication by leading to cell cycle arrest or the premature apoptotic death of the host cell (20, 23, 74, 100, 101). In the late phase, E1B-55K functions together with the E4-orf6 protein to stimulate the preferential nucleocytoplasmic transport and translation of the viral late mRNAs (2, 3, 6, 14, 47, 48, 67, 94, 95, 131, 140).
E1B-55K counteracts the effects of p53 during viral infection through at least two distinct mechanisms. E1B-55K can (i) bind the amino-terminal acidic activation domain of p53 and directly repress p53-mediated transcriptional activation (61, 78, 133, 134) and (ii) function together with E4-orf6 to stimulate the degradation of p53 by proteasomes (11, 18, 44, 83, 98, 103, 104, 122, 130). E1B-55K possesses a generalized transcription repression activity that can inhibit expression from several promoters when targeted by fusion with the Gal4 DNA binding domain (134). This activity has also been shown to inhibit transcription initiation in vitro and, in the context of an infection, is recruited upstream of p53-responsive cellular promoters by direct interaction with DNA-bound p53 (78). E4-orf6 has also been shown to contribute to the repression of transcriptional activation by p53 (18, 27, 84).
The in vivo E1B-55K-mediated inhibition of p53-activated promoters may also involve the recruitment of histone deacetylase complexes. In a recent study by Punga et al., E1B-55K was shown to bind histone deacetylase 1 and the transcriptional corepressor protein mSin3A in both Ad-transformed and lytically infected cells (96). Though the functional significance of these interactions was not demonstrated in the context of a viral infection, a complex containing E1B-55K in 293 cells was shown to catalyze the deacetylation of a histone substrate peptide (96). It was suggested that the association of E1B-55K with this activity may play a role in one or more of the functions attributed to E1B-55K in the infected cell (96).
As mentioned above, E1B-55K also interferes with p53 function by cooperating with E4-orf6 to cause its accelerated proteolytic degradation (83, 84, 86, 98, 104, 122, 130). In infections with the wild-type virus in which both of these proteins are present, p53 levels within the infected cell are markedly reduced (44). In the absence of either of these proteins, however, a dramatic stabilization of p53 is seen (44, 98, 103, 122). The E1B-55K and E4-orf6 proteins appear to be the only viral proteins required to destabilize p53, as even their transient expression in the absence of other adenoviral proteins results in a decrease in p53 half-life (11, 18, 99, 104, 122, 130). The functions of the 26S proteasome were implicated in this degradative process since it was eliminated upon treatment with proteasome inhibitors (97, 104).
The accelerated turnover of p53 also appears to involve additional cellular factors of 84, 19, 16, and 14 kDa (97, 99). In recent studies by Querido et al. (97, 99), the ability of E4-orf6 to cooperate with E1B-55K in enhancing p53 degradation was found to correlate with its ability to associate with these proteins. Using ion trap mass spectrometry, a subset of these factors was later identified to be Cullin-5, Elongin B, and Elongin C (97). These cellular factors, together with Rbx1/ROC1/Hrt1, E1B-55K, and E4-orf6 were also shown to weakly ubiquitinate p53 in vitro (97).
Querido et al. further demonstrated that the degradation of p53 was dependent on an efficient interaction between E1B-55K and p53 (99, 104, 116). Indeed, mutant forms of p53 that could not efficiently interact with E1B-55K were not degraded (99). The enhanced turnover of p53 did not appear to require the ubiquitin ligase activity of mdm2, or the action of p19ARF, however, as E1B-55K and E4-orf6 could mediate the destruction of p53 in both p53/MDM2-null and p19ARF-null mouse embryo fibroblasts (97).
In the late phase of infection, the E1B-55K protein performs additional functions important to viral replication. During the late phase, viral mRNAs are preferentially transported to the cytoplasm to the exclusion of most cellular mRNAs (3, 6, 67, 94, 131). Several studies have shown that the E1B-55K and E4-orf6 proteins play a critical role in modulating this process (2, 3, 47, 95, 131). Indeed, in HeLa cells infected with mutant viruses that fail to express either of these proteins, the late viral mRNAs fail to accumulate efficiently in the cytoplasm, resulting in the impaired synthesis of the corresponding late viral protein products (1, 3, 6, 14, 47, 67, 95, 109, 128, 131). The exact mechanism by which the E1B-55K/E4-orf6 complex (105, 109) mediates this event is not well understood. However, new insight into this process was provided by the identification of a new E1B-55K binding factor, E1B-AP5 (34).
E1B-AP5 (E1B-associated protein 5) is a nuclear RNA-binding protein of the hnRNP (heterogeneous nuclear ribonucleoprotein particle) protein family and has been shown to specifically bind E1B-55K both in vitro and in vivo (34). The stable overexpression of this protein was shown to stimulate the export of late viral transcripts and simultaneously delay the E1B-55K-dependent inhibition of host cell mRNA export in infected cells (34). These findings are consistent with the model presented by Ornelles and Shenk (91), in which the E1B-55K protein is proposed to sequester one or more host factor(s) that may be limiting for mRNA export, and thereby redirect the cellular mRNA export machinery to promote the nucleocytoplasmic transport of viral mRNAs.
Despite our increasing knowledge of the many ways in which E1B-55K impacts the virus life cycle, the molecular mechanisms by which it exerts many of its effects are not well understood. To gain insight into these mechanisms, we sought to identify E1B-55K's molecular partners in these processes. To this end, E1B-55K-containing complexes were immunopurified from Ad-infected HeLa cells, and the components were subsequently identified by a combination of matrix-assisted laser desorption ionization mass spectrometry (MALDI) and nanoelectrospray tandem mass spectrometry (nano-ES MS-MS). Our analysis revealed that E1B-55K associates in vivo with a number of cellular proteins proposed to function in the ubiquitin-proteasome-mediated destruction of proteins. These proteins were further found to reside together with E1B-55K in a high-molecular-mass complex of approximately 800 to 900 kDa. The functional consequence of E1B-55K's association with these putative ubiquitin ligase components was then assessed in an in vitro ubiquitination assay. Our findings support a model in which E1B-55K functions as the substrate binding subunit of a ubiquitin ligase of the Skp1-Cullin-F-box (SCF) type (reviewed in reference 24).
MATERIALS AND METHODS
Cells and viruses.
293 (43) and HeLa suspension cultures were maintained in minimal essential medium modified for suspension culture, supplemented with 5% fetal bovine serum. 293 and HeLa monolayer cultures were maintained in Dulbecco's modified essential medium with 10% fetal bovine serum. The wild-type Ad5 and the E1B-55K defective Ad mutant dl1520 have been described by Barker and Berk (5). The dl1520 virus harbors a deletion within the E1B-55K open reading frame spanning nucleotides 2496 to 3323, in addition to a point mutation at nucleotide 2022 that results in the termination of translation (5). Viruses were propagated in 293 suspension cultures.
Antisera.
The Elongin C protein was detected using the SIII p15 monoclonal antibody from BD Transduction Labs, and the Elongin B, importin-α1, and NEDD8 proteins were detected with goat polyclonal antibodies from Santa Cruz Biotechnologies. The Rbx1/ROC1/Hrt1 rabbit polyclonal antibody was obtained from LabVision. The IVa2 (75) and Cullin-5 antibodies were kind gifts of Claude Kedinger (ESBS, Ecole Européenne des Universités du Rhin Supérieur) and Phil Branton (McGill University), respectively (97). An additional Elongin B antibody was obtained from Joan Conaway of the University of Oklahoma Health Sciences Center (37). The monoclonal NuMA (35) and anti-E-MAP-115/ensconsin (guinea pig) antibodies (30) were generously provided by Duane Compton of Dartmouth Medical School and J. Chloe Bulinski of Columbia University. A rabbit polyclonal antibody directed against the amino terminus of Cullin-5 was also made using a synthetic peptide (Genemed Synthesis, Inc., Covance) (15). Immunoblots for the E1B-55K and E4-orf6 proteins were performed using the anti-E1B-55K 2A6 hybridoma supernatant (111) and the M45 mouse monoclonal antibody that recognizes the amino-terminal portion of the E4-orf6 and E4-orf6/7 proteins (generously provided by Patrick Hearing, SUNY Stony Brook [87]). Immunoprecipitations for the E1B-55K protein were performed using the 2A6 hybridoma supernatant (111), and control immunoprecipitations were performed with the anti-72K DNA binding protein (M186)B6 monoclonal antibody (102).
E32 rabbit antipeptide antiserum was used to detect pp32 in immunoblotting experiments. This antiserum was raised against a 28-residue peptide (DP49) corresponding to a highly conserved 27-amino-acid sequence in murine pp32 and its homologs, plus an added carboxy-terminal cysteine for coupling to keyhole limpet hemocyanin. The amino acid sequence of DP49 corresponds to residues 91 to 117 of human pp32 except for a single mismatch at residue 92. DP49 was conjugated to keyhole limpet hemocyanin using the Pierce Imject conjugation kit, and the conjugate was used as an immunogen. Preimmune serum from the same rabbit did not detect pp32 in control experiments. Of note, the specific sequences obtained from microsequencing the E1B-55K-associated 31-kDa protein contained sequences specific only to pp32.
In vivo labeling of cells.
HeLa (2 × 107) cells were either mock infected or infected with the Ad5 or dl1520 viruses at a multiplicity of infection (MOI) of 100. Cells were labeled with 250 μCi of Tran35S-label (ICN) from 10 to 14 h postinfection as described previously (48).
Preparation of nuclear extracts from uninfected and infected cells.
293 and HeLa suspension cultures were either mock infected or infected with the Ad5 or dl1520 viruses at an MOI of 100. At ∼14 h postinfection, cells were harvested and washed once with phosphate-buffered saline. Nuclear extracts were then prepared by a method similar to that described by Dignam et al. (25, 66). In brief, cells were resuspended in 1 packed cell volume of buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) and incubated 10 min on ice. Cells were then mechanically disrupted by passing five times through a syringe affixed with a 25-gauge needle. Cell nuclei were then collected by centrifugation and subsequently resuspended in 1 packed cell volume of buffer C (20 mM HEPES [pH 7.9], 25% [vol/vol] glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10 mM β-mercaptoethanol, 1 mM PMSF). Nuclear proteins were then extracted by rotating lysates 30 min at 4°C. Upon centrifugation of the lysates for 30 min at 4°C, the supernatants were collected and set aside for immunoprecipitation analysis.
Immunoprecipitation and Western blot analysis.
Immunoprecipitation reactions were prepared by combining nuclear extracts with a concentrated reaction buffer for final reaction conditions of 10 mM Tris-HCl, pH 8.0 (4°C)-150 mM NaCl-1.5 mM MgCl2-1 mM PMSF-0.1% NP-40. The prepared reaction mixtures were precleared by incubating 1 h in the presence of 15 μl of protein A-Sepharose beads at 4°C. Antibody-cross-linked protein A beads (49) (25 μl) were then added to the precleared supernatants, and the resulting reaction mixtures were incubated overnight at 4°C with rotation. Protein-bead complexes were recovered by centrifugation and washed five times with reaction buffer. Precipitated complexes were eluted from the beads by adding an equal volume of 2× Laemmli sample buffer and boiling 15 min (108). The immunoprecipitates were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and either stained with silver nitrate according to the method of Sambrook et al. (108) or transferred to nitrocellulose membranes for Western blot analysis. The nitrocellulose filters were blocked over an hour at room temperature in Tris-buffered saline (TBS) with 10% nonfat dry milk and then incubated overnight at 4°C with the appropriate antibody in TBS-3% milk. The filters were then washed three times in TBS-0.1% Tween 20, incubated with the appropriate horseradish peroxidase-conjugated secondary antibody diluted in TBS-0.1% Tween 20-3% milk, and washed three times in TBS-0.1% Tween 20. Protein bands were then visualized by enhanced chemiluminescence (Pierce).
Mass spectrometry analysis.
Silver-stained bands present specifically in 2A6 anti-E1B-55K immunoprecipitates were excised and digested in-gel with trypsin as described previously (119). Proteins were identified by MALDI peptide mapping and nano-ES MS-MS(132) combined in a layered approach (118).
In vitro p53 ubiquitination assay.
The in vitro p53 ubiquitination activity contained within 2A6 anti-E1B-55K immunoprecipitates was evaluated by a method similar to that described by Scheffner et al. (113). A combined in vitro transcription and translation system (TnT; Promega) was utilized for the generation of 35S-labeled substrate (p53). In brief, pTM.1-ep53 (134) was combined with T7 polymerase, rabbit reticulocyte lysate, and Tran35S-label (ICN), and the reactions were carried out according to the manufacturer's specifications. The products were then resolved by SDS-PAGE and visualized by autoradiography to confirm the efficiency of the reaction. Ubiquitination assays were performed in a 30-μl volume combining 1 μl of radioactively labeled p53, 8 μl of agarose beads of 2A6 anti-E1B-55K immunoprecipitate prepared as described above, and 10 μl of unprogrammed reticulocyte lysate. Where appropriate, the indicated amounts of purified ubiquitin K48R and glutathione S-transferase (GST)-conjugated ubiquitin (Boston Biochem) were also added to the reaction mixtures. Reactions were performed in a buffer containing 25 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 3 mM dithiothreitol. Upon incubation for 4 h at 25°C, the reactions were stopped through the addition of an equal volume of 2× Laemmli sample buffer (108). The resultant products were resolved by SDS-10% PAGE and visualized by autoradiography.
Chromatographic analysis of E1B-55K-containing complexes.
Nuclear extracts were prepared by a modified Dignam method (25). All steps were performed as described previously (25), with the exception that the final dialysis step was performed in a solution containing 10 mM Tris-HCl (pH 8.0) (4°C), 150 mM NaCl, 5% (vol/vol) glycerol, 1.5 mM MgCl2, 0.04 mM EDTA, 2.5 mM β-mercaptoethanol, 1 mM PMSF, and 0.1% Tween 20. Dialyzed nuclear extracts were then centrifuged 10 min at 14,000 rpm (in a Sorvall SA-600 rotor) to pellet precipitants, and the supernatant was additionally centrifuged through a filter (pore size, 0.45 μm; Amicon). One milliliter of the dialyzed and filtered sample was then applied to a 23.76-ml Superose 6 column (HR 10/30; Pharmacia) preequilibrated with a buffer identical to the dialysis buffer described above. Samples were isocratically eluted in the same buffer, and 1-ml fractions were collected. A portion of each fraction was then immunoprecipitated with the 2A6 monoclonal antibody (111) as described above.
E4-orf6 transfection of 293 cells.
293 monolayer cultures (70 to 80% confluent) in 10-cm-diameter tissue culture dishes were transfected with 10 μg of pcDNA3-E4orf6-Flu (85), which was kindly provided by Thomas Dobner (University of Regensburg). Transfections were performed using the Lipofectamine Plus (Invitrogen) reagent as prescribed by the manufacturer. At 42 h posttransfection, nuclear extracts were prepared as described above. The subsequent immunoprecipitation and immunoblotting analyses were also performed as detailed above.
RESULTS
Identification of novel E1B-55K-associated proteins.
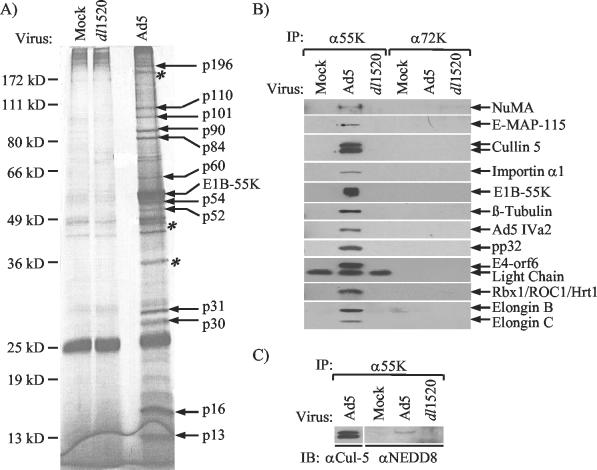
The mechanism(s) by which E1B-55K performs its many functions has not been clearly elucidated. To better understand these processes, we employed a proteomics-based approach to identify new E1B-55K interacting partners. Nuclear extracts were first prepared from Ad5-infected HeLa cells labeled with 35S-labeled methionine and cysteine from 10 to 14 h postinfection. The extracts were immunoprecipitated with 2A6 anti-E1B-55K antibody (111), and the precipitated proteins were analyzed by SDS-PAGE. Several polypeptides coprecipitated with E1B-55K that were not present in parallel reactions from uninfected HeLa nuclear extracts or nuclear extracts prepared from cells infected with a virus defective for E1B-55K expression, dl1520 (data not shown). Such species included proteins that migrated at approximate molecular masses of 196, 110, 101, 90, 84, 60, 54, 52, 31, 30, 16, and 13 kDa (data not shown). Though other species were also detected, these appeared to nonspecifically coprecipitate with E1B-55K, as they were not consistently observed in anti-E1B-55K immunoprecipitates (data not shown).
To permit the identification of the E1B-55K-associated proteins by mass spectrometry, immunoprecipitates from Ad5-infected HeLa cells were obtained in sufficient quantities to visualize by SDS-PAGE followed by silver staining (Fig. 1A). The E1B-55K-associated proteins were then identified through a combination of MALDI mass spectrometry peptide mass mapping and nano-ES MS-MS sequencing (Table 1). A subset of these newly identified E1B-55K binding partners have also been shown to interact with the E4-orf6 protein (Table 1) (9, 97, 99).
FIG. 1.
Identification and characterization of novel E1B-55K-associated proteins. (A) HeLa suspension cultures were either mock infected or infected with the Ad5 or dl1520 viruses at an MOI of 100. At 14 h postinfection, nuclear extracts were prepared and immunoprecipitated with the 2A6 anti-E1B-55K antibody (111). Immunoprecipitates were resolved by SDS-PAGE, and the E1B-55K coprecipitating proteins were visualized by staining with silver nitrate. Asterisks are used to indicate where E1B-55K aggregates and breakdown products were identified. The sizes of the molecular mass markers used are indicated at left. (B and C) Nuclear extracts prepared from mock-, Ad5-, or dl1520-infected cells were immunoprecipitated with antibodies directed against the E1B-55K (B and C) or Ad 72K (B) DNA-binding proteins. Immunoprecipitates were resolved by SDS-PAGE, and upon transfer to a nitrocellulose support, Western analyses for the indicated proteins were performed using the appropriate antibodies.
TABLE 1.
Putative E1B-55K interacting proteins identified by MALDI-nano-ES MS-MS
| Banda | Protein name | Description | Reference(s) |
|---|---|---|---|
| p196 | NuMAb,c | Nuclear mitotic apparatus protein | 137 |
| p110 | E-MAP-115b,c | Microtubule-associated protein | 81 |
| p101 | Ad5 100Kd | Adenovirus late 100K protein | 9, 50 |
| p90 | VACM1/CUL-5b,c,d,e | Vasopressin-activated Ca2+ mobilizing receptor/Cullin homolog 5 | 15, 16 |
| p84 | VACM1/CUL-5b,c,d,e | Vasopressin-activated Ca2+ mobilizing receptor/Cullin homolog 5 | 15, 16 |
| p60 | Importin α-1b,c,e | Nuclear import receptor α subunit | 56, 65, 129 |
| p54 | Tubulin-βb,c | Human tubulin beta-4 chain | 17 |
| p52 | Ad5 IVa2b,c | Adenovirus IVa2 maturation protein | 82, 123 |
| p31 | pp32b,c,e | Potent heat-stable PP2A inhibitor | 77, 127 |
| p30 | E4-orf6b,c,e | Adenovirus early region 34-kDa protein | 110 |
| p16 | Elongin Bb,c,d,e | RNA polymerase II transcription factor SIII p18 subunit | 37 |
| p13 | Elongin Cb,c,d,e | RNA polymerase II transcription factor SIII p15 subunit | 38 |
Molecular weights determined using Kodak 1D image analysis software.
Interactions verified by immunoprecipitation with 2A6 followed by immunoblot analysis in Ad5-infected HeLa cells.
Proteins present in αE1B-55K immunoprecipitates from Ad5-infected 293 cells.
Interactions detected in αE1B-55K immunoprecipitations from E4-orf6-Flu-transfected 293 cells.
The analysis of E1B-55K eluates yielded E4-orf6, as expected (110). Additionally however, a number of cellular proteins that have been implicated in the ubiquitin-proteasome-mediated destruction of proteins were found. These included the Elongins B and C, which together with Cullin-2 and the von Hippel-Lindau gene product have been shown to form an SCF-like E3 ubiquitin ligase (22, 24, 29, 55, 59, 60, 63, 70, 92, 121), and Cullin-5, a member of the Cullin family of proteins (15, 16, 64). Based on their sequence similarity, each member of the Cullin family has been predicted to form an integral component of an SCF-type E3 complex (24, 32, 64). The RING-H2 protein Rbx1/ROC1/Hrt1 has also been shown to be an essential subunit of the SCF-type ubiquitin ligases (59, 88, 116, 120). Rbx1/ROC1/Hrt1 has been shown to interact with a number of the Cullins and appears to enhance the rate at which various E2 ubiquitin-conjugating enzymes ubiquitinate target molecules (32, 58, 59, 60, 88, 97, 116).
Immunoblots of 2A6 immunoprecipitates (111) from Ad5-infected HeLa cells confirmed that the Elongins B and C, Cullin-5, and Rbx1/ROC1/Hrt1 were indeed present in association with E1B-55K (Fig. 1B). These proteins were not found in 2A6 immunoprecipitates (111) from uninfected or dl1520-infected HeLa cells or in parallel immunoprecipitation reactions performed with the (M186)B6 anti-72K DNA binding protein monoclonal antibody (102). Additional immunoblot analyses for the NuMA, E-MAP-115 (30), importin-α1, Ad IVa2 (75), tubulin-β, and pp32 proteins were also performed to validate their specific association with E1B-55K in Ad5-infected HeLa cells (Fig. 1B; Table 1).
The immunoblot analysis for Cullin-5 revealed a closely migrating doublet in the 80- to 90-kDa range. The Cullin proteins have each been shown to be modified by the ubiquitin-like protein NEDD8 (52, 126). Although the role of NEDD8 in regulating the activity of the SCF ubiquitin ligases is not yet clear, several lines of evidence suggest that the NEDDylation of the Cullin is required for the optimal assembly and function of the SCF-type ubiquitin ligases (33, 52, 62, 69, 126). To confirm that the higher-molecular-weight Cullin-5 form was indeed the result of NEDD8 addition, 2A6 immunoprecipitates from uninfected and Ad5- or dl1520-infected HeLa cells were immunoblotted using an anti-NEDD8 antibody. As seen in Fig. 1C, the NEDD8 moiety could be specifically detected in anti-E1B-55K immunoprecipitates from Ad5-infected extracts, and the NEDD8-containing band comigrated with the higher-molecular-weight form of Cullin-5.
E1B-55K assembles into a multiprotein complex with E4-orf6, cellular SCF-type E3 ubiquitin ligase components, and additional cellular proteins.
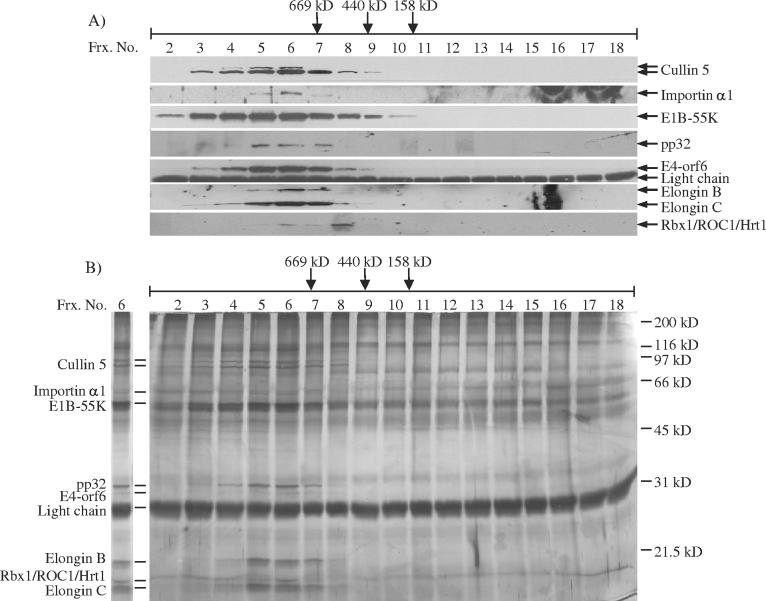
To determine if the cellular SCF-type E3 ubiquitin ligase components Cullin-5, E4-orf6, Rbx1/ROC1/Hrt1, and the Elongins B and C are associated with E1B-55K in a single complex in Ad5-infected cells, nuclear extract was resolved over a Superose 6 gel filtration column. Column fractions were immunoprecipitated with the 2A6 anti-E1B-55K monoclonal antibody (111), and the immunopurified complexes were subsequently resolved by SDS-PAGE and analyzed by Western blotting and silver stain analyses (Fig. 2A and B). E1B-55K eluted in a broad, high-molecular-mass range (Fig. 2A), and E4-orf6, Cullin-5, and the Elongins B and C were associated with a portion of the E1B-55K in a single peak eluting at 800 to 900 kDa (Fig. 2A). Rbx1/ROC1/Hrt1 was additionally detected in this peak, though it appeared that it may also form a distinct, lower-molecular-mass complex with E1B-55K (Fig. 2A). The silver stain analysis of fractions prepared in parallel (Fig. 2B) revealed other E1B-55K-associated polypeptides that coeluted with the proposed ubiquitin ligase components. The identities of some of these cofractionating polypeptides were subsequently verified by Western analysis to be importin-α1 and pp32 (Fig. 2A). The silver-stained gel also showed 2A6 light chain in every fraction, as well as 2A6 heavy chain at 55K, and bands at ∼85K and ∼130K that likely represent cross-linked 2A6 heavy and light chains.
FIG. 2.
Gel filtration chromatographic analysis of E1B-55K-containing complexes. (A and B) Nuclear extract prepared from Ad5-infected HeLa cells at 14 h postinfection was resolved on a Superose 6 column. The isocratically eluted fractions were then immunoprecipitated with the 2A6 anti-E1B-55K monoclonal antibody (111). Immunoprecipitates were resolved by SDS-PAGE and either transferred to a nitrocellulose membrane for Western blot analysis (A) or stained with silver nitrate (B). Western blot analyses were performed using the indicated antibodies (A). (A and B) Fraction numbers are listed at top, with the elution positions of the molecular mass standards thyroglobulin (669 kDa), ferritin (440 kDa) and aldolase (158 kDa) denoted by arrows above the fraction numbers. (B) Fraction 6 is duplicated at left. The major proteins bands coeluting with E1B-55K in the 800- to 900-kDa range are indicated at far left, and SDS-PAGE molecular mass markers are indicated at right.
The E1B-55K complex in Ad5-infected cells polyubiquitinates p53 in vitro.
The E1B-55K and E4-orf6 proteins are required to counter the E1A-induced stabilization of p53 in both virus-infected and -transformed cells (20, 23, 44, 74, 83, 84, 98, 101, 103, 122). Numerous studies have shown that one mechanism by which this is achieved is through the accelerated proteasome-mediated degradation of p53 (83, 84, 86, 98, 104, 122, 130). In Saos-2 osteosarcoma cells transiently expressing p53, for example, p53 levels are dramatically reduced depending on the presence of both the E1B-55K and E4-orf6 proteins (11, 18, 104). The proteasome-mediated degradation pathway was implicated in the rapid turnover of p53, as the treatment of cells coexpressing E1B-55K and E4-orf6 with 26S proteasome inhibitors inhibited this process (97, 104).
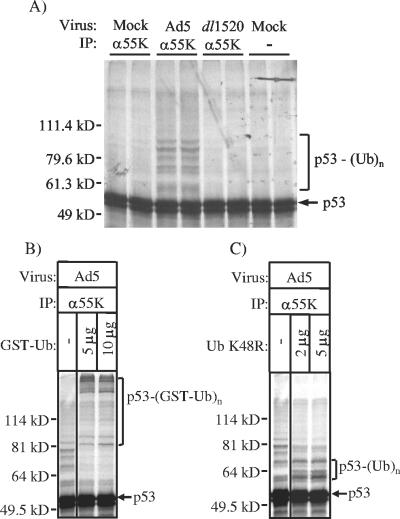
To probe the functional relationship between E1B-55K and the putative ubiquitin ligase components identified in our analysis, we performed ubiquitination assays in vitro. The conjugation of ubiquitin to a target protein typically requires the sequential action of three enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (21, 51). The E3s are primarily responsible for conferring specificity to ubiquitin conjugation and interact with both E2 and substrate (21, 51). Ubiquitination reactions were thus prepared by combining 2A6 anti-E1B-55K immunoprecipitates (111) from mock-, Ad5-, or dl1520-infected HeLa cells and in vitro-translated 35S-labeled p53, with unprogrammed rabbit reticulocyte lysate as a source of ubiquitin, E1, and E2. In vitro p53 ubiquitination was evaluated by resolving reaction products by SDS-PAGE and detecting higher-molecular-weight conjugates of radiolabeled p53 by fluorography.
Significant polyubiquitination activity, as measured by the laddering of 35S-labeled p53, was observed in reactions containing anti-E1B-55K immunoprecipitate from Ad5-infected HeLa cells that contain the 800- to 900-kDa complex with SCF-type E3 ubiquitin ligase components (Fig. 3A). No such activity was detected in similar control reactions containing 2A6 immunoprecipitates (111) from uninfected or dl1520-infected cells that lack the complex. To validate that the higher-molecular-weight labeled species resulted from the conjugation of ubiquitin moieties to p53, ubiquitination reactions were also performed in the presence of GST-tagged ubiquitin (Fig. 3B). The addition of GST-ubiquitin to the reaction mixture produced p53 conjugates with a higher molecular weight than that observed in reactions without added GST-ubiquitin, where p53-ubiquitin conjugates are presumably the result of endogenous ubiquitin in the reticulocyte lysate. These results establish that the 2A6-anti E1B-55K immunoprecipitate containing the 800- to 900-kDa complex has a p53-directed ubiquitin-ligase activity.
FIG. 3.
An E1B-55K-containing complex ubiquitinates p53 in vitro. (A) Nuclear extracts were prepared at 14 h postinfection and immunoprecipitated with the 2A6 anti-E1B-55K antibody. In vitro ubiquitination reactions were prepared by combining the immunoprecipitates with in vitro-transcribed and -translated radiolabeled p53 and an excess of unprogrammed rabbit reticulocyte lysate, as described in Materials and Methods. The reactions were incubated for 4 h at 25°C and then analyzed by SDS-PAGE and autoradiography. Duplicate reactions are shown. Reactions in the right two lanes contained protein A-Sepharose beads without bound antibody that were incubated with nuclear extract from mock-infected cells. Where appropriate, the indicated amounts of purified GST-conjugated ubiquitin (B) and ubiquitin K48R (C) were also added to the reactions. Conjugates of p53 with the various ubiquitin derivatives are denoted by the brackets at the right of each panel, and molecular mass markers are indicated at left.
Lastly, to evaluate if the higher-order p53 species resulted from the addition of a polyubiquitin chain required to signal degradation, as opposed to monoubiquitination at multiple lysines (a functionally distinct signal) (93), additional reactions were performed in the presence of ubiquitin K48R. This ubiquitin mutant terminates the polymerization of polyubiquitin chains by replacing lysine 48, the major site to which ubiquitins are ligated during polymerization, with arginine. The addition of ubiquitin K48R decreased the size of derivatized p53 (Fig. 3C), demonstrating that the laddering of p53 observed in the presence of the E1B-55K immunoprecipitate is due in part to the addition of polyubiquitin chains to the p53 molecule. However, in addition to the presumed monoubiquitin form, a second major p53 species was observed in the presence of high concentrations of ubiquitin K48R. This result suggests that the E1B-55K complex can ubiquitinate p53 at two lysines or cause the covalent attachment of other ubiquitin-like moieties onto the p53 molecule in addition to ubiquitin (42, 73). Altogether, these findings demonstrate that the proteins associated with E1B-55K in Ad5-infected cells have p53 E3 ubiquitin ligase activity, as anticipated from the composition of the 800- to 900-kDa complex.
E4-orf6 is required for assembly of the ubiquitin ligase complex.
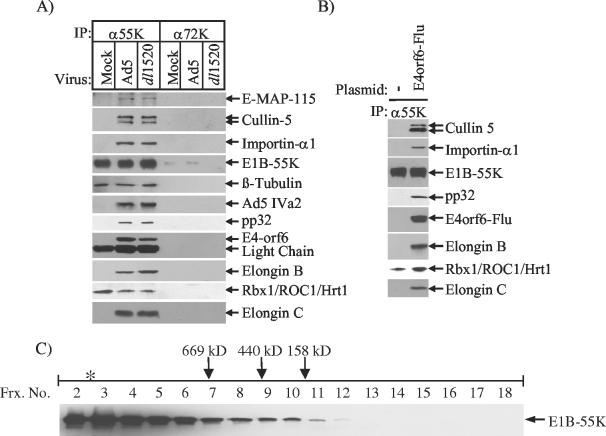
To determine if E1B-55K associates with Cullin-5, Rbx1/ROC1/Hrt1, and the Elongins B and C in Ad5-transformed 293 cells which lack E4-orf6, nuclear extract was immunoprecipitated with the 2A6 anti-E1B-55K monoclonal antibody (111), and the immunoprecipitate was subjected to Western blot analysis (Fig. 4A). Cullin-5 and the Elongins B and C did not coimmunoprecipitate with E1B-55K in these cells. However, an interaction between E1B-55K and Rbx1/ROC1/Hrt1 was observed, as was an interaction with β-tubulin. Upon infection with either wild-type Ad5 or dl1520, both E4-orf6 and the full complement of these SCF-type ubiquitin ligase components (Cullin-5, Rbx1/ROC1/Hrt1, and Elongins B and C) were found to coimmunoprecipitate with E1B-55K (Fig. 4A). NuMA, E-MAP-115, importin-α1, pp32, and Ad IVa2 also associated with E1B-55K in Ad5- and dl1520-infected 293 cells (Fig. 4A, data not shown). These findings indicate that the association of E1B-55K with these factors is an infection-specific event and suggest that one or more viral proteins are required to facilitate their interactions.
FIG. 4.
E4-orf6 induces the assembly of E1B-55K with Cullin-5, Rbx1/Roc1/Hrt1, and the Elongins B and C. (A) 293 cells were either mock infected or infected with wild-type Ad5 or dl1520 at an MOI of 100. At 14 h postinfection, nuclear extracts were prepared and immunoprecipitated with either the 2A6 anti-E1B-55K hybridoma supernatant (111) or (M186)B6 anti-72K DBP monoclonal antibody (102). The immunoprecipitates were subsequently resolved by SDS-PAGE and Western blotted for the indicated proteins. (B) 293 cells were either mock transfected or transfected with pcDNA3-E4orf6-Flu (85). Nuclear extracts prepared at 42 h posttransfection were immunoprecipitated with the 2A6 anti-E1B-55K hybridoma supernatant (111). Immunoprecipitates were resolved by SDS-PAGE and Western blot analyses performed with the indicated antibodies. (C) Nuclear extract from 293 cells was fractionated on Superose 6, and E1B-55K-containing complexes were immunopurified using the 2A6 anti-E1B-55K antibody (111) as described in the legend to Fig. 2. E1B-55K was detected by Western blotting with the 2A6 hybridoma supernatant (111). Superose 6 fraction numbers are indicated at top. The void volume is denoted with an asterisk, and the elution positions of the molecular mass standards thyroglobulin (669 kDa), ferritin (440 kDa), and aldolase (158 kDa) are indicated by the arrows.
Previous studies by Moore et al. demonstrated that p53 levels are substantially reduced in a 293-derived cell line stably expressing E4-orf6 compared to the parental cell line in which the E1A and E1B proteins are the only Ad proteins expressed (83). Similarly, Grifman et al. showed that in the 293-based 10-3 cell line, which expresses E4-orf6 under the control of the sheep metallothionein promoter, the zinc-induced expression of E4-orf6 caused the rapid and specific degradation of p53 (45). Together these findings suggested that E4-orf6 is required to assemble the p53-directed ubiquitin ligase in uninfected 293 cells.
To test this hypothesis, 293 cells were transiently transfected with pcDNA3-E4orf6-Flu (85). 2A6 immunoprecipitations (111) were performed on nuclear extracts prepared from these cells, and the immunopurified complexes were subsequently analyzed by Western blotting. As shown in Fig. 4B, the transient expression of E4-orf6 led to the coimmunoprecipitation of Cullin-5, Elongins B and C, pp32, and importin-α with E1B-55K. This finding demonstrates that E4-orf6 is the only other viral protein required to induce the assembly of E1B-55K with these ubiquitin ligase complex components in 293 cells.
When examined by Superose 6 gel filtration chromatography, the E1B-55K protein in 293 nuclear extracts eluted as heterogeneous high-molecular-weight complexes extending into the void volume of the column (Fig. 4C). The subsequent Superose 6 fractionation of nuclear extracts prepared from E4-orf6-Flu-transfected 293 cells revealed an 800- to 900-kDa complex containing E1B-55K, E4-orf6, Cullin-5, and Elongins B and C, similar or identical to the complex in Ad5-infected HeLa cells (data not shown; Fig. 2A). Thus, in the presence of E4-orf6, a portion of the E1B-55K protein was redistributed to much lower molecular weight fractions in comparison to the E1B-55K in untransfected 293 cells (Fig. 4C). Based on these observations, it is likely that the elution profile observed for E1B-55K from Ad5-infected HeLa cells (Fig. 2A) is the result of its presence both in an 800- to 900-kDa complex with E4-orf6, Cullin-5, Rbx1/ROC1/Hrt1, and the Elongins B and C and in heterogeneous high-molecular-weight complexes like those observed in 293 cells.
DISCUSSION
To further elucidate the mechanisms underlying the early and late phase functions of E1B-55K, we sought to identify new molecular partners of the protein via a proteomics-based approach. A multiprotein, E1B-55K-associated complex of 800 to 900 kDa was immunopurified from Ad5-infected HeLa cells and shown to contain a set of proteins predicted to function in an SCF-type E3 ubiquitin ligase (Cullin-5, Rbx1/ROC1/Hrt1, and Elongins B and C), as well as a number of other proteins of both viral and cellular origin not previously known to associate with E1B-55K. We show here that this complex is capable of directing the polyubiquitination of p53 in vitro. We further demonstrate that these components exist together in a single, high-molecular-weight complex in vivo. In addition to the established SCF-type ubiquitin ligase components, we show that two additional cellular proteins, pp32 and importin-α1, copurify with this complex and may therefore also contribute to its function.
A large body of evidence indicates that E1B-55K, in complex with E4-orf6 (105, 110), induces the degradation of p53 via a ubiquitin- and proteasome-dependent mechanism (83, 84, 86, 98, 104, 122, 130). In the present study, we gain insight into the mechanism underlying this process by demonstrating that at least four cellular proteins previously implicated in the ubiquitination of proteins interact with E1B-55K (Fig. 1; Table 1). These include Rbx1/ROC1/Hrt1 and Elongins B and C, which have previously been shown to participate in an SCF-type E3 ubiquitin ligase with the Cullin-2 and von Hippel-Lindau proteins (22, 24, 29, 55, 59, 60, 63, 70, 92, 121), and Cullin-5, a member of the Cullin family of proteins (15, 16, 64). The Cullins are a recently identified family of proteins classified according to their sequence similarity to the Saccharomyces cerevisiae cdc53 protein and are each proposed to participate in the ubiquitin-dependent proteolysis of proteins (24, 32, 63). Indeed, in a recent study by Kamura et al., the Cullin-5 protein was shown to function together with MUF1, Rbx1/ROC1/Hrt1, and Elongins B and C to reconstitute a ubiquitin ligase in vitro (57).
In a work complementary to ours by Querido et al., these same four cellular proteins were found in association with E4-orf6 (97). Querido et al. further found, using a series of E4-orf6 in-frame deletion mutants, that the ability of the E4-orf6 mutants to mediate the accelerated degradation of p53 correlated with their binding to E1B-55K, Cullin-5, and Elongins B and C (97, 99). We now provide further evidence that these proteins act together as part of a single complex in vivo to ubiquitinate p53, by demonstrating that they can be isolated together with E1B-55K and E4-orf6 from Ad5-infected HeLa nuclear extracts by gel filtration chromatography (Fig. 2). Surprisingly, these components (E1B-55K, E4-orf6, Cullin-5, Elongins B and C, and Rbx1/ROC1/Hrt1) were found in a complex of 800 to 900 kDa, much larger than the sum of their molecular masses. Additionally, we found that the formation of this high-molecular-weight, SCF-type E3 complex could be induced in the E1-expressing 293 cells upon the introduction of an E4-orf6 expression construct (Fig. 4B and data not shown).
We further demonstrated that this high-molecular-weight complex can induce the polyubiquitination of p53 in vitro. When combined with unprogrammed rabbit reticulocyte lysate, E1B-55K-containing complexes immunopurified from Ad5-infected HeLa cells were found to possess a robust p53-directed ubiquitination activity, as indicated by the laddering of p53 (Fig. 3A). The higher-molecular-weight p53 species that resulted were in part the result of polyubiquitin chain addition (Fig. 3B and C). Additionally however, it appeared that p53 could be monoubiquitinated or otherwise posttranslationally modified at more than one site (Fig. 3C) (42, 73).
In a previous study, Querido et al. failed to observe the ubiquitination and degradation of p53 under similar reaction conditions, under which in vitro-translated E1B-55K, E4-orf6, and p53 were combined with an excess of rabbit reticulocyte lysate (99). It is possible that the E1B-55K immunopurified complexes utilized in our in vitro reactions contained additional factors that enabled the ubiquitination of p53 to occur more efficiently. In a later work, Querido et al. were able to reconstitute a weak, p53-directed ubiquitinating activity using E4-orf6-immunopurified complexes from insect cells coinfected with baculovirus expression vectors encoding E1B-55K, E4-orf6, Cullin-5, Rbx1/ROC1/Hrt1, and the Elongins B and C (97). The authors additionally noted, however, that the ubiquitination of p53 in this assay occurred at similar levels both in the presence and absence of E1B-55K, suggesting that E1B-55K was not absolutely required in this process (97).
This result is in contrast to previous observations that the binding of p53 to E1B-55K, but not to E4-orf6, is required for p53 degradation (99, 104, 117). Indeed, Querido et al. have shown that the p53 mutant p53KEEK fails to bind to E4-orf6 but can still be targeted for degradation by the E1B-55K/E4-orf6 complex by virtue of its ability to bind E1B-55K (99). Conversely, the p53 22/23 point mutant that fails to interact with E1B-55K, but likely retains binding to E4-orf6, remains stabilized in the presence of E1B-55K and E4-orf6 (99, 104, 130). Lastly, Shen et al. have shown that the E1B-55K point mutant R240A, retains binding to E4-orf6 and the ability to mediate late-phase functions together with E4-orf6 but fails to efficiently interact with p53 and accelerate its decay (117). Collectively, these findings indicate that the interaction between E1B-55K and p53 is critical for the recruitment of p53 to the E3 complex and suggest that the E1B-55K protein constitutes the principal substrate recognition component of this SCF-type ubiquitin ligase complex.
The E1B-55K immunoprecipitates utilized in our reactions were not, however, sufficient to direct the degradation of p53 in our in vitro assay. This is in contrast to the in vitro E3 ligase activity of the human papillomavirus (HPV) E6 protein (54, 113). Under the conditions used in our study, the HPV type 18 E6 protein is additionally able to stimulate the degradation of p53 (113). These findings suggest that the degradation of p53 induced by the E1B-55K/E4-orf6 complex occurs via a mechanism distinct from that established for the HPV E6 protein (97, 112).
The failure of the E1B-55K immunoprecipitate to facilitate the degradation of p53 in rabbit reticulocyte lysates may be due to the absence of critical factors required to mediate this process, as such lysates are prepared from enucleated cells (97). Alternatively, it is possible that the degradation of p53 may be coupled to another function of E1B-55K that may not be recapitulated in reticulocyte lysates (41). Lastly, the E1B-55K/E4-orf6-mediated degradation of p53 may in fact rely on the activity of nuclear proteasomes (85, 97). Indeed, the mdm2-induced degradation of p53 has been proposed to occur by both nuclear and cytoplasmic proteasomes (10, 31, 39). In support of a role for nuclear proteasomes in the degradation of p53 by E1B-55K and E4-orf6, Querido et al. have found that leptomycin B, a CRM1 nuclear export receptor inhibitor that blocks the export of E1B-55K/E4-orf6 complexes (28), had little effect on E1B-55K/E4-orf6-mediated p53 degradation (85, 99). Further investigation is clearly required to determine where the E1B-55K/E4-orf6-induced degradation of p53 occurs, and whether other cofactors or activities of the E1B-55K and E4-orf6 proteins are required.
It will be of additional interest to determine if the E1B-55K and E4-orf6 proteins are capable of directing the polyubiquitination and degradation of cellular proteins other than p53. Recent studies by Grifman et al. suggest that this may in fact be the case (45). Indeed, in the 293-derived 10-3 cells, the zinc-induced expression of E4-orf6 was followed by a rapid reduction in both p53 and cyclin A levels (45). The effect on these proteins was confirmed to be a specific event and not a general phenomenon, as the levels of other proteins involved in cell cycle control were not affected by the presence of E4-orf6 (45). Additionally, it was found that the regulation occurred at a posttranscriptional step (45). It will be interesting to determine if the degradation of cyclin A in this context occurs via a ubiquitin-proteasome-mediated pathway. It is possible that the targeted degradation of additional cellular factors by the E1B-55K/E4-orf6 ubiquitin ligase complex contributes to the other functions ascribed to these viral proteins, such as the stimulation of late viral mRNA nucleocytoplasmic transport and translation.
Our analysis of E1B-55K-containing complexes in Ad5-infected HeLa cells by gel filtration chromatography revealed that two additional cellular proteins—importin-α1 and pp32—appeared to reside in a single complex together with E1B-55K, E4-orf6, and the other major SCF-like ubiquitin ligase components. These factors were also found in E1B-55K immunoprecipitates from Ad5-infected 293 cells. Though the functional significance of these factors was not established with respect to p53-directed ubiquitination activity, their specific recruitment to the complex in Ad5-infected cells and in 293 cells transfected with an E4-orf6 expression vector suggests that they may participate in the various functions attributed to this complex.
Importin-α1 is a nuclear import factor that has been shown to initiate the nuclear entry of target proteins bearing a nuclear localization signal (NLS) (reviewed in references 56, 65, and 129). Importin-α1 binds specifically and directly to the NLS of cargo proteins and together with importin-β facilitates their movement across the nuclear pore (reviewed in reference 56, 65, and 129). The recruitment of importin-α1 to E1B-55K in E4-orf6-transfected 293 cells may be explained by the observation that E4-orf6 can direct the subcellular localization of E1B-55K (26, 41). When expressed in the absence of other adenoviral proteins in HeLa cells, E1B-55K is primarily restricted to the cytoplasm and adopts a distribution similar to that observed in Ad-transformed cells (8, 26, 41, 125, 135, 136). Upon the cointroduction of E4-orf6, however, the localization of E1B-55K is shifted to the nuclear compartment, which results in a largely diffuse, nucleoplasmic staining pattern (26, 41). It is believed that an amphipathic arginine-rich alpha-helix in the C terminus of E4-orf6 functions as an NLS that is responsible for the targeting and retention of E4-orf6 and E4-orf6-containing complexes to the nucleus (26, 89, 90). Indeed, E1B-55K/E4-orf6 complexes are known to undergo dynamic, nucleocytoplasmic shuttling (26, 28, 41). The association of importin-α1 with the nuclear complex described here may result from its binding to the E4-orf6 NLS during the import phase of nucleocytoplasmic shuttling.
pp32 is a member of a family of acidic, leucine-rich nuclear phosphoproteins found in cells capable of self-renewal (77, 127). It has been demonstrated to have a number of activities, including the suppression of transformation in vitro by various oncogene pairs and a potent and specific inhibition of protein phosphatase 2A (PP2A), a major serine/threonine phosphatase involved in the regulation of a variety of cellular events (4, 19, 68). The association of pp32 with E1B-55K may reflect a common theme among the DNA tumor viruses to modulate the activities of PP2A (106). Indeed, PP2A is inhibited by SV40 small t antigen and polyomavirus middle t antigen, thus contributing to transformation by these viruses (106). Alternatively, since phosphorylation stimulates the association of a number of substrates with their corresponding E3 ubiquitin ligases (24), the association of a phosphatase inhibitor with the Ad-specific ubiquitin ligase complex may ensure that PP2A does not remove such signals.
pp32 has additionally been shown to be a ligand for the HuR protein, a member of the ELAV (embryonic lethal abnormal vision) family of RNA-binding proteins that regulates the stability and nucleocytoplasmic trafficking of a subset of cellular mRNAs (12, 40, 76). It has been proposed that pp32 modulates the activities of HuR, by mediating HuR export from the nucleus, and enhancing its affinity for target mRNAs (12, 13, 36). Consistent with this type of function, pp32 itself has been shown to shuttle between the nucleus and cytoplasm in a CRM1-dependent fashion (12).
Like HuR, E1B-55K has been proposed to modulate the preferential nucleocytoplasmic transport of viral mRNAs in the late phase of infection. Indeed, genetic studies have shown that in the absence of a functional E1B-55K protein, the late viral messages fail to preferentially accumulate in the cytoplasm and host cell mRNAs continue to be trafficked and translated into the late phase of infection (2, 3, 6, 47, 67, 95, 131). A similar defect has been noted for viruses harboring mutations in E4-orf6 (14, 47, 109, 128). In support of a role for E1B-55K in this type of RNA export function, E1B-55K has been shown to bind to RNA, and the E1B-55K/E4-orf6 complex exhibits a nucleocytoplasmic shuttling activity (26, 28, 41, 53). Perhaps pp32 modulates the RNA binding and export functions of the E1B-55K/E4-orf6 complex as it does those of HuR (12, 13, 36). Our finding that pp32 is associated with a ubiquitin ligase complex raises the question of whether protein ubiquitination plays a role in pp32-regulated transport of cellular mRNAs.
pp32 has also been shown to be a part of the INHAT complex (inhibitor of acetyltransferases), a cellular complex biochemically purified on the basis of its ability to inhibit histone acetylation by p300 (115). The subunits of this complex include human template activating factor 1α (TAF-1α), Set/TAF-1β (myeloid leukemia-associated oncoprotein), and two phosphoforms of pp32 (115). Each member of this four-subunit complex has been demonstrated to inhibit histone acetylation by both p300/CBP and PCAF, by binding to their substrate histones and preventing them from serving as substrates (114, 115). Indeed, in vitro-labeled TAF-1α, Set/TAF-1β, and pp32 all efficiently associate with purified histones, and these INHAT subunits have also been found to associate with histones on chromatin in vivo (115). The “masking” of histones by this complex has additionally been shown to block histone acetylase (HAT)-dependent transcriptional activation by CBP in transfection studies (115). It was therefore broadly proposed that the INHAT complex mediates a key event in chromatin remodeling and transcriptional regulation, by either inhibiting or enhancing the access of histones to the acetylases (115).
It would be interesting to determine if the various INHAT components can also modulate the acetylation of other HAT substrates, such as p53. p53 has been shown to be acetylated by p300, CBP, and PCAF, and its DNA-binding ability seems to vary according to its acetylation status (46, 71, 107). In this context, it is noteworthy that the E1B-55K protein appears to specifically inhibit the acetylation of p53 by PCAF both in vivo and in vitro (72). In cells stably expressing E1B-55K for example, the levels of acetylated p53 are reduced, and a corresponding decrease in the sequence-specific DNA binding ability of p53 is observed (72). It has additionally been shown that the E1B-55K protein interferes with the physical association between PCAF and p53, suggesting that the principal mode of inhibition is the prevention of an enzyme-substrate interaction (72). It remains possible, however, that E1B-55K recruits pp32 to acetylation sites on p53 and thereby prevents its acetylation by occluding their access to PCAF. E1B-55K may thus repress transcription from p53-responsive promoters during the early phase of infection by a dual mechanism, involving the recruitment of both HAT inhibitors and transcriptional corepressors to p53 (79).
Our analysis of E1B-55K coprecipitating proteins also produced the L4-100K late nonstructural protein, which has been shown to be required for the efficient translation of the late viral mRNAs (50). Though this protein was also initially detected in complex with E4-orf6 in Ad5-infected cells, the reverse immunoprecipitation performed with an anti-L4-100K antibody failed to coprecipitate either the E4-orf6 or E1B-55K proteins (9). Furthermore, as the L4-100K protein was detected in control anti-E4-orf6 immunoprecipitates from cells infected with a virus that did not express E4-orf6, it was concluded that the association between E4-orf6 and L4-100K was nonspecific (9).
The analysis of the remaining E1B-55K copurifying proteins identified factors whose presence cannot be readily understood in the context of the known functions of E1B-55K. These include the NuMA, E-MAP-115, β-tubulin, and Ad IVa2 proteins. NuMA and E-MAP-115 have both been implicated in the modulation of microtubule dynamics (reviewed in references 17, 80, 81, and 137). NuMA has been shown to be required for the organization of the mitotic spindle (reviewed in reference 137), and E-MAP-115 appears to be involved in the stabilization and reorganization of microtubules (reviewed in references 17, 80, and 81). Interestingly, pp32 (also known as mapmodulin) has also been shown to bind a number of microtubule-associated proteins and appears to slow their rate of association with microtubules (124). The only previously identified functions of the Ad IVa2 protein are in the transactivation of the major late promoter and in viral DNA packaging (75, 82, 123, 138, 139). It is therefore not clear at present if or how these factors may impact E1B-55K function. What is clear, however, is that the interactions made by the E1B-55K protein are far more numerous and complex than previously thought.
Finally, the fact that the known E1B-55K interactors E1B-AP5 (34), PCAF (72), histone deacetylase 1 (96), and mSin3A (96) did not appear to coprecipitate with E1B-55K in our analysis (Fig. 1A) suggests that the E1B-55K-anchored proteome is substantially more complex than revealed in this work. It remains formally possible that these factors were in fact present in substoichiometric amounts in our E1B-55K immunoprecipitates prepared from Ad5-infected HeLa cells (Fig. 1A) but were not detected under the conditions utilized in our study. Alternatively, it is possible that complexes containing E1B-55K and these factors were enriched in the cytoplasmic fraction generated during the preparation of nuclear extracts from Ad-infected cells. This fraction, which we did not analyze, contained ∼50% of the total cellular E1B-55K. (J. N. Harada, unpublished results). The extracted nuclear fraction that we analyzed contained the remaining ∼50%, while the insoluble nuclear fraction contained <∼5%.
In this work we have described the immunopurification of proteins associated with the E1B-55K protein of Ad5. Though our work has increased our knowledge of the mechanism by which adenovirus counters the effects of p53, it has also made it clear that we are just beginning to understand the many ways in which the E1B-55K protein can impact the virus's life cycle. Additional studies are clearly required to understand how these factors may contribute to the many viral processes mediated by E1B-55K. Better understanding of these molecular contacts may eventually provide insight for the design of an improved oncolytic vector that is more effective for tumor therapy than the existing 55K deletion mutant, dl1520 (ONYX-015) (7).
Acknowledgments
We thank Carol Eng for providing excellent technical assistance, Xing Xian Yu for assistance in making the E32 antiserum, and Sergei Sikora and Helen Brown for helpful discussions. We also acknowledge Sergei Sikora's contributions to the production of the anti-Cullin-5 polyclonal serum. We are grateful to Claude Kedinger, Philip Branton, Joan Conaway, Duane Compton, J. Chloe Bulinski, and Patrick Hearing for their kind gifts of antisera directed against the Ad IVa2, Cullin-5, Elongin B, NuMA, E-MAP-115/ensconsin, and E4-orf6 proteins, respectively. We also thank Arnold Levine for providing us with both the 2A6 anti-E1B-55K and (M186)B6 anti-Ad 72K DBP hybridoma supernatants and Thomas Dobner for providing us with pcDNA3-E4orf6-Flu.
This work was supported by Public Health Service grant CA 64799 to A.J.B. and was supported in part by a Public Health Service National Research Award GM07185 to J.N.H. and NIH grant CA57327 to D.C.P.
REFERENCES
- 1.Babich, A., L. T. Feldman, J. R. Nevins, J. E. Darnell, Jr., and C. Weinberger. 1983. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translational discrimination. Mol. Cell. Biol. 3:1212-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss, L. E., and H. S. Ginsberg. 1984. Adenovirus type 5 early region 1B gene product is required for efficient shutoff of host protein synthesis. J. Virol. 50:202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, J., J. R. Brody, S. S. Kadkol, and G. R. Pasternack. 2001. Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene 20:2153-2160. [DOI] [PubMed] [Google Scholar]
- 5.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. [DOI] [PubMed] [Google Scholar]
- 6.Beltz, G. A., and S. J. Flint. 1979. Inhibition of HeLa cell protein synthesis during adenovirus infection. J. Mol. Biol. 131:353-373. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff, J. R., D. H. Kirn, A. Williams, C. Heise, S. Horn, M. Muna, L. Ng, J. A. Nye, A. Sampson-Johannes, A. Fattaey, and F. McCormick. 1996. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 274:373-376. [DOI] [PubMed] [Google Scholar]
- 8.Blair Zajdel, M. E., M. D. Barker, S. C. Dixon, and G. E. Blair. 1985. The use of monoclonal antibodies to study the proteins specified by the transforming region of human adenoviruses. Biochem. J. 225:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boivin, D., M. R. Morrison, R. C. Marcellus, E. Querido, and P. E. Branton. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 73:1245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd, S. D., K. Y. Tsai, and T. Jacks. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563-568. [DOI] [PubMed] [Google Scholar]
- 11.Boyer, J. L., and G. Ketner. 2000. Genetic analysis of a potential zinc-binding domain of the adenovirus E4 34k protein. J. Biol. Chem. 275:14969-14978. [DOI] [PubMed] [Google Scholar]
- 12.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 15.Burnatowska-Hledin, M. A., W. S. Spielman, W. L. Smith, P. Shi, J. M. Meyer, and D. L. Dewitt. 1995. Expression cloning of an AVP-activated, calcium-mobilizing receptor from rabbit kidney medulla. Am. J. Physiol. 268:F1198-F1210. [DOI] [PubMed] [Google Scholar]
- 16.Byrd, P. J., T. Stankovic, C. M. McConville, A. D. Smith, P. R. Cooper, and A. M. Taylor. 1997. Identification and analysis of expression of human VACM-1, a cullin gene family member located on chromosome 11q22-23. Genome Res. 7:71-75. [DOI] [PubMed] [Google Scholar]
- 17.Cassimeris, L., and C. Spittle. 2001. Regulation of microtubule-associated proteins. Int. Rev. Cytol. 210:163-226. [DOI] [PubMed] [Google Scholar]
- 18.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, T. H., J. R. Brody, F. E. Romantsev, J. G. Yu, A. E. Kayler, E. Voneiff, F. P. Kuhajda, and G. R. Pasternack. 1996. Structure of pp32, an acidic nuclear protein which inhibits oncogene-induced formation of transformed foci. Mol. Biol. Cell 7:2045-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiou, S.-K., and E. White. 1997. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J. Virol. 71:3515-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22:442-451. [DOI] [PubMed] [Google Scholar]
- 22.Cockman, M. E., N. Masson, D. R. Mole, P. Jaakkola, G. W. Chang, S. C. Clifford, E. R. Maher, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275:25733-25741. [DOI] [PubMed] [Google Scholar]
- 23.Debbas, M., and E. White. 1993. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7:546-554. [DOI] [PubMed] [Google Scholar]
- 24.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell. Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 25.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. [DOI] [PubMed] [Google Scholar]
- 26.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 28.Dosch, T., F. Horn, G. Schneider, F. Kratzer, T. Dobner, J. Hauber, and R. H. Stauber. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan, D. R., A. Pause, W. H. Burgess, T. Aso, D. Y. Chen, K. P. Garrett, R. C. Conaway, J. W. Conaway, W. M. Linehan, and R. D. Klausner. 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269:1402-1406. [DOI] [PubMed] [Google Scholar]
- 30.Faire, K., C. M. Waterman-Storer, D. Gruber, D. Masson, E. D. Salmon, and J. C. Bulinski. 1999. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J. Cell Sci. 112:4243-4255. [DOI] [PubMed] [Google Scholar]
- 31.Freedman, D. A., and A. J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18:7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furukawa, M., T. Ohta, and Y. Xiong. 2002. Activation of UBC5 ubiquitin conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J. Biol. Chem. 277:15758-15765. [DOI] [PubMed] [Google Scholar]
- 33.Furukawa, M., Y. Zhang, J. McCarville, T. Ohta, and Y. Xiong. 2000. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20:8185-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabler, S., H. Schutt, P. Groitl, H. Wolf, T. Shenk, and T. Dobner. 1998. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 72:7960-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaglio, T., A. Saredi, and D. A. Compton. 1995. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 131:693-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 37.Garrett, K. P., T. Aso, J. N. Bradsher, S. I. Foundling, W. S. Lane, R. C. Conaway, and J. W. Conaway. 1995. Positive regulation of general transcription factor SIII by a tailed ubiquitin homolog. Proc. Natl. Acad. Sci. USA 92:7172-7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrett, K. P., D. Haque, R. C. Conaway, and J. W. Conaway. 1994. A human cDNA encoding the small subunit of RNA polymerase II transcription factor SIII. Gene 150:413-414. [DOI] [PubMed] [Google Scholar]
- 39.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569-573. [DOI] [PubMed] [Google Scholar]
- 40.Good, P. J. 1995. A conserved family of elav-like genes in vertebrates. Proc. Natl. Acad. Sci. USA 92:4557-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodrum, F. D., T. Shenk, and D. A. Ornelles. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol. 70:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characterization of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-72. [DOI] [PubMed] [Google Scholar]
- 44.Grand, R. J. A., M. L. Grant, and P. H. Gallimore. 1994. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology 203:229-240. [DOI] [PubMed] [Google Scholar]
- 45.Grifman, M., N. N. Chen, G. P. Gao, T. Cathomen, J. M. Wilson, and M. D. Weitzman. 1999. Overexpression of cyclin A inhibits augmentation of recombinant adeno-associated virus transduction by the adenovirus E4orf6 protein. J. Virol. 73:10010-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 47.Halbert, D. N., and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harada, J. N., and A. J. Berk. 1999. p53-independent and -dependent requirements for E1B-55K in adenovirus type 5 replication. J. Virol. 73:5333-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 52.Hori, T., F. Osaka, T. Chiba, C. Miyamoto, K. Okabayashi, N. Shimbara, S. Kato, and K. Tanaka. 1999. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18:6829-6834. [DOI] [PubMed] [Google Scholar]
- 53.Horridge, J. J., and K. N. Leppard. 1998. RNA-binding activity of the E1B 55-kilodalton protein from human adenovirus type 5. J. Virol. 72:9374-9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Izaurralde, E., and S. Adam. 1998. Transport of macromolecules between the nucleus and the cytoplasm. RNA 4:351-364. [PMC free article] [PubMed] [Google Scholar]
- 57.Kamura, T., D. Burian, Q. Yan, S. L. Schmidt, W. S. Lane, E. Querido, P. E. Branton, A. Shilatifard, R. C. Conaway, and J. W. Conaway. 2001. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276:29748-29753. [DOI] [PubMed] [Google Scholar]
- 58.Kamura, T., M. N. Conrad, Q. Yan, R. C. Conaway, and J. W. Conaway. 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 60.Kamura, T., S. Sato, K. Iwai, M. Czyzyk-Krzeska, R. C. Conaway, and J. W. Conaway. 2000. Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA 97:10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kao, C. C., P. R. Yew, and A. J. Berk. 1990. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology 179:806-814. [DOI] [PubMed] [Google Scholar]
- 62.Kawakami, T., T. Chiba, T. Suzuki, K. Iwai, K. Yamanaka, N. Minato, H. Suzuki, N. Shimbara, Y. Hidaka, F. Osaka, M. Omata, and K. Tanaka. 2001. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20:4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kibel, A., O. Iliopoulos, J. A. DeCaprio, and W. G. Kaelin, Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269:1444-1446. [DOI] [PubMed] [Google Scholar]
- 64.Kipreos, E. T., L. E. Lander, J. P. Wing, W. W. He, and E. M. Hedgecock. 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85:829-839. [DOI] [PubMed] [Google Scholar]
- 65.Komeili, A., and E. K. O'Shea. 2001. New perspectives on nuclear transport. Annu. Rev. Genet. 35:341-364. [DOI] [PubMed] [Google Scholar]
- 66.Lee, K. A., A. Bindereif, and M. R. Green. 1988. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal. Tech. 5:22-31. [DOI] [PubMed] [Google Scholar]
- 67.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1B 55 kD protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li, M., A. Makkinje, and Z. Damuni. 1996. Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry 35:6998-7002. [DOI] [PubMed] [Google Scholar]
- 69.Liakopoulos, D., T. Busgen, A. Brychzy, S. Jentsch, and A. Pause. 1999. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc. Natl. Acad. Sci. USA 96:5510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lisztwan, J., G. Imbert, C. Wirbelauer, M. Gstaiger, and W. Krek. 1999. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 13:1822-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu, Y., A. L. Colosimo, X. J. Yang, and D. Liao. 2000. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol. Cell. Biol. 20:5540-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lohrum, M. A., D. B. Woods, R. L. Ludwig, E. Balint, and K. H. Vousden. 2001. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21:8521-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus E1A and accompanies apoptosis. Genes Dev. 7:535-545. [DOI] [PubMed] [Google Scholar]
- 75.Lutz, P., and C. Kedinger. 1996. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J. Virol. 70:1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 77.Malek, S. N., A. I. Katumuluwa, and G. R. Pasternack. 1990. Identification and preliminary characterization of two related proliferation-associated nuclear phosphoproteins. J. Biol. Chem. 265:13400-13409. [PubMed] [Google Scholar]
- 78.Martin, M. E., and A. J. Berk. 1998. Adenovirus E1B 55K represses p53 activation in vitro. J. Virol. 72:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin, M. E. D., and A. J. Berk. 1999. Corepressor required for adenovirus E1B 55,000-molecular-weight protein repression of basal transcription. Mol. Cell. Biol. 19:3403-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masson, D., and T. E. Kreis. 1995. Binding of E-MAP-115 to microtubules is regulated by cell cycle-dependent phosphorylation. J. Cell Biol. 131:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Masson, D., and T. E. Kreis. 1993. Identification and molecular characterization of E-MAP-115, a novel microtubule-associated protein predominantly expressed in epithelial cells. J. Cell Biol. 123:357-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mondesert, G., C. Tribouley, and C. Kedinger. 1992. Identification of a novel downstream binding protein implicated in late-phase-specific activation of the adenovirus major late promoter. Nucleic Acids Res. 20:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 1997. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc. Natl. Acad. Sci. USA 94:1206-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 2000. Two distinct activities contribute to the oncogenic potential of the adenovirus type 5 E4orf6 protein. J. Virol. 74:5168-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nevels, M., T. Spruss, H. Wolf, and T. Dobner. 1999. The adenovirus E4orf6 protein contributes to malignant transformation by antagonizing E1A-induced accumulation of the tumor suppressor protein p53. Oncogene 18:9-17. [DOI] [PubMed] [Google Scholar]
- 87.Obert, S., R. J. O'Connor, S. Schmid, and P. Hearing. 1994. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol. Cell. Biol. 14:1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 89.Orlando, J. S., and D. A. Ornelles. 1999. An arginine-faced amphipathic alpha helix is required for adenovirus type 5 E4orf6 protein function. J. Virol. 73:4600-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Orlando, J. S., and D. A. Ornelles. 2002. E4orf6 variants with separate abilities to augment adenovirus replication and direct nuclear localization of the E1B 55-kilodalton protein. J. Virol. 76:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pause, A., B. Peterson, G. Schaffar, R. Stearman, and R. D. Klausner. 1999. Studying interactions of four proteins in the yeast two-hybrid system: structural resemblance of the pVHL/elongin BC/hCUL-2 complex with the ubiquitin ligase complex SKP1/cullin/F-box protein. Proc. Natl. Acad. Sci. USA 96:9533-9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pickart, C. M. 2000. Ubiquitin in chains. Trends Biochem. Sci. 25:544-548. [DOI] [PubMed] [Google Scholar]
- 94.Pilder, S., K. Leppard, J. Logan, and T. Shenk. 1986. Functional analysis of the adenovirus E1B-55K polypeptide. Cancer Cells 4:285-290. [Google Scholar]
- 95.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Punga, T., and G. Akusjarvi. 2000. The adenovirus-2 E1B-55K protein interacts with a mSin3A/histone deacetylase 1 complex. FEBS Lett. 476:248-252. [DOI] [PubMed] [Google Scholar]
- 97.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Querido, E., M. R. Morrison, H. Chu-Pham-Dang, S. W. Thirlwell, D. Boivin, P. E. Branton, and M. R. Morisson. 2001. Identification of three functions of the adenovirus E4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Querido, E., J. G. Teodoro, and P. E. Branton. 1997. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J. Virol. 71:3526-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rao, L., M. Debbas, D. Sabbatini, D. Hockenberry, S. Korsmeyer, and E. White. 1992. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc. Natl. Acad. Sci. USA 89:7742-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480-484. [DOI] [PubMed] [Google Scholar]
- 103.Ridgway, P. J., A. R. Hall, C. J. Myers, and A. W. Braithwaite. 1997. p53/E1b58kDa complex regulates adenovirus replication. Virology 237:404-413. [DOI] [PubMed] [Google Scholar]
- 104.Roth, J., C. Konig, S. Wienzek, S. Weigel, S. Ristea, and M. Dobbelstein. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 72:8510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rubenwolf, S., H. Schutt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 71:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rundell, K., and R. Parakati. 2001. The role of the SV40 ST antigen in cell growth promotion and transformation. Semin. Cancer Biol. 11:5-13. [DOI] [PubMed] [Google Scholar]
- 107.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 109.Sandler, A. B., and G. Ketner. 1989. Adenovirus early region 4 is essential for normal stability of late nuclear RNAs. J. Virol. 63:624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sarnow, P., P. Hearing, C. W. Anderson, D. N. Halbert, T. Shenk, and A. J. Levine. 1984. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early-region 4 25,000-dalton protein in productively infected cells. J. Virol. 49:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510-517. [DOI] [PubMed] [Google Scholar]
- 112.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 113.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 15 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 114.Seo, S. B., T. Macfarlan, P. McNamara, R. Hong, Y. Mukai, S. Heo, and D. Chakravarti. 2002. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J. Biol. Chem. 277:14005-14010. [DOI] [PubMed] [Google Scholar]
- 115.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 116.Seol, J. H., R. M. Feldman, W. Zachariae, A. Shevchenko, C. C. Correll, S. Lyapina, Y. Chi, M. Galova, J. Claypool, S. Sandmeyer, K. Nasmyth, and R. J. Deshaies. 1999. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13:1614-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shen, Y., G. Kitzes, J. A. Nye, A. Fattaey, and T. Hermiston. 2001. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J. Virol. 75:4297-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shevchenko, A., O. N. Jensen, A. V. Podtelejnikov, F. Sagliocco, M. Wilm, O. Vorm, P. Mortensen, H. Boucherie, and M. Mann. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 93:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 120.Skowyra, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 121.Stebbins, C. E., W. G. Kaelin, Jr., and N. P. Pavletich. 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284:455-461. [DOI] [PubMed] [Google Scholar]
- 122.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 123.Tribouley, C., P. Lutz, A. Staub, and C. Kedinger. 1994. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 68:4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ulitzur, N., M. Humbert, and S. R. Pfeffer. 1997. Mapmodulin: A possible modulator of the interaction of microtubule-associated proteins with microtubules. Proc. Natl. Acad. Sci. USA 94:5084-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van den Heuvel, S. J., T. van Laar, W. M. Kast, C. J. Melief, A. Zantema, and A. J. van der Eb. 1990. Association between the cellular p53 and the adenovirus 5 E1B-55kd proteins reduces the oncogenicity of Ad-transformed cells. EMBO J. 9:2621-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wada, H., E. T. Yeh, and T. Kamitani. 1999. Identification of NEDD8-conjugation site in human cullin-2. Biochem. Biophys. Res. Commun. 257:100-105. [DOI] [PubMed] [Google Scholar]
- 127.Walensky, L. D., D. S. Coffey, T. H. Chen, T. C. Wu, and G. R. Pasternack. 1993. A novel Mr 32,000 nuclear phosphoprotein is selectively expressed in cells competent for self-renewal. Cancer Res. 53:4720-4726. [PubMed] [Google Scholar]
- 128.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weis, K. 1998. Importins and exportins: how to get in and out of the nucleus. Trends Biochem. Sci. 23:185-189. [DOI] [PubMed] [Google Scholar]
- 130.Wienzek, S., J. Roth, and M. Dobbelstein. 2000. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J. Virol. 74:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Williams, J., B. D. Karger, Y. S. Ho, C. L. Castaglia, T. Mann, and S. J. Flint. 1986. The adenovirus E1B 495R protein plays a role in regulating the transport and stability of the viral late messages. Cancer Cells 4:275-284. [Google Scholar]
- 132.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 133.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]
- 134.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]
- 135.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44-58. [DOI] [PubMed] [Google Scholar]
- 136.Zantema, A., P. I. Schrier, A. Davis-Olivier, T. van Laar, R. T. Vaessen, and E. A. van der Eb. 1985. Adenovirus serotype determines association and localization of the large E1B tumor antigen with cellular tumor antigen p53 in transformed cells. Mol. Cell. Biol. 5:3084-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zeng, C. 2000. NuMA: a nuclear protein involved in mitotic centrosome function. Microsc. Res. Tech. 49:467-477. [DOI] [PubMed] [Google Scholar]
- 138.Zhang, W., and M. J. Imperiale. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang, W., J. A. Low, J. B. Christensen, and M. J. Imperiale. 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75:10446-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang, Y., D. Feigenblum, and R. J. Schneider. 1994. A late adenovirus factor induced eIF-4E dephosphorylation and inhibition of cell protein synthesis. J. Virol. 68:7040-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]