Abstract
The small nonstructural NS2 proteins of parvovirus minute virus of mice (MVMp) were previously shown to interact with the nuclear export receptor Crm1. We report here the analysis of two MVM mutant genomic clones generating NS2 proteins that are unable to interact with Crm1 as a result of amino acid substitutions within their nuclear export signal (NES) sequences. Upon transfection of human and mouse cells, the MVM-NES21 and MVM-NES22 mutant genomic clones were proficient in synthesis of the four virus-encoded proteins. While the MVM-NES22 clone was further able to produce infectious mutant virions, no virus could be recovered from cells transfected with the MVM-NES21 clone. Whereas the defect of MVM-NES21 appeared to be complex, the phenotype of MVM-NES22 could be traced back to a novel distinct NS2 function. Infection of mouse cells with the MVM-NES22 mutant led to stronger nuclear retention not only of the NS2 proteins but also of infectious progeny MVM particles. This nuclear sequestration correlated with a severe delay in the release of mutant virions in the medium and with prolonged survival of the infected cell populations compared with wild-type virus-treated cultures. This defect could explain, at least in part, the small size of the plaques generated by the MVM-NES22 mutant when assayed on mouse indicator cells. Altogether, our data indicate that the interaction of MVMp NS2 proteins with the nuclear export receptor Crm1 plays a critical role at a late stage of the parvovirus life cycle involved in release of progeny viruses.
The minute virus of mice prototype strain (MVMp) is an autonomously replicating parvovirus that encodes two types of nonstructural proteins, NS1 and NS2, which are required at various steps of the parvovirus replication cycle. The 83-kDa nuclear phosphoprotein NS1 is a multifunctional protein that exhibits site-specific DNA binding, ATPase, helicase, and nickase activities. These activities account for the role played by NS1 during viral DNA replication (for a review, see reference 22). NS1 also controls transcription in trans from both parvovirus promoters P4 and P38, which drive the expression of nonstructural and capsid proteins, respectively, and from at least one cellular promoter (21, 28, 29, 86). Furthermore, NS1 is described as the major effector of parvovirus-induced cytotoxicity (10, 13, 50; for a review, see reference 87).
The small nonstructural NS2 proteins of MVMp consist of three isoforms, NS2-P, -Y, and -L, that differ at their carboxy termini as a result of alternative splicing events (19, 59). They have a molecular mass of about 25 kDa, and all three isoforms share a common amino-terminal domain with NS1 which comprises the first 85 amino acids (aa) of each protein (21, 23, 43). NS2 proteins exist in phosphorylated and unphosphorylated forms that are mainly located in the cytoplasm of infected cells; however, nonphosphorylated NS2 can also be found in the nucleus (15, 19). NS2 proteins are the predominant virus-encoded proteins detected early in the S phase of infected cells, but their accumulation rapidly diminishes as, on the one hand, NS2 proteins exhibit a relatively short half-life (about 90 min) and, on the other hand, activity of the P4 promoter declines later in infection (19, 58, 74).
It has been previously reported that NS2 proteins from MVMp play a critical role in viral capsid assembly and, as a probable consequence, in the generation of viral single-stranded DNA (17, 61, 62). NS2 proteins have also been implicated in the production of the other viral replicative forms (14, 61, 62). Furthermore, NS2 proteins seem to enhance NS1-associated parvovirus-induced cell killing in some but not all cell lines tested (10, 13, 50). Although the molecular mechanisms behind NS2 functioning have not been elucidated yet, MVM NS2 proteins were shown to be strictly required for productive infection in cells from their natural host species, i.e., from the mouse, both in tissue cultures and in animals (12, 14, 17, 61, 62). Similarly, the small nonstructural proteins NS2 from the closely related H-1 virus are absolutely required for efficient virus production in rat cells but are dispensable in cell lines from other origins (51). In this case, NS2 proteins appeared to play a role not only in viral DNA synthesis but also in expression of all viral proteins. It was indeed suggested that NS2 proteins from H-1 virus modulate viral mRNA translation via a sequence located in the 3′ untranslated region of the transcripts (52). In contrast to NS2 from MVMp and H-1 virus, the small nonstructural proteins from canine parvovirus (CPV) appeared to act in a host-independent manner, as a CPV-NS2-null mutant efficiently replicates in both canine and feline cell lines (88).
The species dependence of murine NS2 activity as well as the diversity of the viral processes in which these proteins appear to be involved led us to speculate that NS2 proteins need to interact with specific cellular factors to achieve some of their functions. Indeed, we previously reported that MVM NS2 proteins are able to interact with two members of the 14-3-3 protein family, the epsilon and the beta or zeta isoforms (11). This interaction seems to play a role in posttranslational modifications of the NS2 proteins (7). More recently we, and others, also found that unphosphorylated NS2 proteins interact with the nuclear export receptor Crm1 and are indeed actively exported out of the nuclei of infected cells via a Crm1-mediated nuclear export pathway (7, 64).
Crm1, also known as exportin-1, is a member of the importin beta family that recognizes and binds to a short, leucine-rich nuclear export signal (NES) and further forms a competent nuclear export complex together with Ran-GTP (4, 33, 36, 63, 65, 78). Studies on a number of exported proteins have led to the definition of a consensus NES as a set of critically spaced hydrophobic, primarily leucine, residues (9), but not all sequences conforming to the consensus are functional, and avid interaction with Crm1 can also occur in the absence of a consensus sequence (38, 41). We previously described that MVMp NS2 proteins contain a functional NES located at the junction of NS2 exon 1 and exon 2, which is required for Crm1 binding (U. Bodendorf, V. Eichwald, M. Fornerod, R. Bischoff, M. Klein, J. Rommelaere, and N. Salomé, abstr. 2es Journées Francophones de Virologie, Virologie 4:179, 2000). Compared to other well-characterized NESs, the MVM NS2 NES showed the highest affinity for Crm1 (3, 49), although the NS2 NES sequence is rather poor in leucine residues (3; Bodendorf et al., Virologie 4:179, 2000) when compared to the proposed NES consensus (9). Several viral proteins, including human immunodeficiency virus type 1 (HIV-1) Rev protein (31), human T-cell lymphotropic virus type 1 Rex protein (9), herpes simplex virus ICP27/IE63 protein (72), adenovirus E1B 55-kDa and E4 34-kDa proteins (27, 46), and hepatitis B virus X protein (32), contain functional NESs. Most of these viral proteins play a critical role in the nuclear export of viral mRNAs or can inhibit cellular RNA export. NESs have also been identified in cellular proteins, many of which are involved in transcription, cell signaling cascades, oncogenic transformation, and cell cycle regulation. Examples include cAMP-dependent protein kinase inhibitor (PKI) (89), mitogen-activated protein kinase kinase (1), TFIIIA (35), Mdm2 (34), p53 (79), IκBα (42), NF-AT (45), cyclin B1 (84), c-Abl (80), and Smad1 (92). The activities of most of these proteins appear to be tightly regulated by their NESs.
Given the facts that MVM NS2 proteins are absolutely required for virus production in a host-cell dependent manner and that they undergo Crm1-dependent nuclear export, it is tempting to speculate that a strong, functional NES might contribute to NS2 activities during the parvovirus replication cycle. To test this hypothesis, we investigated whether the nuclear export function of NS2 contributes to the efficiency of virus production and, if so, which stages of the infectious cycle depend on NS2 export. To this end, two MVM mutant genomic clones generating NES-deficient NS2 proteins were constructed and further assayed for their ability to sustain efficient progeny virion production. Although both mutants were proficient in viral capsid protein synthesis, infectious mutant virions could be recovered for only one of these mutant genomic clones after transfection of human or mouse cells. In this case, disruption of the NES site within the NS2 proteins resulted in nuclear retention not only of these viral products but also of infectious progeny MVM particles. These results indicate a possible role for the NES-dependent nuclear export and/or cytoplasmic localization of NS2 in the efficient release of progeny virions from infected cell nuclei. Our data also show that nuclear export or proper subcellular localization of NS2 contributes to the efficiency of parvovirus infection and cytotoxicity.
MATERIALS AND METHODS
Construction of MVM-NES(−) mutant expression plasmids.
The infectious parvovirus molecular clone pdBMVp (kindly provided by P. Tattersall) (44), which contains the MVMp genome with an extension of its 3′ end in order to constitute the so-called minimal origin of DNA replication as defined previously (18, 20), was used to generate the two MVM-NES(−) genomes summarized in Fig. 1.
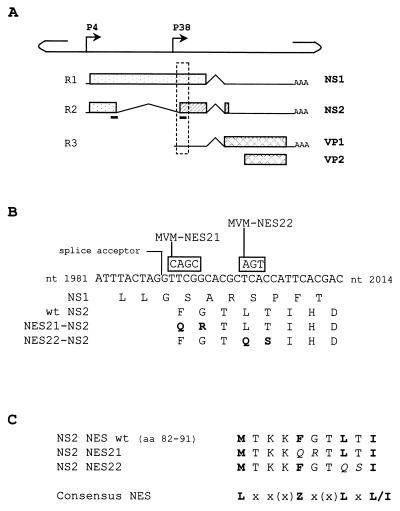
FIG. 1.
Mutagenesis strategy in the NES site of NS2. (A) The line scheme of the single-stranded MVMp genome shows the positions of the nonstructural gene promoter (P4) and the capsid gene promoter (P38). The three classes of RNA transcripts encoded by MVM, denoted R1 through R3, with introns (∧), polyadenylation sites (AAA), and open reading frames (boxes) encoding NS1, NS2, VP1, and VP2 proteins, are aligned below. The two black bars below the R2 transcript indicate the position of the NES site within the NS2 coding sequence. The stippled frame indicates the region that is shown in greater detail in panel B. (B) DNA sequence between MVMp nt 1981 and 2014 and amino acid sequence for this region of open reading frames 3 (for NS1) and 2 (for NS2). NS2 exon 2 initiates at the indicated large intron splice acceptor site. Mutations introduced into this sequence to create the two MVM-NES(−) genomes are shown in boxes above the wild-type sequence, and the resulting substitutions within the NS2 coding sequence are indicated in the last two lines, with the modified amino acids highlighted in bold. (C) Alignment of the wild-type and mutated NS2 NES sites with the NES consensus sequence (9). Leucine-like hydrophobic residues are shown in bold, and amino acid substitutions within the mutated NS2 NES sequences are shown in italics. x, any amino acid; Z, Met, Leu, Ile, Phe, or Val.
(i) pdBMV-NES21.
Two synthetic oligonucleotide primers, 5′ CCTATAAATTTACTAGGCAGCGCACGCTCACCATTCACG 3′ and 5′ CGTGAATGGTGAGCGTGCGCTGCCTAGTAAATTTATAGG 3′, which harbor the desired nucleotide changes (underlined in primer sequences) within the NS2 NES coding region and are complementary to opposite strands of the pdBMVp template, were extended using the QuickChange site-directed mutagenesis kit (Stratagene). The resulting mutated DNA was further transformed into the Escherichia coli strain SURE and isolated according to the manufacturer's protocol. The BstEII-XhoI (nucleotide [nt] 1884 to 2070) fragment, containing the modified fraction of the region encoding the NS2 NES site, was excised from the mutated DNA and subsequently substituted for the equivalent wild-type BstEII-XhoI fragment of pdBMVp, thus generating the pdBMV-NES21 plasmid.
(ii) pdBMV-NES22.
Two oligonucleotide primers, 5′ CTAGGTTCGGCACGCAGTCCATTCACGACACCG 3′ and 5′ CGGTGTCGTGAATGGACTGCGTGCCGAACCTAG 3′, which harbor the desired nucleotide changes (underlined in primer sequences) within the NS2 NES coding region, were introduced into the pdBMVp plasmid as described for the pdBMV-NES21 construction.
Subcloning steps were performed according to standard procedures (71). Both the pdBMV-NES21 and pdBMV-NES22 constructions were sequenced across the entire substituted BstEII-XhoI fragment to ensure that no other mutation than those expected had been introduced. Plasmids containing wild-type and mutated MVMp genomes were propagated in the E. coli strain SURE (Stratagene). Isolation and purification of plasmid DNA were performed using the plasmid Maxi kit (Qiagen) according to the manufacturer's instructions.
Mammalian cell lines.
Mouse A9 fibroblasts (81) were grown in Eagle's minimum essential medium (MEM; Sigma) supplemented with 5% heat-inactivated fetal bovine serum, 1% nonessential amino acids, 2 mM l-glutamine, and 100 μg of gentamicin per ml. Human 293T cells, which were shown to be highly transfectable (67), were propagated in Dulbecco's modified Eagle's medium (Sigma) supplemented with the same additives except the nonessential amino acids.
Transfection, virus stock, and infection.
The pdBMVp, pdBMV-NES21, and pdBMV-NES22 plasmids were transfected in 293T cells using the standard calcium phosphate precipitation protocol (39) with 10 μg of plasmid DNA per 106 cells and in A9 cells using the Lipofectamine transfection reagent as recommended by the supplier (Gibco BRL) with 4 μg of DNA per 106 cells.
For virus production, cells were diluted 1:10 at day 2 posttransfection and further incubated until the appearance of cytopathic effects or until otherwise specified. Cells were then harvested and lysed in 50 mM Tris (pH 8.7)-0.5 mM EDTA by means of three freeze-thaw cycles. Cell debris was removed by centrifugation and virus stocks were stored at 4°C. Virus stocks were titered by DNA hybridization assays as previously described (44). Briefly, A9 and NB-324K indicator cells were inoculated with serial dilutions of virus, incubated for 48 h, and transferred to nitrocellulose filters (NC45; Schleicher and Schuell). The filters were treated with 0.5 M NaOH-1.5 M NaCl to denature DNA and subsequently neutralized with 0.5 M Tris (pH 7.5)-1.5 M NaCl. The membranes were backed at 80°C, hybridized with a 703-bp 32P-labeled MVM DNA probe corresponding to the EcoRV (nt 384)-EcoRI (nt 1084) fragment from the NS coding region, and exposed to X-ray films (Kodak) to detect DNA replication-positive cells. Virus titers were expressed as replication units (RU) per milliliter of virus suspension. Wild-type and mutant viruses were also tested for the ability to generate plaques on A9 and NB-324K indicator cells as described elsewhere (81).
To estimate the single-stranded DNA (ssDNA) content in the virus stocks, equivalent RU of wild-type and mutant viruses extracted in 50 mM Tris (pH 8.7)-0.5 mM EDTA were lysed in Hirt buffer (10 mM Tris [pH 7.4], 10 mM EDTA, 0.6% sodium dodecyl sulfate [SDS]) and treated with proteinase K (0.5 mg/ml; Boehringer Mannheim) to remove viral capsid proteins. The samples were DpnI digested, run on a 0.8% agarose gel, and subjected to Southern blot analysis (77) using the above-mentioned 32P-labeled MVM DNA probe. Blots were exposed to a Phosphor screen and ssDNA was quantitated using a Molecular Dynamics PhosphorImager and ImageQuant 5.1 software. Asynchronous A9 and 293T cells were then infected at a multiplicity of infection (MOI) of 10 RU per cell for wild-type MVMp and 30 RU per cell for the MVM-NES22 mutant in order to compensate for the difference in the ratio of ssDNA-containing particles versus replicative centers observed between wild-type and mutant viruses (see text). Where indicated, neuraminidase (type V, from Clostridium perfringens; Sigma) at 0.1 U per ml of culture medium was added at 4 h postinfection in order to limit infection to a single round by preventing readsorption of virus to cell receptors.
Antibodies.
The following antibodies were used: for Crm1, the commercially available polyclonal goat antiserum SC-7825 (Santa Cruz Biotechnology); for NS2, the polyclonal rabbit antiserum SP6 directed against the C-terminal region of MVMp NS2 (11); for assembled capsids, the monoclonal antibody (MAb) B7, kindly provided by C. Parrish (Cornell, Ithaca, N.Y.) and purified by J. M. Almendral (Madrid, Spain), that specifically recognizes intact MVM capsids (53); for capsid proteins, the polyclonal rabbit antiserum VP2-Pas, a generous gift from J. M. Almendral (Madrid, Spain) that recognizes linear epitopes from viral structural VP1 and VP2 proteins and, to a lesser extent, intact capsids (E. Hernando and J. M. Almendral, manuscript in preparation), or a polyclonal rabbit antiserum directed against a region common to H-1 VP1 and VP2 (44). This last antiserum was used in immunoprecipitation and immunoblotting experiments whereas MAb B7 and VP2-Pas were used in the immunofluorescence assays.
For secondary antibodies, horseradish peroxidase-conjugated goat anti-rabbit or donkey anti-goat antibodies (Santa Cruz Biotechnology) were used in immunoblotting assays and Oregon green 488-conjugated goat anti-rabbit or goat anti-mouse antibodies (Molecular Probes) were used in immunofluorescence assays.
Metabolic radiolabeling and cell extracts.
At 24 h postinfection or 48 h posttransfection, cells (106 cells per 10-cm dish) were metabolically labeled for 2 h with 200 μCi of Tran35S-label (1,000 Ci/mmol; ICN Pharmaceuticals) in Met- and Cys-free Eagle's MEM supplemented with 5% dialyzed fetal calf serum. Cells were subsequently lysed in 1 ml of the so-called Raf buffer (20 mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 1% Nonidet P-40) containing a mixture of proteinase inhibitors (Complete; Roche Molecular Biochemicals), and proteins were harvested after removing cell debris by centrifugation in an Eppendorf centrifuge at full speed for 15 min.
Immunoprecipitation.
Equal amounts (107 cpm) of labeled protein extracts were immunoprecipitated with 5 μl of appropriate antiserum and 60 μl of a 50% suspension of protein A-Sepharose beads (Pharmacia) for about 2 h at 4°C. The immunoprecipitates were washed four times with Raf buffer and resuspended in loading dye buffer (47). Dissociated immunocomplexes were separated in SDS-containing bipartite 8 and 12% polyacrylamide gels and visualized by autoradiography.
For immunoprecipitation-Western blot assays, equal amounts (ca. 4 × 105 equivalent cells) of cold protein extracts were immunoprecipitated with 20 μl of appropriate antiserum and 100 μl of a 50% suspension of protein A-Sepharose beads for 2 h at 4°C, washed as described above, and further processed for immunoblotting.
Immunoblotting.
Total or immunoprecipitated protein extracts were fractionated by SDS-polyacrylamide gel electrophoresis with the indicated percentages of polyacrylamide and were transferred to a nitrocellulose membrane (Schleicher and Schuell). Membranes were blocked by an overnight incubation with 10% low-fat milk powder and 0.5% Tween 20 in phosphate-buffered saline (PBS) at 4°C, incubated with the indicated primary antibodies for 1 h at room temperature, washed three times in PBS containing 0.5% Tween 20, and further incubated with appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Immunoreactive proteins were detected by enhanced chemiluminescence as recommended by the supplier (Perkin-Elmer Life Sciences, Inc.).
Immunofluorescence.
Cells were grown on coverslips, fixed in 3.7% formaldehyde for 10 min, dehydrated in cold methanol for 5 min and cold acetone for 2 min, and permeabilized with 0.2% Triton X-100 for 10 min. Cells were washed first with 0.2% Tween and 0.2% Nonidet P-40 in PBS and then with 2 mM MgCl2 in PBS and were further preincubated with 1% normal goat serum in PBS for 30 min at room temperature. Cells were then successively incubated with primary and secondary antibodies for 1 h each at room temperature, followed by the two washing steps described above. After quick staining with DAPI (4′,6-diamidino-2-phenylindole), coverslips were dried with ethanol and mounted onto glass slides in the presence of Elvanol (polyvinyl alcohol; molecular weight, 77,000 to 79,000; ICN Biomedicals Inc.). Samples were examined with a conventional epifluorescence microscope (Leica DMRBE; ×63 objective with immersion oil). Images were captured using a Hamamatsu Orca digital camera and processed using Openlab 2 (Improvision) and Micrografx Picture Publisher 7a software. Some samples were analyzed by a Leica TCS SP laser scanning confocal device fitted to a Leica IMRBE microscope. In the latter case, Oregon green 488 fluorochrome was excited by the 488-nm excitation line of an argon-krypton laser and detected using a 500- to 540-nm emission window. Single optical sections of about 0.5 μm in depth were performed at the level of the cytoplasm and the nucleus, and images were recorded using Leica TCS-NT version 1 and Adobe Photoshop 4.4 software.
Measurement of cell growth, virus release, and VP protein accumulation.
A9 cells were seeded at 2 × 105 cells per 10-cm-diameter dish and infected with wild-type MVMp (MOI = 10 RU/cell) or the MVM-NES22 mutant (MOI = 30 RU/cell). At daily intervals postinfection, cells were harvested and counted. Viable cell numbers were determined by trypan blue exclusion. At each time point, virus was recovered from supernatants after a clear spin and analyzed by plaque assay on A9 indicator cells to determine virus production. At each time point, cells were lysed in 1 ml of RIPA buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing a mixture of proteinase inhibitors (Complete; Roche Molecular Biochemicals), proteins were harvested after a clear spin, and equivalent volumes of protein extracts were further analyzed by immunoblotting.
RESULTS
Mutations within the NES site of NS2 abolish the interaction between NS2 and Crm1.
The MVM NS2 proteins contain a leucine-rich-like NES that binds with high affinity to Crm1 (3, 49; Bodendorf et al., Virologie 4:179, 2000). In conformity to the NES consensus (9), the NS2 NES sequence (amino acids 82 to 91) contains four properly spaced large hydrophobic residues (Fig. 1C). This NES sequence is located at the junction of NS2 exons 1 and 2, and therefore the first half of this motif (amino acids 82 to 85) is also present in the major nonstructural protein NS1 (Fig. 1A).
To test whether the NS2-Crm1 complex plays a critical role during parvovirus replication, we introduced mutations affecting the NS2 NES site into an infectious clone of MVMp. Substitutions of alanine residues for the critical hydrophobic amino acids have been shown to disrupt NES function in a number of proteins, including HIV-1 Rev (55), human T-cell lymphotropic virus Rex (9), or PKI (89). We indeed observed that by mutating Phe86, Leu89, or Ile91 to alanine in the context of the full-length NS2 protein, the interaction between NS2 and Crm1 was abolished (Bodendorf et al., Virologie 4:179, 2000). However, in the context of the full-length MVM genome, these mutations will also affect the NS1 coding sequence. Furthermore, it should be stated that Met82 and Ile91 cannot be replaced by alanine or another nonhydrophobic amino acid without affecting NS1 as well. Therefore, the two following MVM-NES(−) mutant genomic clones were generated: pdBMV-NES21, which encodes NS2 proteins with substitutions of Gln and Arg for the amino acids Phe86 and Gly87, respectively, and pdBMV-NES22, which expresses NS2 proteins with substitutions of Gln and Ser for the amino acids Leu89 and Thr90, respectively (Fig. 1C). Both mutants were designed so that the primary sequence of the NS2 NES site is modified without altering the sequence of the major nonstructural protein NS1 (Fig. 1B). Although the NES21 and NES22 mutations were introduced in the vicinity of the NS2 large intron splice acceptor, none of them was expected to dramatically impair the splicing of NS2 mRNAs according to a previous report which described the viral elements required for proper MVM pre-mRNA processing (37). In agreement with this expectation, accumulation of NS2 as measured by Western blotting (see below; Fig. 2) was comparable in cells transfected with wild-type and NES mutant genomic clones.
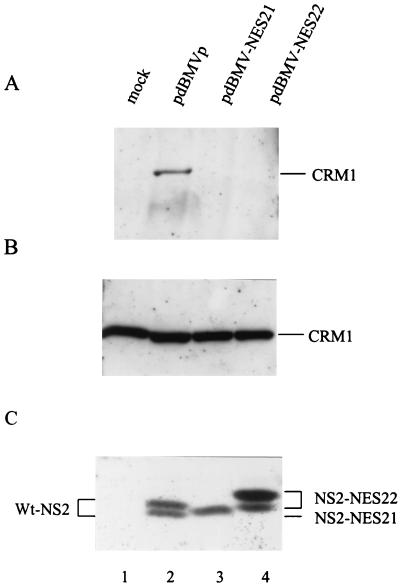
FIG. 2.
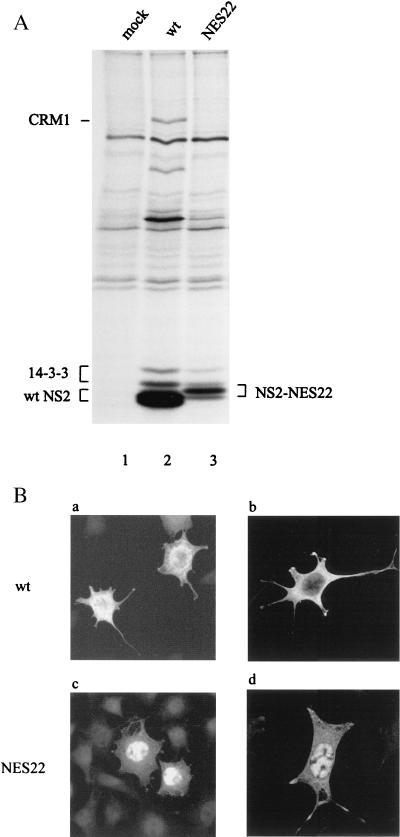
The NES21 and NES22 substitutions within the NS2 NES site prevent the interaction of NS2 with Crm1 in vivo. 293T cells were transfected with pdBMVp, pdBMV-NES21, pdBMV-NES22, or no DNA (mock) and further incubated for 48 h. (A) Whole-cell lysates were first immunoprecipitated with an NS2-specific antiserum and then immunoblotted with anti-Crm1 antibodies. As controls, nonimmunoprecipitated lysates (10% of the amount used for panel A) were analyzed by immunoblotting with either the anti-Crm1 (B) or anti-NS2 (C) antibodies. Proteins were separated on SDS-containing bipartite 8 and 12% polyacrylamide gels. Immunoblots were visualized by chemiluminescence.
We first checked whether the NS2 proteins produced by the MVM-NES(−) mutant genomic clones were indeed impaired in their ability to interact with the nuclear export factor Crm1 as a result of the amino acid substitutions within their NES sites. Human 293T cells were transfected with wild-type, MVM-NES21, or MVM-NES22 mutant DNA clones, and coimmunoprecipitation reactions were performed, using whole-cell extracts and NS2-specific antibodies. As shown in Fig. 2, endogenous Crm1 was coprecipitated with wild-type NS2 (Fig. 2A, lane 2), in agreement with previous reports (7, 64). In contrast, no interaction of Crm1 with either the NS2-NES21 or NS2-NES22 mutant could be detected (Fig. 2A, lanes 3 and 4), although both Crm1 and NS2 proteins were present in similar amounts in all three cell extracts (Fig. 2B and C, respectively). Our results therefore indicated that the 2-aa substitutions in the NS2 NES site were sufficient to inhibit NS2-Crm1 complex formation.
It is worth noting that both mutant proteins were still able to interact with members of the 14-3-3 protein family as efficiently as wild-type NS2 (data not shown), as was previously demonstrated in the case of wild-type MVMp-infected cell extracts (11). This result suggests that the 2-aa substitutions introduced into the NS2 NES site did not drastically modify the conformation of the viral protein. Since the interaction with 14-3-3 proteins requires a phosphorylated amino acid at the 14-3-3 binding site in the target protein (60), our data also indicated that phosphorylation of at least one critical NS2 site was not affected in either of the NES-modified NS2 proteins. Yet, minor alterations in NS2 structure or other posttranslational modifications cannot be excluded, as both NS2-NES21 and NS2-NES22 proteins migrated somewhat slower than wild-type NS2 in SDS-polyacrylamide gels (Fig. 2C, lanes 3 and 4 versus lane 2).
NS2-Crm1 interaction is not required for MVM protein expression in human and mouse cells.
We then tested the ability of the mutated MVM genomes to properly express viral proteins, including the NES-modified NS2 proteins, in human as well as mouse cell lines. Viral proteins were immunoprecipitated from [35S]methionine-cysteine-labeled cells that had been previously transfected with either wild-type or MVM-NES(−) mutant genomic clones. As illustrated in Fig. 3A, the MVM-NES22 mutant genome was able to support the synthesis of NES-modified NS2 proteins in both human 293T (lane 4) and mouse A9 (lane 8) cells. As expected, NS2-NES22 mutant proteins were not able to interact with the nuclear export receptor Crm1, although they could still bind to members of the 14-3-3 protein family. The NES22 substitutions did not appear to interfere with the expression of the other viral polypeptides, as NS1, VP1, and VP2 proteins were produced with similar efficiency upon transfection of human or mouse cells with either wild-type or NES22-mutated DNA clones (Fig. 3B and C). The lack of a significant effect of NES22 substitutions L89Q and T90S on parvovirus gene expression was confirmed by Western blotting assays (data not shown). Altogether, our results indicate that NS2 interaction with the nuclear export factor Crm1 is not required for an efficient production of the virus-encoded proteins.
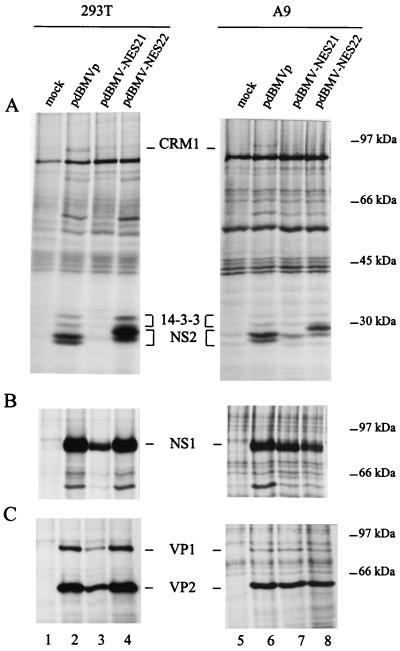
FIG. 3.
Effect of the NES21 and NES22 substitutions within the NS2 NES site on viral protein synthesis. Lysates of 35S-labeled human 293T and mouse A9 cells, transfected with pdBMVp, pdBMV-NES21, pdBMV-NES22, or no DNA (mock), were precipitated with antisera directed against the NS2 C terminus (A), the NS1 C terminus (B), or the VP1 and VP2 C termini (C). The autoradiograms show immunoprecipitated proteins after separation by electrophoresis through SDS-containing bipartite 8 and 12% polyacrylamide gels. The positions of Crm1 (110 kDa), 14-3-3 (30 and 32 kDa), NS2 (ca. 25 kDa), NS1 (83 kDa), VP1 (83 kDa), and VP2 (65 kDa) proteins are indicated. Molecular masses of prestained standard proteins are shown on the right.
The MVM-NES21 mutant genome was also able to support synthesis of NS1, VP1, and VP2 proteins in both human and mouse cells (Fig. 3B and C, lanes 3 and 7). In contrast, amounts of immunoprecipitated NS2 varied from barely detectable (Fig. 3A, lanes 3 and 7) to nearly wild-type-like levels (data not shown), suggesting an altered instability and/or synthesis of these mutant viral proteins. There is so far no clear explanation for this variation, but it is possible that the NES21 mutations, which gave rise to the F86Q and G87R substitutions within the NS2 sequence, may have other effects besides impairment of the NS2 NES motif.
During the parvovirus replication cycle, the viral nonstructural proteins NS1 and NS2 are expressed after conversion of the input single-stranded genome into a double-stranded transcription template, driving the subsequent steps of viral DNA amplification, expression, and possibly packaging. By analogy with other viral regulatory proteins such as the HIV-1 Rev protein (8, 31, 54), the NS2 proteins may conceivably contribute, through their association with Crm1, to the export of viral mRNAs to the cytoplasm and the ensuing synthesis of viral proteins. The phenotype of the MVM-NES(−) mutant genomic clones described above argued against this possibility and led us to analyze other events of the parvovirus life cycle.
The MVM-NES22 genomic clone is able to generate progeny viruses in transfected human and mouse cells.
It was previously described that expression of MVM NS2 protein is not essential for the production of infectious virions in human cells but is absolutely required for a productive infection of mouse cells (17, 61, 62). Given the similar proficiency in the production of at least NS1, VP1, and VP2 proteins, wild-type and MVM-NES21 or MVM-NES22 genomic clones were compared for their ability to give rise to virus production in human and mouse cells. To this end, human 293T and mouse A9 cells were transfected with the respective molecular clones and further incubated for 6 days, with a 1:10 dilution performed at day 2 posttransfection. At this time cytopathic effects became apparent in the wild-type but not MVM-NES(−) DNA-transfected cultures. Virus was recovered from all transfected cell cultures by repeated freezing and thawing cycles and was titered by DNA replication centers and plaque assays, using A9 or NB-324K indicator cell lines. Similar results were obtained with both cell lines and are illustrated in Fig. 4 for A9 indicator cells.
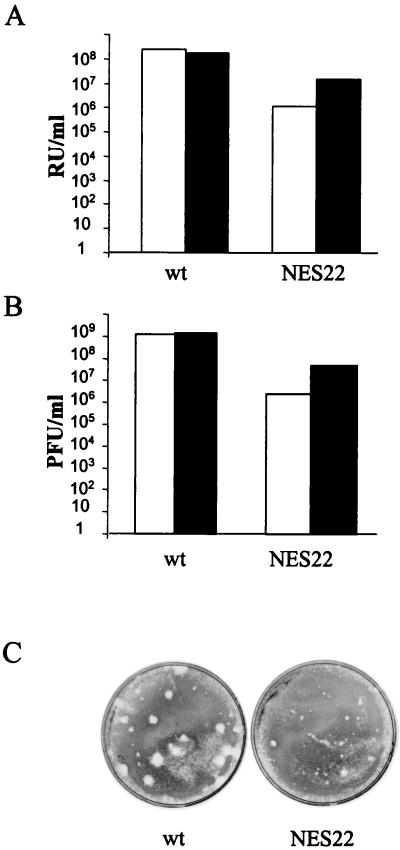
FIG. 4.
The MVM-NES22 genomic clone generates the production of infectious virions in human 293T and mouse A9 cells. Cells were transfected with either pdBMVp or pdBMV-NES22 plasmid DNA and viruses were harvested in the same final volume 6 days after transfection. Virus stocks were used to infect A9 indicator cells, and virus yields were determined by performing either hybridization assays, for which titers are given in RU per milliliter of crude extract (A), or plaque assays, for which titers are given in PFU per milliliter of crude extract (B). White bars, virus stocks generated in A9 cells; black bars, virus stocks generated in 293T cells. (C) Typical size appearance of plaques obtained upon infection of A9 indicator cells with viruses generated in A9 cells. Similar patterns were observed upon infection of A9 indicator cells with viruses generated in 293T cells (data not shown). wt, wild-type MVMp; NES22, MVM-NES22 mutant.
As shown in Fig. 4A and B, the MVM-NES22 mutant genomic clone was competent for infectious virus production not only in human 293T but also in mouse A9 cell lines. Yet, the progeny virus yields were about 12- to 460-fold lower in cultures transfected with the mutant than in those transfected with the wild-type clone. The largest difference concerned production in A9 cells, in agreement with the greater dependence of MVM replication on NS2 proteins in mouse versus human cells. Interestingly, viruses generated by the MVM-NES22 genomic clone gave rise to tiny plaques in comparison with wild-type virions (Fig. 4C). Altogether, these results indicate that the association of NS2 with Crm1 is not required for the formation of infectious particles; however, the presence of NS2 proteins, which harbored the amino acid substitutions L89Q and T90S in their NES site, impaired, at least to some extent, the intracellular production, release, and/or infectivity of MVM virions.
In marked contrast, the MVM-NES21 mutant genomic clone failed to support infectious virion production not only in mouse A9 but also human 293T cell cultures, even 8, 11, and 17 days after transfection as measured by plaque assay using either A9 or NB-324K as indicator cell lines (data not shown). This phenotype of the MVM-NES21 mutant, which can be distinguished from both the MVM-NES22 and MVM-NS2null mutant phenotypes (17, 61, 62), might be ascribed to secondary effects of NS2 related to changes not only in the expression (see above) but also in the quality of the NS2 products.
Altogether, our results led us to focus on the MVM-NES22 mutant to further analyze the consequences of the disruption of NS2-Crm1 interaction on the parvovirus replication cycle. To this end, wild-type MVMp and MVM-NES22 mutant virus stocks were prepared in A9 cells as described above, except that each virus was collected at the appearance of cytopathic effects. The ratio of full particles to replicative centers was determined for both the wild-type and mutant virus stocks by hybridization measurement of ssDNA in proteinase K-treated samples and of viral DNA-amplifying cells. This ratio was somewhat (about threefold) higher for the mutant virus, indicating a slightly lower infectivity of the MVM-NES22 mutant than of the wild-type virus (data not shown). However, it is unlikely that the lower yields and smaller plaques generated from MVM-NES22 could only be ascribed to the lower infectivity of the mutant. These results prompted us to carry out further experiments to analyze MVM-NES22 mutant virus production, in particular in relation to nuclear export processes.
Deficiency of NS2 nuclear export in MVM-NES22 mutant-infected mouse cells.
It was first ascertained that the viruses produced in cells transfected with MVM-NES22 mutant genomic DNA were not revertants and encoded NS2 proteins displaying the original NES(−) phenotype. A9 cells were infected with either wild-type or MVM-NES22 mutant virions, labeled de novo with [35S]methionine-cysteine, and processed for NS2 protein immunoprecipitation using NS2-specific antibodies. As illustrated in Fig. 5, the NS2 proteins produced by MVM-NES22 viruses were unable to bind to Crm1 while still interacting with 14-3-3 proteins (lane 3), whereas wild-type NS2 proteins precipitated both Crm1 and 14-3-3 proteins (lane 2). Furthermore, the NS2 proteins from MVM-NES22 mutant-infected cells showed the mobility shift noticed above for NS2 produced in cells transfected with the MVM-NES22 mutant genomic clone (Fig. 2). Therefore, MVM-NES22 viruses appeared to encode the expected mutant form of NS2.
FIG. 5.
NS2 proteins expressed from the MVM-NES22 mutant exhibit an NES(−) phenotype. (A) Coimmunoprecipitation assays from infected mouse cell extracts. Lysates of 35S-labeled A9 cells infected with wild-type MVMp (MOI = 10 RU/cell), MVM-NES22 mutant (MOI = 30 RU/cell), or no virus were immunoprecipitated with an NS2-specific antiserum. The autoradiogram shows immunoprecipitated proteins after separation by electrophoresis through an SDS-containing bipartite 8 and 12% polyacrylamide gel. (B) Subcellular distribution of wild-type and NES22-modified NS2 proteins. A9 cells were infected as described above and further analyzed by indirect immunofluorescence assay 24 h after infection, using an NS2-specific antiserum. Cells were examined by either epifluorescence (a and c) or confocal (b and d) microscopy. mock, no virus; wt, wild-type MVMp; NES22, MVM-NES22 mutant.
We and others previously demonstrated that in the presence of leptomycin B, a drug that specifically inhibits Crm1-mediated nuclear export (33, 66, 91), the cytoplasmic NS2 proteins predominantly relocalized to the nuclei of MVMp-infected cells (7, 64). This prompted us to analyze the subcellular localization of NS2 proteins in MVM-NES22-infected cells compared to wild-type virus-infected cells by indirect immunofluorescence using NS2-specific antibodies. NS2 proteins expressed from wild-type MVMp-infected cells predominantly accumulated in the cytoplasm, although they were also present to some extent in the nucleus (Fig. 5B, panels a and b), in agreement with previous reports (15, 19). In marked contrast, the inverse pattern was observed in MVM-NES22 mutant-infected cells, with a stronger accumulation of NS2 in the nucleus than in the cytoplasm (Fig. 5B, panels c and d). Altogether, these results confirmed that the MVM-NES22 mutant encoded NS2 proteins that were impaired in Crm1-mediated nuclear export.
Retention of MVM-NES22 progeny particles in the nucleus of infected mouse cells.
Given the facts that infection of mouse cells with the MVM-NES22 mutant, on the one hand, led to a predominantly nuclear localization of the NES-modified NS2 proteins and, on the other hand, gave rise to the formation of tiny plaques, it was tempting to speculate that abolition of the NS2-Crm1 interaction may affect the intracellular distribution of other viral products. The subcellular localization of the VP proteins and capsids was determined in wild-type MVMp and MVM-NES22 mutant virus-infected cells by indirect immunofluorescence using specific polyclonal antibodies (VP2-Pas) or MAbs. The polyclonal antiserum VP2-Pas was previously shown to be able to recognize both VP1 and VP2 capsid proteins and, to a lesser extent, assembled capsids (Hernando and Almendral, manuscript in preparation), while the MAb B7 can only detect assembled capsids (53). After infection with wild-type MVMp or the MVM-NES22 mutant, mouse A9 cells were incubated for various times and further processed for immunofluorescence staining. In agreement with the above-mentioned immunoprecipitation experiments (Fig. 3), a similar proportion of cells in cultures infected with either virus was positive for the presence of structural proteins and capsids (data not shown). In contrast, wild-type MVMp and MVM-NES22 virus-infected cells could be distinguished by the intracellular distribution of VP proteins and capsids.
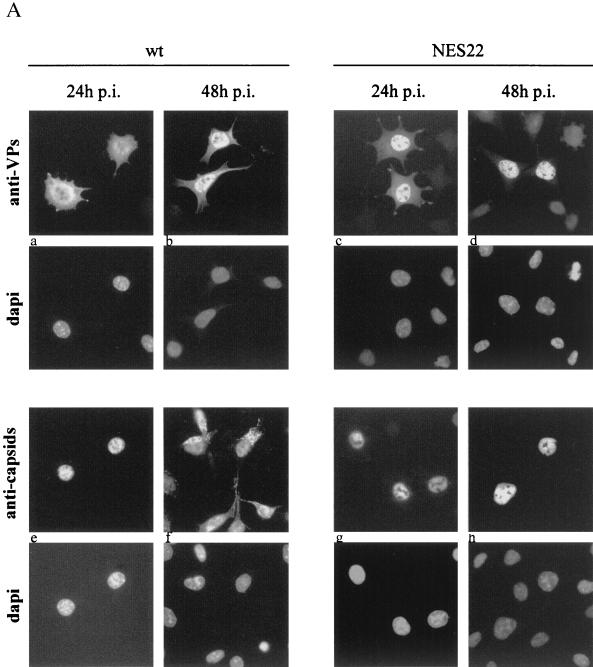
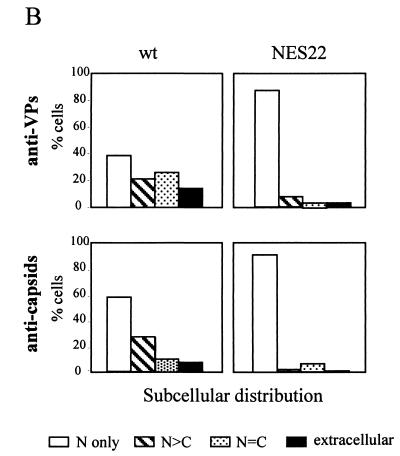
A9 cells infected with wild-type MVMp showed both cytoplasmic and nuclear staining for VP proteins, which became predominantly nuclear 48 h after infection (Fig. 6A, panels a and b, and 6B, top left). Assembled particles could be detected almost exclusively in the nuclei of infected cells 24 h after infection (Fig. 6A, panel e), while at later times, the capsid-specific antibodies gave rise to additional punctate signals in the cytoplasm and at the cell periphery (Fig. 6A, panel f, and 6B, bottom left). Altogether, these results were consistent with the current view that upon infection with the wild-type virus, de novo-synthesized VP proteins migrate to the nucleus, in which capsid assembly takes place (53, 73, 85), followed by virus release from the cells and further reinfection of the surrounding cells (69, 76). It is worth noting that in neuraminidase-treated cultures, cells that were exclusively labeled in their cytoplasm could not be detected, indicating that second rounds of infection were prevented. However, a small fraction of cells showing capsid staining in the nucleus were also able to exhibit a cytoplasmic capsid-specific signal at 48 h postinfection (Fig. 6B, bottom left), although this cytoplasmic labeling essentially appeared diffuse rather than punctate (data not shown). These data suggest that at least part of the cytoplasmic signal may correspond to nucleus-released rather than reinfecting virions.
FIG. 6.
MVM-NES22 particles are retained in the nucleus of infected mouse cells. (A) A9 cells, infected with either wild-type MVMp (MOI = 10 RU/cell) or the MVM-NES22 mutant (MOI = 30 RU/cell), were analyzed by indirect immunofluorescence assay at 24 and 48 h postinfection, using antisera directed against the structural proteins (anti-VPs) or intact capsids (anti-capsids), and were examined by epifluorescence microscopy. The images shown are representative of the most abundant phenotypes found in the wild-type and mutant infected cells. Panels: a, VP staining in both the nucleus and cytoplasm; b and c, VP staining predominantly in the nucleus; d, VP staining in the nucleus only; e, g, and h, capsid staining in the nucleus only; f, capsid staining in both the nucleus and cytoplasm. Nuclei were visualized within each field by staining the DNA with DAPI. (B) Subcellular distribution of the structural protein (top) and capsid (bottom) antigens. Cells were treated as for panel A except for the addition of neuraminidase 4 h after virus inoculation to prevent secondary rounds of infection. The values are the percentages of at least 90 immunofluorescence-positive cells counted 48 h after infection. Scored cells were classified into four categories as follows: immunofluorescent staining that was exclusively nuclear (N), mainly nuclear (N > C), comparably nuclear and cytoplasmic (N = C), and at the periphery of infected cells (extracellular). wt, wild-type MVMp; NES22, MVM-NES22 mutant.
At 24 h postinfection, MVM-NES22 mutant-infected A9 cells showed a staining pattern similar to that of wild-type virus-infected cells, namely VP staining over both the nucleus and the cytoplasm and a capsid-specific signal confined to the nucleus (Fig. 6A, panels c and g). In contrast, MVM-NES22 differed from wild-type MVMp at later times postinfection in that the VP staining became mostly nuclear and no significant capsid-specific signal outside the nucleus could be detected (Fig. 6A, panels d and h, and 6B, top and bottom right). It was concluded from these observations that MVM-NES22 mutant viruses are competent for VP production, nuclear translocation, and assembly into capsids but are deficient in the export of virions out of the nucleus. This phenotype can be assumed to impair virus release and may explain, at least in part, the above-mentioned tiny size of the plaques generated by MVM-NES22 upon infection of the indicative cells. It is worth noting that the effect of disrupting the NS2 NES motif on the intracellular distribution of viral products was specific for VP proteins and capsids besides the NS2 proteins themselves, as the localization of the major nonstructural protein NS1 was identical, i.e., predominantly nuclear, in both wild-type MVMp and MVM-NES22 virus-infected cells (data not shown).
Delayed virus release and cell death after infection with the MVM-NES22 mutant.
The impaired export of progeny MVM-NES22 virions out of infected cell nuclei led us to compare the mutant and wild-type viruses for other late events in the parvovirus life cycle, namely cell death, particle release, and ensuing secondary infections. To this end, asynchronous A9 cell cultures were infected with wild-type MVMp or the MVM-NES22 mutant and were assayed in parallel for cell growth, virus release in the medium, and further internalization at daily intervals postinfection.
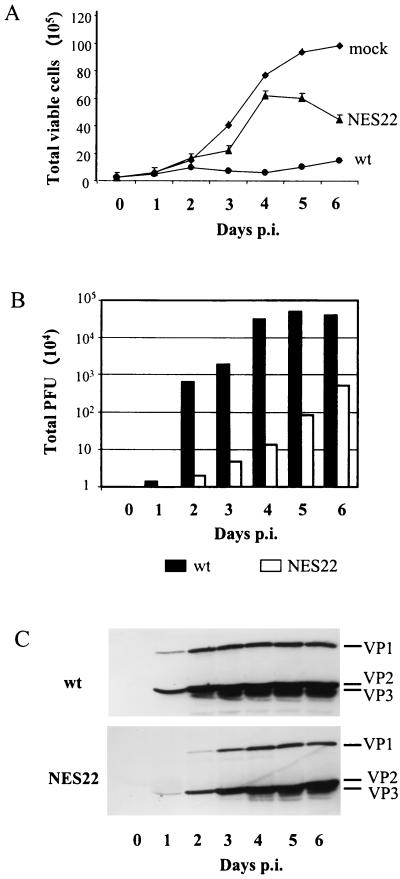
As shown in Fig. 7B, increasing amounts of infectious particles were detected in the medium of wild-type MVMp-infected cultures until day 4 postinfection, at which time virus production reached a plateau. As expected, the continuous virus production taking place during the first 4 days postinfection was accompanied by a drastic decrease in the viability of the wild-type MVMp-infected cells compared with that of mock-treated ones, resulting in a lack of significant growth of infected cultures (Fig. 7A). This growth inhibition was the result of repeated infection cycles within the cell cultures, as evidenced by the increased accumulation of VP3 polypeptides, besides the VP1 and VP2 capsid proteins, in whole-cell extracts from infected cells (Fig. 7C, upper panel).
FIG. 7.
Delayed cell killing, virus release, and subsequent internalization in MVM-NES22 mutant-infected mouse cell cultures. (A) Cell survival kinetics of A9 cell cultures infected with wild-type MVMp (MOI = 10 RU/cell), the MVM-NES22 mutant (MOI = 30 RU/cell), or no virus. The number of viable cells in each culture was determined by multiplying the total number of cells by the percentage of living cells assayed by trypan blue exclusion. (B) Time course of virion release in infected mouse cell cultures. A9 cells were infected with wild-type MVMp or the MVM-NES22 mutant as mentioned for panel A. The amounts of infectious particles released in the medium over time were determined by plaque assays using A9 indicator cells and were expressed as total PFU after subtraction of the day 0 value. (C) Time course appearance of VP1, VP2, and VP3 proteins in infected mouse cell extracts. A9 cells were infected with either wild-type MVMp or the MVM-NES22 mutant as mentioned for panel A. Whole-cell lysates, prepared at days 1 to 6 after infection, were analyzed by immunoblotting with an antiserum directed against the common C terminus of the VP polypeptides following protein separation on SDS-containing 10% polyacrylamide gels. Immunoblots were visualized by chemiluminescence. The experiments represented in all three panels were done in parallel. mock, no virus; wt, wild-type MVMp; NES22, MVM-NES22 mutant; p.i., postinfection.
The MVM-NES22 mutant was distinguishable from wild-type MVMp by a delayed and slower virus release which continued during the whole time interval studied (Fig. 7B). In agreement with these data, MVM-NES22 mutant-infected cell cultures kept growing, albeit at a slower rate than mock-treated controls, until day 4 to 5 postinfection, at which time the growth of the cultures became overwhelmed by cell death (Fig. 7A). This decrease in cell survival could be correlated with a progressive appearance of VP3 polypeptides in the corresponding whole-cell extracts (Fig. 7C, lower panel), indicating that second rounds of infection were taking place in the MVM-NES22 mutant-infected cultures, though at later times than with wild-type MVMp-infected cells. Altogether, these results showed that MVM-NES22 viruses were able to induce a productive infection in A9 cells, leading to the release of a burst of infectious progeny particles, but that their replication cycle is slower, thereby allowing host cells to survive longer than after infection with wild-type viruses. From the above-mentioned impairment of MVM-NES22 mutant-infected cells in the nuclear export of virions, it may be assumed that the MVM-NES22 replication cycle is delayed at late stages involved, in particular, in the egress of viral DNA-containing assembled particles and in the subsequent internalization of these infectious virions.
DISCUSSION
We and others previously described that MVMp NS2 proteins are able to interact with the nuclear export receptor Crm1, leading to their active export out of the nucleus of infected cells (7, 64). In the present study, the 2-aa substitutions F86Q-G87R (mutation NES21) and L89Q-T90S (mutation NES22) were introduced within the NS2 NES site, using an MVMp genomic clone. The mutated genomes both generated NS2 proteins that had lost the capacity for interacting with Crm1 but behaved differently upon transfection of human or mouse cells. The MVM-NES22 genomic clone gave rise to a productive infection in both cell lines, whereas the MVM-NES21 genomic clone failed to support infectious virion production in either of them. Both genomic clones expressed NS2 proteins that ran more slowly than wild-type NS2 in denaturing polyacrylamide gels. Yet NES21-NS2 proteins could be further distinguished from both wild-type and NES22-NS2 polypeptides in that they migrated predominantly as a single band and showed some variability in their expression. This led us to conclude that the 2-aa substitution F86Q-G87R might affect other functions of NS2 besides the one related to the functionality of the NS2 NES site. A further characterization of the MVM-NES21 genomic clone is in progress. On the other hand, experiments using the MVM-NES22 mutant demonstrated that in the absence of interaction between NS2 and Crm1, infectious virions could be generated in mouse cells. However, the egress of MVM-NES22 viruses from the host cell nuclei was severely retarded. This delay in the completion of the parvovirus replication cycle correlated with a slower release of the mutant virus than of the wild-type virus from infected cells and could account, at least in part, for the longer survival of infected cell cultures and the generation of tiny plaques on indicator cells.
Altogether, these data indicate that the association between NS2 and Crm1 plays an important role in MVMp propagation, in particular at later stages of the viral replication cycle. This phenotype distinguishes MVM-NES22 from the other NS2 mutants described so far. It has been previously reported that various MVM-NS2 mutants encoding either C-terminally modified or no NS2 proteins were unable to productively replicate in mouse cells as a result of major defects in both viral DNA replication and capsid assembly (14, 17, 61, 62). In addition, no C-terminally modified or truncated NS2 proteins could be detected in mutant virus-infected mouse cells, although some of these proteins were expressed to wild-type levels in transformed human or rat cells (11, 17, 61). The MVM-NES22 mutant described in this study shows a phenotype that is clearly different from the one exhibited by MVM-NS2null mutants, since it proved able to both express mutated NS2 proteins and generate fully infectious particles in mouse cells. In proliferating mouse fibroblasts infected with MVMp, the viral NS2 proteins are localized in the cytoplasmic and, to a much lesser extent, nuclear compartments (19). As shown in this study, disruption of the NS2 NES site leads to both an increased nuclear accumulation of the viral NS2 proteins and a nuclear retention of fully assembled MVMp capsids in infected mouse cells. It therefore appears that efficient nuclear exit of the virions depends on NS2 activities linked to either the cytoplasmic localization of the protein or its transport out of the nucleus. Whether nuclear accumulation of NS2 interferes with other steps in the parvovirus replication cycle cannot be excluded either. Indeed, a recent report by Miller and Pintel (57) shows that abolishment of NS2-Crm1 interaction not only affects the nuclear export of MVM infectious particles but also causes a significant reduction in the accumulation of ssDNA in infected mouse cells. According to this report, the primary defect in the Crm1 interaction mutant is most likely at the level of capsid egress from the nucleus and the authors proposed a possible feedback mechanism, direct or indirect, between the nuclear exit of full virions and continued production of progeny ssDNA (57).
In the present study, we showed that the release of MVM-NES22 virions from the nucleus of infected cells was delayed, suggesting that the particles were only temporarily blocked in the nuclear compartment. The mechanisms by which MVM particles are released from infected cells remain enigmatic. Parvovirus release was previously considered to result from the degeneration of both the nucleus and the cytoplasm, leading to the disruption of the plasma membrane (69, 76). Ultra-histological studies, however, reported the presence of virus particles within the cytoplasm in the vicinity of nuclear pores at late phases of infection, suggesting that nuclear exit of progeny virions might result from an active transport process. On the other hand, the presence of virus in the cytoplasm could also reflect a passive release of progeny virions from a degenerating nucleus (75). Nonetheless, active and passive mechanisms for parvovirus nuclear exit are both compatible with virion release into the culture medium following lysis of infected cells. Such a cytoplasmic passage prior to cell lysis to release nonenveloped virus in the culture medium has been recently demonstrated for adenovirus particles (68). Although we cannot exclude the fact that nuclear degeneration is delayed in MVM-NES(−)-infected cells, the nuclear retention of mutant virions reported in this study may reflect a defective transport of MVM-NES(−) particles as a consequence of the nuclear accumulation of NES-modified NS2 proteins. The late egress of the mutant may then be related to the fact that a small but significant proportion of NES-modified NS2 proteins was still localized in the cytoplasm. This cytoplasmic subpopulation of NS2 proteins may either fail to transit through the nucleus or be released from the nucleus through an NES-independent pathway. It is noteworthy that NS2 proteins are small proteins of about 25 kDa and could therefore go in and out of the nucleus by passive diffusion through the nuclear membrane, since the cutoff size for facilitated transport versus diffusion across the nuclear pore complex is considered to be 40 to 50 kDa (25). Assuming that the nuclear export of NS2 controls virus exit from the nucleus, passive diffusion of NS2 alone or bound to other small molecules through the nuclear membrane may participate in the release of virions, albeit at a lower rate than with active NS2 transport.
The reason why suppression of the Crm1-mediated export of NS2 out of the nucleus impairs the nuclear egress of infectious virions is currently a matter of speculation. As stated above, one possible explanation would be that NS2 controls the latter process by acting in the cytoplasm, for instance, by modulating posttranslational modifications of the VP proteins prior to their import in the nucleus. On the other hand, NS2 may work as cargo to coexport from the nucleus either the viral particles themselves or factors that interfere with the nuclear release of assembled virions. The MVMp capsid is composed of 60 protein subunits, 90% of which consist of VP2 polypeptides and 10% of which consist of VP1 polypeptides (82). It has also been shown that VP2 is necessary and sufficient for the encapsidation of virus progeny ssDNA (85). Parvovirus capsid formation is a multistep process which is still not fully understood. The two capsid proteins VP1 (83 kDa) and VP2 (64 kDa) are first synthesized in the cytoplasm of infected cells, where they cooperatively interact to further migrate to the nucleus (53). In the nucleus, VP1- and VP2-containing complexes are assembled into icosahedral virions which are able to package single-stranded viral DNA (2, 24, 85) and are released from the infected cells, most likely after passage through the cytoplasm (69, 75, 76). In full virions, the N terminus (ca. 20 to 25 amino acids) of some VP2 molecules becomes exposed at the particle surface and further cleaved during release and/or subsequent uptake of infectious particles into the host cells, leading to the appearance of VP3 (60 kDa) (2, 73, 83). The nuclear export of NS2 might be coupled with that of assembled virions in a direct way. It should be stated, however, that attempts to detect interactions between the viral NS2 and VP2 proteins using the yeast two-hybrid system were not successful (U. Bodendorf and N. Salomé, unpublished results). In addition, no interaction between NS2 and fully assembled capsids has been described so far. Alternatively, NS2 may act indirectly, for instance, by participating in the modification of VP components before or after their assembly into capsids.
Interestingly, it has recently been reported that phosphorylation of the VP2 N terminus plays an important role in the parvovirus life cycle. Indeed, MVMp mutants lacking VP2 N-terminal phosphorylation were found to form tiny plaques on human indicator cells (56), which appeared to result from an impaired release of mutant virions from infected cells (B. Maroto and J. M. Almendral, Abstr. VIIIth Parvovirus Workshop, p. 32, 2000). This phenotype is similar to the one previously described for MVMp-NS2null mutants in human cells (17) as well as to the one observed here for the MVMp-NES(−) mutant. Altogether, these data lead us to speculate that the viral NS2 proteins contribute to the regulation of VP2 phosphorylation. The dependence of this effect may tentatively be assigned to the fact that the VP2 phosphorylation events concerned take place in the cytoplasm prior to the migration of VP complexes into the nucleus and/or require the NS2-mediated intracellular delocalization of a kinase, phosphatase, or associated regulating factor(s).
Several steps of the parvovirus replication cycle, including viral DNA amplification, capsid assembly, and viral ssDNA packaging, are taking place in the nucleus of infected cells. It has been recently demonstrated that MVM DNA replication proceeds in specific virus-induced nuclear structures termed autonomous parvovirus-associated replication (APAR) bodies (6). These APAR bodies might also correspond to active sites of de novo capsid assembly and/or viral ssDNA packaging (93). It is relevant to note that the generation of MVM ssDNA is tightly connected to the availability of assembled MVM capsids (22, 24). In mouse cells, MVM NS2 proteins are needed for capsid assembly and generation of progeny ssDNA but not for the synthesis of the viral capsid proteins VP1 and VP2 and their subsequent migration to the nucleus, most likely as preassembled particles (17, 53). Interestingly, nuclear accumulation of NS2 does not dramatically impair assembly of capsid proteins into infectious particles, as demonstrated in the present study. Therefore, one can speculate that the NS2 function(s) would be to improve the interaction between (pre)assembled capsids and viral DNA cleavage and packaging machinery. This could be achieved by modulating cellular or viral factors that are involved in the nuclear formation or stabilization of MVM particles and/or in the positioning of these particles in the nuclear parvovirus replication centers, i.e., the APAR bodies. This model would be consistent with a nuclear localization of the NS2 proteins and might explain why NS2-NES(−) mutant proteins are still efficient for progeny virion production. On the other hand, either cytoplasmic NS2 or NS2 transport through the nuclear envelope seems to be required to facilitate the release of infectious MVM particles. It is worth noting that a role in proper localization of assembled capsids and in viral DNA cleavage and packaging reactions has been proposed for other viral regulatory proteins, including the herpes simplex virus type 1 UL32 gene product (48). The molecular mechanisms that are behind NS2 activities in both virion formation and release still remain to be elucidated. However, NS2 is probably acting indirectly, since specific cellular determinants, provided, for example, in transformed human cells, can complement for NS2 function(s) although to a limited extent, as suggested by a delayed release of MVM-NS2null mutant and a higher nuclear accumulation of MVM-NES(−) mutant in infected NB-324K cells (17, 57).
Some members of the parvovirus family, including MVMp and H-1 virus, displayed interesting oncosuppressive properties which, together with the ability of these viruses to cause clinically asymptomatic infections, point to the use of these agents as vectors for the transfer and preferential expression of anticancer genes in tumors (5, 16, 26). Indeed, several laboratories including ours have developed H-1 and MVMp-based vectors in which the early P4 promoter and the sequence encoding the NS1 protein have been retained but the genes coding for the capsid proteins have been partially deleted and replaced by different transgenes (30, 40, 70, 90). At present, a limiting step to the assessment of these vectors lies in the relatively low titers of the recombinant virus stocks obtained. The present work shows that NS2 needs to be considered among the parameters that determine the efficiency of parvovirus production. Therefore, a better knowledge of NS2 functioning during the late steps of the parvovirus life cycle will be of great interest for optimizing parvovirus-based vector production and infectivity.
Acknowledgments
We are indebted to P. Tattersall (Yale University, New Haven, Conn.) for kindly providing the pdBMVp infectious plasmid and to C. Parrish (Cornell University, Ithaca, N.Y.) and J. M. Almendral (Universidad Autonoma, Madrid, Spain) for the generous gift of MVM monoclonal anti-capsid antibody and polyclonal anti-VP antiserum. We thank J. M. Almendral for valuable comments, C. Cziepluch and C. Dinsart for critical reading of the manuscript, and the members of the laboratory for helpful discussions.
V.E. was supported by a fellowship from the Deutsches Krebsforschungszentrum and L.D. was supported first by an Alexander von Humboldt fellowship and further by a Marie Curie fellowship from the European Community.
REFERENCES
- 1.Adachi, M., M. Fukuda, and E. Nishida. 2000. Nuclear export of MAP kinase (ERK) involves a MAP kinase kinase (MEK)-dependent active transport mechanism. J. Cell Biol. 148:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 3.Askjaer, P., A. Bachi, M. Wilm, F. R. Bischoff, D. L. Weeks, V. Ogniewski, M. Ohno, C. Niehrs, J. Kjems, I. W. Mattaj, and M. Fornerod. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askjaer, P., T. H. Jensen, J. Nilsson, L. Englmeier, and J. Kjems. 1998. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 273:33414-33422. [DOI] [PubMed] [Google Scholar]
- 5.Avalosse, B., F. Dupont, and A. Burny. 1995. Gene therapy for cancer. Curr. Opin. Oncol. 7:94-100. [PubMed] [Google Scholar]
- 6.Bashir, T., J. Rommelaere, and C. Cziepluch. 2001. In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies. J. Virol. 75:4394-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodendorf, U., C. Cziepluch, J. C. Jauniaux, J. Rommelaere, and N. Salomé. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 73:7769-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogerd, H. P., A. Echarri, T. M. Ross, and B. R. Cullen. 1998. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 72:8627-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogerd, H. P., R. A. Fridell, R. E. Benson, J. Hua, and B. R. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandenburger, A., D. Legendre, B. Avalosse, and J. Rommelaere. 1990. NS-1 and NS-2 proteins may act synergistically in the cytopathogenicity of parvovirus MVMp. Virology 174:576-584. [DOI] [PubMed] [Google Scholar]
- 11.Brockhaus, K., S. Plaza, D. J. Pintel, J. Rommelaere, and N. Salomé. 1996. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J. Virol. 70:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brownstein, D. G., A. L. Smith, E. A. Johnson, D. J. Pintel, L. K. Naeger, and P. Tattersall. 1992. The pathogenesis of infection with minute virus of mice depends on expression of the small nonstructural protein NS2 and on the genotype of the allotropic determinants VP1 and VP2. J. Virol. 66:3118-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cater, J. E., and D. J. Pintel. 1992. The small non-structural protein NS2 of the autonomous parvovirus minute virus of mice is required for virus growth in murine cells. J. Gen. Virol. 73:1839-1843. [DOI] [PubMed] [Google Scholar]
- 15.Clemens, K. E., D. R. Cerutis, L. R. Burger, C. Q. Yang, and D. J. Pintel. 1990. Cloning of minute virus of mice cDNAs and preliminary analysis of individual viral proteins expressed in murine cells. J. Virol. 64:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelis, J. J., A. Haag, C. Kornfeld, G. Balboni, A. Y. Dege, B. Neeb, C. Dinsart, and J. Rommelaere. 2000. Autonomous parvovirus vectors, p. 97-115. In A. Cid-Arregui and A. Garcia-Carranca (ed.), Viral vectors: basic science and gene therapy. Eaton Publishing, Natick, Mass.
- 17.Cotmore, S. F., A. M. D'Abramo, Jr., L. F. Carbonell, J. Bratton, and P. Tattersall. 1997. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231:267-280. [DOI] [PubMed] [Google Scholar]
- 18.Cotmore, S. F., J. P. Nuesch, and P. Tattersall. 1993. Asymmetric resolution of a parvovirus palindrome in vitro. J. Virol. 67:1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotmore, S. F., and P. Tattersall. 1990. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology 177:477-487. [DOI] [PubMed] [Google Scholar]
- 20.Cotmore, S. F., and P. Tattersall. 1994. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 13:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91-174. [DOI] [PubMed] [Google Scholar]
- 22.Cotmore, S. F., and P. Tattersall. 1995. DNA replication in the autonomous parvoviruses. Semin. Virol. 6:271-281. [Google Scholar]
- 23.Cotmore, S. F., and P. Tattersall. 1986. Organization of nonstructural genes of the autonomous parvovirus minute virus of mice. J. Virol. 58:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotmore, S. F., and P. Tattersall. 1996. Parvovirus DNA replication, p. 799-813. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Dingwall, C., and R. A. Laskey. 1986. Protein import into the cell nucleus. Annu. Rev. Cell Biol. 2:367-390. [DOI] [PubMed] [Google Scholar]
- 26.Dinsart, C., J. J. Cornelis, and J. Rommelaere. 1996. Recombinant autonomous parvoviruses: new tools for the gene therapy of cancer? Chem. Today 14:32-38. [Google Scholar]
- 27.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doerig, C., B. Hirt, J. P. Antonietti, and P. Beard. 1990. Nonstructural protein of parvoviruses B19 and minute virus of mice controls transcription. J. Virol. 64:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doerig, C., B. Hirt, P. Beard, and J. P. Antonietti. 1988. Minute virus of mice non-structural protein NS-1 is necessary and sufficient for trans-activation of the viral P39 promoter. J. Gen. Virol. 69:2563-2573. [DOI] [PubMed] [Google Scholar]
- 30.Dupont, F., B. Avalosse, A. Karim, N. Mine, M. Bosseler, A. Maron, A. V. Van den Broeke, G. E. Ghanem, A. Burny, and M. Zeicher. 2000. Tumor-selective gene transduction and cell killing with an oncotropic autonomous parvovirus-based vector. Gene Ther. 7:790-796. [DOI] [PubMed] [Google Scholar]
- 31.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 32.Forgues, M., A. J. Marrogi, E. A. Spillare, C. G. Wu, Q. Yang, M. Yoshida, and X. W. Wang. 2001. Interaction of the hepatitis B virus X protein with the Crm1-dependent nuclear export pathway. J. Biol. Chem. 276:22797-22803. [DOI] [PubMed] [Google Scholar]
- 33.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 34.Freedman, D. A., and A. J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18:7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fridell, R. A., U. Fischer, R. Luhrmann, B. E. Meyer, J. L. Meinkoth, M. H. Malim, and B. R. Cullen. 1996. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1. Rev. Proc. Natl. Acad. Sci. USA 93:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 37.Gersappe, A., and D. J. Pintel. 1999. CA- and purine-rich elements form a novel bipartite exon enhancer which governs inclusion of the minute virus of mice NS2-specific exon in both singly and doubly spliced mRNAs. Mol. Cell. Biol. 19:364-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 39.Graham, F. L., and A. J. van der Eb. 1973. Transformation of rat cells by DNA of human adenovirus 5. Virology 54:536-539. [DOI] [PubMed] [Google Scholar]
- 40.Haag, A., P. Menten, J. Van Damme, C. Dinsart, J. Rommelaere, and J. J. Cornelis. 2000. Highly efficient transduction and expression of cytokine genes in human tumor cells by means of autonomous parvovirus vectors; generation of antitumor responses in recipient mice. Hum. Gene Ther. 11:597-609. [DOI] [PubMed] [Google Scholar]
- 41.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell. Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 42.Huang, T. T., N. Kudo, M. Yoshida, and S. Miyamoto. 2000. A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc. Natl. Acad. Sci. USA 97:1014-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jongeneel, C. V., R. Sahli, G. K. McMaster, and B. Hirt. 1986. A precise map of splice junctions in the mRNAs of minute virus of mice, an autonomous parvovirus. J. Virol. 59:564-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kestler, J., B. Neeb, S. Struyf, J. Van Damme, S. F. Cotmore, A. D'Abramo, P. Tattersall, J. Rommelaere, C. Dinsart, and J. J. Cornelis. 1999. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum. Gene Ther. 10:1619-1632. [DOI] [PubMed] [Google Scholar]
- 45.Klemm, J. D., C. R. Beals, and G. R. Crabtree. 1997. Rapid targeting of nuclear proteins to the cytoplasm. Curr. Biol. 7:638-644. [DOI] [PubMed] [Google Scholar]
- 46.Kratzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850-857. [DOI] [PubMed] [Google Scholar]
- 47.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 48.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee, S. H., and M. Hannink. 2001. The N-terminal nuclear export sequence of IkappaBalpha is required for RanGTP-dependent binding to CRM1. J. Biol. Chem. 276:23599-23606. [DOI] [PubMed] [Google Scholar]
- 50.Legrand, C., J. Rommelaere, and P. Caillet-Fauquet. 1993. MVM(p) NS-2 protein expression is required with NS-1 for maximal cytotoxicity in human transformed cells. Virology 195:149-155. [DOI] [PubMed] [Google Scholar]
- 51.Li, X., and S. L. Rhode, 3rd. 1991. Nonstructural protein NS2 of parvovirus H-1 is required for efficient viral protein synthesis and virus production in rat cells in vivo and in vitro. Virology 184:117-130. [DOI] [PubMed] [Google Scholar]
- 52.Li, X., and S. L. Rhode, 3rd. 1993. The parvovirus H-1 NS2 protein affects viral gene expression through sequences in the 3′ untranslated region. Virology 194:10-19. [DOI] [PubMed] [Google Scholar]
- 53.Lombardo, E., J. C. Ramirez, M. Agbandje-McKenna, and J. M. Almendral. 2000. A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74:3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 55.Malim, M. H., D. F. McCarn, L. S. Tiley, and B. R. Cullen. 1991. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 65:4248-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maroto, B., J. C. Ramirez, and J. M. Almendral. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J. Virol. 74:10892-10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller, C. L., and D. J. Pintel. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 76:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller, C. L., and D. J. Pintel. 2001. The NS2 protein generated by the parvovirus minute virus of mice is degraded by the proteasome in a manner independent of ubiquitin chain elongation or activation. Virology 285:346-355. [DOI] [PubMed] [Google Scholar]
- 59.Morgan, W. R., and D. C. Ward. 1986. Three splicing patterns are used to excise the small intron common to all minute virus of mice RNAs. J. Virol. 60:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muslin, A. J., J. W. Tanner, P. M. Allen, and A. S. Shaw. 1996. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889-897. [DOI] [PubMed] [Google Scholar]
- 61.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J. Virol. 64:6166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naeger, L. K., N. Salomé, and D. J. Pintel. 1993. NS2 is required for efficient translation of viral mRNA in minute virus of mice-infected murine cells. J. Virol. 67:1034-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 64.Ohshima, T., T. Nakajima, T. Oishi, N. Imamoto, Y. Yoneda, A. Fukamizu, and K. Yagami. 1999. CRM1 mediates nuclear export of nonstructural protein 2 from parvovirus minute virus of mice. Biochem. Biophys. Res. Commun. 264:144-150. [DOI] [PubMed] [Google Scholar]
- 65.Ossareh-Nazari, B., F. Bachelerie, and C. Dargemont. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278:141-144. [DOI] [PubMed] [Google Scholar]
- 66.Otero, G. C., M. E. Harris, J. E. Donello, and T. J. Hope. 1998. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 72:7593-7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puvion-Dutilleul, F., S. Besse, E. Pichard, and C. Cajean-Feroldi. 1998. Release of viruses and viral DNA from nucleus to cytoplasm of HeLa cells at late stages of productive adenovirus infection as revealed by electron microscope in situ hybridization. Biol. Cell 90:5-38. [DOI] [PubMed] [Google Scholar]
- 69.Richards, R., P. Linser, and R. W. Armentrout. 1977. Kinetics of assembly of a parvovirus, minute virus of mice, in synchronized rat brain cells. J. Virol. 22:778-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russell, S. J., A. Brandenburger, C. L. Flemming, M. K. Collins, and J. Rommelaere. 1992. Transformation-dependent expression of interleukin genes delivered by a recombinant parvovirus. J. Virol. 66:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 72.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santaren, J. F., J. C. Ramirez, and J. M. Almendral. 1993. Protein species of the parvovirus minute virus of mice strain MVMp: involvement of phosphorylated VP-2 subtypes in viral morphogenesis. J. Virol. 67:5126-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 181:22-34. [DOI] [PubMed] [Google Scholar]
- 75.Siegl, G. 1984. Biology and pathogenicity of autonomous parvoviruses, p. 297-362. In K. I. Berns (ed.), The parvoviruses. Plenum Press, New York, N.Y.
- 76.Singer, I. I., and S. L. Rhode, III. 1978. Electron microscopy and cytochemistry of H-1 parvovirus intracellular morphogenesis, p. 479-504. In D. C. Ward and P. Tattersall (ed.), Replication of mammalian parvoviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 77.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 78.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 79.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taagepera, S., D. McDonald, J. E. Loeb, L. L. Whitaker, A. K. McElroy, J. Y. Wang, and T. J. Hope. 1998. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc. Natl. Acad. Sci. USA 95:7457-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tattersall, P., P. J. Cawte, A. J. Shatkin, and D. C. Ward. 1976. Three structural polypeptides coded for by minute virus of mice, a parvovirus. J. Virol. 20:273-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tattersall, P., and S. F. Cotmore. 1988. The nature of parvoviruses, p. 5-41. In J. R. Pattison (ed.), Parvoviruses and human disease. CRC Press, Boca Raton, Fla.
- 84.Toyoshima, F., T. Moriguchi, A. Wada, M. Fukuda, and E. Nishida. 1998. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 17:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J. Virol. 67:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vanacker, J. M., R. Corbau, G. Adelmant, M. Perros, V. Laudet, and J. Rommelaere. 1996. Transactivation of a cellular promoter by the NS1 protein of the parvovirus minute virus of mice through a putative hormone-responsive element. J. Virol. 70:2369-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vanacker, J. M., and J. Rommelaere. 1995. Non-structural proteins of autonomous parvoviruses: from cellular effects to molecular mechanisms. Semin. Virol. 6:291-297. [Google Scholar]
- 88.Wang, D., W. Yuan, I. Davis, and C. R. Parrish. 1998. Nonstructural protein-2 and the replication of canine parvovirus. Virology 240:273-281. [DOI] [PubMed] [Google Scholar]
- 89.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 90.Wetzel, K., P. Menten, G. Opdenakker, J. Van Damme, H. J. Grone, N. Giese, A. Vecchi, S. Sozzani, J. J. Cornelis, J. Rommelaere, and C. Dinsart. 2001. Transduction of human MCP-3 by a parvoviral vector induces leukocyte infiltration and reduces growth of human cervical carcinoma cell xenografts. J. Gene Med. 3:326-337. [DOI] [PubMed] [Google Scholar]
- 91.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 92.Xiao, Z., N. Watson, C. Rodriguez, and H. F. Lodish. 2001. Nucleocytoplasmic shuttling of Smad1 conferred by its nuclear localization and nuclear export signals. J. Biol. Chem. 276:39404-39410. [DOI] [PubMed] [Google Scholar]
- 93.Young, P. J., K. T. Jensen, L. R. Burger, D. J. Pintel, and C. L. Lorson. 2002. Minute virus of mice NS1 interacts with the SMN protein, and they colocalize in novel nuclear bodies induced by parvovirus infection. J. Virol. 76:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]