Abstract
Theiler's virus is a neurotropic murine picornavirus which, depending on the strain, causes either acute encephalitis or persistent demyelinating disease. Persistent strains of Theiler's virus (such as DA) produce an 18-kDa protein called L* from an open reading frame overlapping that encoding the viral polyprotein. Neurovirulent strains (such as GDVII) are thought not to produce the L* protein, as the alternative open reading frame of these strains starts with an ACG codon instead of an AUG codon. However, we observed that both persistent and neurovirulent strain derivatives can produce two forms of the L* protein through unusual type II internal ribosome entry site-mediated translation. A full-length 18-kDa protein can be expressed from an ACG or an AUG initiation codon, whereas an N-terminally truncated 15-kDa product can be translated from a downstream AUG initiation codon. The expression of the 18-kDa form is required for efficient persistence of DA virus derivatives in the central nervous system.
Theiler's murine encephalomyelitis virus (TMEV or Theiler’s virus) belongs to the Cardiovirus genus of the Picornaviridae family. Strains of this virus are divided into two subgroups on the basis of the diseases that they induce in the central nervous system (CNS) of the mouse. Neurovirulent strains, such as GDVII, replicate permissively in the neurons of the brain and cause acute fatal encephalitis. Persistent strains, such as DA or BeAn, induce a biphasic disease (4, 11, 18). During the early phase, the virus replicates in the gray matter of the brain, causing subclinical encephalitis. At 2 to 3 weeks postinfection, the virus migrates from the brain to the spinal cord, where it persists in the white matter during the entire life of the mouse. At this stage, the virus is predominantly found in macrophages (or microglial cells), oligodendrocytes, and possibly astrocytes (1, 2, 12, 33). Viral persistence in the CNS is associated with inflammation and with demyelinating lesions. In this regard, TMEV is considered an experimental model for studying multiple sclerosis (23).
The Theiler's virus genome is a single-stranded positive-sense RNA molecule containing a 7-kb-long open reading frame (ORF) encoding a polyprotein which is cleaved to produce 12 mature viral proteins. Persistent strains of TMEV contain an additional ORF which overlaps the main ORF and codes for a 156-amino-acid-long protein called L* (with a calculated molecular mass of 18 kDa). This overlapping ORF, first identified by Kong and Roos (8), is unique among picornaviruses.
The ORF coding for the L* protein starts with two AUG codons in the same frame, separated by 9 nucleotides (nt). The first codon has been suggested to initiate translation of the L* protein because it has a strong Kozak context (A in −3 and G in +4). The translation of both the main ORF and the L* ORF is driven by an internal ribosome entry site (IRES) found in the 5′ noncoding region of the genome. Cardioviruses (such as TMEV) contain type II IRESs that are thought to allow exact positioning of the ribosome on the AUG used for initiation of translation of the polyprotein (6, 7, 22). Translation initiation of the alternative ORF encoding the L* protein has been proposed to occur through leaky scanning (32). In this hypothesis, the ribosome which initiates translation at the L* protein AUG would first land at the polyprotein AUG and then scan through the stem-loop present between these AUGs.
Neurovirulent strains of TMEV are believed not to produce the L* protein, as the AUG initiating the L* ORF and the second AUG located 5 codons downstream in the same reading frame have been specifically mutated to ACG during the course of evolution of these strains (17, 24).
The role of the L* protein is still poorly understood. This protein has been shown to increase the infection of macrophage cell lines (20, 28). Macrophage infection enhancement is due to the L* protein and not merely to competition for translation of the two overlapping ORFs of the virus (30).
Enhancement of macrophage infection could be linked to an antiapoptotic role reported for the L* protein (5). Recently, L* was shown to associate with microtubules within infected cells (19). The relationship between these phenotypes is still unclear.
In vivo, the role of the L* protein is controversial. Using a mutant DAFL3 virus in which an ACG codon was substituted for the L* AUG initiation codon, Ghadge et al. showed that L* was essential for persistence of the virus in the CNS (5). However, a similar AUG-to-ACG L* mutant virus obtained in our laboratory from another molecular clone (DA1) of the DA virus persisted almost as well as the wild-type virus (30). Moreover, recombinant virus R2, formed from the DA and GDVII viruses, persisted in the CNS of the mouse in spite of the fact that this virus had the GDVII 5′ noncoding region and the ACG initiation codon for L* (13).
To gain insight into the influence of the L* protein on macrophage infection and viral persistence, we analyzed the phenotypes of a series of L* mutant viruses. We observed that two forms of the L* protein, of 15 and 18 kDa, can be expressed by TMEV, even by neurovirulent virus derivatives which were thought not to produce this protein. The expression of these L* products involves unusual translation initiation mechanisms for a type II IRES, including initiation at a non-AUG codon. The expression of the 18-kDa form is required for efficient persistence of DA virus derivatives in the CNS.
MATERIALS AND METHODS
Construction of mutant viruses.
Viruses mutated in the L* region were obtained by site-directed mutagenesis. Mutations were generated by the method of Kunkel (9) by using subclones carrying the appropriate region. Restriction fragments carrying the mutations were sequenced on one strand to ensure that no unwanted mutation had occurred and were subsequently cloned back into plasmids containing the full-length viral cDNA.
Plasmids pOV23, pOV41, and pOV42 were described previously (30). pOV46 was obtained by cloning the BsiWI-SphI fragment (nt 1265 to 2162 of DA1) of pOV42 into pOV23. The recombinant plasmid obtained, pOV46, thus carries the genome of a virus (OV46) combining the AUG-to-ACG mutation of OV23 and the stop codon mutation of OV42.
To confirm the surprising persistence of AUG-to-ACG mutant OV23, we constructed an identical recombinant from nonpersistent virus OV46. The BsiWI-MscI fragment (nt 1265 to 1705 of DA1) of pTM410 was used to replace the corresponding fragment of pOV46. pTM410 is a pTZ19R (Pharmacia) derivative containing the 5′ end (nt 1 to 1729) of the DA1 genome. The new AUG-to-ACG mutant was called OV48. Plasmid pOV48 is identical to plasmid pOV23.
The AUG-to-ACG mutation at codon 41 of the L* ORF was introduced by site-directed mutagenesis with oligonucleotide TM338 (GCC AGA AGA CGT CGT CGT CCA GGT CCA). The mutation was introduced into the pTM410 and pOV20 subclones, carrying nt 1 to 1730 of the DA1 and OV23 genomes, respectively. The BbrPI-BsiWI fragments (nt 804 to 1265 of DA1) carrying the mutation (nt 1199) were then used to replace the corresponding fragment of pTM598 (31). The recombinant plasmids obtained, pOV82 and pOV90, contain the full-length genomes of mutant viruses carrying the AUG41-to-ACG mutation alone (pOV82) or in combination with the AUG1-to-ACG and AUG5-to-ACG mutations (pOV90).
A stop codon was introduced at codon 13 of the L* ORF of pTM410 with oligonucleotide TM337 (CGG GAG TAA CGT CTA CGG CTG TGC AAA T). The BbrPI-BsiWI fragment of the mutated plasmid (nt 804 to 1265 of DA1) was then cloned into pTM598. The recombinant plasmid obtained, carrying the Leu-to-stop codon mutation at codon 13 of the L* ORF, was called pOV84. The virus produced from this construct was called OV84.
A stop codon at codon 13 was also introduced into the genome of a virus carrying the AUG1-to-ACG and AUG5-to-ACG mutations. As described above, pOV20 was used as an intermediate subclone to perform mutagenesis. The recombinant plasmid obtained, called pOV89, thus combines the stop codon 13 mutation with the AUG1-to-ACG and AUG5-to-ACG mutations of pOV23.
A premature stop codon was introduced into the L* ORF of GDVII. Mutagenesis was performed with oligonucleotide TM244 (TAT GCG ATA CTG TGA TGG AAC CCC AGG) by using pTM427, a pTZ18R derivative containing the 5′ end (nt 1 to 1733) of the GDVII genome. The BbrPI-AocI fragment (nt 807 to 1334 of GDVII) was then ligated to the AocI-SgrAI and SgrAI-BbrPI fragments of pTMGDVII derivatives to form pOV47. This plasmid contains the full-length genome of the GDVII virus with the Ser-to-stop codon mutation at codon 68 of the L* ORF. The virus produced from this construct was called OV47.
The premature stop codon of pOV42 (stop codon 93) was introduced into the L* ORF of recombinant virus R2 (13). The BsiWI-Van91 fragment of pOV42 (nt 1265 to 2983) was cloned back into pTMR2 (13). Recombinant virus R2 carrying the Leu-to-stop codon mutation at codon 93 of the L* ORF was called OV57 (on plasmid pOV57).
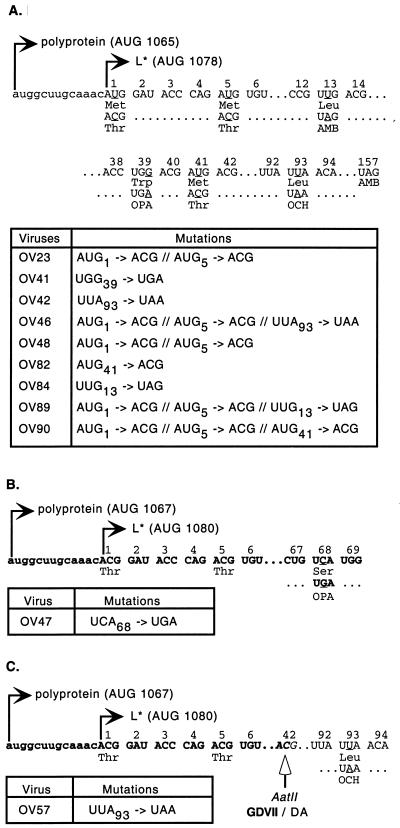
The characteristics of the L* mutants used in this study are presented in Fig. 1.
FIG. 1.
L* mutant viruses. The nucleotide and amino acid sequences of L* mutations are shown. The various mutations are indicated under the corresponding sequence segments of the parental strain. Mutated nucleotides are underlined. Nucleotides of the main ORF upstream of the L* ORF are in lowercase. Numbering corresponds to the codons of the L* protein. The nucleotide sequence of the neurovirulent genome is indicated in bold. For each parental strain, a table summarizes the mutations present in the corresponding mutant viruses. (A) DA1 derivatives. (B) GDVII and OV47. (C) R2 and OV57. Note that OV23 and OV48 are identical. OV48 was derived from OV46 to confirm data obtained with OV23.
Cell cultures and virus production.
BHK-21 and RAW264.7 cells were cultured as described previously (30). Standard fetal calf serum was used in this study for RAW264.7 cells to maintain macrophage susceptibility to TMEV infection (27).
Viruses DA1, GDVII, and R2 and the various OV viruses were produced as described previously (16) by transfection of BHK-21 cells with genomic RNAs transcribed in vitro from plasmids carrying the corresponding cDNAs: pTMDA1 (14, 16); pTMGDVII (29); pTMR2 (13); pOV23, pOV41, and pOV42 (30); and pOV46, pOV47, pOV48, pOV57, pOV82, pOV84, pOV89, and pOV90 (this work).
Culture supernatants were collected after completion of the cytopathic effect (generally between 48 and 72 h after transfection). Supernatants were frozen, thawed, and centrifuged at 4,000 × g for 10 min. Supernatants were then collected and stored in aliquots at −70°C. Viruses were titrated by standard plaque assays with BHK-21 cells.
Analysis of mixed infections.
The proportions of the two viruses in a mixture were estimated by restriction analysis of reverse transcription (RT)-PCR products. To this end, RNA was extracted from infected cells or tissues, and a 1.1-kb fragment of the viral genome spanning the leader region was amplified by RT-PCR with primers TM4 and TM132 (26). A diagnostic restriction enzyme was then used to digest the PCR product. Upon gel electrophoresis, the proportions of the two viruses were estimated from the relative intensities of bands specific for the viruses. This approach is very sensitive for detecting small differences between two viruses, since it is independent of sample-to-sample variations and of any technical bias related to virus quantification, RNA extraction, cDNA synthesis, or PCR amplification.
Endonuclease TruI was used to discriminate between AUG-to-ACG (OV23) and stop codon 93 (OV42) mutant genomes and for R2-OV57 mixtures. Endonuclease Hsp92II was used to discriminate between GDVII and OV47 genomes.
Infection of mice.
Three-week-old female SJL mice (Iffa-Credo) were inoculated intracranially in the right hemisphere with 40 μl of a viral suspension containing the indicated virus dose.
For dot blot or RT-PCR analysis, RNA was prepared from the brains and the spinal cords by the technique of Chomczynski and Sacchi (3). Dot blot and RT-PCR analyses to detect viral RNA were performed as described previously (26).
Real-time PCR.
Real-time PCR was performed by using a GeneAmp 5700 system (Applied Biosystems) with a Platinum quantitative PCR kit (Invitrogen Life Technologies) and Sybr green detection. Primers used for the detection of viral RNA were TM346 (GCC GCT CTT CAC ACC CAT) and TM347 (AGC AGG GCA GAA AGC ATC AC). After an initial cycle of 10 min at 95°C and 1 min at 60°C, cycling was performed for 40 cycles of 95°C for 15 s and 60°C for 1 min.
Luciferase assay.
Equivalent amounts of DNA from different bicistronic constructs were transfected into BHK-21 cells with FuGene6 reagent (Roche) according to the manufacturer's recommendations. At 24 h posttransfection, luciferase activity was measured by using a dual luciferase assay (Promega).
Detection of the L* protein in infected cells.
BHK-21 cells infected in a serum-free medium with different mutant viruses were collected after completion of the cytopathic effect. Cells were harvested and lysed directly in sample buffer (62.5 mM Tris [pH 6.8], 2% β-mercaptoethanol, 3% sodium dodecyl sulfate, 10% glycerol, 0.1% bromophenol blue). Samples were then run on Tris- Tricine- sodium dodecyl sulfate- 11% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Amersham-Pharmacia Biotech). Western blotting was performed with polyclonal rabbit antiserum raised against a peptide corresponding to amino acids 70 to 88 of L* from strain DA (21), kindly provided by Y. Ohara (Kanazawa Medical University, Kanazawa, Japan). Detection was performed with a horseradish peroxidase-conjugated anti-rabbit antibody (Dako) and chemoluminescence (ECL+ kit; Amersham Pharmacia Biotech).
RESULTS
Influence of the L* protein on viral persistence.
We compared infection of mice by a series of L* mutant viruses carrying either a stop codon mutation in the L* ORF or an AUG-to-ACG replacement of the L* ORF initiation codon (Fig. 1). Viruses OV41 and OV42 carry a stop codon in the L* ORF at codons 39 and 93, respectively. Viruses OV23 and OV48 are identical constructs generated independently by substituting two ACG codons for the two AUG codons (codons 1 and 5 of L*) potentially used for L* protein translation initiation. Virus OV46 was obtained by combining the AUG-to-ACG mutations of OV23 and the stop codon of OV42. None of the mutations introduced into the L* ORF affected the amino acid sequences of the L and VP4 proteins encoded by the overlapping ORF.
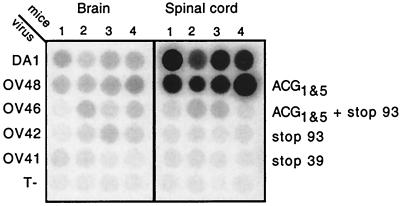
Groups of four SJL mice were inoculated with 105 PFU of viruses DA1, OV48, OV41, OV42, and OV46. Four control mice were inoculated with virus-free culture medium. At 45 days after inoculation, the amounts of viral RNA present in the brains and spinal cords of these mice were compared by RNA dot blot hybridization (Fig. 2). As observed previously for OV23, the AUG-to-ACG mutant virus (OV48) persisted almost as well as the wild-type virus. In contrast, viruses carrying a stop codon mutation in L*, either without (OV41 or OV42) or in combination with (OV46) AUG-to-ACG mutations, were strongly affected in their ability to persist in the CNS. Phosphorimager analysis of the dot blots showed RNA levels of stop codon mutant viruses to be about 30-fold lower than RNA levels of the wild-type virus.
FIG. 2.
Detection of viral RNA in the brains and spinal cords of infected SJL mice 45 days postinoculation. Viral RNA was detected by dot blot hybridization of total RNA extracted from the brains and spinal cords of four mice inoculated with 105 PFU of the indicated viruses. Control mice (T−) were mock infected. Viruses and corresponding L* ORF mutations are shown to the left and right, respectively.
In spite of the low level of persistence of stop codon mutants, RT-PCR allowed us to amplify viral cDNA from spinal cord RNA samples of mice infected for 45 days with OV41, OV42, and OV46. Sequencing of the PCR products confirmed the identities of these viruses.
The surprising difference between stop codon and AUG-to-ACG mutant viruses was confirmed in an independent experiment involving viruses OV23 and OV42. The occurrence of a spontaneous point mutation in the genome of stop codon mutants cannot account for the nonpersistence of these viruses. Indeed, the OV48 (AUG-to-ACG) mutant persisted, although it was derived from one of these nonpersistent constructs (OV46).
To examine whether the L* protein plays a role in the early phase of infection or whether this protein is required only for persistence in the spinal cord white matter during the late phase of disease, viral loads in the brains of infected SJL mice 5 days after inoculation were examined. At this time, the viral loads of stop codon mutant viruses (OV41, OV42, and OV46) were already three times lower than that of the DA1 virus (data not shown). This threefold reduction in viral RNA amounts was unlikely to have resulted merely from altered RNA replication of stop codon mutant viruses, since these viruses formed wild-type-size plaques and replicated like wild-type viruses in BHK-21 cells, as shown by dot blot experiments and by mixed-infection assays (30) (data not shown). Thus, it appears that the L* protein can play a role during the early phase of disease.
Influence of the L* protein on infection of a macrophage cell line.
As L* protein expression is known to increase macrophage infection (20, 28), we examined whether the difference between stop codon and AUG-to-ACG mutants was also detectable during infection of a macrophage cell line. To do so, we used a mixed-infection assay described previously (30). Briefly, BHK-21 cells (fibroblasts) and RAW264.7 cells (macrophages) were infected with a 1:1 mixture of OV48 and OV42 viruses. OV42 virus produces a truncated L* protein, whereas OV48 virus has the AUG-to-ACG mutations. After serial passage of the virus mixture on either cell type, the proportion of the two viruses was evaluated by RT-PCR and restriction endonuclease digestion analysis.
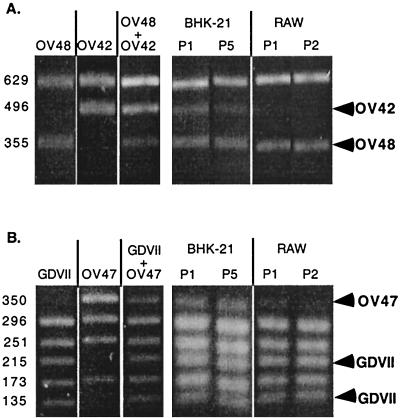
As shown in Fig. 3A, the proportion of the two viruses remained 1:1 in BHK-21 cells, even after five passages. In contrast, in macrophages, the infectivity of the stop codon mutant (OV42) was lower than that of the AUG-to-ACG mutant (OV48). After one passage on RAW264.7 cells, the OV42 virus almost disappeared from the mixture. Thus, as observed in vivo, the mutant with the stop codon mutation was more severely affected than the AUG-to-ACG mutant.
FIG. 3.
Mixed infections of fibroblast and macrophage cell lines. A mixture of two viruses (1:1) was prepared and used to infect BHK-21 and RAW264.7 (RAW) cells. RNA was extracted from infected cells after one to five passages of the virus mixture. As a control, RNA was extracted from the virus mixture before infection and from the parental virus stocks. The L* region was amplified by RT-PCR and digested with a restriction enzyme, allowing discrimination between the two viruses present in the mixture. (Left panels) Control analysis of the parental viruses and of the mixture used. (Right panels) Analysis of the virus mixture contained in the infected cells after one (P1) to five (P5) passages. Arrowheads indicate the fragments that are diagnostic of a given virus. (A) Competition between OV48, an AUG-to-ACG mutant, and OV42, a stop codon 93 mutant of DA1. TruI was used to digest the PCR fragments. The sizes of the fragments are indicated in base pairs on the left. (B) Competition between GDVII and its stop codon derivative, OV47. Hsp92II was used to digest the PCR fragments.
Several explanations can be put forward concerning the difference in the behavior of stop codon and AUG-to-ACG mutants.
First, the truncated form of the L* protein produced by OV42 could have a dominant-negative effect on viral replication. This possibility is unlikely, as the OV46 mutant, which harbors both the AUG-to-ACG and the stop codon mutations (and which is thus not expected to produce the truncated form of the L* protein), has the same phenotype as the stop codon mutants OV41 and OV42.
Second, the introduction of a stop codon in the alternative ORF encoding the L* protein, although not modifying the amino acid sequence of the polyprotein, could influence the translation of this overlapping ORF, either by modifying the secondary structure of RNA or by perturbing codon usage. However, we think that these two hypotheses are unlikely, because stop codon mutant viruses OV41 and OV42 contain a stop codon at different positions of the genome but display the same phenotype.
A third possibility is that, in spite of the absence of the AUG codons, the OV23 virus could initiate a low level of L* protein translation by an alternative mechanism, involving either initiation at an ACG codon or leaky scanning to a downstream AUG codon. This scenario could explain the surprising phenotype of AUG-to-ACG mutants OV23 and OV48. In this hypothesis, neurovirulent strains could also produce small amounts of the L* protein, as they carry the entire L* ORF, but with ACG codons instead of AUG codons for translation initiation.
Influence of the alternative ORF on neurovirulent strains.
To test whether neurovirulent strains can produce small amounts of the L* protein from the ACG codon of the putative L* ORF, we constructed a GDVII mutant carrying a premature stop codon at codon 68 of the L* ORF and called it OV47 (Fig. 1B). Again, this point mutation in L* did not modify the amino acid sequence of the polyprotein encoded by the overlapping ORF.
These viruses, GDVII and OV47, were compared for their ability to infect BHK-21 and RAW264.7 cells in a mixed-infection experiment (Fig. 3B). As observed for the persistent strain derivatives, the stop codon mutant virus OV47 was impaired in its ability to infect macrophages. In contrast, both viruses infected BHK-21 cells equally well. These results suggest that although this L* ORF starts with an ACG codon, neurovirulent viruses can translate small amounts of the L* protein.
To analyze whether this L* mutation could affect the acute encephalomyelitis induced by GDVII, we inoculated mice with 103 PFU of the GDVII-OV47 virus mixture. At 5 days postinoculation, we evaluated the proportions of the two viruses in the brains and spinal cords of infected mice. However, we failed to detect a difference in viral propagation between the two viruses (data not shown).
Taken together, the results obtained with persistent and neurovirulent strains suggest that small amounts of the L* protein can be translated from the alternative ORF of OV23 and GDVII viruses, possibly via the use of ACG codons as initiation codons. However, the influence of the L* protein in the GDVII background was not detected in mice with acute encephalitis.
Bicistronic constructs for testing translation initiation from ACG codons.
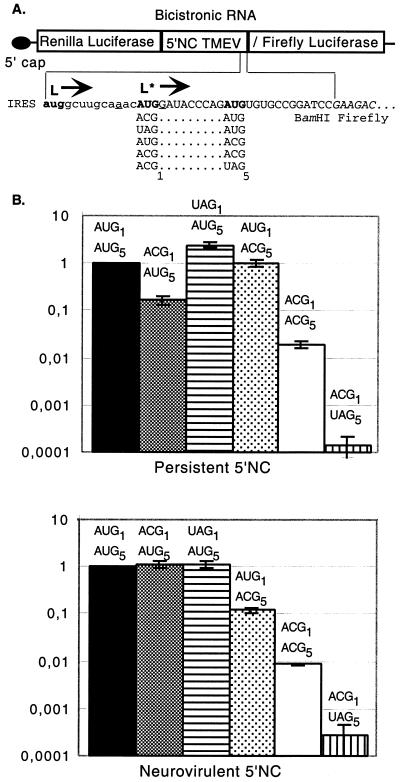
To examine the possibility of translation initiation from ACG codons, we constructed a bicistronic vector in which the first seven codons of L* are fused in frame with the firefly luciferase gene. The hybrid protein is translated from the L* initiation codon by an IRES-mediated mechanism, as in the viral context. Constructs were made with the 5′ noncoding regions of both DA1 and GDVII viruses. In these constructs, L* codons 1 and 5 were replaced by combinations of AUG, ACG, or UAG (stop) codons (Fig. 4A). The Renilla luciferase gene was cloned as the first cistron to allow the control of transfection efficiency. This gene is thus translated by the conventional cap-dependent mechanism, while translation of the firefly luciferase gene is IRES mediated. These constructs, shown in Fig. 4A, were transfected into BHK-21 cells. Firefly and Renilla luciferase activities were determined 24 h after transfection by using the dual luciferase assay.
FIG. 4.
Bicistronic constructs and IRES-mediated expression of luciferase from AUG or ACG initiation codons. (A) Bicistronic constructs in which translation of the firefly luciferase gene is under the control of the TMEV IRES. Constructs were made in the pHMG vector background (15). The 5′ end of the L* ORF, followed by a BamHI restriction site, is fused in frame with the second codon of the firefly luciferase gene. The polyprotein and L* AUG initiation codons are shown in bold. The Kozak context sequence of the first L* AUG is underlined. The sequence of the firefly luciferase gene is italicized. The different codon combinations replacing AUG1 and AUG5 of L* are shown under the sequence. NC, noncoding region. Nucleotides of the L*-luciferase gene fusion are in uppercase. Nucleotides of the main ORF upstream of the L* ORF are in lowercase. (B) Luciferase activity expressed by the different constructs in BHK-21 cells. The bicistronic constructs shown in panel A were transfected into BHK-21 cells. At 24 h posttransfection, the level of expression of both luciferase genes was measured. The firefly/Renilla luciferase expression ratio (construct:control) was calculated and normalized to the value obtained for the AUG1-AUG5 combination. The histograms show means and standard deviations for at least three transfection experiments.
As shown in Fig. 4B, the level of expression of the firefly luciferase gene from AUG initiation codons was very high (about 1 × 103-fold to 5 × 103-fold the background) for constructs carrying the DA1 and GDVII 5′ noncoding regions, suggesting that both IRESs can promote efficient translation of the L* ORF. In our constructs, both codons 1 and 5 could serve as initiation codons, since AUG-ACG as well as ACG-AUG or stop codon-AUG combinations yielded high levels of luciferase activity.
For both IRESs, translation from the ACG-ACG combination was about 50- to 100-fold higher than that of the negative control (AUG-stop codon combination), suggesting that translation from ACG codons occurred. However, this translation from ACG codons was about 100-fold lower than that of the positive control (AUG-AUG combination).
To confirm that ACG-initiated firefly luciferase gene expression was indeed IRES mediated in these experiments, Northern blot experiments were performed with the luc gene as a probe to detect the bicistronic mRNA in cells transfected with the ACG-ACG constructs. For the constructs containing the DA1 IRES, only one band of the expected size (4 kb) was detected, confirming that the mRNA was indeed bicistronic. For the constructs with the GDVII IRES, however, an additional band appeared, compatible with splicing or cryptic promoter activity (data not shown). To determine whether this smaller mRNA was a potential source of IRES-independent luciferase gene translation, we constructed control bicistronic constructs carrying either a 4-nt insertion (at coordinate 932 of DA1) or a 130-nt deletion (coordinates 800 to 932 of DA1) in the IRES region. Unexpectedly, these constructs still expressed rather high levels of firefly luciferase. The mean reductions in luciferase activity due to IRES alteration were only 1.7-fold for constructs with the IRES of GDVII and 3.7-fold for constructs with the IRES of DA1. These controls, together with the Northern blot results, indicate that at least part of the firefly luciferase activity shown in Fig. 4 represents IRES-mediated translation, as expected. However, the values should be interpreted with some caution, as they may partly represent IRES-independent translation, especially for the GDVII IRES.
L* protein expression in infected cells.
We observed above that in artificial constructs, the ACG codons of the alternative ORF could serve as initiation codons in IRES-driven translation. We were thus interested in testing, in the viral context, whether such alternative initiation could yield detectable amounts of the L* protein in cells infected with ACG mutant viruses. We thus performed Western blotting with infected BHK-21 cell extracts by using a very sensitive anti-L* protein antibody kindly provided by Y. Ohara. This polyclonal antibody was raised against a synthetic peptide corresponding to amino acids 70 to 88 of the DA L* protein. Due to sequence polymorphism between DA and GDVII in that region, this antibody cannot be used to detect L* protein production from neurovirulent virus derivatives.
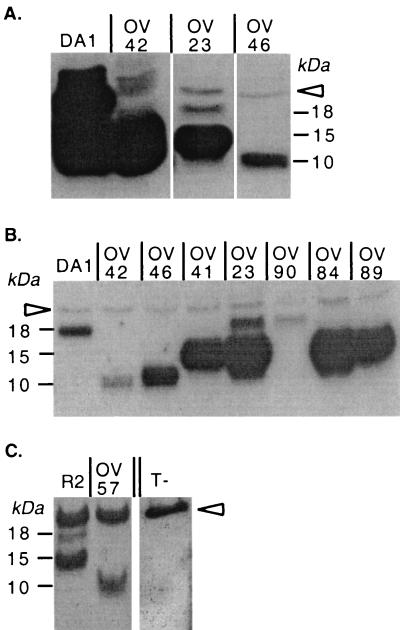
As shown in Fig. 5, the entire (18-kDa) and the truncated (10-kDa) L* proteins were readily detected in extracts from cells infected with DA1 and OV42 viruses, respectively. Interestingly, cells infected with the OV23 mutant virus also produced proteins detected with the anti-L* protein antibody. However, only a minor band could be detected at the expected molecular mass (18 kDa). The major band detected in the OV23-infected cell extracts had a molecular mass of 15 kDa and corresponded to a minor band detected in the DA1-infected cell extracts (Fig. 5A and B). The two bands detected in the OV23-infected cell extracts were tentatively identified as full-size L* protein initiated at the level of the ACG initiation codon and a shorter protein initiated at the level of the AUG41 codon in the L* sequence, possibly by a (leaky) scanning mechanism. OV46 virus produced small amounts of a truncated L* protein with the same molecular mass (10 kDa) as that produced by OV42 virus (Fig. 5A and B). This protein corresponded to the expected C-terminally truncated protein initiated at the level of the ACG codon.
FIG. 5.
Detection by Western blotting of L* protein expression in infected BHK-21 cells. (A) Comparison of the amounts of L* protein produced by viruses with AUG or ACG: wild type virus DA1 and mutant viruses OV23, OV42, and OV46. (B) Comparison of the viruses in panel A and OV41, OV84, OV89, and OV90 mutant viruses. Note that samples from DA1- and OV42-infected cells were diluted 100-fold to allow a better molecular mass comparison. (C) Comparison of recombinant virus R2 and the corresponding stop codon mutant virus, OV57. The arrowhead points to a nonspecific band detected in both infected and mock-infected cell extracts. T-, mock-infected cell extract.
This Western blotting experiment demonstrates that viruses harboring the AUG-to-ACG mutation of the initiation codon can still produce small amounts of the L* protein in infected cells.
Alternative sites for L* protein translation initiation.
To examine whether the 18- and 15-kDa proteins expressed by OV23 indeed corresponded to proteins initiated at the levels of the ACG1 and AUG41 codons, respectively, we constructed additional mutant viruses (Fig. 1). First, we introduced a premature stop codon at codon 13 of the L* ORF in the genome of DA1 (wild-type) or OV23 (AUG-to-ACG mutant) viruses to construct two mutant viruses, called OV84 or OV89, respectively. Second, we replaced the AUG41 codon with an ACG codon. The corresponding mutants, derived from the DA1 and OV23 virus genomes, were called OV82 and OV90, respectively.
Viruses OV82 (data not shown) and OV90, carrying the AUG-to-ACG replacement at codon 41, produced no detectable 15-kDa product, confirming that this form of the L* protein indeed corresponded to a protein initiated at the level of AUG41 (Fig. 5B). As also shown in Fig. 5B, viruses with a stop codon at position 13 (OV84 and OV89) or at position 39 (OV41) failed to produce the 18-kDa form of the L* protein, confirming that this protein could be initiated at the level of ACG1. These viruses produced the 15-kDa form of the L* protein initiated downstream, at the level of AUG41.
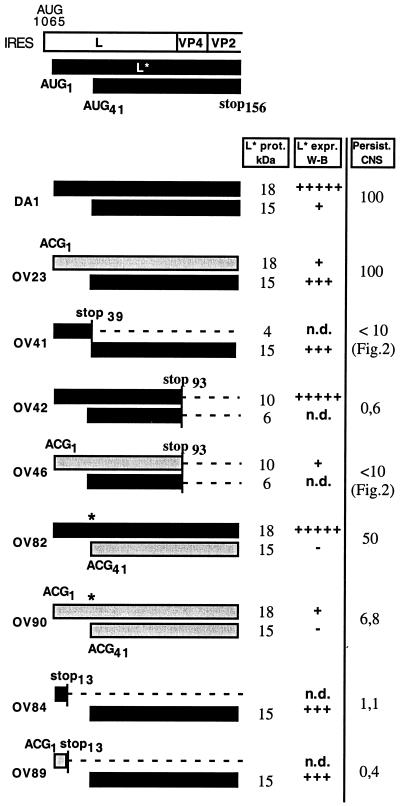
To examine whether the 15-kDa form of the L* protein could influence viral persistence in the CNS, we inoculated mice with 105 PFU of new mutant viruses OV82, OV84, OV89, and OV90. Mice were inoculated in parallel with viruses OV23, OV42, and DA1 or were mock infected. Real-time PCR was used to compare the amounts of viral RNA present in the spinal cords of the mice at 6 weeks postinfection (Table 1 and Fig. 6). The data showed a good correlation between the expression of the full-length form of the L* protein (from AUG or ACG) and efficient viral persistence. The AUG1-to-ACG and AUG5-to-ACG mutations of OV23 had no effect on viral persistence, although Western blot experiments indicated that the amount of the L* protein produced in vitro was small. The AUG41-to-ACG (Met41-Thr) mutation of the L* protein found in OV82 slightly affected viral persistence, suggesting that the amino acid replacement due to this mutation partly affected the function of the L* protein. In the context of the Met41-Thr mutation, the effect of the AUG1-to-ACG and AUG5-to-ACG mutations became clear (OV90 versus OV82). We believe that for the Met41-Thr mutant, higher levels of L* protein expression are necessary for function, in view of the weak activity of the protein.
TABLE 1.
Comparison, by real-time RT-PCR, of viral RNA amounts present in the spinal cords of infected mice 6 weeks postinfection
| Virus | Δ CTa
|
Relative no. of viral RNA copiesb | |
|---|---|---|---|
| Mean | SD | ||
| DA1 | 0 | 0.3 | 100 |
| OV23 | −0.1 | 0.55 | 107 |
| OV42 | 8.98 | 1.94 | 0.19 |
| OV82 | 1.93 | 1.15 | 26.24 |
| OV90 | 5.55 | 0.95 | 2.13 |
| OV84 | 8.24 | 1.64 | 0.33 |
| OV89 | 9.8 | 1.69 | 0.11 |
| Nonec | 19.05 | 0.77 | <0.01 |
CT, number of PCR cycles required to reach an arbitrary PCR product amount threshold during the logarithmic phase of the amplification; Δ CT, difference between a given CT and the CT obtained for DA1.
Data were normalized to the value for positive control DA1, which was taken as 100.
Control mice that were mock infected.
FIG. 6.
L* protein expression and viral persistence. The expected L* protein products for the various L* mutants are represented by bars. The black and gray bars represent the L* protein products initiated from AUG and ACG codons, respectively. An asterisk symbolizes the Met41-to-Thr mutation of the L* protein introduced to mutate the AUG41 codon. For each L* product, the expected molecular mass (L* prot. kDa) and the estimated amount of protein detected by Western blotting in BHK-21 cells (L* expr. W-B) are shown. +++++, wild-type expression level; +++, intermediate expression level; +, low expression level; −, not detectable; n.d., not determined. The persistence of the various mutants in the CNS, as deduced from dot blot (Fig. 2) or real-time PCR (Table 1) measurements, is indicated (Persist. CNS).
L* protein expression by neurovirulent viruses.
We next wanted to examine whether the 18- and 15-kDa forms of the L* protein could also be produced by viruses derived from neurovirulent virus GDVII. As the anti-L* protein antibody failed to detect the L* protein of viruses derived from GDVII, we used virus R2, constructed by McAllister et al., in which the capsid-coding region of virus DA replaces the capsid-coding region of virus GDVII (13). This virus has the IRES and the 5′ end of the L* ORF (with the ACG1 and ACG5 codons) from neurovirulent virus GDVII, but the region of the L* protein recognized by the antibody is from virus DA. We constructed a mutant of virus R2 by introducing a stop codon at position 93 of the L* ORF, as in OV42. This new virus was called OV57 (Fig. 1C).
When tested with R2 in mixed-infection experiments, OV57 had a decreased ability to infect macrophages in vitro or to persist in the CNS of SJL mice (data not shown), suggesting that L* protein translation could indeed be driven by the GDVII IRES and the ACG initiation codon. Accordingly, both the 18-kDa and the 15-kDa L* products could be detected by Western blotting in extracts of R2-infected BHK-21 cells (Fig. 5C). As expected, the OV57 stop codon mutant produced a 10-kDa truncated form of the L* protein (Fig. 5C).
DISCUSSION
Role of the L* protein in viral persistence.
The genome of Theiler's virus contains two overlapping ORFs. The main ORF encodes a polyprotein processed to form the 12 mature proteins necessary for the life cycle of the virus. A second ORF, overlapping the main one, encodes an 18-kDa protein called L*, first identified by Kong and Roos (8). Neurovirulent strains of TMEV were thought not to produce this protein, as in these strains, the L* ORF started with an ACG codon instead of an AUG codon.
The involvement of the L* ORF in viral persistence has been somewhat controversial. A role was suggested for the L* ORF because this ORF was conserved in all the persistent sequenced strains of TMEV (17). Through the analysis of a mutant DA virus in which an ACG codon was substituted for the AUG start codon, Ghadge et al. showed that the L* ORF was required for persistence of the virus (5). However, a similar AUG-to-ACG mutant constructed in our laboratory from another molecular clone of the DA strain (DA1) persisted to almost wild-type levels (30). In addition, a recombinant virus (R2) formed from viruses DA and GDVII was reported to persist in the spinal cord in spite of the fact that the IRES of the virus and the 5′ end of the L* ORF (including the ACG1 and ACG5 codons) originated from neurovirulent virus GDVII (13).
Our results obtained with L* stop codon mutants confirm the data of Ghadge et al. showing that the L* protein is required for persistence of the virus (5). Our data show that viral persistence was influenced by the L* protein itself and not merely by a competition mechanism for the translation of the two overlapping ORFs of the viral genome.
Persistence of mutants carrying an L* protein ACG initiation codon.
We observed that some L* protein could be produced by mutant viruses in which the AUG initiation codon was replaced by the ACG codon. Moreover, there was a perfect correlation between the expression of the 18-kDa L* protein in ACG mutants and viral persistence. This observation explains the persistence of the previously analyzed OV23 and R2 viruses, which both express some level of L* protein from the ACG codon. Interestingly, persistence was correlated only with expression of the full-length L* protein. Mutant viruses carrying a stop codon at position 13 (OV84 and OV89) or at position 39 (OV41) and thus lacking the 18-kDa form of the L* protein but readily expressing the truncated 15-kDa form of the protein from codon 41 were dramatically affected in their ability to persist. Recently, Obuchi et al. reported the binding of the L* protein to microtubules; N-terminally truncated L* proteins lacked such binding activity (19). The fact that viruses expressing the 15-kDa form of the L* protein showed impaired persistence could be related to this inability to bind microtubules, although the relationship between microtubule binding and persistence remains to be established.
It is worth noting that none of the L* protein mutants was completely cleared from the CNS at 6 weeks after infection. Although viral RNA levels were reduced more than 30-fold for the mutants (Table1; Fig. 2), viral RNA could still be detected in all the samples by the very sensitive RT-PCR procedure.
The exact mechanism by which the L* protein can influence viral persistence in the CNS is still poorly understood. Lin et al. suggested that the L* protein can inhibit the cytotoxic T-lymphocyte response (10). Ghadge et al. proposed that this protein can have an antiapoptotic effect during viral infection of a macrophage cell line (5). However, there is no evidence yet to correlate these effects with the role of the L* protein in establishing viral persistence in the CNS.
Strain variations.
The results presented in this report allow a reconciliation of views concerning the requirement for L* protein in viral persistence. However, we are still puzzled by the phenotype discrepancy existing for the AUG-to-ACG mutant viruses constructed from the DA1 (OV23) and the DAFL3 (DAL*-1) molecular clones of the DA virus (5, 30). We believe that nucleotide differences between the two cloned viruses are responsible for the difference in persistence ability. One possibility is that a point mutation in the IRES of DAFL3 prevents the initiation of the L* protein from an ACG codon. Another possibility is that the two viruses have different requirements (threshold) of L* protein expression due to subtle differences in replication levels.
Expression of the L* protein by neurovirulent viruses.
We postulated that the neurovirulent GDVII strain which contains the entire L* ORF but which naturally possesses ACG1 and ACG5 codons could also produce the L* protein. Several lines of evidence support this conclusion. (i) The IRES of GDVII could promote the translation of luciferase from ACG codons in a bicistronic construct. (ii) Mixed-infection experiments showed that a GDVII mutant virus carrying a stop codon in the L* ORF (OV47) infected macrophages less efficiently than the wild-type GDVII virus. (iii) The 18-kDa form of the L* protein was detected by Western blotting in BHK-21 cells infected with virus R2, which contains the GDVII IRES as well as the 5′ end of the L* ORF, including the ACG1 and ACG5 codons.
We observed that L* mutant OV47 had a decreased ability to infect macrophages in vitro compared to GDVII. Accordingly, we showed previously that increasing the expression of the L* protein from GDVII by creating an AUG initiation codon also increased the ability of the virus to infect macrophages. However, no clear phenotype difference could be demonstrated between GDVII and the mutants during acute neuronal cell infection in vivo. We believe that the high level of neurovirulence of these viruses may conceal the role of the L* protein. On the other hand, during viral propagation in the brain, GDVII mainly infects neurons. If the effect of the L* protein is specific for macrophages, as reported, one could envision that the L* protein of neurovirulent strains is not critical for neurovirulence after intracerebral inoculation but that this protein may participate in neuroinvasion or at any stage of the fecal-oral transmission of the virus.
IRES-mediated translation of L* protein products.
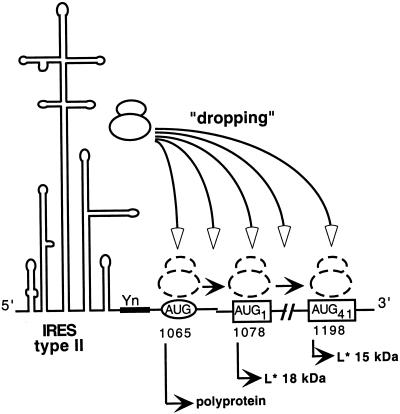
How does translation initiation of the L* ORF occur? The L* initiation codon is located 13 nt downstream from the AUG1065 codon initiating the translation of the main ORF. The translation of both ORFs is dependent on the TMEV IRES, which is classified as a type II IRES. Type II IRESs, found in cardioviruses and aphthoviruses, are believed to promote direct ribosome recognition of the initiator codon. In TMEV, translation initiation occurs at least at the level of three different codons: AUG1065 (polyprotein), AUG1 or ACG1 of L*, and AUG41 of L*. Leaky scanning could be responsible for initiation at codons 1 and 41 of L* but is unlikely because AUG1065 is in a good Kozak context (AxxAUGG). Furthermore, when a mini-L* ORF is created by the insertion of a stop codon at L* position 13 or 39, translation of the downstream 15-kDa form of the L* protein from codon 41 is not inhibited, indicating either that reinitiation occurred or that leaky scanning occurred over two potential initiation codons. It is tempting to speculate that the IRES “drops” the ribosome near nt 1065 but that, on the basis of the RNA secondary or tertiary structure and of cellular factors associated with the IRES, dropping occurs either at 1065 or after that point. Scanning would then occur from the place where the ribosome was dropped to the next potential initiation codon (Fig. 7). We do not know at the present time whether the relative amounts of initiation at the various start codons are controlled. Yamasaki et al. (32) suggested that L* protein translation could be regulated by cellular factors. It will be of interest to determine whether parameters such as the kinetics of infection or the cell type influence the rates of translation initiation at the various start codons.
FIG. 7.
Dropping model for type II IRES-mediated multiple translation initiation. Yn, oligopyrimidine tract conserved in picornavirus IRESs. The circled AUG is the start codon for translation of the polyprotein. Boxed AUG codons are codons that would initiate translation of the 18- and 15-kDa L* products. Instead of positioning the small ribosomal subunit at a fixed position corresponding to the initiation codon, the IRES would be more “flexible” and drop the ribosome at various positions. From there, the ribosome could scan to find the first available initiator codon.
To our knowledge, non-AUG start codons are usually decoded as methionine codons. Only in picornavirus-like insect viruses, such as Plautia stali intestine virus, has translation been shown to be initiated with non-Met (glutamine) amino acids (25). This very unusual initiation mechanism involves the formation of a pseudoknot in the RNA structure. For TMEV, we have no evidence that such a structure can be formed. We have no reason to believe that ACG is not decoded as methionine, although we have no proof for this belief.
Acknowledgments
We are indebted to M. Obuchi and Y. Ohara for efforts to provide us with the anti-L* protein antibody. We are grateful to Pierre Rensonnet and Muriel Minet for expert technical assistance. We thank Vincent van Pesch for helpful discussions. We thank Jean-Luc Vaerman (SANG Unit, University of Louvain) for help in setting up real-time PCR.
O.V. is a fellow of the Belgian FRIA (Fonds pour la Recherche dans l'Industrie et l'Agriculture). T.M. is a senior research associate with the FNRS. This work was supported by convention 3.4573.94F from the FRSM, by research grant 1.5.095.00 from the FNRS (Belgian Fund for Scientific Research), and by the Fonds de Développement Scientifique (FSR) of the University of Louvain.
REFERENCES
- 1.Aubert, C., M. Chamorro, and M. Brahic. 1987. Identification of Theiler's virus infected cells in the central nervous system of the mouse during demyelinating disease. Microb. Pathog. 3:319-326. [DOI] [PubMed] [Google Scholar]
- 2.Brahic, M., W. G. Stroop, and J. R. Baringer. 1981. Theiler's virus persists in glial cells during demyelinating disease. Cell 26:123-128. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 4.Drescher, K. M., L. R. Pease, and M. Rodriguez. 1997. Antiviral immune responses modulate the nature of central nervous system (CNS) disease in a murine model of multiple sclerosis. Immunol. Rev. 159:177-193. [DOI] [PubMed] [Google Scholar]
- 5.Ghadge, G. D., L. Ma, S. Sato, J. Kim, and R. P. Roos. 1998. A protein critical for a Theiler's virus-induced immune system-mediated demyelinating disease has a cell type-specific antiapoptotic effect and a key role in virus persistence. J. Virol. 72:8605-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 7.Kaminski, A., G. J. Belsham, and R. J. Jackson. 1994. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J. 13:1673-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong, W. P., and R. P. Roos. 1991. Alternative translation initiation site in the DA strain of Theiler's murine encephalomyelitis virus. J. Virol. 65:3395-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, X., R. P. Roos, L. R. Pease, P. Wettstein, and M. Rodriguez. 1999. A Theiler's virus alternatively initiated protein inhibits the generation of H-2K-restricted virus-specific cytotoxicity. J. Immunol. 162:17-24. [PubMed] [Google Scholar]
- 11.Lipton, H. L. 1975. Theiler's virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect. Immun. 11:1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipton, H. L., G. Twaddle, and M. L. Jelachich. 1995. The predominant virus antigen burden is present in macrophages in Theiler's murine encephalomyelitis virus-induced demyelinating disease. J. Virol. 69:2525-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAllister, A., F. Tangy, C. Aubert, and M. Brahic. 1990. Genetic mapping of the ability of Theiler's virus to persist and demyelinate. J. Virol. 64:4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAllister, A., F. Tangy, C. Aubert, and M. Brahic. 1989. Molecular cloning of the complete genome of Theiler's virus, strain DA, and production of infectious transcripts. Microb. Pathog. 7:381-388. [DOI] [PubMed] [Google Scholar]
- 15.Mehtali, M., M. LeMeur, and R. Lathe. 1990. The methylation-free status of a housekeeping transgene is lost at high copy number. Gene 91:179-184. [DOI] [PubMed] [Google Scholar]
- 16.Michiels, T., V. Dejong, R. Rodrigus, and C. Shaw-Jackson. 1997. Protein 2A is not required for Theiler's virus replication. J. Virol. 71:9549-9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michiels, T., N. Jarousse, and M. Brahic. 1995. Analysis of the leader and capsid coding regions of persistent and neurovirulent strains of Theiler's virus. Virology 214:550-558. [DOI] [PubMed] [Google Scholar]
- 18.Monteyne, P., J. F. Bureau, and M. Brahic. 1997. The infection of mouse by Theiler's virus: from genetics to immunology. Immunol. Rev. 159:163-176. [DOI] [PubMed] [Google Scholar]
- 19.Obuchi, M., T. Odagiri, K. Asakura, and Y. Ohara. 2001. Association of L* protein of Theiler's murine encephalomyelitis virus with microtubules in infected cells. Virology 289:95-102. [DOI] [PubMed] [Google Scholar]
- 20.Obuchi, M., Y. Ohara, T. Takegami, T. Murayama, H. Takada, and H. Iizuka. 1997. Theiler's murine encephalomyelitis virus subgroup strain-specific infection in a murine macrophage-like cell line. J. Virol. 71:729-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obuchi, M., J. Yamamoto, T. Odagiri, M. N. Uddin, H. Iizuka, and Y. Ohara. 2000. L* protein of Theiler's murine encephalomyelitis virus is required for virus growth in a murine macrophage-like cell line. J. Virol. 74:4898-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilipenko, E. V., A. P. Gmyl, S. V. Maslova, G. A. Belov, A. N. Sinyakov, M. Huang, T. D. Brown, and V. I. Agol. 1994. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J. Mol. Biol. 241:398-414. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez, M. 1992. Central nervous system demyelination and remyelination in multiple sclerosis and viral models of disease. J. Neuroimmunol. 40:255-263. [DOI] [PubMed] [Google Scholar]
- 24.Roos, R. P., W. P. Kong, and B. L. Semler. 1989. Polyprotein processing of Theiler's murine encephalomyelitis virus. J. Virol. 63:5344-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki, J., and N. Nakashima. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. USA 97:1512-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw-Jackson, C., and T. Michiels. 1999. Absence of internal ribosome entry site-mediated tissue specificity in the translation of a bicistronic transgene. J. Virol. 73:2729-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw-Jackson, C., and T. Michiels. 1997. Infection of macrophages by Theiler's murine encephalomyelitis virus is highly dependent on their activation or differentiation state. J. Virol. 71:8864-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takata, H., M. Obuchi, J. Yamamoto, T. Odagiri, R. P. Roos, H. Iizuka, and Y. Ohara. 1998. L* protein of the DA strain of Theiler's murine encephalomyelitis virus is important for virus growth in a murine macrophage-like cell line. J. Virol. 72:4950-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tangy, F., A. McAllister, and M. Brahic. 1989. Molecular cloning of the complete genome of strain GDVII of Theiler's virus and production of infectious transcripts. J. Virol. 63:1101-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Eyll, O., and T. Michiels. 2000. Influence of the Theiler's virus L* protein on macrophage infection, viral persistence, and neurovirulence. J. Virol. 74:9071-9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamasaki, K., C. C. Weihl, and R. P. Roos. 1999. Alternative translation initiation of Theiler's murine encephalomyelitis virus. J. Virol. 73:8519-8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, L., M. A. Calenoff, and M. C. Dal Canto. 2001. Astrocytes, not microglia, are the main cells responsible for viral persistence in Theiler's murine encephalomyelitis virus infection leading to demyelination. J. Neuroimmunol. 118:256-267. [DOI] [PubMed] [Google Scholar]