Abstract
Immediate-early protein IE1 is a principal regulator of viral transcription and a contributor to origin-specific DNA replication of the baculovirus Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV). Since these viral functions involve interaction of dimeric IE1 with palindromic homologous region (hr) enhancer-origin elements of the AcMNPV genome within the nucleus, it is presumed that proper nuclear transport of IE1 is essential for productive infection. To investigate the mechanisms of IE1 nuclear import, we analyzed the effect of site-directed mutations on IE1 subcellular distribution. As demonstrated by fluorescence microscopy and biochemical fractionation of plasmid-transfected cells, wild-type IE1 localized predominantly to the nucleus. Substitution or deletion of amino acid residues within a positively charged domain (residues 534 to 538) adjacent to IE1's oligomerization motif impaired nuclear import and caused loss of transactivation. Moreover, upon coexpression, these import-defective mutations prevented nuclear entry of wild-type IE1. In contrast, double-mutated IE1 defective for both nuclear import and dimerization failed to block nuclear entry or transactivation by wild-type IE1. Thus, import-defective IE1 dominantly interfered with wild-type IE1 by direct interaction and cytosolic trapping. Collectively, our data indicate that the small basic domain encompassing residues R537 and R538 constitutes a novel nuclear localization element that functions only upon IE1 dimerization. These findings support a model wherein IE1 oligomerizes within the cytosol as a prerequisite for nuclear entry and subsequent high-affinity interaction with the symmetrical binding sites comprising AcMNPV hr enhancer-origin elements.
Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is the best-characterized member of Baculoviridae, a family of large, double-stranded DNA viruses pathogenic to invertebrates (for a recent review, see reference 11). AcMNPV replication occurs within the nucleus of infected cells, necessitating timely nuclear import of viral proteins involved in transcription, DNA replication, and virus assembly. Among these required nuclear factors is IE1, an immediate-early protein that regulates viral transcription and contributes to DNA replication (for reviews, see references 10 and 38). IE1 is a highly conserved, 67-kDa DNA binding protein that transactivates baculovirus early gene promoters and supports late gene expression in plasmid transfection assays (4, 7, 14, 16, 25, 26, 28, 31, 36, 43, 50-53, 56). In similar assays, IE1 is one of six viral factors required for plasmid DNA replication (24, 39). The involvement of IE1 in these AcMNPV replicative processes and its association with viral DNA replication factories within the nucleus (44) suggest that proper IE1 nuclear transport is essential for productive infection.
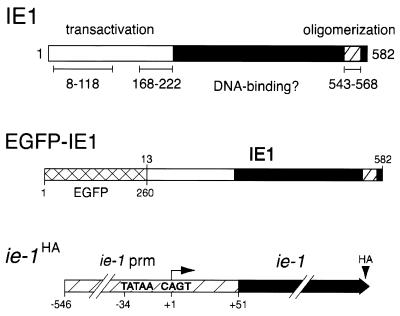
AcMNPV IE1 possesses separable domains that contribute to promoter transactivation and DNA binding (Fig. 1). The N-terminal half of this 582-residue phosphoprotein contains transcriptional stimulatory domains from residue 8 to 118 and 168 to 222 (25, 54, 56). The C-terminal three-fourths of IE1 is required for DNA binding, but the residues involved in direct DNA interaction are unknown. IE1 binds to the ∼28-bp imperfect palindrome (28-mer) that constitutes repetitive sequences within multiple homologous regions (hrs) found dispersed throughout the AcMNPV genome (13, 15, 28, 52). The hrs function as transcriptional enhancers and probable origins of DNA replication (12, 13, 31, 43, 48, 52, 53). IE1-mediated stimulation of promoters cis-linked to the hr enhancer is correlated with an increase in the level of transcription initiation (15). The hr 28-mer is the minimal sequence motif required for IE1-mediated enhancer and origin-specific replication functions. IE1 interacts with the 28-mer as a dimer, presumably by making direct contact with the palindromic half sites, both of which are required for hr enhancer activity (12, 28, 32, 53, 54). A helix-loop-helix (HLH)-like domain (residues 543 to 568) at the C terminus of IE1 mediates homo-oligomerization (Fig. 1). Our previous studies indicated that amino acid substitutions within the HLH-like domain disrupt IE1 oligomerization, thereby eliminating 28-mer binding and causing loss of promoter transactivation (46, 54). Thus, oligomerization is required for IE1 replicative functions. In this report, we show that oligomerization is also a prerequisite for proper import of IE1 into the nucleus.
FIG. 1.
Structure of IE1 and EGFP-IE1 fusions. Top panel: IE1 functional domains. The 582-residue IE1 protein possesses transcriptional transactivation domains (open) from residue 8 to 118 and 168 to 222 and an oligomerization domain (hatched) from residue 543 to 568. The DNA binding domain is unknown. Middle panel: EGFP-IE1. Residues 1 to 260 of EGFP (crosshatched) were fused to residues 13 to 582 of IE1 (shaded). Bottom panel: genetic organization of ie-1HA expression plasmids. Sequences encoding the HA epitope were inserted into the ie-1 open reading frame (solid) after residue 579. The resulting ie-1HA gene or mutations thereof were placed under control of the full-length ie-1 promoter (nucleotides −546 to +51) for transient expression assays. Transcription initiates (arrow) from the CAGT (+1) initiator motif.
Nuclear transport is a highly regulated, energy-dependent process by which selected proteins traffic in and out of the nucleus (for reviews, see references 20 and 23). Regulated import of larger proteins (>40 kDa) usually involves the interaction of shuttle proteins, including importin α, with a nuclear localization signal (NLS) embedded within the transported protein. The classical monopartite NLS, such as that found in large T antigen of simian virus 40 (SV40), consists of a stretch of positively charged arginines or lysines flanked by helix-breaking residues (20, 23). Nuclear entry of several baculovirus proteins has been examined, and putative NLS elements have been identified (2, 3, 9, 19, 21, 22, 27, 35, 37, 44, 49). However, the molecular mechanisms of baculovirus nuclear targeting are poorly understood.
To define the NLS of AcMNPV IE1 and investigate the mechanism of nuclear import of an early viral protein, we mapped IE1 domains required for nuclear entry by using site-directed mutagenesis. Consistent with nuclear localization of IE1 during infection (44), wild-type IE1 was concentrated in the nucleus after plasmid transfection of cultured Spodoptera frugiperda SF21 cells. Substitutions and deletion of residues within a positively charged domain preceding the C-terminal HLH-like domain of IE1 disrupted nuclear import and caused loss of transactivation. Furthermore, nuclear import-defective IE1 dominantly interfered with wild-type IE1 by prohibiting nuclear entry through direct interaction and cytosolic trapping. The failure of heterodimers consisting of wild-type and import-defective IE1 to enter the nucleus indicated that each monomer of the IE1 dimer contributes to nuclear import. Thus, analogous to basic-HLH transcription activators (for reviews, see references 34, 41, and 47), the positively charged motif located immediately adjacent to the IE1 oligomerization domain is required for nuclear import and likely functions as the IE1 NLS. However, this nuclear localization element (NLE) is distinguished by its trans-acting, bipartite structure, which must dimerize for activity.
MATERIALS AND METHODS
Plasmids. (i) IE1 basic domain II alterations.
Plasmid pIE1 encodes the wild-type AcMNPV ie-1 gene under control of its own full-length promoter (43). The influenza virus hemagglutinin (HA) epitope was inserted after IE1 residue 579 of pIE1BS (50) by using PCR mutagenesis to generate pIE1HA/BS (46) (Fig. 1). IE1 amino acid substitutions R524A/K526A, R537A/R538A, and L550D/I554E were generated by site-directed mutagenesis of pIE1BS and subsequently inserted into pIE1HA/BS to generate pIE1B1/HA, pIE1B2/HA, and pIE1H1/HA. The IE1 substitutions R524A/K526A and R537A/R538A were combined to generate pIE1B1:B2/HA by inserting the SwaI-SfaNI fragment of pIE1B1/HA into the corresponding sites of pIE1B2/HA. Sequences encoding residues 534 to 538 were deleted from within pIE1HA/BS by using the oligonucleotides 5′-G CAT CAC ATG TTT GTA ATC GGT GAG AGC ACT AC-3′ and 5′-CAA ATT ATT GTG CAA TGT AGT GCT CTC ACC GAT TAC-3′ to generate pIE1ΔB. The IE1 substitution K534N/R537G/R538N was engineered into pIE1HA/BS by using complementary oligonucleotides 5′-C GGT AAT GTT AAC GGA AAT GAG AGC ACT ACA TTG CAC-3′ and 5′-GTG CAA TGT AGT GCT CTC ATT TCC GTT AAC ATT ACC G-3′ to generate pIE1B3/HA. We noted that pIE1B3/HA contained a second site mutation, I532T, that had no effect on IE1 intracellular localization. The substitution K534N/R537G/R538N was also engineered into pIE1H1/HA by site-directed mutagenesis using complementary oligonucleotides 5′-C GGT AAT GTT AAC GGA AAT GAG AGC ACT AC-3′ and 5′-GT AGT GCT CTC ATT TCC GTT AAC ATT ACC G-3′ to generate pIE1B3:H1/HA. To remove the HA epitope from pIE1B3/HA and pIE1B3:H1/HA, the BstBI-NdeI fragment was replaced with the corresponding fragment of pIE1BS. All site-directed mutations were verified by nucleotide sequence determination.
(ii) IE1 basic domain I alterations.
Substitutions K152A/K154A, R156A/K158A, and K160A/K161A were generated by site-directed mutagenesis of pIE1HA/BS by using the complementary oligonucleotide pairs 5′-GTG GTG GGC CAG TTT AAC GCA ATT GCA TTG AGG CC-3′ and 5′-GG CCT CAA TGC AAT TGC GTT AAA CTG GCC CAC CAC-3′, 5′-G GGC CAG TTT AAC AAA ATT AAG CTT GCG CCT GCA TAC AAG-3′ and 5′-CTT GTA TGC AGG CGC AAG CTT AAT TTT GTT AAA CTG GCC C-3′, and 5′-GG CCT AAA TAC GCG GCT AGC ACA ATT CAA AGC TGT GC-3′ and 5′-GC ACA GCT TTG AAT TGT GCT AGC CGC GTA TTT AGG CC-3′ to generate pIE1BI1/HA, pIE1BI2/HA, and pIE1BI3/HA, respectively. Sequences encoding residues 150 to 162 were deleted within pIE1HA/BS by using the oligonucleotides 5′-GGC CAG TTT AAC AGC ACA ATT CAA AGC TGT GCA ACC-3′ and 5′-TTG AAT TGT GCT GTT AAA CTG GCC CAC CAC ACC TTG-3 to generate pIE1ΔBI/HA. All site-directed mutations were verified by nucleotide sequence determination.
(iii) EGFP-IE1 fusions.
To generate plasmid pEGFP-IE1, sequences encoding residues 1 to 260 of the enhanced green fluorescent protein (EGFP) were fused to those of full-length IE1 (residues 13 to 582) (Fig. 1, middle panel). To this end, the EGFP-encoding NheI-SmaI fragment of pEGFP-C2 (Clontech) was fused to the ie-1-containing Eco47III-XbaI fragment of pIE1BS and inserted into the SpeI site of pIE1hr/PA (6), generating pIE1hr/EGFP-IE1/PA. A plasmid encoding deletion EGFP-IE1Δ515-580 was engineered by inserting the NdeI-SacI fragment of pIE1ΔAccI-BstBI/BS (54) into the corresponding sites of pIE1hr/EGFP-IE1/PA. The PshAI-HpaI fragment of pIE1hr/EGFP-IE1/PA was isolated and intramolecularly ligated to generate a plasmid encoding deletion EGFP-IE1Δ70-515. Plasmid pEGFP was constructed by inserting the NheI-XbaI fragment of pEGFP-C2 into the SpeI site of pIE1hr/PA.
(iv) Other plasmids.
The hr-dependent reporter pBAS35K-Luc/28mer-up+/PA, which carries a single copy of the AcMNPV hr5 28-mer linked to the basal p35 promoter upstream from the luciferase gene, was described previously (46). Plasmid pIAPHA, encoding HA epitope-tagged Op-IAP under control of the ie-1 promoter, was also described previously (40).
Cells and plasmid transfections.
Spodoptera frugiperda IPLB-SF21 (58) cells were propagated in TC100 growth medium (GIBCO Laboratories) supplemented with 2.6 mg of tryptose broth/ml and 10% heat-inactivated fetal bovine serum. For transfections, SF21 monolayers (2 × 106 cells per 60-mm-diameter plate) were overlaid with transfection mix containing N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate-l-phosphatidylethanolamine, dioleoyl (C18:1, [cis]-9) (DOTAP-DOPE) and plasmid DNA in TC100 medium. After 4 h at room temperature, the mix was replaced with supplemented TC100 medium. For transfections in suspension, SF21 cells were incubated with transfection mix containing DOTAP-DOPE and plasmid DNA in TC100 medium and mixed gently by rotation. After 4 h, the cells were plated and overlaid with supplemented TC100 medium.
Fluorescence microscopy.
SF21 cells were transfected in suspension with plasmid DNA (4 μg) encoding EGFP or EGFP-IE1 and plated in 8-chambered glass slides (Nalge Nunc International). After 48 h, the transfected cells were fixed with 2% formaldehyde-0.2% glutaraldehyde in phosphate-buffered saline (PBS) (30) at 4°C for 5 min. The fixed cells were washed with PBS and overlaid with Hoechst stain (0.5 μg/ml) (bisbenzimide H33342, fluorochrome, trihydrochloride; Calbiochem) for 2 min at 27°C. After washing with PBS, EGFP and Hoechst stain fluorescence was monitored by excitation at λ488 and λ346, respectively. Photos (×100 magnification) were taken using a Zeiss Axiovert 135 TV fluorescence microscope equipped with a digital camera and prepared using Adobe Photoshop.
Biochemical fractionation.
SF21 cells (2 × 106/60-mm-diameter plate) were transfected with plasmid DNA encoding IE1HA and IAPHA (4 and 0.5 μg, respectively, or the amounts indicated in the figure legends). Cells were collected 48 h later, washed with PBS, and lysed by suspension in 170 μl of TBN buffer (10 mM Tris [pH 6.5], 140 mM NaCl, 0.5% NP-40, 3 mM MgCl2, protease inhibitors) for 15 min on ice (22). After clarification by centrifugation (5,220 × g, 5 min), the supernatant was retained as the cytosolic fraction. The pellet was washed with TBN buffer (250 μl), lysed by suspension for 15 min on ice with 170 μl of lysis buffer (62.5 mM Tris [pH 6.8], 40% glycerol, 4% sodium dodecyl sulfate [SDS], 3% dithiothreitol), and forced 10 to 20 times through a 25-gauge needle. After clarification by centrifugation (16,000 × g, 10 min), the supernatant was retained as the nuclear fraction. To analyze total cell proteins, a sample of the cell suspension was removed prior to cell lysis and mixed with an equal volume of 2% β-mercaptoethanol-2% SDS.
Reporter transactivation assays.
SF21 cells (2 × 106/60-mm-diameter plate) were transfected with the hr-dependent luciferase reporter pBAS35K-Luc/28mer-up+/PA (2 μg) either alone or with the indicated IE1HA-encoding plasmids (0.5 μg). Cells were collected 48 h later, washed with PBS, and lysed by suspension in 250 μl of cell culture lysis reagent (Promega). After clarification by centrifugation, luciferase activity in the cell lysate (20 μl) was determined using a Monolight 3010 luminometer according to the manufacturer's instructions. The assay was linear over a range from 104 to 108 relative light units (RLU) when using purified luciferase.
Immunoprecipitations.
SF21 cells (7 × 106/100-mm-diameter plate) were transfected with plasmid DNA (11 μg) encoding untagged wild-type IE1 and IE1HA. Cells were collected 24 h later, washed with PBS, and lysed by suspension in 900 μl of E1A buffer (50 mM HEPES [pH 7.0], 250 mM NaCl, 0.1% NP-40, protease inhibitors) for 45 min on ice. After clarification by centrifugation (16,000 × g, 10 min), cell lysate (300 μl) was mixed with 918 μl of WCE buffer (10 mM HEPES, [pH 7.0], 400 mM NaCl, 0.10 mM EGTA, protease inhibitors) containing ∼10 μg (2 μl) of HA.11 monoclonal antibody (α-HA) (BAbCO). Immunoprecipitates were collected using protein G Sepharose beads (Sigma) as described previously (46) and eluted by boiling in 1% SDS-2.5% β-mercaptoethanol.
Immunoblot analysis.
Protein samples in SDS and β-mercaptoethanol were electrophoresed on SDS-7.5% polyacrylamide gels. After protein transfer, nitrocellulose membranes were incubated with a 1:20,000 dilution of α-IE1 (46) or a 1:3,000 dilution of α-FKBP46 (1) followed by alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories). HA epitope-tagged proteins were detected using a 1:1,000 dilution of α-HA (HA.11) followed by alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories). Signal development was performed using nitroblue tetrazolium chloride-BCIP (5-bromo-4-chloro-3-indolylphosphate p-toluidine salt) colorimetric detection as described previously (18) or the Western-Star chemiluminescent detection system (Tropix). To quantify protein levels, immunoblots were scanned at a resolution of 300 dots/in. using a UMAX PowerLook III and analyzed using the program ImageQuant (IQMac v1.2; Molecular Dynamics). Images were printed using Adobe Photoshop and an Epson Stylus Pro 5000 printer.
RESULTS
Nuclear localization of IE1.
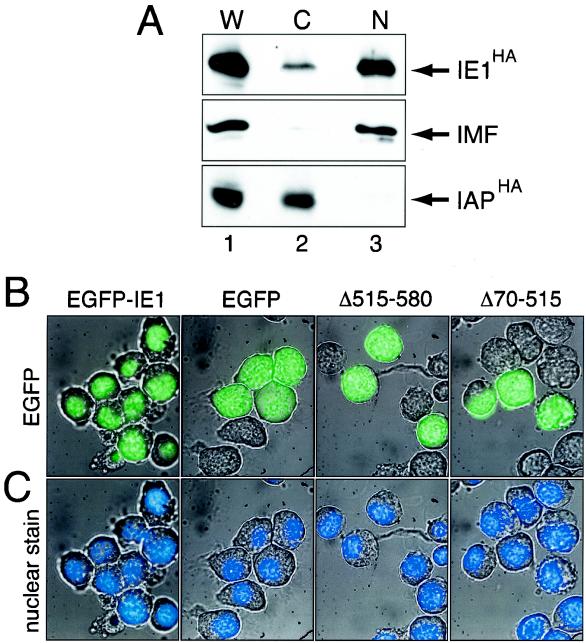
As determined by plasmid transfection assays, IE1 is required for viral gene expression and DNA replication (for reviews, see references 10 and 38). Thus, it is to be expected that efficient transport of IE1 into the nucleus is required for proper IE1 function. Previous immunostaining studies indicated that during infection, IE1 localizes to the nucleus and associates with viral DNA replication factories (44). To assess the intracellular distribution of IE1 in the absence of viral proteins, we biochemically fractionated SF21 cells that were transfected with a plasmid encoding C-terminal epitope-tagged IE1 (IE1HA) under control of the ie-1 promoter (Fig. 1). As determined by cellular fractionation using nonionic detergent and differential centrifugation, IE1HA localized predominantly to the nucleus (Fig. 2A, lane 3). Lesser amounts of IE1HA were detected in the cytosolic fraction (Fig. 2A, lane 2). The integrity of the cytosolic and nuclear fractions was verified by monitoring the distribution of immunophilin FKBP46 (IMF) and Op-IAP (IAPHA), representing nuclear and cytoplasmic proteins, respectively (1, 40). As expected, endogenous IMF was detected in the nuclear fraction, with little or no accumulation in the cytosolic fraction (Fig. 2A, lanes 2 and 3). In contrast, IAPHA, which was synthesized upon plasmid transfection, was detected exclusively in the cytosolic fraction (Fig. 2A, lane 2). Thus, there was little or no contamination of the nuclear or cytosolic fractions.
FIG. 2.
Intracellular distribution of IE1. (A) Biochemical fractionation. SF21 cells were transfected with plasmids encoding IE1HA and IAPHA and fractionated 48 h later. Samples (2 × 105 cell eq) of whole-cell lysate (W) (lane 1), cytosolic (C) (lane 2), and nuclear (N) (lane 3) fractions were subjected to immunoblot analyses. IE1HA and IAPHA were detected using HA-specific monoclonal antibody (α-HA), and immunophilin (IMF) was detected using FKBP46-specific antiserum (α-FKBP46). (B) EGFP fluorescence. SF21 cells were transfected with plasmids encoding EGFP, EGFP-IE1, or the indicated deletions thereof. Cells were fixed and treated with Hoechst stain 48 h later. EGFP and Hoechst signals were visualized by fluorescence microscopy. Representative fields are shown. Magnification, ×100.
To confirm the nuclear distribution of IE1 and define the protein domains involved, we transfected SF21 cells with plasmids encoding EGFP-IE1 fusion proteins and monitored protein distribution in intact cells by fluorescence microscopy. To this end, the gene encoding the 260-residue EGFP was fused to sequences encoding IE1 residues 13 to 582 and placed under control of the ie-1 promoter. The resulting EGFP-IE1 fusion (Fig. 1) was fully functional for transactivation (data not shown). In transfected cells, EGFP-IE1 localized selectively to the nucleus, as indicated by EGFP-specific fluorescence (Fig. 2B) that was coincident with Hoechst staining of the nucleus (Fig. 2C). In contrast, EGFP alone was evenly distributed within the nucleus and cytoplasm of transfected cells (Fig. 2B), indicating a lack of specific localization. C-terminal deletion EGFP-IE1Δ515-580 was also evenly distributed between cytoplasm and nucleus (Fig. 2B). This finding suggested that the C-terminal 65 residues of IE1 contribute to nuclear targeting. However, EGFP-IE1Δ70-515, which lacks internal IE1 residues 70 to 515, was also distributed throughout the cell (Fig. 2B). These data suggested that residues comprising the C terminus of IE1 are required but are not sufficient for nuclear accumulation.
Identification of IE1 residues required for nuclear localization.
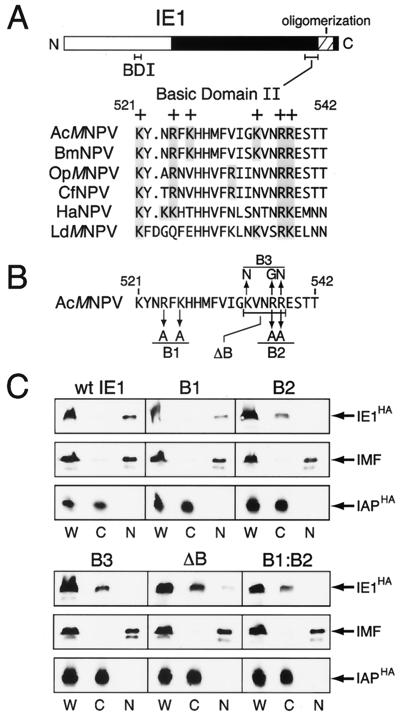
NLSs are usually composed of basic amino acid residues (for reviews, see references 20 and 23). Inspection of the amino acid sequence of AcMNPV IE1 revealed two regions with a high proportion of arginine and lysine: that of residues 152 to 161 (basic domain I, pI of 11.5) within the transactivation domain and that of residues 521 to 538 (basic domain II, pI of 12.3) preceding the C-terminal oligomerization domain (Fig. 3A). In particular, basic domain II is highly conserved among baculovirus IE1 proteins, suggesting a critical and conserved role in IE1 function.
FIG. 3.
Intracellular distribution of IE1 with basic domain II mutations. (A) Basic domain II. The locations of basic domain I (BDI) and II (BDII) within IE1 are shown. AcMNPV IE1 residues 521 to 542 were aligned with corresponding residues of IE1 from the indicated baculoviruses. Conserved basic (+) residues are shaded. (B) Mutagenesis. Amino acid substitutions within basic domain II are indicated (arrows) for IE1 mutations B1, B2, and B3. Residues deleted from IE1 mutation ΔB are bracketed. (C) Biochemical fractionation. SF21 cells were transfected with plasmids encoding the indicated IE1HA mutation and IAPHA, fractionated, and subjected to immunoblot analysis as described in the legend to Fig. 2A. W, whole-cell lysate; C, cytosolic fraction; N, nuclear fraction.
To assess participation of basic domain II in nuclear entry, we constructed a series of mutations in this region and determined their effect on IE1 intracellular distribution. Replacement of R524 and K526 with alanines (Fig. 3B) had little or no effect, since accumulation of mutated IE1B1 within the nucleus was comparable to that of wild-type IE1 (Fig. 3C). In contrast, substitutions of alanines for R537 and R538 caused accumulation of mutated IE1B2 in the cytosol instead of the nucleus (Fig. 3C). The contribution of residues R537 and R538 was dominant, since the cytoplasmic restriction of double-mutated IE1B1:B2 was comparable to that of IE1B2 (Fig. 3C). When K534, R537, and R538 were replaced with nonpolar residues within IE1B3 (Fig. 3B), nuclear entry was also compromised (Fig. 3C). Likewise, deletion of residues 534 to 538 within IE1ΔB caused loss of nuclear entry without affecting total IE1 levels. In all of these fractionations, cytoplasmic IAPHA and nuclear IMF were distributed normally (Fig. 3C). We concluded that basic domain II residues R537, R538, and probably K534 are required for IE1 nuclear localization. Moreover, since nearby residues R524 and K526 contributed little to nuclear import of IE1, it is unlikely that basic domain II functions as a bipartite NLS, as is found to be the case in some transcriptional activators (20, 23).
Minimal role of basic domain I in IE1 localization.
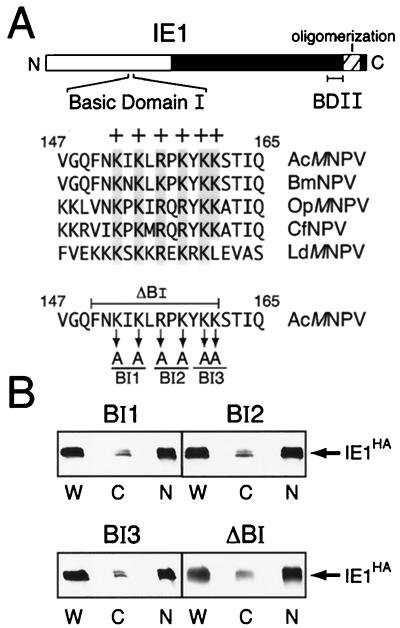
We used a similar strategy of site-directed mutagenesis and plasmid transfection assays to assess the role of basic domain I (residues 152 to 161) in IE1 nuclear transport. Conserved among baculovirus IE1s and positioned within the transactivation domain, basic domain I has the highest concentration of lysine and arginine within IE1. Pairwise substitutions of alanine for basic residues (Fig. 4A) had no measurable effect on nuclear localization. Upon transfection, mutated IE1BI1, IE1BI2, and IE1BI3 exhibited a nuclear and cytoplasmic distribution (Fig. 4B) that was indistinguishable from that of wild-type IE1 (Fig. 2A). Likewise, deletion of residues 150 to 162, comprising the entire domain, failed to alter the normal nuclear targeting of IE1ΔBI (Fig. 4B). We concluded that basic domain I contributes little to nuclear entry of AcMNPV IE1 in the absence of other viral proteins.
FIG. 4.
Intracellular distribution of IE1 with basic domain I mutations. (A) Basic domain I. AcMNPV IE1 residues 147 to 165 were aligned with corresponding IE1 residues from the indicated baculoviruses. Conserved basic (+) residues are shaded. Alanine substitutions are indicated (arrows) for IE1 mutations BI1, BI2, and BI3. Residues deleted from IE1 mutation ΔBI are bracketed. (B) Biochemical fractionation. SF21 cells were transfected with plasmids encoding the indicated IE1HA mutation, fractionated 48 h later, and analyzed using α-HA as described in the legend to Fig. 2A.
Requirement of basic domain II for IE1 transactivation.
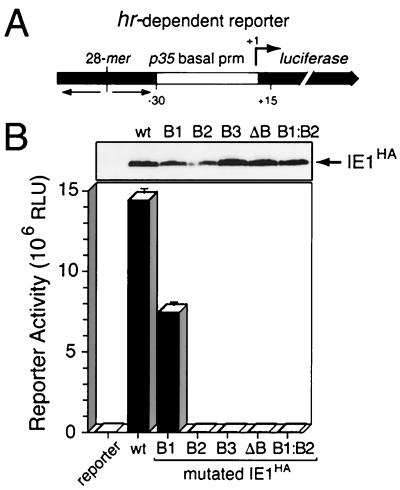
To further define the role of basic domain II encompassing residues 534 to 538, we determined the effect of mutations within this domain on promoter transactivation by IE1. As a reporter for plasmid transfection assays, we used the luciferase gene under control of the basal promoter of the AcMNPV p35 gene that was cis linked to a single copy of the 28-mer of the hr5 enhancer (Fig. 5A). The p35 basal promoter is highly sensitive to transactivation by IE1 when linked to a 28-mer binding site (52, 53). All mutated IE1s defective for nuclear localization, including IE1B2, IE1B3, IE1ΔB, and IE1B1:B2, failed to transactivate the hr-dependent reporter (Fig. 5B). Since each of these mutated IE1s was readily detected in transfected cells, the loss of transactivation was not due to protein instability. In contrast, IE1B1 was transactivation competent, exhibiting 50% of the activity of wild-type IE1HA (Fig. 5B). Immunoblot analysis suggested that the decrease in transactivation was due to reduced IE1B1 stability. These data indicated that basic domain II residues 534 to 538 are required for IE1 transactivation and are consistent with an NLS function of this domain.
FIG. 5.
hr-dependent transactivation by IE1 basic domain II mutations. (A) Luciferase reporter. The luciferase gene under control of the p35 basal promoter (TATA element to the +1 RNA start site) (arrow) was linked to a single copy of the palindromic 28-mer from the hr5 enhancer. (B) Reporter transactivation. SF21 cells (2 × 106/plate) were transfected with reporter plasmid (2 μg) alone and plasmid (2 μg) encoding wild-type (wt) IE1HA or the indicated basic domain II-IE1HA mutations. Cell lysates prepared 48 h later were subjected to immunoblot analysis (1.2 × 105 cell eq) using α-HA (top panel) or assayed for luciferase activity (bottom panel). Values shown are averages ± standard deviations of triplicate transfections and are reported as RLU.
Oligomerization of nonnuclear IE1 mutations.
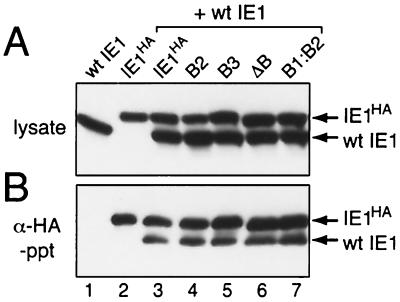
IE1 oligomerization is required for promoter transactivation (46, 53). Since the residues required for IE1 nuclear entry immediately precede the HLH-like oligomerization domain, disruption of oligomerization by basic domain II mutations may cause loss of IE1 function. To test this possibility, we assessed the oligomeric state of mutated IE1 in cells simultaneously transfected with plasmids encoding untagged, wild-type IE1 and IE1HA, each under control of the ie-1 promoter. Wild-type and mutated IE1s were detected in lysates of plasmid-transfected cells (Fig. 6A). Untagged, wild-type IE1 oligomerized with IE1HA, as demonstrated by the detection of both proteins in a complex after immunoprecipitation with α-HA (Fig. 6B, lane 3). Untagged IE1 alone was not immunoprecipitated with α-HA (Fig. 6B, lane 1). The less-than-equimolar ratio of untagged IE1 to tagged IE1HA was due to the capacity of IE1HA to homodimerize, allowing only one-third of the available IE1HA to heterodimerize with untagged IE1. Each of the mutated IE1s, including IE1B2, IE1B3, IE1ΔB, and IE1B1:B2, formed an immunoprecipitable complex with wild-type IE1 (Fig. 6B, lanes 4 to 7) and thus retained the capacity to oligomerize in vivo. These data indicated that the loss of transactivation caused by basic domain II mutations was not due to disruption of oligomerization.
FIG. 6.
Oligomerization of IE1HA basic domain II mutations with wild-type IE1. (A) Intracellular levels of IE1. SF21 cells were transfected with plasmids encoding wild-type untagged IE1 and wild-type IE1HA or the indicated IE1HA mutations. Plasmid levels were adjusted for comparable protein synthesis. A constant level of plasmid DNA was maintained by supplementation with pIE1-lacZ (5). Nonionic detergent lysates were prepared 24 h after transfection. Samples (105 cell eq) representing 10% of that used for immunoprecipitation (panel B) were subjected to immunoblot analysis using IE1 antiserum (α-IE1). (B) Immunoprecipitations. HA-tagged proteins from transfected-cell lysates (106 cell eq) were immunoprecipitated with α-HA and subjected to immunoblot analysis using α-IE1. IE1HA and wild-type (wt) IE1 proteins are indicated.
Dominant interference of wild-type IE1 by nonnuclear IE1 mutations.
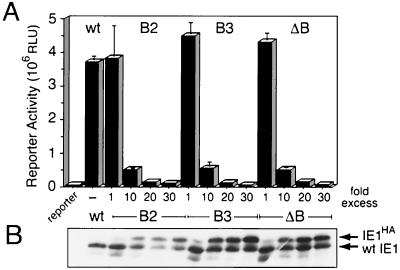
The finding that basic domain II-mutated IE1s retained the capacity to oligomerize suggested that these nonfunctional mutations might dominantly interfere with wild-type IE1 and thus provide insight into mechanisms of IE1 nuclear transport. To test this possibility, we determined the effect of these cytoplasm-restricted IE1s on wild-type IE1 transactivation of the hr-dependent luciferase reporter (Fig. 5A). SF21 cells were transfected with a constant level of plasmid encoding wild-type IE1 and increasing levels of plasmid encoding mutated IE1. With increasing levels of IE1B2, IE1B3, or IE1ΔB, reporter activity decreased dramatically (Fig. 7A). At a 20-fold excess of IE1HA mutation-encoding plasmid, IE1 transactivation was reduced >25-fold. Double-mutated IE1B1:B2 exhibited a similar inhibitory effect (data not shown). Immunoblot analysis indicated that the level of each basic domain II-mutated IE1HA increased with that of the increasing plasmid (Fig. 7B). However, the mutated IE1s exhibited disparate levels of accumulation, which was likely due to differences in protein stability. In contrast, wild-type IE1 levels decreased or remained stable in cotransfected cells (Fig. 7B). Since the levels of ie-1 promoter were held constant, it is unlikely that the reduction in wild-type IE1 upon cotransfection with IE1B2-encoding plasmid was due to decreased ie-1 transcription. Collectively, these data suggested that the decrease in wild-type IE1 transactivation was due to direct inhibition, protein destabilization, or both.
FIG. 7.
Dominant interference of wild-type IE1 transactivation. (A) Reporter assays. SF21 cells were transfected with the hr-dependent luciferase (Fig. 5A) plasmid (2 μg) and plasmid encoding wild-type IE1 (0.1 μg) either alone (−) or with increasing amounts of plasmid (0.1 to 3 μg) encoding the indicated IE1HA basic domain II mutation. Plasmid DNA levels were normalized using pIE1-lacZ. Cell lysates were prepared 48 h later and assayed for luciferase activity. The values are averages ± standard deviations of triplicate transfections and are reported as RLU. (B) Immunoblots. Levels of wild-type IE1 (wt) and the indicated IE1HA mutations were determined by immunoblot analysis using α-IE1 of whole-cell lysates (2.4 × 105 cell eq).
Mechanism of dominant inhibition by nonnuclear IE1 mutations.
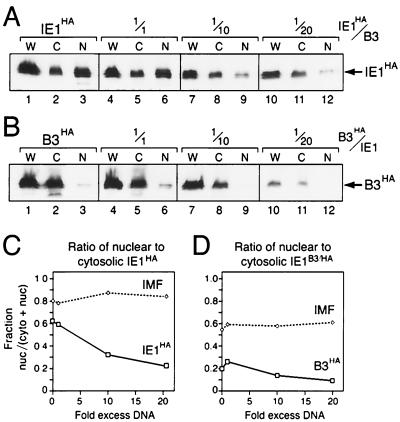
To investigate the possibility that nonnuclear IE1 inhibited wild-type IE1 by trapping it within the cytoplasm, we determined the effect of mutated IE1B3 on nuclear import of wild-type IE1. To this end, SF21 cells were transfected with a constant level of plasmid encoding wild-type IE1HA and increasing levels of untagged IE1B3. As IE1B3 levels increased, total intracellular IE1HA levels decreased (Fig. 8A, lanes 1, 4, 7, and 10). However, the intracellular distribution of wild-type IE1HA progressively switched from the nucleus to the cytosol (Fig. 8A, lanes 3, 6, 9, and 12). As shown by a graph plotting the ratio of nuclear IE1 to cytosolic IE1 (Fig. 8C), nuclear import of IE1HA decreased threefold over the range of IE1B3 plasmid levels used. In contrast, the nuclear distribution of cellular IMF remained constant.
FIG. 8.
IE1 distribution upon dominant inhibition. (A) Localization of wild-type IE1HA. SF21 cells were transfected with plasmids encoding wild-type IE1HA (0.5 μg), IAPHA (0.25 μg), and untagged IE1B3 (0.5 to 10 μg). Plasmid DNA levels were normalized using pIE1-lacZ. Cells were fractionated 48 h later and subjected to immunoblot analysis using α-HA. The integrity of cell fractions was monitored by immunoblot analysis of IMF and IAPHA (data not shown). W, whole-cell lysate; C, cytosolic fraction; N, nuclear fraction. (B) Localization of IE1B3/HA. SF21 cells were transfected with plasmid encoding IE1B3/HA (0.5 μg), IAPHA (0.25 μg), and untagged wild-type IE1 (0.5 to 10 μg), fractionated, and subjected to immunoblot analysis using α-HA as described for panel A. (C and D) Change in intracellular distribution of IE1s. The relative levels of wild-type IE1HA and IE1B3/HA, as determined by α-HA immunoblots (panels A and B, respectively), were quantified for nuclear and cytosolic fractions. The level of nuclear IE1 was divided by the sum of the levels of nuclear and cytosolic IE1 and plotted as a function of the severalfold excess of plasmid DNA encoding either untagged IE1B3 (panel C) or wild-type IE1 (panel D). Nuclear and cytosolic immunophilin (IMF) were quantified and plotted similarly.
The capacity of mutated IE1B3 to block nuclear entry of wild-type IE1 (Fig. 8A) predicted that wild-type IE1 would not affect the cytoplasmic restriction of IE1B3 despite its capacity to heterodimerize. We tested this premise by determining the subcellular distribution of HA-tagged IE1B3 upon cotransfection with increasing levels of plasmid encoding untagged wild-type IE1. Despite an observed reduction in IE1B3/HA synthesis (Fig. 8B, lanes 1, 4, 7, and 10), IE1B3/HA remained predominantly in the cytosol (lanes 2, 5, 8, and 11). The plotted ratio of nuclear to cytosolic IE1B3/HA was low and remained unchanged as the level of wild-type IE1 increased (Fig. 8D). Taken together, these data suggested that the dominant inhibition of IE1 transactivation (Fig. 7) by nonnuclear IE1 mutations was due to cytoplasmic sequestration of wild-type IE1.
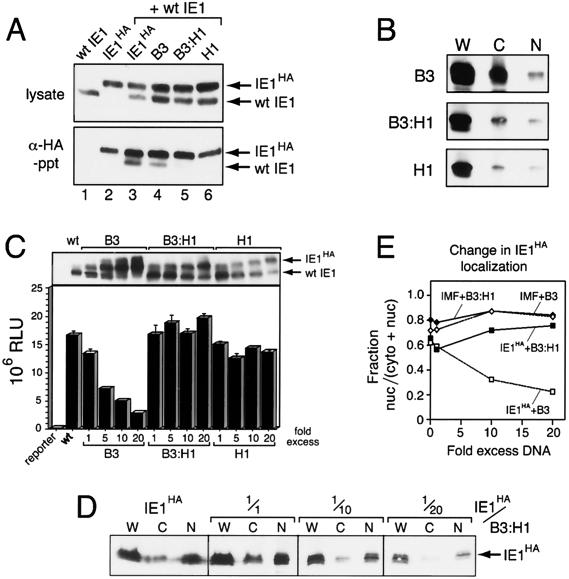
Loss of dominant interference by disrupting oligomerization.
If interaction with nonnuclear IE1 prevents wild-type IE1 from nuclear entry, then eliminating hetero-oligomerization should alleviate dominant inhibition. We tested this prediction by constructing a plasmid encoding IE1B3:H1, which carries the nuclear import-defective mutation (B3) and a substitution mutation, L550D/I554E (H1), that disrupts oligomerization mediated by HLH interactions (46). As determined by immunoprecipitations of extracts from plasmid-transfected cells, IE1B3:H1 exhibited little if any oligomerization with wild-type IE1 (Fig. 9A, lane 5). As expected, wild-type IE1 interacted with IE1HA and IE1B3 (lanes 3 and 4) but not IE1H1 (lane 6). Also, IE1B3:H1 accumulated predominantly in the cytoplasm (Fig. 9B) despite its instability (46) and failed to transactivate the hr-dependent reporter (data not shown). Thus, IE1B3:H1 is impaired for oligomerization, nuclear localization, and transactivation.
FIG. 9.
Lack of dominant inhibition by IE1B3:H1. (A) Immunoprecipitations. SF21 cells were transfected with plasmids encoding untagged IE1 (wt IE1) and IE1HA or the indicated IE1HA mutation. Plasmid levels were adjusted to provide comparable levels of each protein. Cell lysates were subjected to immunoblot analysis with α-IE1 (top panel) or immunoprecipitation with α-HA and detection using α-IE1 (bottom panel) as described in the legend to Fig. 6. (B) IE1B3:H1 intracellular distribution. SF21 cells were transfected with plasmid encoding IE1B3/HA, IE1B3:H1/HA, or IE1H1/HA, fractionated, and subjected to immunoblot analysis using α-HA. W, whole-cell lysate; C, cytosolic fraction; N, nuclear fraction. (C) Reporter assays. SF21 cells were transfected with the hr-dependent luciferase reporter (2 μg) and plasmid encoding wild-type (wt) IE1 (0.1 μg) either alone or with increasing amounts of plasmid (0.1 to 2 μg) encoding the indicated IE1HA mutation as described in the legend to Fig. 7. Cell lysates prepared 48 h later were subjected to immunoblot analysis using α-IE1 (top panel) or assayed for luciferase activity (bottom panel). The values are averages ± standard deviations of triplicate transfections and are reported as RLU. (D) Localization of wild-type IE1HA. SF21 cells were transfected with plasmids encoding wild-type IE1HA (0.5 μg), IAPHA (0.25 μg), and untagged IE1B3:H1 (from 0.5 to 10 μg), fractionated, and subjected to immunoblot analysis using α-HA. Cell fraction integrity was monitored by immunoblot analysis of IMF and IAPHA (data not shown). (E) Change in intracellular distribution of IE1HA. The relative levels of wild-type IE1HA and immunophilin (IMF) in nuclear and cytosolic fractions (panel D) were quantified and plotted as described in the legend of Fig. 8. The effect of IE1B3 on wild-type IE1HA and IMF localization as shown in Fig. 8C is included for comparison (line with open squares).
To test for dominant interference by IE1B3:H1, we transfected SF21 cells with plasmids encoding the hr-dependent reporter, wild-type IE1, and increasing amounts of plasmid encoding IE1B3, IE1B3:H1, or IE1H1. All proteins were produced upon transfection (Fig. 9C). As expected, nonfunctional IE1B3 dominantly interfered with reporter transactivation by wild-type IE1. However, even with a 20-fold excess of IE1B3:H1-encoding plasmid, the presence of IE1B3:H1 exerted little if any effect on wild-type IE1 activity (Fig. 9C). A similar lack of interference was exhibited by IE1H1. Thus, the loss of dominant interference coincided with the failure to oligomerize. To determine the effect, if any, on intracellular distribution of wild-type IE1, we fractionated cells transfected with plasmids encoding wild-type IE1HA and untagged IE1B3:H1. Despite decreased levels of synthesis, IE1HA accumulated predominantly in the nucleus in the presence of excess IE1B3:H1 (Fig. 9D). Over a 20-fold increase in IE1B3:H1 plasmid, the ratio of nuclear IE1HA to total IE1HA remained steady and paralleled that of nuclear IMF (Fig. 9E). Moreover, the cytoplasmic localization of IE1B3:H1/HA was not affected by increasing levels of wild-type IE1 (data not shown). We concluded that nonnuclear IE1 interferes with wild-type IE1 function by a mechanism involving direct oligomerization, which impedes nuclear entry and subsequent transactivation.
DISCUSSION
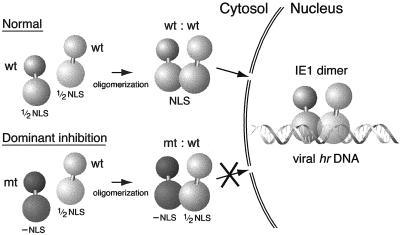
Normal function of AcMNPV IE1, including transactivation of early viral promoters, requires proper transport to the nucleus. In this study, we have identified IE1 residues necessary for nuclear import of IE1. These residues within basic domain II, including R537 and R538, comprise a positively charged motif, designated here as an NLE. Positioned adjacent to the HLH-like oligomerization domain at the C terminus of IE1, the NLE alone is insufficient for IE1 nuclear import. Rather, two copies of the motif, each located in a separate monomer of an IE1 dimer, are required for nuclear entry. Thus, the functional NLS of IE1 likely consists of a novel dimeric arrangement of basic residues. Our study suggests that oligomerization of IE1 after synthesis in the cytoplasm is a prerequisite for nuclear entry and thus ensures proper assembly of IE1 for high-affinity, stable interaction with the palindromic binding sites comprising the AcMNPV hr elements for transcriptional enhancement and DNA replication (Fig. 10).
FIG. 10.
Model for IE1 nuclear import and transactivation. (Top panel) Upon synthesis in the cytoplasm, wild-type IE1 monomers (wt) assemble into dimers (wt : wt), thereby constituting a fully functional NLS for nuclear import. Upon entering the nucleus, dimeric IE1 interacts with symmetrical IE1 binding sites within the hr enhancer-origin elements to stimulate transcription of early viral genes. (Bottom panel) When an NLS-defective (−NLS), oligomerization-competent IE1 mutation (mt) is coproduced with wild-type IE1, heterodimers (mt : wt) assemble. As a consequence, wild-type IE1 is trapped in the cytosol and prohibited from nuclear entry. Thus, IE1 transactivation is dominantly inhibited.
The NLE is required for IE1 nuclear entry and transactivation.
IE1 shares multiple characteristics with eukaryotic transcriptional activators (for reviews, see references 34, 41, and 47). The positively charged, basic domain that precedes the oligomerization domain of basic-HLH transactivators often functions in nuclear import and DNA binding. Our investigations here indicated that basic domain II (residues 534 to 538), which precedes the HLH-like oligomerization motif of IE1, is required for efficient nuclear entry and constitutes an NLE. Although basic domain I (residues 152 to 161) also contains a high proportion of positively charged residues, we found no evidence that this region contributes to nuclear entry or nuclear retention (Fig. 4). In contrast, site-specific substitutions of positively charged residues within basic domain II impeded nuclear entry and caused cytoplasmic accumulation of IE1 (Fig. 3). Mutagenesis also caused loss of hr-dependent transactivation by IE1 without disrupting dimerization (Fig. 5 and 6), which is also required for IE1 function (46). Regions of IE1 involved in oligomerization, including the HLH-like domain, indirectly contribute to nuclear transport, since dimerization is required for nuclear entry (see below). Thus, IE1 depends on the NLE within basic domain II as well as on its oligomerization domain(s) for nuclear import. To formally define the NLE as a true NLS, it will be necessary to demonstrate that it can direct a heterologous cytosolic protein to the nucleus. Analogous to other basic-HLH transcriptional activators, the IE1 NLE may also contribute to DNA binding. Our previous in vitro assays demonstrated that DNA (28-mer) binding by IE1 was disrupted upon substitution of basic domain II residues R537 and R538 (54). Subsequent studies (45) have indicated that a primary determinant of 28-mer binding also lies within an IE1 domain N-terminal to basic domain II.
NLE-defective IE1 prevents nuclear import of wild-type IE1.
The basic domain II loss-of-function mutations IE1B2, IE1B3, and IE1ΔB each dominantly interfered with wild-type IE1 in dose-response assays (Fig. 7). Wild-type IE1 accumulated within the cytosol, along with the inhibitory, import-defective IE1 (Fig. 8). However, upon disruption of the capacity of import-defective IE1 to oligomerize, dominant inhibition was alleviated and wild-type IE1 trafficked unimpeded into the nucleus (Fig. 9). In these transfection assays, we noted that wild-type IE1 levels were sometimes reduced in the presence of import-defective IE1 (Fig. 7 and 8). However, since a similar reduction was also observed in the presence of oligomer-defective and import-defective IE1, which failed to inhibit IE1 activity (Fig. 9), the reduced IE1 production was not responsible for the inhibitory effect. We concluded that dominant inhibition was the result of an HLH-mediated interaction between import-defective IE1 and wild-type IE1 that trapped wild-type IE1 in the cytosol and blocked nuclear entry. This mechanism of IE1 inactivation should prove useful in defining the function(s) of wild-type IE1 during AcMNPV infection.
IE1 requires a dimeric NLE.
The observation that nuclear import-defective IE1 dominantly interfered with wild-type IE1 by binding and trapping it in the cytosol suggested that a single NLE is insufficient to mediate nuclear transport of IE1 oligomers. This conclusion was supported by the finding that nuclear import-defective IE1 (IE1B3) was not transported to the nucleus even in the presence of a large excess of functional IE1 (Fig. 8B). Also as predicted, oligomerization-defective IE1 (IE1H1), with its monomeric NLE, exhibited reduced nuclear import (Fig. 9B). Since the H1 mutation (L550D/I554E) severely impairs but does not eliminate dimerization (46), it is not surprising that low levels of nuclear IE1H1 were detected. Collectively, our data indicated that efficient nuclear import requires that each IE1 monomer of the dimer possess a functional NLE (Fig. 10). Furthermore, oligomerization is necessary for nuclear entry. This requirement of oligomerization for nuclear import explains for the first time why dimerization is necessary for transactivation of both hr-dependent and -independent promoters by IE1 in transfection assays (46).
IE1's dependence on a dimeric NLE is strikingly similar to that of the mammalian signal transducers STAT1 and STAT2, which heterodimerize for transport from their normal residence in the cytosol to the nucleus, where they bind DNA and activate interferon-responsive genes (42). Each STAT contains a nonclassical, arginine- and lysine-rich NLE that must cooperate with the analogous NLE in its partner STAT during dimerization to achieve nuclear import. Like NLE-deficient IE1, import-defective STAT dominantly interferes with nuclear import of wild-type STAT (42).
Mechanism for IE1 nuclear transport and transactivation.
The high conservation of residues comprising basic domain II among baculovirus IE1s (and IE0s) (Fig. 3) argues that IE1 uses a common mechanism for nuclear import. Nuclear entry is often controlled by modulating the accessibility or affinity of the NLS to the importins (for reviews, see references 20 and 23). The NLE residues of IE1 lie adjacent to the HLH-like oligomerization domain. Thus, it is likely that dimerization juxtaposes the NLE of each monomer, thereby doubling the net positive charge of the region. This union may enhance IE1's affinity for nuclear shuttle proteins, including importin α. Consistent with this possibility, the IE1 NLE contains relatively few basic residues (K534, R537, and R538) when compared to that of known nuclear proteins (20), including the NLS of SV40 large T antigen (PKKKRKV), papillomavirus E2 (KCYRFRVKKNHRHR), and AcMNPV polyhedrin (KRKK) (22, 29, 55). Alternatively, IE1 oligomerization may induce an undefined conformational change that activates or unmasks the NLE. For some proteins, conformational changes mediated by homo- or hetero-oligomerization contribute to nuclear transport (17, 33, 57, 59). Lastly, phosphorylation can affect the affinity of various transcription factors for their transport receptors (20, 23). Although IE1 is phosphorylated during infection (7, 8, 56), the functional role of this modification is unknown.
Our present model (Fig. 10) for IE1-mediated stimulation of viral replication events first involves oligomerization of newly synthesized IE1 in the cytosol. By virtue of the juxtaposition of NLE residues upon dimerization mediated by the C-terminal HLH, IE1 interacts with nuclear import factors and is efficiently transported into the infected cell nucleus. Within the nucleus, oligomeric IE1 makes contact with both half sites of the palindromic 28-mer comprising hr enhancer-origin elements of the viral genome to stimulate transcription and initiate DNA replication events (28, 32, 53). For transcriptional activation, simultaneous interaction with both 28-mer half sites by the IE1 dimer likely induces an alteration in either IE1 or the bound DNA that facilitates the recruitment of transcription coactivators or the basal transcriptional machinery (53). Thus, a mechanism that requires IE1 oligomerization prior to nuclear entry ensures that this transactivator is properly configured for immediate high-affinity binding to viral DNA. The critical molecular events occurring after IE1 binding to viral DNA require further study.
Acknowledgments
We thank Emad Alnemri (Thomas Jefferson University) for the gift of α-FKBP46 antiserum.
This work was supported in part by Public Health Service grant AI25557 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Alnemri, E. S., T. Fernandes-Alnemri, K. Pomerenke, N. M. Robertson, K. Dudley, G. C. DuBois, and G. Litwack. 1994. FKBP46, a novel Sf9 insect cell nuclear immunophilin that forms a protein-kinase complex. J. Biol. Chem. 269:30828-30834. [PubMed] [Google Scholar]
- 2.Braunagel, S. C., H. He, P. Ramamurthy, and M. D. Summers. 1996. Transcription, translation, and cellular localization of three Autographa californica nuclear polyhedrosis virus structural proteins: ODV-E18, ODV-E35, and ODV-EC27. Virology 222:100-114. [DOI] [PubMed] [Google Scholar]
- 3.Broussard, D. R., L. A. Guarino, and D. L. Jarvis. 1996. Mapping functional domains in AcMNPV pp31. Virology 222:318-331. [DOI] [PubMed] [Google Scholar]
- 4.Carson, D. D., M. D. Summers, and L. A. Guarino. 1991. Transient expression of the Autographa californica nuclear polyhedrosis virus immediate-early gene, IE-N, is regulated by three viral elements. J. Virol. 65:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartier, J. L. 1994. Master’s thesis. University of Wisconsin—Madison, Madison.
- 6.Cartier, J. L., P. A. Hershberger, and P. D. Friesen. 1994. Suppression of apoptosis in insect cells stably transfected with baculovirus p35: dominant interference by N-terminal sequences p351-76. J. Virol. 68:7728-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, J., and L. A. Guarino. 1995. The baculovirus transactivator IE1 binds to viral enhancer elements in the absence of insect cell factors. J. Virol. 69:4548-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, J., and L. A. Guarino. 1995. Expression of the IE1 transactivator of Autographa californica nuclear polyhedrosis virus during viral infection. Virology 209:99-107. [DOI] [PubMed] [Google Scholar]
- 9.Eason, J. E., R. H. Hice, J. J. Johnson, and B. A. Federici. 1998. Effects of substituting granulin or a granulin-polyhedrin chimera for polyhedrin on virion occlusion and polyhedral morphology in Autographa californica multinucleocapsid nuclear polyhedrosis virus. J. Virol. 72:6237-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friesen, P. D. 1997. Regulation of baculovirus early gene expression, p. 141-166. In L. K. Miller (ed.), The baculoviruses. Plenum Publishing Corporation, New York, N.Y.
- 11.Friesen, P. D., and L. K. Miller. 2001. Insect viruses, p. 599-628. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Guarino, L. A., and W. Dong. 1994. Functional dissection of the Autographa californica nuclear polyhedrosis virus enhancer element hr5. Virology 200:328-335. [DOI] [PubMed] [Google Scholar]
- 13.Guarino, L. A., M. A. Gonzalez, and M. D. Summers. 1986. Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J. Virol. 60:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarino, L. A., and M. D. Summers. 1986. Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J. Virol. 57:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarino, L. A., and M. D. Summers. 1986. Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J. Virol. 60:215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarino, L. A., and M. D. Summers. 1987. Nucleotide sequence and temporal expression of a baculovirus regulatory gene. J. Virol. 61:2091-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heerklotz, D., P. Doring, F. Bonzelius, S. Winkelhaus, and L. Nover. 2001. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 21:1759-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershberger, P. A., D. J. LaCount, and P. D. Friesen. 1994. The apoptotic suppressor P35 is required early during baculovirus replication and is targeted to the cytosol of infected cells. J. Virol. 68:3467-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, T., M. D. Summers, and S. C. Braunagel. 1997. N-terminal sequences from Autographa californica nuclear polyhedrosis virus envelope proteins ODV-E66 and ODV-E25 are sufficient to direct reporter proteins to the nuclear envelope, intranuclear microvesicles and the envelope of occlusion-derived virus. Proc. Natl. Acad. Sci. USA 94:4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jans, D. A., C. Y. Xiao, and M. H. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis, D. L., D. A. Bohlmeyer, and A. Garcia, Jr. 1992. Enhancement of polyhedrin nuclear localization during baculovirus infection. J. Virol. 66:6903-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis, D. L., D. A. Bohlmeyer, and A. Garcia, Jr. 1991. Requirements for nuclear localization and supramolecular assembly of a baculovirus polyhedrin protein. Virology 185:795-810. [DOI] [PubMed] [Google Scholar]
- 23.Kaffman, A., and E. K. O'Shea. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15:291-339. [DOI] [PubMed] [Google Scholar]
- 24.Kool, M., C. H. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl. Acad. Sci. USA 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacs, G. R., J. Choi, L. A. Guarino, and M. D. Summers. 1992. Functional dissection of the Autographa californica nuclear polyhedrosis virus immediate-early 1 transcriptional regulatory protein. J. Virol. 66:7429-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovacs, G. R., L. A. Guarino, and M. D. Summers. 1991. Novel regulatory properties of the IE1 and IE0 transactivators encoded by the baculovirus Autographa californica multicapsid nuclear polyhedrosis virus. J. Virol. 65:5281-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krappa, R., R. Roncarati, and D. Knebel-Morsdörf. 1995. Expression of PE38 and IE2, viral members of the C3HC4 finger family, during baculovirus infection: PE38 and IE2 localize to distinct nuclear regions. J. Virol. 69:5287-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kremer, A., and D. Knebel-Morsdorf. 1998. The early baculovirus he65 promoter: on the mechanism of transcriptional activation by IE1. Virology 249:336-351. [DOI] [PubMed] [Google Scholar]
- 29.Lanford, R. E., and J. S. Butel. 1984. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell 37:801-813. [DOI] [PubMed] [Google Scholar]
- 30.Lee, H. H., and L. K. Miller. 1978. Isolation of genotypic variants of Autographa californica nuclear polyhedrosis virus. J. Virol. 27:754-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leisy, D. J., C. Rasmussen, H. T. Kim, and G. F. Rohrmann. 1995. The Autographa californica nuclear polyhedrosis virus homologous region 1a: identical sequences are essential for DNA replication activity and transcriptional enhancer function. Virology 208:742-752. [DOI] [PubMed] [Google Scholar]
- 32.Leisy, D. J., and G. F. Rohrmann. 2000. The Autographa californica nucleopolyhedrovirus IE-1 protein complex has two modes of specific DNA binding. Virology 274:196-202. [DOI] [PubMed] [Google Scholar]
- 33.Liang, S. H., and M. F. Clarke. 1999. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. J. Biol. Chem. 274:32699-32703. [DOI] [PubMed] [Google Scholar]
- 34.Littlewood, T. D., and G. I. Evan. 1995. Transcription factors 2: helix-loop-helix. Protein Profile 2:621-702. [PubMed] [Google Scholar]
- 35.Liu, G., and E. B. Carstens. 1999. Site-directed mutagenesis of the AcMNPV p143 gene: effects on baculovirus DNA replication. Virology 253:125-136. [DOI] [PubMed] [Google Scholar]
- 36.Lu, A., and E. B. Carstens. 1993. Immediate-early baculovirus genes transactivate the p143 gene promoter of Autographa californica nuclear polyhedrosis virus. Virology 195:710-718. [DOI] [PubMed] [Google Scholar]
- 37.Lu, A., and E. B. Carstens. 1991. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology 181:336-347. [DOI] [PubMed] [Google Scholar]
- 38.Lu, A., P. J. Krell, J. M. Vlak, and G. F. Rohrmann. 1997. Baculovirus DNA replication, p. 171-191. In L. K. Miller (ed.), The baculoviruses. Plenum Publishing Corporation, New York, N.Y.
- 39.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manji, G. A., R. R. Hozak, D. J. LaCount, and P. D. Friesen. 1997. Baculovirus inhibitor of apoptosis functions at or upstream of the apoptotic suppressor P35 to prevent programmed cell death. J. Virol. 71:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melen, K., L. Kinnunen, and I. Julkunen. 2001. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem. 276:16447-16455. [DOI] [PubMed] [Google Scholar]
- 43.Nissen, M. S., and P. D. Friesen. 1989. Molecular analysis of the transcriptional regulatory region of an early baculovirus gene. J. Virol. 63:493-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olson, V. A. 2001. Ph.D. thesis. University of Wisconsin—Madison, Madison.
- 46.Olson, V. A., J. A. Wetter, and P. D. Friesen. 2001. Oligomerization mediated by a helix-loop-helix-like domain of baculovirus IE1 is required for early promoter transactivation. J. Virol. 75:6042-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patikoglou, G., and S. K. Burley. 1997. Eukaryotic transcription factor-DNA complexes. Annu. Rev. Biophys. Biomol. Struct. 26:289-325. [DOI] [PubMed] [Google Scholar]
- 48.Pearson, M. N., R. M. Bjornson, G. D. Pearson, and G. F. Rohrmann. 1992. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science 257:1382-1384. [DOI] [PubMed] [Google Scholar]
- 49.Prikhod'ko, E. A., and L. K. Miller. 1999. The baculovirus PE38 protein augments apoptosis induced by transactivator IE1. J. Virol. 73:6691-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pullen, S. S., and P. D. Friesen. 1995. Early transcription of the ie-1 transregulator gene of Autographa californica nuclear polyhedrosis virus is regulated by DNA sequences within its 5′ noncoding leader region. J. Virol. 69:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodems, S. M., and P. D. Friesen. 1993. The hr5 transcriptional enhancer stimulates early expression from the Autographa californica nuclear polyhedrosis virus genome but is not required for virus replication. J. Virol. 67:5776-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodems, S. M., and P. D. Friesen. 1995. Transcriptional enhancer activity of hr5 requires dual-palindrome half sites that mediate binding of a dimeric form of the baculovirus transregulator IE1. J. Virol. 69:5368-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodems, S. M., S. S. Pullen, and P. D. Friesen. 1997. DNA-dependent transregulation by IE1 of Autographa californica nuclear polyhedrosis virus: IE1 domains required for transactivation and DNA binding. J. Virol. 71:9270-9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skiadopoulos, M. H., and A. A. McBride. 1996. The bovine papillomavirus type 1 E2 transactivator and repressor proteins use different nuclear localization signals. J. Virol. 70:1117-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slack, J. M., and G. W. Blissard. 1997. Identification of two independent transcriptional activation domains in the Autographa californica multicapsid nuclear polyhedrosis virus IE1 protein. J. Virol. 71:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szilvay, A. M., K. A. Brokstad, S. O. Boe, G. Haukenes, and K. H. Kalland. 1997. Oligomerization of HIV-1 Rev mutants in the cytoplasm and during nuclear import. Virology 235:73-81. [DOI] [PubMed] [Google Scholar]
- 58.Vaughn, J. L., R. H. Goodwin, G. L. Thompkins, and P. McCawley. 1977. Establishment of two insect cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae). In Vitro 13:213-217. [DOI] [PubMed] [Google Scholar]
- 59.Xia, Y.-P., C.-T. Yeh, J.-H. Ou, and M. M. C. Lai. 1992. Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J. Virol. 66:914-921. [DOI] [PMC free article] [PubMed] [Google Scholar]