Abstract
Latency-associated nuclear antigen 1 (LANA1) of Kaposi's sarcoma-associated herpesvirus (KSHV) is implicated in the maintenance of the viral genome during latent infection. LANA1 colocalizes with KSHV episomes on the host chromosome and mediates their maintenance by attaching these viral structures to host chromosomes. Data from long-term selection of drug resistance in cells conferred by plasmids containing the terminal repeat (TR) sequence of KSHV revealed that KSHV TRs and LANA1 act as cis and trans elements of viral latent replication, respectively. In this study, we further characterized the cis- and trans-acting elements of KSHV latent replication by using a transient replication assay with a methylation-sensitive restriction enzyme, DpnI. Transient reporter and replication assays disclosed that the orientation and basal transcriptional activity of TR constructs did not significantly affect the efficiency of replication. However, at least two TR units were necessary for efficient replication. The N-terminal 90 amino acids comprising the chromosome-binding domain of LANA1 were required for the mediation of LANA1 C-terminal DNA-binding and dimerization domains to support the transient replication of KSHV TRs. LANA1 interacted with components of the origin recognition complexes (ORCs), similar to Epstein-Barr virus nuclear antigen 1. Our data suggest that LANA1 recruits ORCs to KSHV TRs for latent replication of the viral genome.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV)/human herpesvirus 8 is a member of the gammaherpesvirus group, along with the closely related herpesvirus saimiri and Epstein-Barr virus (EBV). KSHV is implicated in KS and several lymphoproliferative diseases, including primary effusion lymphoma or body cavity-based lymphoma and some cases of multicentric Castleman's disease (9, 10, 36). KSHV DNA is found in the endothelial and spindle cells of KS lesions, as well as lymphocytes and lymphoid tissues, and establishes a predominantly latent state of infection. During latent infection, only a small subset of viral genes is expressed (47), and the circularized viral genome is maintained as a multicopy episome (8, 13). Following chemical treatment or overexpression of KSHV open reading frame 50, the lytic cycle is induced, resulting in virus particle production (30, 31, 35, 37, 44, 51).
Latency-associated nuclear antigen 1 (LANA1), originally identified by use of an immunofluorescence assay with the sera of KS patients (16, 24), is one of the latent genes encoded by open reading frame 73 of the KSHV genome (40, 46). This 222- to 234-kDa nuclear protein, composed of 1,162 amino acids, associates with the mitotic chromosome (5, 12, 38) and binds to sites within the terminal repeat (TR) sequence of the KSHV genome encompassing the putative origin of plasmid replication (oriP) (6, 17), thereby allowing the stable maintenance of plasmids containing this region (5, 6). Therefore, it has been suggested that LANA1 plays an important role in the maintenance of the KSHV genome during host cell mitosis by tethering the viral episome to host chromosomes. These characteristics imply that KSHV LANA1 is a functional analog of EBV nuclear antigen 1 (EBNA-1) in the latent replication of the viral genome. Additionally, LANA1 acts as a transcriptional repressor when attached to promoters via the heterologous DNA-binding domain (DBD) (27, 49). The protein is involved in the regulation of several viral and cellular promoters, including the cyclin E promoter (CCNE1p) (39), the telomerase reverse transcriptase promoter (hTERTp) (26), two EBV latent promoters (Cp and LMP1p) (20, 27), long TRs of human immunodeficiency virus type 1 (23, 45), the interleukin-6 promoter (4), KSHV TRs (17), and its own promoter (45). LANA1 also interacts directly with several cellular and viral factors and regulates their transcriptional activities (15, 28, 29, 39).
The maintenance of the viral genome in host cells during latent infection involves two distinct steps, specifically, replication of the viral genome by cellular machinery during the S phase and the equal segregation or partition of the replicated viral genome to daughter cells after mitosis of the host cell. Previous studies on the latent replication of KSHV were based on the long-term selection in cells of drug resistance conferred by putative oriP-containing plasmids (5, 6). However, this assay does not discriminate between the events of replication and equal segregation and detects only the result of their cooperation. In this study, we describe for the first time a transient DNA replication assay for KSHV that uses a methylation-sensitive enzyme, DpnI, which is unable to digest plasmids replicated in eukaryotic cells. We investigate the roles of viral trans- and cis-acting elements during latent infection in the replication and transcription of KSHV TRs, which encompass the putative oriP.
MATERIALS AND METHODS
Plasmids.
The pcDNA3 LANA1 and pFLAG-CMV2 LANA1 vectors and deletion mutants of LANA1 were constructed as described previously (28, 29). A BglII/EcoRI fragment of pM or pVP16 (Clontech, Palo Alto, Calif.) was inserted into BamHI/EcoRI sites of pcDNA3 (Invitrogen, Carlsbad, Calif.) or its derivatives to express the GAL4 DBD- or VP16 activation domain (AD)-fused protein driven by a cytomegalovirus (CMV) promoter, respectively. cDNA corresponding to amino acids 1 to 90 of EBNA-1 was amplified with appropriate primers from pCEP4 (Invitrogen) and inserted into the EcoRI site of pFLAG LANA1 C to express an EBNA-1-LANA1 C hybrid protein.
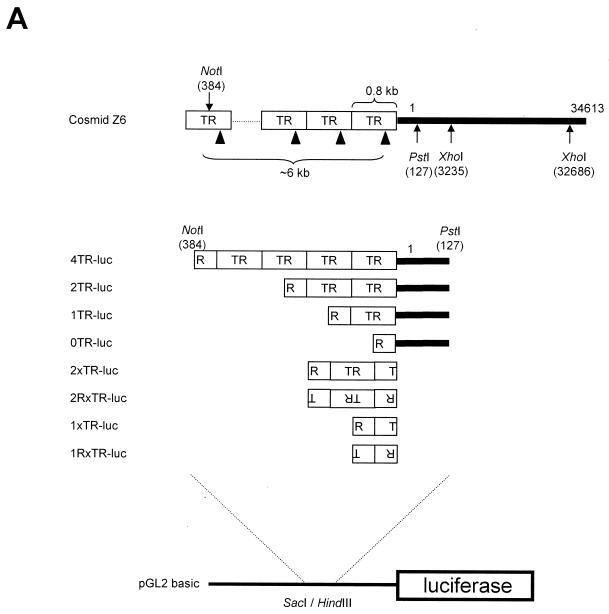
The EcoRI/XhoI fragment of cosmid Z6 spanning from KSHV left-end TRs to nucleotide 3235 of the KSHV long unique region (LUR) was cloned between the corresponding restriction sites of pBluescript II KS(−) (Stratagene, San Diego, Calif.) and designated pTR-3K. Partial NotI digestion of pTR-3K was performed to construct vectors p4TR-3K, p2TR-3K, p1TR-3K, and p0TR-3K (designated according to the number of TRs). The SacI/PstI fragments of these constructs were inserted into the corresponding sites of pBluescript II KS(−) to remove most of the KSHV LUR and designated pBS-4TR, pBS-2TR, pBS-1TR, and pBS-0TR, respectively. Digested fragments (SacI/HindIII) of these constructs were cloned into the corresponding sites of pGL2-basic (Promega, Madison, Wis.) to generate p4TR-luc, p2TR-luc, p1TR-luc, and p0TR-luc, respectively. From these plasmids, SacI/XhoI fragments were cloned into pGL3-promoter (Promega) to generate p4TRSV-luc, p2TRSV-luc, p1TRSV-luc, and p0TRSV-luc, respectively. One or two units of TRs were generated by partial NotI digestion of p4TR-luc and inserted into the corresponding restriction site of pBluescript II KS(-). The orientation of the TR sequence was confirmed by restriction enzyme mapping, followed by insertion into pGL2-basic to generate plasmids p2xTR-luc, p2RxTR-luc, p1xTR-luc, and p1RxTR-luc, respectively. See Fig. 2A for a schematic representation of various TR constructs.
FIG. 2.
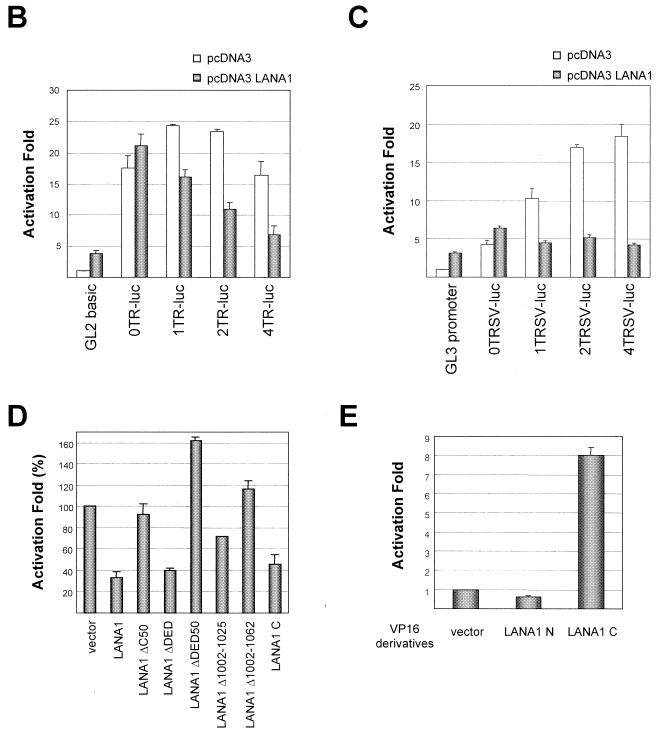
Transcriptional activity of KSHV TRs is down-regulated by the C terminus of LANA1. (A) Schematic diagram of plasmids containing KSHV TR variants used in transient reporter and replication assays. LBSs within TRs are indicated by black triangles. (B) 293T cells in a 60-mm dish were cotransfected with 1 μg of the indicated reporter and 4 μg of pcDNA3 or pcDNA3 LANA1. Activation was calculated with respect to the normalized luciferase activity of pGL2-basic in the presence of pcDNA3, which was set as 1. (C) 293T cells were cotransfected with 0.1 μg of the indicated reporter and 4 μg of pcDNA3 or pcDNA3 LANA1. Activation was calculated with respect to the normalized luciferase activity of pGL3-promoter in the presence of pcDNA3, which was set as 1. (D) 293T cells were cotransfected with 1 μg of p4TR-luc and 4 μg of the indicated pcDNA3 derivatives. Activation was calculated with respect to the normalized luciferase activity of p4TR-luc in the presence of pcDNA3, which was set as 100%. (E) 293T cells were cotransfected with 1 μg of p4TR-luc and 4 μg of the indicated VP16 derivatives. Activation was calculated with respect to the normalized luciferase activity of p4TR-luc in the presence of VP16, which was set as 1. The results represent the averages of at least three independent experiments, and standard deviations are depicted as error bars.
The pGEX-hORC1, pGEX-hORC2, pGEX-hORC4, and pGEX-hORC5 constructs were generous gifts from H. Ariga (Hokkaido University, Hokkaido, Japan). cDNA fragments from pGEX-hORC1 and pGEX-hORC2 were inserted into BamHI/NotI sites of pcDNA3. N- or C-terminal deletion mutants of pGEX-hORC1 and pGEX-hORC2 fused to glutathione S-transferase (GST) were generated by PCR with appropriate primers, followed by cloning into pGEX4T-1 (Amersham Pharmacia Biotech, Uppsala, Sweden).
Cell culture, transfection, and reporter assay.
293T cells were maintained and transfected as described previously (28). BJAB cells, KSHV- and EBV-negative B-cell lymphoma lines derived from EBV-negative Burkitt lymphoma biopsy samples, were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and transfected by electroporation (31). A constant amount of total DNA for transfection was maintained by using an appropriate blank vector. The transient reporter assay was performed as described in an earlier study (28).
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed as described previously, with minor modifications (6). Synthetic oligonucleotides TR-13 (5′-TCCCGCCCGGGCATGGGGCC-3′) and TR-12 (5′-CGGGCATGGGGCCGCGCGCCGCC-3′) with 5′-GATC overhangs were labeled with [γ-32P]ATP by using T4 polynucleotide kinase and annealed to complementary strands. LANA1 and derivatives were synthesized by using the TNT T7 coupled transcription-translation reticulocyte lysate system according to the manufacturer's instructions (Promega). Labeled probe (approximately 200 fmol per reaction) was incubated with 3 μl of in vitro-translated LANA1 or its derivatives in binding buffer 1 [20 mM Tris (pH 7.5), 10% glycerol, 50 mM KCl, 0.1 mM dithiothreitol, 10 mM MgCl2, 1 mM EDTA, 1 μg of poly(dI-dC)] at room temperature for 30 min. Unlabeled specific or nonspecific competitors were included in the reaction mixtures (at a 12.5- to 50-fold excess) when needed. Reactions were terminated by loading onto a 3.5% native polyacrylamide gel in Tris-borate-EDTA running buffer. The gel was dried and subjected to autoradiography.
Transient replication assay.
293T cells in 100-mm dishes were cotransfected with 2 μg of oriP-containing plasmid, 2 μg of pGL2-basic as an internal control, and 8 μg of trans-acting element expression plasmid. At ∼36 h posttransfection, 293T cells were trypsinized, collected by centrifugation, and serially split to make equal amounts of cells (∼5 × 106 cells per sample) at harvesting time. BJAB cells were cotransfected with 7.5 μg of oriP-containing plasmid, 7.5 μg of pGL2-basic as an internal control, and 15 μg of trans-acting element expression plasmid. Following electroporation, BJAB cells were serially split to make equal amounts of cells (∼107 cells per sample) at harvesting time. Cells were washed twice with phosphate-buffered saline, and low-molecular-weight DNA was extracted by the Hirt lysis method (21). Supernatants were successively extracted with phenol-chloroform-isoamyl alcohol and chloroform. Ethanol-precipitated DNA was dissolved in 50 μl of distilled water containing RNase. Thirty percent of Hirt-extracted DNA from ∼5 × 106 293T cells and ∼107 BJAB cells was digested with DpnI and Alw44I to remove unreplicated DNA and linearize pGL2-basic derivatives, respectively. The completion of DpnI digestion was routinely checked by monitoring the complete digestion of the unreplicated internal control, pGL2-basic. Digested DNA was separated by 0.8% agarose gel electrophoresis and analyzed by Southern blot hybridization. Probes specific for the luciferase gene of pGL2-basic were synthesized with an NEBlot Phototope kit and detected with a Phototope-Star detection kit according to the manufacturer's instructions (New England Biolabs, Beverly, Mass.).
In vitro binding assay.
Escherichia coli expressing GST fusion proteins was harvested and stored at −70°C until use. Cell pellets were resuspended in binding buffer 2 (20 mM Tris [pH 7.5], 50 mM NaCl, 2 mM MgCl2, 1 mM dithiothreitol, 0.5% Nonidet P-40) with brief sonication. After removal of cell debris, the supernatants were incubated with glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) at 4°C for 1 h. The resin was washed twice, and in vitro-translated 35S-labeled protein was added. After a further 3 h of incubation at 4°C, the resin was washed four times with binding buffer 2, and bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. When needed, 1 mM ATP and 50 pmol of double-stranded oligonucleotide were included in the binding reaction mixtures. Purification of His-tagged LANA1 and LANA1 N by using a recombinant baculovirus expression system and pull-down assays with Ni+-nitrilotriacetic acid-agarose were performed as described previously (29).
RESULTS
Integration of the C terminus of KSHV LANA1 is important for binding to the TR region of the KSHV genome.
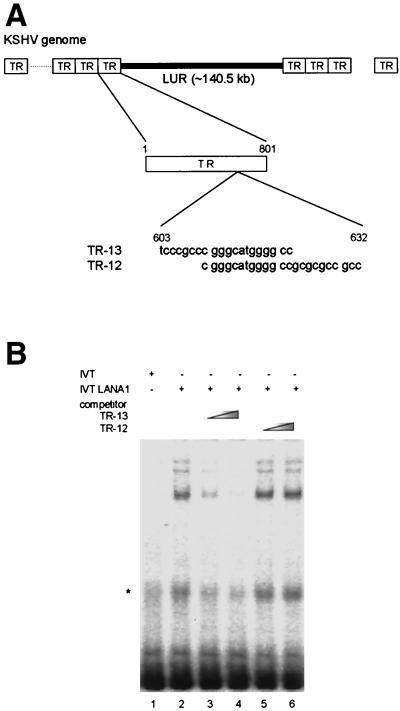
Previous reports showed that LANA1 binds the imperfect 20-nucleotide palindromic sequence within the TR region of the KSHV genome (TR-13) via its C-terminal domain (6, 17). The location of the LANA1-binding sequence within the TR region is depicted in Fig. 1A. As described previously, in vitro-translated LANA1 interacted with TR-13 (Fig. 1B, lane 2). The addition of excess, unlabeled TR-13 (lanes 3 and 4) but not unlabeled TR-12 (lanes 5 and 6), spanning another portion of the TR region, resulted in competition for binding to LANA1, further confirming the specificity of this interaction. Similar results were obtained with nuclear extracts from 293T cells transfected with a FLAG-tagged LANA1 expression vector (data not shown).
FIG. 1.
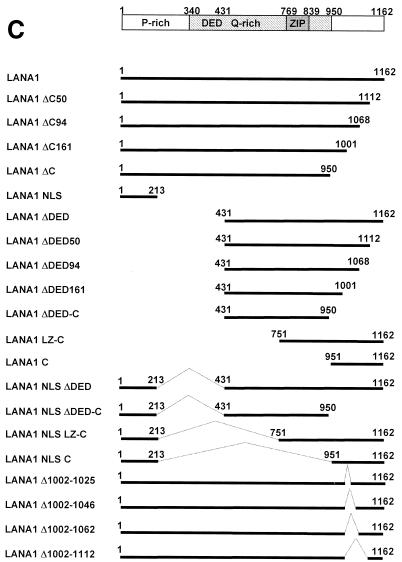
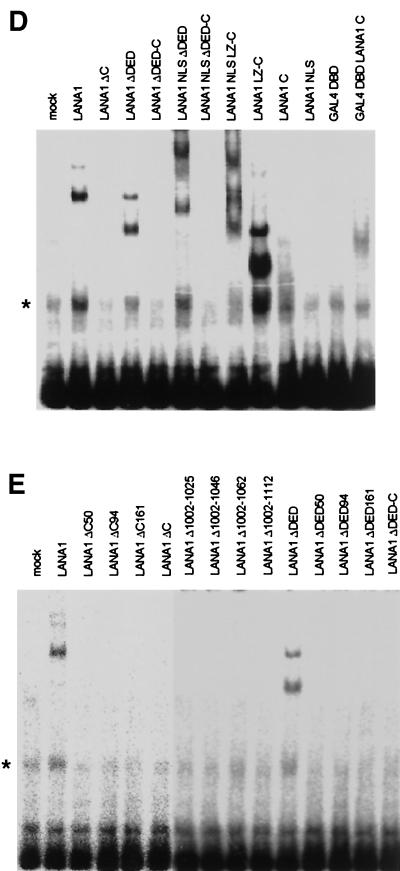
EMSAs of in vitro-translated LANA1 and derivatives. (A) Locations of LANA1-binding sequence, TR-13, and adjacent TR-12 within the TR region of the KSHV genome. (B) Control reticulocyte lysate (IVT) (lane 1) or in vitro-translated LANA1 (IVT LANA1) (lanes 2 to 6) was incubated with a radiolabeled TR-13 probe in an EMSA. Unlabeled competitors, such as TR-13 (lanes 3 and 4) or TR-12 (lanes 5 and 6), were included at a 12.5- to 50-fold molar excess in the binding reaction mixture. (C) Schematic diagram of LANA1 deletion mutants used in EMSAs. DED, aspartate/glutamate-rich repeat region. (D) In vitro-translated LANA1 deletion mutants containing the C terminus of LANA1 were used in EMSAs. (E) In vitro-translated LANA1 deletion mutants containing small truncations or internal deletions within the LANA1 C terminus were used in EMSAs. Nonspecific bands are marked by asterisks.
For fine mapping of the LANA1 DBD, a series of deletion mutants were constructed and tested for TR-13-binding ability in an EMSA. A schematic diagram of the LANA1 deletion mutants used in EMSA analyses is depicted in Fig. 1C. The expression of these mutant proteins was confirmed by their sizes upon in vitro translation in the presence of [35S]methionine (data not shown). As shown in Fig. 1D, the C terminus of LANA1 was necessary and sufficient for binding to TR-13, although the bands shifted by LANA1 C were weak and not discrete. However, the GAL4 DBD fused to the C terminus of LANA1 (but not the GAL4 DBD alone) clearly bound to TR-13, suggesting that the C terminus of LANA1 acts independently as a DNA-binding module, similar to the GAL4 DBD. In addition, a small C-terminal truncation or internal deletion within the C terminus of LANA1 completely abolished TR-13 binding ability (Fig. 1E), indicating that the overall structure of the C terminus of LANA1 is required for DNA binding.
Binding of LANA1 via the C terminus results in the transcriptional repression of TRs.
From cosmid Z6 spanning the region between the left-end TRs and the ∼33-kb LUR of the KSHV genome, we subcloned a series of KSHV TR variants into pGL2-basic (Fig. 2A) and examined their transcriptional promoter activities in the absence or presence of LANA1. TR sequences including the 127-bp left end of the LUR of KSHV led to more than 20-fold transcriptional activation of pGL2-basic, which was repressed by LANA1 (Fig. 2B). We also subcloned KSHV TR variants into pGL3-promoter containing the simian virus 40 (SV40) early promoter and examined their transcriptional enhancer activities in the absence or presence of LANA1. As shown in Fig. 2C, TR variants also induced the transcriptional activation of pGL3-promoter containing the SV40 early promoter. Interestingly, their transcriptional enhancer activities were increased in proportion to the number of TR units and repressed by LANA1, consistent with earlier data (17). However, reporter plasmids containing only TRs without the left end of the LUR, such as p2xTR-luc, p2RxTR-luc, p1xTR-luc, and p1RxTR-luc, did not show any promoter activities in a similar transient reporter assay, although transcriptional enhancer activities were seen with pGL3-promoter (data not shown).
Next, we determined the specific domain of LANA1 responsible for the transcriptional repression of KSHV TRs by using a series of LANA1 deletion mutants in a transient reporter assay. The results confirmed earlier data showing that the C terminus of LANA1 alone was sufficient for the transcriptional repression of KSHV TRs (17). Moreover, small deletions within this region abolished activity (Fig. 2D), suggesting that direct binding via the C terminus of LANA1 induced the transcriptional repression of KSHV TRs. Since the C terminus of LANA1 alone was sufficient for the DNA-binding module in the EMSA and the transcriptional repression of TRs in a transient reporter assay, we assumed that a transcriptional activator tethered to TRs via the C terminus of LANA1 would result in the transcriptional activation of TRs, although the LANA1 C terminus itself repressed transcription from the TRs. As shown in Fig. 2E, the herpes simplex virus VP16 AD fused to LANA1 C efficiently activated the transcription of KSHV TRs. This activation was not due to the complicated transcriptional response of TRs to VP16 AD, since VP16 AD activated the transcription of p4TR-luc only when fused to LANA1 C (Fig. 2E) and VP16 AD fused to LANA1 C did not activate the transcription of pGL2-basic, a backbone reporter of p4TR-luc (data not shown). Taken together, these results further validate the theory that the C terminus of LANA1 acts independently as a DNA-binding module.
KSHV TRs replicate only in the presence of LANA1 in a transient DNA replication assay.
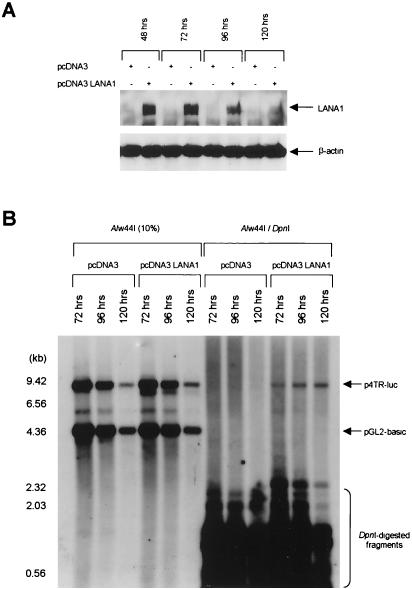
For the transient DNA replication assay, TR variants used in the transient reporter assay along with pGL2-basic as an internal control were cotransfected with a blank or an LANA1 expression vector into 293T or BJAB cells. At ∼36 h posttransfection, cells were serially split to make equal amounts of cells harvested at various times. The levels of expression of LANA1 and the numbers of transfected 293T cells were monitored by Western blotting with rabbit anti-LANA1 polyclonal antiserum and anti-β-actin monoclonal antibody, respectively (Fig. 3A). As shown in Fig. 3B, DpnI-resistant replicated plasmids containing TR units (but not pGL2-basic) were detectable only in the presence of LANA1 at 72 h posttransfection. Similar results were obtained when BJAB cells were used instead of 293T cells (Fig. 3C).
FIG. 3.
Transient DNA replication of p4TR-luc in the presence of LANA1. (A) 293T cells in a 100-mm dish were cotransfected with 2 μg of p4TR-luc, 2 μg of pGL2-basic, and 8 μg of pcDNA3 or pcDNA3 LANA1. At ∼36 h posttransfection, cells were split and harvested at the indicated times. LANA1 expression levels and cell numbers were monitored by Western blotting of total cell lysates with rabbit anti-LANA1 polyclonal antiserum (upper panel) and anti-β-actin monoclonal antibody (lower panel), respectively. (B) Hirt-extracted DNA from transfected 293T cells was digested with Alw44I/DpnI (right), and 10% of the samples were digested with Alw44I alone as input (left). Digested DNA was separated by 0.8% agarose gel electrophoresis and analyzed by Southern blot hybridization with a probe specific for the luciferase gene. DNA size markers are depicted on the left. (C) About 107 BJAB cells were cotransfected with 7.5 μg of p4TR-luc, 7.5 μg of pGL2-basic, and 15 μg of pcDNA3 or pcDNA3 LANA1. After electroporation, cells were split and harvested at the indicated times. Hirt-extracted DNA was digested with Alw44I alone (left) or Alw44I/DpnI (right) and analyzed by Southern blotting. (D) 293T cells in a 100-mm dish were cotransfected with 2 μg of p4TR-luc or pFR-luc and 8 μg of cytomegalovirus G4 or pcDNA3 derivatives. At ∼36 h posttransfection, cells were split and harvested at the indicated times. Hirt-extracted DNA from transfected 293T cells was digested with Alw44I/DpnI (right). DNA extracted from cells harvested at 96 h posttransfection was digested with Alw44I as input (left). Southern blotting was performed with a luciferase gene-specific probe.
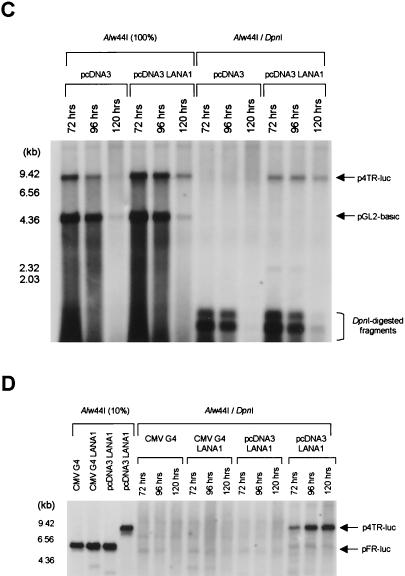
The EMSA and the transient reporter assay revealed that LANA1 exerts transcriptional repressor activity on TRs by direct binding. LANA1 also acts as a transcriptional repressor when attached to the promoter via the heterologous DBD (27, 49) Therefore, we examined whether this protein could facilitate the replication of plasmid DNA when artificially tethered to plasmid DNA via the GAL4 DBD. For this purpose, a pFR-luc reporter plasmid containing five tandem GAL4-binding sites was cotransfected with a GAL4 DBD, a GAL4 DBD fused to LANA1, or a LANA1 expression vector into 293T cells, and a transient reporter assay was performed like that described above. As shown in Fig. 3D, the pFR-luc reporter plasmid was not replicated in the presence of either LANA1 alone or LANA1 fused to the GAL4 DBD. However, LANA1 fused to the GAL4 DBD but not LANA1 alone resulted in the transcriptional repression of the pFR-luc reporter plasmid in the transient reporter assay (data not shown), consistent with previous reports (27, 49). These results reveal that the artificial access of LANA1 in the vicinity of DNA is not sufficient for supporting the initiation of DNA replication and that only specific interactions between the viral trans- and cis-acting elements of KSHV latent replication result in KSHV oriP activity.
A minimum of two LANA1-binding sites (LBSs) in TRs are required for efficient replication.
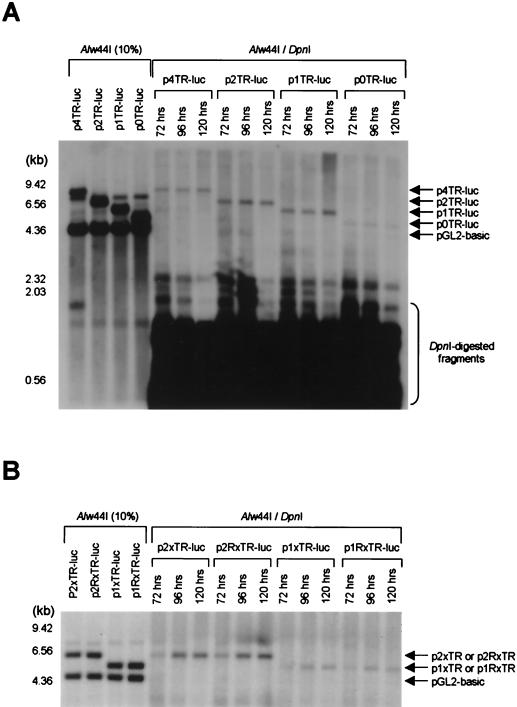
To examine the role of cis-acting elements in KSHV oriP activity, the various TR constructs used in the transient reporter assay (Fig. 2A) were used in a transient replication assay. As shown in Fig. 4A, TR constructs, such as p4TR-luc, p2TR-luc, and p1TR-luc, which contain more than two LBSs were efficiently replicated, while p0TR-luc, with one LBS, was only weakly replicated. Similar results were obtained when TR constructs containing only the TR region without the left end of the KSHV LUR and displaying no promoter activity were used in the replication assay. TR orientation had no effect on the efficiency of replication (Fig. 4B). These results suggest that basal transcription and replication activities at KSHV oriP are uncoupled and that at least two LBSs are required for efficient KSHV oriP activity.
FIG. 4.
Transient replication of KSHV TR variants in the presence of LANA1. The indicated plasmids (2 μg) containing variable numbers of KSHV TR units (A) or reverse-oriented TR units (B) were cotransfected with 2 μg of pGL2-basic and 8 μg of pcDNA3 LANA1 into 293T cells. After cells were harvested at the indicated times, Hirt-extracted DNA was digested and analyzed by Southern blotting with a luciferase gene-specific probe as described in the legend to Fig. 3.
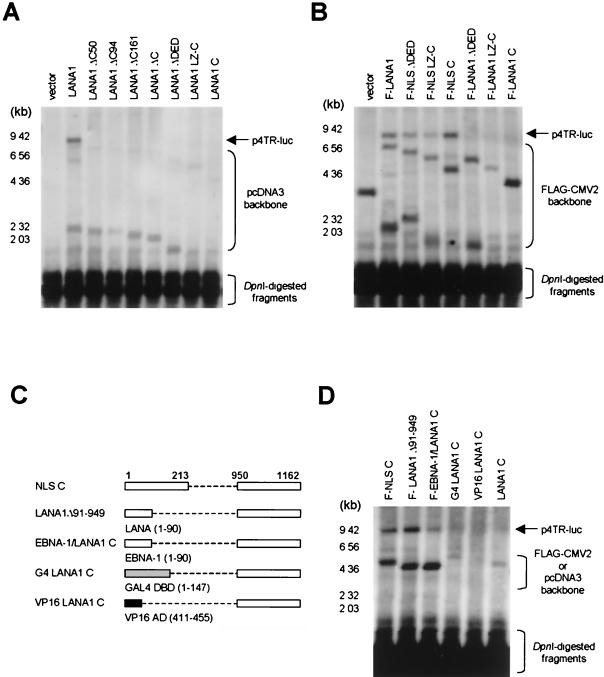
Both N and C termini of LANA1 are required for KSHV oriP activity.
The EMSA and the transient reporter assay revealed that the C-terminal domain of LANA1 is necessary and sufficient for binding to the TR region, thereby resulting in the transcriptional repression of KSHV TRs. To determine whether the C terminus of LANA1 alone is sufficient to induce KSHV oriP activity, various LANA1 deletion mutants were used in a transient DNA replication assay. As expected, LANA1 ΔC50, containing a small deletion at the LANA1 C terminus, was unable to support KSHV oriP activity, indicating that the direct binding of LANA1 to TRs is necessary for this process (Fig. 5A). However, none of the N-terminal deletion mutants of LANA1 which bound KSHV TRs and repressed its transcription was also able to facilitate the replication of plasmids containing KSHV TRs. A transient DNA replication assay with FLAG-tagged internal deletion mutants of LANA1 revealed that both the N and the C termini of LANA1 are required for the efficient replication of the TR region (Fig. 5B). It is unlikely that the inability of LANA1 mutants to support DNA replication was due to reduced expression levels in transfected cells, since titration of increasing amounts of a LANA1 expression vector and Western blotting data showed that the transient DNA replication assay was performed under conditions in which trans-acting element concentrations were largely in excess (data not shown). Various sizes of faint DpnI-resistant bands in Fig. 5A and darker ones in Fig. 5B were pcDNA3 and pFLAG-CMV2 derivatives, respectively, because they unexpectedly cross-reacted with the probe that we used. They contained the SV40 origin of replication and replicated in 293T cells expressing SV40 large T antigen. Therefore, they exhibited various sizes of DpnI-resistant bands according to LANA1 deletion mutants.
FIG. 5.
Determination of LANA1 domains required for p4TR-luc replication in a transient replication assay. Plasmids p4TR-luc and pGL2-basic (2 μg) were cotransfected with 8 μg of pcDNA3 LANA1 derivatives, including the N-terminal or C-terminal deletion mutants of LANA1 (A), or pFLAG-CMV2 LANA1 derivatives, including the N-terminal or internal deletion mutants of LANA1 (B), into 293T cells. Cells were harvested at 72 h posttransfection, and Hirt-extracted DNA was digested with Alw44I/DpnI. Southern blotting with a luciferase gene-specific probe was performed as described above. (C) Schematic diagram of LANA1 hybrid proteins used in a transient replication assay with p4TR-luc. (D) Plasmids (8 μg) expressing the indicated hybrid proteins were cotransfected with 2 μg of p4TR-luc and 2 μg of pGL2-basic into 293T cells. Hirt-extracted DNA from cells harvested at 72 h posttransfection was digested with Alw44I/DpnI and probed with a luciferase gene-specific probe. F, FLAG-tagged.
A previous report demonstrated that the extreme N terminus of LANA1 is the chromosome-binding domain (38). Therefore, we focused on this region and performed a DNA replication assay with hybrid proteins containing various N-terminal domains fused to the C terminus of LANA1. These included amino acids 1 to 90 of KSHV LANA1 and EBNA-1, corresponding to the chromosome-binding domain, the GAL4 DBD, and the VP16 AD (Fig. 5C). As shown in Fig. 5D, the N-terminal 90 amino acids of LANA1 fused to the C terminus efficiently facilitated KSHV TR replication. Interestingly, KSHV TRs were also replicated in the presence of the EBNA-1-LANA1 C hybrid, albeit relatively weakly. Western blotting with an anti-FLAG antibody revealed similar expression levels for LANA1 Δ91-949 and EBNA-1-LANA1 C in transfected 293T cells (data not shown). This specificity of the N-terminal domains of hybrid proteins was further confirmed by experiments with GAL DBD-LANA1 C and VP16 AD-LANA1 C, which failed to support the efficient replication of KSHV TRs.
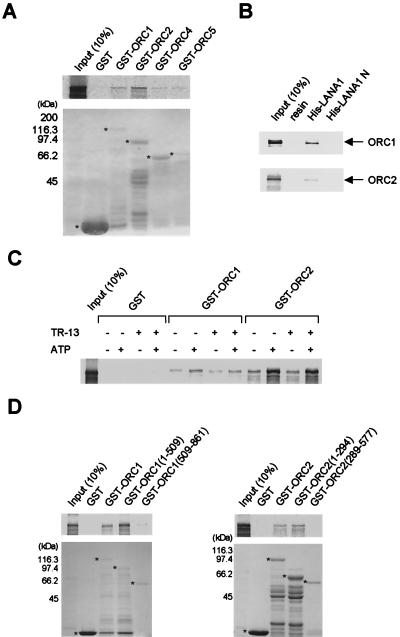
KSHV LANA1 interacts with components of the ORCs.
EBNA-1 recruits cellular origin recognition complexes (ORCs) to EBV oriP for latent replication of the genome (11, 14, 48). In theory, KSHV should also utilize cellular replication machinery for the maintenance of the viral genome, since viral proteins expressed during latent infection do not display enzymatic activity for DNA replication. LANA1, as an analog of EBNA-1 that functions in the latent DNA replication of KSHV, is a good candidate as an adaptor molecule linking the viral origin of replication and cellular replication machinery. Therefore, we used an in vitro binding assay to examine the interactions between ORCs and KSHV LANA1. As shown in Fig. 6A, in vitro-translated LANA1 was efficiently precipitated in the presence of bacterially expressed GST-ORC1 and GST-ORC2. GST-ORC4 and GST-ORC5 (but not GST alone) also precipitated in vitro-translated LANA1 very weakly. The addition of 200 μg of ethidium bromide/ml in binding buffer 2 did not affect the binding affinity of LANA1 for GST-ORC1 and GST-ORC2, confirming that these interactions are not mediated by nonspecific DNA contaminants (data not shown).
FIG. 6.
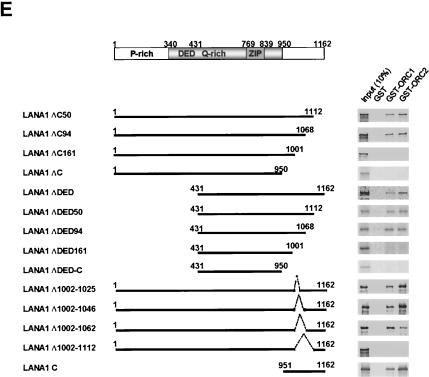
Interactions between KSHV LANA1 and ORCs. (A) In vitro-translated LANA1 in the presence of 35S-methionine was incubated with glutathione-Sepharose 4B precoated with the indicated GST fusion proteins. Bound proteins were subjected to autoradiography (upper panel); Coomassie blue staining of GST fusion proteins used in a pull-down assay is also depicted (lower panel). (B) In vitro- translated ORC1 or ORC2 in the presence of 35S-methionine wasincubated with His-tagged LANA1 or the LANA1 N purified from insect cells infected with the appropriate recombinant baculovirus. After precipitation with Ni2+-nitrilotriacetic acid-agarose, bound proteins were subjected to autoradiography. (C) ATP (1 mM) and a double-stranded oligonucleotide (50 pmol) containing the LANA1-binding sequence (TR-13) were included in a GST pull-down assay. (D) ORC1 (left panel) or ORC2 (right panel) deletion mutants fused to GST were used in a binding assay. Bound proteins were subjected to autoradiography (upper panels); Coomassie blue staining of GST fusion proteins used in a pull-down assay is also shown (lower panels). Protein markers are shown to the left of Coomassie blue stain panels. GST proteins with the correct size are indicated by asterisks. (E) (Left panel) Schematic diagram of LANA1 deletion mutants used in GST pull-down assays. DED, aspartate/glutamate-rich repeat region. (Right panel) Each LANA1 deletion mutant was in vitro translated in the presence of 35S-methionine and then used in GST pull-down assays. The results obtained with GST, GST-ORC1, and GST-ORC2 are shown. Input, 10% in vitro-translated proteins used in the binding assay.
Reciprocal pull-down experiments with His-tagged LANA1 purified with a baculovirus expression system and in vitro-translated ORC1 and ORC2 disclosed similar results (Fig. 6B). Interestingly, the presence of ATP but not oligonucleotides containing LBSs enhanced the binding affinity between ORC1 or ORC2 and LANA1 (Fig. 6C). This observed augmentation of binding affinity by ATP disappeared when lysates used in the in vitro translation assay were frozen before inclusion in the binding reaction (data not shown). The LANA1-binding regions of ORC1 and ORC2, determined by similar GST pull-down assays with deletion mutants of these complexes, were mapped to the relatively nonconserved N-terminal regions, although very weak interactions between the C terminus of ORC1 and in vitro-translated LANA1 were additionally observed (Fig. 6D). On the other hand, the ORC-binding region of LANA1, determined by including in vitro-translated proteins of various LANA1 deletion mutants in a similar binding assay, was mapped to a small region of the C terminus of LANA1 (Fig. 6E).
DISCUSSION
The viral genome of EBV is maintained as an episome by cis- and trans-acting elements during latent infection. This activity involves the semiconservative replication of the genome once per cell cycle and equal segregation and partition of the viral genome to daughter cells following mitosis of the host cell (1, 53). The two distinct events are mediated, respectively, by two components of EBV oriP, i.e., dyad symmetry and family of repeats, which are separated by ∼1 kbp within the viral genome (34, 42). The dyad symmetry and family-of-repeats components contain multiple EBNA-1-binding sites with low and high affinities, respectively (3, 41). EBNA-1 is the only viral trans-acting element required for stable maintenance of the viral genome (32, 42), although it was previously reported that oriP-containing plasmids undergo at least two rounds of DNA replication in the absence of this element (2). Another member of the gammaherpesviruses family, KSHV, displays similar characteristics of latent replication. KSHV oriP corresponds to an ∼800-bp TR sequence of the viral genome containing an LBS. Moreover, LANA1 is the only viral trans-acting element required for the maintenance of TR-containing plasmids, as verified by the long-term selection of drug resistance in cells harboring oriP-containing plasmids (5, 6). Using a transient DNA replication assay with a methylation-sensitive enzyme, we additionally show here that KSHV TRs and LANA1 function as viral cis- and trans-acting elements of KSHV latent replication, respectively.
Interestingly, KSHV LANA1 and EBV EBNA-1 (functional analogs of the viral latent replication factor) have similar domain structures, despite differences in their primary amino acid sequences. Both proteins contain an N-terminal chromosome-binding domain, an internal repeat sequence, and a C-terminal DNA-binding and dimerization domain. The DNA-binding and dimerization domain of EBNA-1 alone is not sufficient for the efficient replication of EBV oriP, and the additional chromosome-binding domain/DNA-linking region is required (22, 25, 33); the latter may be substituted for by the chromosome-binding domain of the high-mobility group 1 protein or histone H1 (22). For KSHV LANA1, similar results were obtained with various LANA1 deletion mutants and hybrid proteins, although the chromosome-binding domain of EBNA-1 could not fully replace that of LANA1 in domain-swapping experiments. It is possible that the viral genomes of KSHV and EBV attach to distinct sites of host chromosomes (via the respective chromosome-binding domains of LANA1 and EBNA-1) where latent replication of the viral genome occurs or that structural differences between these regions result in different efficiencies of replication. Other possibilities, including unknown functions of the N terminus of LANA1 that may contribute to efficient replication mediated by the C terminus of LANA1, cannot be excluded.
LANA1 and EBNA-1 do not display enzymatic activities, such as helicase or ATPase, required for replication events, unlike SV40 large T antigen, bovine papillomavirus 1 E1, and origin-binding protein of herpes simplex virus type 1, which are involved in viral replication (7, 50, 52). Since viral oriP is replicated by cellular machinery and LANA1 and EBNA-1 are the only trans-acting elements supporting the latent replication of the cis-acting element, it is reasonable to assume that these proteins target cellular replication machinery to the viral origin of replication and synchronize viral replication with that of host cells. Recent data suggest that ORCs are recruited to EBV oriP by EBNA-1 (11, 14, 48). We additionally reveal that LANA1 directly interacts with ORC components. The LANA1-binding domains of ORC1 and ORC2 were identified in the relatively nonconserved N termini (19), a result which may explain the host range that is permissive to viral genome replication. It is noteworthy that the ORC-binding region of LANA1 was mapped to the C terminus of the protein in a GST pull-down assay (Fig. 6E). Therefore, the inability of the N-terminal deletion mutants of LANA1 to support KSHV TR replication is not due to the loss of an interaction with ORCs.
KSHV displays distinct latent replication characteristics. Sequence analyses have revealed that a GC-rich sequence within KSHV TRs contains various zinc finger transcription factor-binding sites. Additionally, the p4TR-luc reporter is responsive to several cellular factors (data not shown), and individual TR units act as enhancer elements (17), like other viral origins of replication, such as those of EBV, SV40, polyomavirus, and bovine papillomavirus. However, the binding of LANA1 induced the repression of the transcriptional activation of KSHV TRs (17), in contrast to that of EBNA-1 to the EBV origin of replication, which resulted in transcriptional activation (42, 43). The results of the transient replication assays with various TR constructs showed that the number of TR units, but not the orientation or basal transcriptional activity of TRs, significantly affects the efficiency of replication, implying that the replication and transcription of KSHV TRs may be somehow uncoupled.
Recently, Garber et al. (18) reported that LANA1 cooperatively binds to two adjacent sites separated by 21 bp within a single unit of KSHV TRs. They demonstrated that LANA1 binds first to LBS 1, corresponding to TR-13, with a high affinity, and then has an increased affinity for LBS 2; they concluded that the binding affinity of LANA1 for the LBS correlates with the transcriptional suppression and replication of KSHV TRs. Since p0TR-luc contains both LBS 1 and LBS 2, its low efficiency of transcriptional repression and replication by LANA1 may be due not to the loss of high-affinity binding sites but to insufficient numbers of LBS 1 and LBS 2. Since Garber et al. (18) manipulated a single unit of TRs and used them in transient reporter and replication assays, they could not show the different efficiencies of transcriptional suppression and replication on the basis of the number of TRs. Further studies are required to determine the relationship between replication and transcription of the KSHV TR region. The mechanism by which LANA1 represses KSHV TRs and its biological significance are currently under investigation.
Acknowledgments
This work was supported in part by grants from the National Research Laboratory Program of the Korea Institute of Science and Technology, Evaluation, and Planning (KISTEP); the Korea Science and Engineering Foundation (KOSEF) through the Protein Network Research Center at Yonsei University; and the BK21 Program of the Ministry of Education, Korea.
REFERENCES
- 1.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar, A., C. Tyree, and B. Sugden. 1998. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 17:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambinder, R. F., W. A. Shah, D. R. Rawlins, G. S. Hayward, and S. D. Hayward.1990. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J. Virol. 64:2369-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An, J., A. K. Lichtenstein, G. Brent, and M. B. Rettig. 2002. The Kaposi sarcoma-associated herpesvirus (KSHV) induces cellular interleukin 6 expression: role of the KSHV latency-associated nuclear antigen and the AP1 response element. Blood 99:649-654. [DOI] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 6.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruckner, R. C., J. J. Crute, M. S. Dodson, and I. R. Lehman. 1991. The herpes simplex virus 1 origin binding protein: a DNA helicase. J. Biol. Chem. 266:2669-2674. [PubMed] [Google Scholar]
- 8.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 9.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 10.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, M. A., and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 13.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 15.Friborg, J., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 16.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among North Americans, Italians, and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 17.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 19.Gavin, K. A., M. Hidaka, and B. Stillman. 1995. Conserved initiator proteins in eukaryotes. Science 270:1667-1671. [DOI] [PubMed] [Google Scholar]
- 20.Groves, A. K., M. A. Cotter, C. Subramanian, and E. S. Robertson. 2001. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 75:9446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 22.Hung, S. C., M.-S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-1 or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyun, T. S., C. Subramanian, M. A. Cotter, R. A. Thomas, and E. S. Robertson. 2001. Latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus interacts with tat and activates the long terminal repeat of human immunodeficiency virus type 1 in human cells. J. Virol. 75:8761-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in Kaposi's sarcoma risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 25.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight, J. S., M. A. Cotter, and E. S. Robertson. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971-22978. [DOI] [PubMed] [Google Scholar]
- 27.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 29.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 30.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 31.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupton, S., and A. J. Levine. 1985. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 5:2533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Middleton, T., and B. Sugden. 1994. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA-1. J. Virol. 68:4067-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, G., M. Rigsby, L. Heston, E. Grogan, R. Sun, C. Metroka, S.-J. Gao, Y. Chang, and P. S. Moore. 1996. Antibodies to butyrate inducible antigens of Kaposi's sarcoma-associated herpesvirus in HIV-1 infected patients. N. Engl. J. Med. 334:1292-1297. [DOI] [PubMed] [Google Scholar]
- 36.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 37.Moore, P. S., S.-J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. M. Knowles, R. Garber, D. J. McGeoch, P. Pellett, and Y. Chang. 1996. Primary characterization of a herpesvirus-like agent associated with Kaposi's sarcoma. J. Virol. 70:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piolot, T., M. Tramier, M. Coppey, J.-C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 40.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42:859-868. [DOI] [PubMed] [Google Scholar]
- 42.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reisman, D., and B. Sudgen. 1986. trans-Activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol. Cell. Biol. 6:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 45.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. X. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stahl, H., P. Droge, and R. Knippers. 1986. DNA helicase activity of SV40 large tumor antigen. EMBO J. 5:1939-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]