Abstract
Studies on hepatitis C virus (HCV) replication have been greatly advanced by the development of cell culture models for HCV known as replicon systems. The prototype replicon consists of a subgenomic HCV RNA in which the HCV structural region is replaced by the neomycin phosphotransferase II (NPTII) gene, and translation of the HCV proteins NS3 to NS5 is directed by the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES). The interferon (IFN)-inducible protein kinase PKR plays an important role in cell defense against virus infection by impairing protein synthesis as a result of eIF-2α phosphorylation. Here, we show that expression of the viral nonstructural (NS) and PKR proteins and eIF-2α phosphorylation are all variably regulated in proliferating replicon Huh7 cells. In proliferating cells, induction of PKR protein by IFN-α is inversely proportional to viral RNA replication and NS protein expression, whereas eIF-2α phosphorylation is induced by IFN-α in proliferating but not in serum-starved replicon cells. The role of PKR and eIF-2α phosphorylation was further addressed in transient-expression assays in Huh7 cells. These experiments demonstrated that activation of PKR results in the inhibition of EMCV IRES-driven NS protein synthesis from the subgenomic viral clone through mechanisms that are independent of eIF-2α phosphorylation. Unlike NS proteins, HCV IRES-driven NPTII protein synthesis from the subgenomic clone was resistant to PKR activation. Interestingly, activation of PKR could induce HCV IRES-dependent mRNA translation from dicistronic constructs, but this stimulatory effect was mitigated by the presence of the viral 3′ untranslated region. Thus, PKR may assume multiple roles in modulating HCV replication and protein synthesis, and tight control of PKR activity may play an important role in maintaining virus replication and allowing infection to evade the host's IFN system.
Infection with hepatitis C virus (HCV) causes chronic liver disease that can lead to hepatocellular carcinoma (35). The virus is a flavivirus with a positive-stranded RNA genome of approximately 9.6 kb (4). The viral RNA consists of 5′ and 3′ untranslated regions (UTRs) and a large open reading frame encoding a polyprotein of 3,010 to 3,033 amino acids that undergoes proteolytic processing by both host signal peptidases and viral proteases to yield 10 mature structural (C, E1, and E2) and nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins. HCV replication is error prone, resulting in continuous generation of viral variants with differences located mainly in the hypervariable region of the E1 and E2 genes. This dynamic variation may play a role in the establishment and maintenance of persistent infection.
The only approved treatments for HCV infection are alpha interferon (IFN-α) alone or in combination with ribavirin (5). The mechanisms utilized by IFN-α to block HCV replication are not well understood, nor are the reasons for the limited effectiveness of IFN therapy known (5). It was hypothesized that differences in HCV genomic sequence may affect the structure and function of the viral RNA and proteins, which in turn can alter the host's response to IFN-α treatment. In fact, comparison of full-length sequences of IFN-α-responsive and nonresponsive viruses from HCV-infected patients showed that patients who responded completely to IFN therapy carried HCV 1b isolates with multiple mutations within a discrete region encoding 40 amino acids in the carboxy-terminal half of NS5A between amino acids 2209 and 2248 of the HCV polyprotein (13, 14). This region, known as the interferon sensitivity determining region (ISDR), has been thought to play an important role in HCV resistance to IFN treatment (34). Although several studies reported conflicting data on the relationship between the ISDR sequence and clinical resistance to IFN, recent statistical analysis of data from these studies confirmed a positive association between the ISDR sequence and clinical resistance to IFN (63).
Binding of IFN-α to its receptor induces many signaling pathways that control transcription of a large number of interferon-inducible genes encoding proteins with antiviral and antiproliferative activities (53). Transcriptional activation of interferon-inducible genes by IFN-α is regulated mainly by activation of the Jak-Stat pathway (53). The importance of Jak-Stat activation in HCV replication may be underscored by the finding that this signaling pathway is impaired in cells expressing the HCV polyprotein (24). Among the many interferon-inducible gene products, the double-stranded RNA (dsRNA)-activated protein kinase PKR has been shown to play an important role in cell defense against infections by many viruses by suppressing protein synthesis (28).
PKR is a serine/threonine kinase that exhibits distinct activities: dimerization upon binding to dsRNA and autophosphorylation at many serine and threonine sites and phosphorylation of the α subunit of translation initiation factor eIF-2, a modification that leads to the inhibition of protein synthesis (28). Through this capacity, PKR is thought to be a mediator of the antiviral and antiproliferative actions of IFN-α (53).
To bypass PKR activation and the inhibition of host protein synthesis, many RNA and DNA viruses have evolved mechanisms to inactivate PKR (26). In the case of HCV, both the NS5A and E2 proteins were shown to block PKR activation. In the case of NS5A, it has been proposed that the ISDR of the viral protein from IFN-resistant but not IFN-sensitive strains binds and blocks PKR activity (18, 20). Consequently, inhibition of PKR activity by NS5A abolishes PKR-dependent apoptosis and induces malignant transformation of NIH 3T3 cells, providing evidence for an important biological function of PKR inactivation by the viral protein (20, 21). Contrary to the inhibitory role of NS5A reported earlier, newer studies suggested that ISDR may not be required to confer virus resistance to IFN-α (15, 46, 47) and that NS5A expression in cultured cells mediates IFN resistance independently of PKR (46).
These contradictory findings have been explained by the pleiotropic effects of NS5A expressed in various cell lines and by the genetic differences and variations in NS5A expression levels between the chosen cell lines (55). On the other hand, E2 protein contains a 12-amino-acid sequence that shows high homology with several putative autophosphorylation sites within the amino terminus of PKR (57, 58). A similar sequence, known as the PKR-eIF-2α phosphorylation homology domain, found within eIF-2α was shown to be required for E2 inhibition of PKR activity in Saccharomyces cerevisiae (57). How E2 blocks PKR activity is not yet clear, but a recent study indicated that E2 functions via a pseudosubstrate mechanism (58). Together, both NS5A and E2 may play an important role in evading IFN response, but they are unlikely to be the only viral genes that contribute to evasion from IFN action (56).
For many years, traditional approaches to studying the mechanisms of HCV replication were unavailable, largely due to the limitations in propagating the virus in cultured cells. In recent years, significant progress in HCV research was initially made by the development of subgenomic dicistronic HCV replicon RNA capable of successfully replicating in the hepatoma cell line Huh7 in vitro (38). This prototype replicon was a subgenomic HCV RNA in which the HCV structural region was replaced by the neomycin phosphotransferase II (NPTII) gene. Synthesis of the NPTII protein was directed by the HCV internal ribosome entry site (IRES), whereas synthesis of HCV proteins NS3 to NS5B was directed by the IRES from the encephalomyocarditis virus (EMCV) (38). A number of adaptive mutations within the viral genome localized within NS5A, including a deletion of the entire ISDR, were later identified and found to enhance the initiation of replication in vitro (6, 37, 44). Most interestingly, the adaptive mutations did not alter the sensitivity of HCV replication to IFN-α treatment, suggesting that the ISDR may not be essential in this process (6, 50).
The availability of a system with which to study the mechanisms of virus replication in vitro prompted us to examine whether and how PKR modulates gene expression from the original subgenomic HCV clone (38). Here, we show that PKR plays a direct role in inhibition of viral protein synthesis from the subgenomic viral clone and provide strong evidence that this effect is mediated at the translational level. We also demonstrate the ability of PKR to induce the HCV IRES and inhibit EMCV IRES activity and the ability of the viral 3′ UTR to mitigate PKR function. These data clearly implicate PKR in HCV gene expression in vitro, raising the possibility of a similar functional role in vivo.
MATERIALS AND METHODS
Cell culture and IFN treatment.
The original wild-type pFKI389-NS3-3′ replicon DNA from genotype 1b and the generation of Huh7 HCV replicon cells were described earlier (38). Huh7 cells were maintained in Dulbecco's modified Eagle's medium (Gibco Life Technologies, Inc., Rockville, Md.) supplemented with 10% heat-inactivated fetal bovine serum, 1% nonessential amino acids, 100 U of penicillin G per ml, and 100 μg of streptomycin per ml at 37°C in a humidified atmosphere of 5% CO2. Cells containing the HCV replicon were maintained in culture in the presence of 500 μg of G418 (Geneticin; Life Technologies)/ml, which was removed 2 days before the experiments. For IFN treatment, 1,000 IU of human IFN-α2b (Intron A; Schering-Plough Corp., Kenilworth, N.J.)/ml was used.
RNA extraction and Northern blot analysis.
Total RNA was purified with Trizol (Gibco) according to the manufacturer's specification. RNA samples (10 μg) were treated with RNase-free DNase (Sigma), run on formaldehyde-containing 1% agarose gels, transferred to a nylon membrane (ICN, Aurora, Ohio), and subjected to Northern blotting with an [α-32P]dCTP-labeled 3.6-kb fragment (HindIII and EcoRI restriction sites) of pFKI389-NS3-3′ DNA or a 0.6-kb fragment of ribosomal 18S DNA as described previously (49). Radioactive signals were exposed on Kodak films, and quantification of the bands was performed by densitometric analysis of films within the linear range of exposure.
HCV plasmid constructions.
Two sets of IRES dicistronic constructs were used. The first set consisted of the EMCV or HCV IRES in the same plasmid DNA under control of the T7 RNA polymerase and was described previously (54). The second set of dicistronic constructs containing either the HCV IRES alone or the HCV IRES and the 3′ viral UTR downstream of the luciferase gene was generated by recovering the HCV IRES from the vector pBC(HCV)L (a kind gift from C. Schultz-Witherell and G. Witherell, RiboGene, Inc.). For the construction of pBC(HCV)L, the HCV IRES was amplified by PCR from vector pKIV (a kind gift from K. F. Tsukiyama-Kohara) (sense primer, 5′-TTATGATCAGGTTACGTTTGGTTTTTCTTTGAGG-3′; antisense primer, 5′-ATAGGATCCGATTGGGGGCGACACTCCACCATAGATC-3′) (60). The amplified region corresponds to the HCV 5′ UTR plus 40 nucleotides corresponding to the core protein coding region (from nucleotides 13 to 383, numbering with respect to the HCV genome). pBC(HCV)L was digested with EcoRI, and the fragment containing the HCV IRES was recovered and cloned into the intercistronic spacer of pcDNA3-CAT/Luc, containing the chloramphenicol acetyltransferase (CAT) gene and firefly luciferase (Luc) gene as reporter genes (a kind gift from S. Pyronnet) previously digested with EcoRI (48).
The HCV IRES dicistronic construct containing the HCV 3′ UTR vector was generated by recovering the HCV 3′ UTR by PCR amplification from vector pFKI389-NS3-3′/wild-type (a kind gift from R. Bartenschlager; EMBL database accession number AJ242654) (sense primer, 5′-CCGCTCGAGTGACGGGGAGCTAAACACTC-3′; antisense primer, 5′-CCGCTCGAGCGGCCGCACTTGATCTGCAGAGAGG-3′) (38). XhoI restriction sites were added for cloning purposes, and a NotI site was added to verify the orientation of the cloned segment. The PCR-amplified 3′ UTR (from nucleotides 7771 to 8001) was inserted in vector HCV IRES previously digested with XhoI upstream from the luciferase gene. The HCV IRES and 3′ UTR sequences in the dicistronic constructs were confirmed by DNA sequencing.
Recombinant vaccinia virus/T7 virus expression system.
One day before transfection, 0.8 × 106 Huh7 parental cells were seeded in 6-cm plates. Cells were infected with recombinant vaccinia virus containing the bacterial T7 RNA polymerase gene (17) for an hour, followed by transfection with 5 μg of Lipofectace reagent (Gibco) and 2 μg of DNA containing the gene of interest in the expression vector under the control of the T7 promoter. Wild-type PKR, PKR mutants, wild-type eIF-2α, and eIF-2α S51A cDNAs as well as K3L, E2, and green fluorescent protein (GFP) DNAs were expressed from the T7 promoter within the pcDNA3 vector (Invitrogen). Cells were incubated in serum-free medium at 37°C for 6 h, followed by the addition of complete medium and incubation for an additional 18 h before RNA or protein extraction.
Protein extraction and immunoblot analysis.
Cells were washed twice with ice-cold 1× phosphate-buffered saline (140 mM NaCl, 15 mM KH2PO4 [pH 7.2], 2.7 mM KCl), and proteins were extracted with 1× lysis buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 2 mM MgCl2, 1% Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 3 μg of aprotinin per ml, 1 μg of leupeptin per ml, and 1 μg of pepstatin per ml. After incubation on ice for 20 min, the lysates were centrifuged at 14,000 × g for 10 min at 4°C. The supernatant was transferred to a fresh tube, the protein concentration was measured by the Bradford assay (Bio-Rad, Hercules, Calif.), and samples were stored at −85°C.
Proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore Corp.). Immunoblottings were performed according to the standard protocol (49). The primary antibodies were as follows: anti-HCV-NS5A monoclonal antibody (MAb) (dilution 1:1,000; Biogenesis, London, United Kingdom); anti-NS3 rabbit polyclonal antibodies (dilution 1:1,000) (59); anti-human PKR MAb (dilution 1:1,000; clone F9 or E8) (36, 41); anti-Flag MAb (dilution 1:1,000; Sigma); anti-NPTII rabbit polyclonal (dilution 1:500; Cortex Biochem); anti-E2 MAb (dilution 1:500, generous gift from J. Dubuisson); rabbit serum to human eIF-2α (dilution 1:1,000; SC-11386, Santa Cruz Biotechnology); anti-mouse eIF-2α MAb (dilution 1:1,000) (36); rabbit serum to phosphoserine 51 of eIF-2α (dilution 1:1,000) (36); chicken polyclonal anti-GFP antibodies (dilution 1:1,000); and antiactin MAb (dilution 1:5,000; ICN). The secondary antibodies were horseradish peroxidase-conjugated anti-mouse immunoglobulin G, horseradish peroxidase-conjugated anti-rabbit immunoglobulin G, horseradish peroxidase-conjugated anti-goat immunoglobulin G, or horseradish peroxidase-conjugated anti-chicken immunoglobulin G (all at 1:1,000; Amersham Pharmacia Biotechnology). Proteins were visualized by the enhanced chemiluminescence detection system according to the manufacturer's instructions (Amersham Pharmacia Biotechnology).
Reporter gene assays.
To measure luciferase activity, transfected cells were lysed in reporter lysis buffer (Promega, Madison, Wis.) according to the manufacturer's recommendations. Twenty micrograms of total protein was mixed with 100 μl of substrate buffer, and luciferase activity was assayed by integrating the total light emission over 10 s with a luminometer (Luman LB9507; EG&G Berthold, Berlin, Germany). CAT activity was determined by incubating 20 μg of total protein with 400 μM acetyl-coenzyme A and [14C]chloramphenicol (50 mCi/mmol) (ICN Biochemicals Corp.) as a substrate, in a 150-μl reaction volume at 37°C for 2 h as described before. The acetylated products were separated by thin-layer chromatography (Whatman Ltd., Kent, England) and visualized by autoradiography. Densitometric analysis of autoradiograms was performed with the Scion Image software (Scion Corporation, Frederick, Md.). Quantification was done in samples with linear CAT activity, and the percent conversion was calculated as acetylated products divided by acetylated plus nonacetylated products.
RESULTS
PKR protein expression and eIF-2α phosphorylation levels in HCV replicon cells.
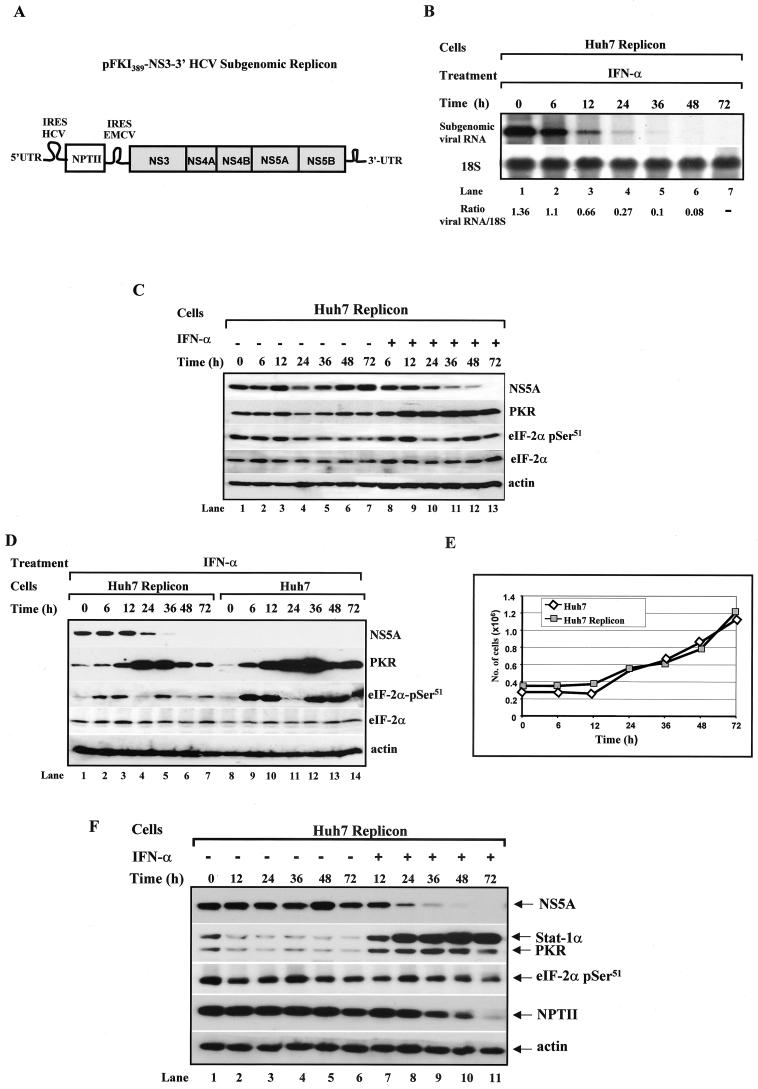
With the HCV replicon system previously described by Lohmann et al. (38) (Fig. 1A), we first verified the ability of IFN-α to inhibit viral RNA synthesis. Northern blot analysis demonstrated the downregulation of the subgenomic HCV RNA levels as early as 6 h after treatment, leading to undetectable RNA levels 48 h after IFN-α treatment (Fig. 1B, upper panel). In parallel, we examined the levels of NS5A and PKR proteins as well as eIF-2α phosphorylation in replicon cells stimulated with IFN-α (Fig. 1C). Replicon cells were maintained in culture in the absence (lanes 1 to 7) or presence (lanes 8 to 13) of IFN-α for up to 72 h. Immunoblot analysis of protein extracts from untreated cells (lanes 1 to 7) showed that NS5A protein (top panel), PKR protein (second from the top panel), and eIF-2α phosphorylation levels (third from the top panel) varied with time, indicating that they were affected by cell proliferation.
FIG. 1.
Regulation of replication and protein synthesis from an HCV subgenomic clone in human Huh7 cells by IFN-α. (A) Schematic representation of the prototype HCV subgenomic clone. The HCV structural region was replaced by the NPTII gene. NPTII expression is under the control of the HCV IRES, whereas expression of proteins NS3 to NS5B is controlled by the EMCV IRES (38). (B) Detection of viral RNA expression by IFN-α. HCV replicon Huh7 cells (∼3 × 105 cells) were stimulated with 1,000 IU of human IFN-α/ml for the indicated periods. Total RNA (10 μg) was isolated and subjected to Northern blot analysis with a 32P-labeled cDNA consisting of either the entire viral subgenomic sequences of pFKI389-NS3-3′ vector (upper panel) or ribosomal 18S DNA (lower panel). The radioactive bands were quantified with NIH Image version 1.54 software, and the ratios of viral to 18S RNA are indicated. (C and D) Expression of viral and host proteins in replicon Huh7 cells in the absence or presence of IFN-α. Replicon cells (B and C) and parental Huh7 cells (D) were seeded at a concentration of ∼3 × 105 cells/10-cm dish. Twenty-four hours later (time zero), the medium was refreshed, and cells were left untreated (lanes 1 to 7) or treated with 1,000 IU of human IFN-α/ml for the indicated times (lanes 8 to 13). Protein extracts (50 μg) were subjected to immunoblot analysis with anti-NS5A MAb (top panel), anti-human PKR MAb (second panel from the top), anti-eIF-2α phosphoserine 51-specific antibody (third panel from the top), rabbit polyclonal anti-eIF-2α antibody (fourth panel from the top), or antiactin MAb (bottom panel). (E) Proliferation of parental and replicon Huh7 cells by IFN-α. Cells cultured as described for panel D were harvested at the indicated times, stained with trypan blue, and counted in a hemacytometer. Values represent the average of two separate experiments. (F) Expression of viral and host proteins in serum-starved replicon Huh7 cells. Huh7 replicon cells were plated at ∼3 × 105 cells/10-cm dish in the absence of serum. Twenty-four hours later (time zero), the cells were refreshed and maintained in serum-free medium in the absence (lanes 1 to 6) or presence of 1,000 IU of human IFN-α/ml. Protein extracts (50 μg) were subjected to immunoblot analysis with anti-NS5A MAb (top panel), anti-human PKR MAb together with anti-human Stat1α MAb (second panel from the top), anti-eIF-2α phosphoserine 51-specific antibody (third panel from the top), anti-NPTII MAb (fourth panel from the top), or antiactin MAb (bottom panel).
Interestingly, NS5A and PKR protein as well as eIF-2α phosphorylation levels decreased at the 24-h time point (Fig. 1C, lane 4), a pattern that was highly consistent in other experiments, particularly for eIF-2α phosphorylation after treatment with IFN-α (Fig. 1C, lane 10; see also Fig. 1D below). On the other hand, immunoblot analysis of protein extracts from IFN-α-treated cells (Fig. 1C, lanes 8 to 13) clearly showed a decrease in NS5A protein (top panel), which coincided with an increase in PKR protein (second from the top panel). The eIF-2α phosphorylation levels varied with the time of IFN-α treatment (Fig. 1C, third panel from the top), whereas total eIF-2α protein levels remained unchanged (Fig. 1C, fourth panel from the top). A clear increase in eIF-2α phosphorylation in IFN-α-treated over untreated cells was observed after prolonged periods of treatment (compare lanes 11 to 13 with lanes 5 to 7). Also, we noticed that high eIF-2α phosphorylation levels did not always correlate with increased PKR protein levels in IFN-α-treated cells (for example, compare lane 10 with lane 9 or 12), possibly indicating that, under these experimental conditions, phosphorylation of eIF-2α may not be modulated solely by PKR.
When we compared PKR protein and eIF-2α phosphorylation levels in IFN-α-treated cells (Fig. 1D), we found that a larger amount of PKR was induced in control cells (second panel from the top, lanes 9 to 14) than in replicon cells (lanes 2 to 7). Since HCV proteins were reported to negatively regulate the activation of the Jak-Stat pathway and the induction of IFN-inducible genes (24), the reduced expression of PKR in replicon cells could be explained by a partial transcriptional inhibition of the pkr gene in response to IFN-α. However, the possibility remains that the different PKR levels induced by IFN-α were caused by clonal variations between replicon and control Huh7 cells.
In regard to eIF-2α (Fig. 1D, third panel from the top), we noticed lower levels of phosphorylation in replicon cells (lanes 2 to 7) than in control cells (lanes 9 to 14) after treatment with IFN-α. As observed in Fig. 1C, eIF-2α phosphorylation was decreased at 24 h after IFN-α treatment in both parental and replicon cells, whereas PKR protein levels were highly induced (Fig. 1D, lanes 4 and 11). This further supports the notion that eIF-2α phosphorylation in response to IFN-α may not be entirely PKR dependent. The total eIF-2α protein levels in IFN-α-treated parental and replicon Huh7 cells remained unchanged (fourth panel from the top). Because eIF-2α phosphorylation levels varied with cell proliferation (Fig. 1C, lanes 1 to 7), we speculated that the phosphorylation differences between the parental and replicon cells were due to differential suppression of cell proliferation by IFN-α. However, we found that IFN-α did not suppress the proliferation of parental and replicon Huh7 cells (Fig. 1E), a result that also indicates that the lower PKR protein levels do not render replicon cells more sensitive to the antiproliferative effects of IFN-α. These findings suggested that protein expression from the subgenomic HCV clone coincides with an overall decrease in eIF-2α phosphorylation, although it was not clear whether this was regulated by PKR.
To bypass the side effects of cell proliferation, we assessed NS5A and PKR protein and eIF-2α phosphorylation levels in serum-starved, growth-arrested replicon cells (Fig. 1F). In this experimental setting, we saw that NS5A protein levels decreased with time after IFN-α treatment (top panel, lanes 7 to 11), and this coincided with an induction of PKR and Stat1 protein, which was used as an additional marker of IFN-α treatment (second panel from the top, lanes 7 to 11). In untreated replicon cells, we noticed that both PKR and Stat1 protein levels were decreased when cells were maintained in the absence of serum (second panel from the top, compare lane 1 with lanes 2 to 6). Unlike in proliferating replicon cells, eIF-2α phosphorylation levels did not vary significantly throughout the experiment (third panel from the top).
These data suggested that inhibition of viral replication and protein synthesis by IFN-α may be independent of eIF-2α phosphorylation status. When we examined whether IFN-α modulates NPTII protein expression, which is under the control of HCV IRES activity, we found that NPTII protein was also decreased in IFN-α-treated replicon cells (fourth panel from the top) but with slower kinetics than the decrease in the NS5A protein (top panel). These differences between the NS5A and NPTII proteins may reflect variations in the stability of the two proteins (see below) and/or differential responses of the HCV and EMCV IRES to IFN-α. These data raised the questions of whether PKR is directly involved in the regulation of gene expression from the subgenomic HCV clone and what the role of eIF-2α phosphorylation is in this process.
PKR directly impairs NS protein expression from the subgenomic HCV clone.
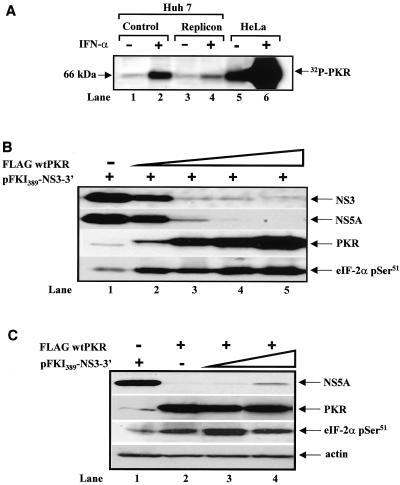
Next, we examined whether PKR in the parental and replicon Huh7 cells could be activated in vitro. To do so, PKR was immunoprecipitated from untreated and IFN-α-treated cells. Activation of PKR was then tested by autophosphorylation in the presence of reovirus activator dsRNA and [γ-32P]ATP (Fig. 2A). In these experiments, we detected PKR autophosphorylation in both parental and replicon cells prior to IFN-α treatment. Stimulation of cells with IFN-α caused the induction of PKR autophosphorylation in both cell types, which was higher for parental than for replicon cells (compare lanes 2 and 4). This difference most likely reflects the different levels of PKR protein induction in both cell types after IFN-α treatment (see Fig. 1D). These findings suggested that Huh7 cells contain a functional PKR.
FIG. 2.
Inhibition of subgenomic NS protein expression by wild-type PKR. (A) PKR autophosphorylation levels in parental and replicon Huh7 cells. Protein extracts (400 μg) from untreated (lanes 1, 3, and 5) or IFN-α-treated (1,000 IU/ml; 24 h) cells were immunoprecipitated with 5 μg of anti-human PKR MAb (clone F9). Immunoprecipitates were subjected to the in vitro kinase assay in the presence of 0.1 μg of reovirus dsRNA per ml and 1 μCi of [γ-32P]ATP as described before (36). Phosphorylated proteins were resolved by SDS-PAGE and visualized by autoradiography. HeLa protein extracts before (lane 5) and after (lane 6) IFN-α treatment were used as positive controls. (B) Wild-type PKR impairs NS protein synthesis from the subgenomic clone. HCV subgenomic DNA (pFKI389-NS3-3′; 1 μg) was transiently expressed in Huh7 cells alone (lane 1) or in the presence of 0.010 μg (lane 2), 0.050 μg (lane 3), 0.1 μg (lane 4), or 0.5 μg (lane 5) of Flag-tagged wild-type PKR cDNA with the vaccinia virus/T7 virus expression system. Eighteen hours later, protein extracts (50 μg) were subjected to immunoblot analysis to detect NS3 (top panel), NS5A (second panel from the top), PKR (third panel from the top), or eIF-2α serine 51 phosphorylation (bottom panel). (C) Overexpression of the subgenomic clone cannot rescue PKR-mediated inhibition of NS protein synthesis. Huh7 cells were treated with recombinant vaccinia virus/T7 virus as above with 1 μg of pFKI389-NS3-3′ DNA alone (lane 1), 0.050 μg of Flag-tagged wild-type PKR cDNA (lane 2), or a constant amount (0.050 μg) of Flag-tagged wild-type PKR together with 0.1 μg (lane 3) or 1 μg of pFKI389-NS3-3′ DNA. Eighteen hours later, protein extracts (50 μg) were subjected to immunoblotting for the detection of NS5A (top panel), PKR (second panel from the top), eIF-2α serine 51 phosphorylation (third panel from the top), or actin levels (bottom panel).
To better address the role of PKR in viral gene expression, we examined NS protein synthesis from the subgenomic clone by wild-type PKR in transient-expression assays in Huh7 cells. To this end, expression of wild-type human PKR and viral proteins was mediated by gene delivery with the vaccinia virus/T7 virus method. In this method, transfected genes under the control of the bacteriophage T7 promoter are efficiently transcribed in the cytoplasm by the T7 RNA polymerase delivered to the cells by infection with recombinant vaccinia viruses (17). This method is suitable for studying the translational functions of PKR (36) and has been used successfully to investigate HCV replication mechanisms in cultured cells (9, 40).
When the HCV subgenomic DNA (i.e., pFKI389-NS3-3′ DNA) was coexpressed with increasing amounts of Flag-tagged human wild-type PKR cDNA (Fig. 2B), expression of both NS3 (top panel) and NS5A proteins (second from the top panel) was suppressed in a PKR-dependent manner (third panel from the top). We also noticed that expression of Flag-tagged wild-type PKR was accompanied by an induction of endogenous eIF-2α phosphorylation (bottom panel), which was proportional to the amount of expressed PKR (third panel from the top), demonstrating that the transfected PKR was functional.
We also tested whether a higher expression of NS proteins was capable of antagonizing the inhibitory action of PKR. That is, we hypothesized that increased NS5A protein levels may relieve the inhibition of NS protein synthesis by blocking PKR, as reported earlier (20). To this end, the Flag-tagged wild-type PKR cDNA was coexpressed with a small (0.1 μg; lane 3) or large (1 μg; lane 4) amount of pFKI389-NS3-3′ DNA, and protein levels were detected by immunoblotting (Fig. 2C). We found that NS5A expression was slightly increased (∼3-fold) when a 10-fold-larger amount of the subgenomic DNA was used (top panel, lane 4). We also saw that PKR protein (second panel from the top) and activity levels, as judged by the endogenous eIF-2α phosphorylation levels (third panel from the top), were both unaffected by the larger amount of viral subgenomic DNA (compare lanes 3 and 4).
Therefore, the increase in NS5A protein in lane 4 was caused by the 10-fold-larger amount of the subgenomic DNA rather than by relief from a PKR-mediated translational block. The data do not, however, rule out the possibility that NS5A negatively regulates PKR activity, since a larger amount of NS5A may be needed to mediate this effect. Nevertheless, this result shows the strong inhibitory effects of PKR on NS protein synthesis (see below), since the induction of viral protein expression (∼3-fold, lane 4) was not proportional to the amount of subgenomic DNA (10-fold, lane 4) used for the expression of the viral proteins.
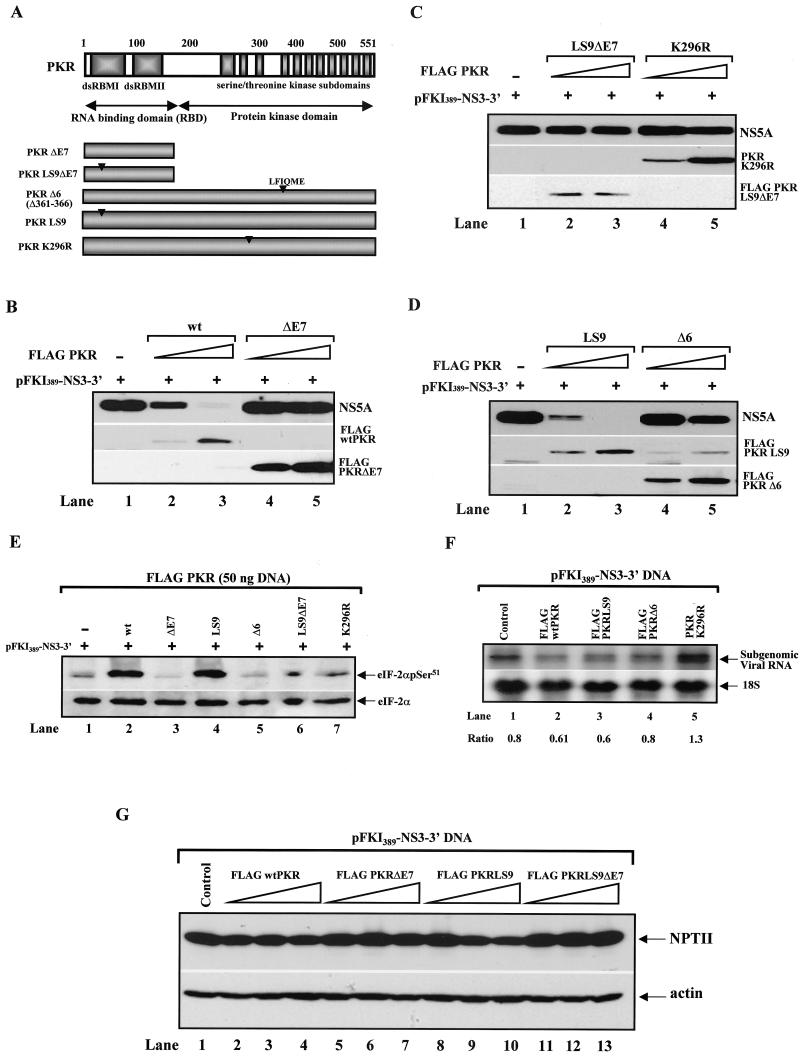
Catalytic activity of PKR is required for suppression of protein expression from the subgenomic HCV clone.
To examine the structural and functional requirements of PKR in viral protein synthesis, we used various catalytically inactive or dsRNA-binding-defective Flag-tagged PKR mutants, which are shown in Fig. 3A, in coexpression assays with the subgenomic HCV DNA. These mutants of PKR were (i) PKRΔE7, a 21-kDa protein, a product of alternative splicing of exon 7 of human PKR with dominant-negative functions (36) (Fig. 3B, lanes 4 and 5); (ii) PKRLS9ΔE7, PKRΔE7 bearing the LS9 mutation (Ala66/Ala68 to Glyc66/Pro68) (22), which completely abolishes binding to dsRNA (36) (Fig. 3C, lanes 2 and 3); (iii) PKRK296R, a catalytically defective mutant with substitution of the invariant Lys296 to Arg (27) (Fig. 3C, lanes 4 and 5); (iv) PKRLS9, an RNA-binding-defective mutant bearing the LS9 mutation (22) (Fig. 3D, lanes 2 and 3); and (v) PKRΔ6, a catalytically defective and dominant-negative mutant of human PKR with a deletion of the Leu-Phe-Ile-Gln-Met-Glu residues between amino acids 361 and 366 (32) (Fig. 3D, lanes 4 and 5).
FIG. 3.
Structure-function analysis of PKR in inhibition of HCV replicon protein synthesis. (A) Schematic representation of the functional domains of wild-type (wt) PKR and various PKR mutants. The N-terminal domain of PKR contains the two dsRNA-binding motifs (dsRBMs) that are essential for dsRNA binding, whereas the C terminus contains the 11 catalytic subdomains that are highly conserved among all serine/threonine kinases and mediate the enzymatic activity. PKRΔE7, an alternatively spliced form of PKR lacking the kinase domain; PKRΔE7LS9, PKRΔE7 containing the LS9 mutation, which completely abolishes dsRNA-binding activity; PKRΔ6, a catalytically inactive and dominant-negative PKR mutant with a 6-amino-acid deletion between amino acids 361 and 366 of human PKR; PKRLS9, a dsRNA-binding-defective mutant of human PKR; PKRK296R, a catalytically inactive form of PKR with a mutation of lysine (K) 296 to arginine (R). (B to D) Control of viral protein synthesis by the PKR mutants. The vaccinia virus/T7 virus system was used to express transiently 1 μg of the subgenomic HCV DNA pFKI389-NS3-3′ in Huh7 cells in the absence (lane 1) or presence of 0.010 μg (lanes 2 and 4) or 0.050 μg (lanes 3 and 5) of the indicated forms of PKR. Protein extracts (50 μg) were used for the immunodetection of NS5A (top panel) and various forms of Flag-tagged PKR (middle and bottom panels). Note that PKRK296R (C, middle panel) and Flag-PKRΔ6 levels (D, bottom panel) were also detected with the anti-human PKR MAb (clone F9), which does not recognize the LS9 mutation of PKR (41). (E) Regulation of eIF-2α phosphorylation by various forms of PKR in Huh7 cells. Protein extracts (50 μg) from Huh7 cells treated with recombinant vaccinia virus/T7 virus to express 1 μg of pFKI389-NS3-3′ DNA and 0.050 μg of each PKR cDNA (as described for B to D) were analyzed by immunoblot analysis for serine 51 phosphorylation of eIF-2α with a phosphoserine-specific antibody (top panel). The protein extracts were normalized to endogenous total eIF-2α protein levels by immunoblot analysis with a rabbit polyclonal antibody specific to human protein (bottom panel). (F) Regulation of subgenomic HCV RNA levels in Huh7 cells expressing various PKR forms. Total RNA (10 μg) from Huh7 cells treated with recombinant vaccinia virus/T7 virus to express 1 μg of pFKI389-NS3-3′ and 0.050 μg of each PKR cDNA (as described for B to D) were subjected to Northern blot analysis for detection of viral and 18S RNA expression as described for Fig. 1A. The ratios of viral to 18S RNA from the radioactive bands are indicated. (G) Control of NPTII protein expression by wild-type PKR and PKR mutants. With the recombinant vaccinia virus/T7 virus system, Huh7 cells were treated to express 1 μg of the pFKI389-NS3-3′ DNA alone (lane 1) or in the presence of 0.010 μg (lanes 2, 5, 8, and 11), 0.050 μg (lanes 3, 6, 9, and 12), or 0.1 μg (lanes 4, 7, 10, and 13) of Flag-tagged wild-type PKR cDNA (lanes 2 to 4), Flag-PKRΔE7 cDNA (lanes 5 to 7), Flag-PKRLS9 cDNA (lanes 8 to 10), or Flag-PKRLS9ΔE7 cDNA (lanes 11 to 13). Protein extracts (50 μg) were subjected to immunoblot analysis with a rabbit polyclonal anti-NPTII antibody and normalized by immunoblotting with antiactin MAb.
PKR protein levels were detected by immunoblotting with either an anti-Flag antibody (Fig. 3B, middle and bottom panels; Fig. 3C, bottom panel; Fig. 3D, middle panel) or anti-human PKR monoclonal antibody (Fig. 3C, middle panel; Fig. 3D, bottom panel). On the other hand, viral protein synthesis was monitored by immunoblotting with anti-NS5A antibody (Fig. 3B to D, top panels). We found that similar to wild-type PKR (Fig. 3B, top panel, lanes 2 and 3), expression of PKRLS9 strongly inhibited NS5A protein expression (Fig. 3D, top panel, lanes 2 and 3). Contrary to this, expression of PKRΔ6 (Fig. 3D, lanes 4 and 5), PKRK296R (Fig. 3C, lanes 4 and 5), PKRΔE7 (Fig. 3B, lanes 4 and 5), or PKRLS9ΔE7 (Fig. 3C, lanes 2 and 3) did not significantly affect NS5A protein levels.
Immunoblot analysis for the detection of endogenous eIF-2α phosphorylation levels (Fig. 3E) demonstrated that wild-type PKR and PKRLS9 induced eIF-2α phosphorylation to equal levels (top panel, compare lanes 2 and 4). Phosphorylation of eIF-2α in cells expressing PKRΔE7 (lane 3) or PKRΔ6 (lane 5) was further diminished compared to that in mock-transfected cells (lane 1) due to the strong dominant-negative effects of these PKR mutants. On the other hand, eIF-2α phosphorylation levels were unaffected in PKRΔE7LS9 (lane 6) or PKRK296R (lane 7). Furthermore, Northern blot analysis showed no significant differences in viral RNA expression levels in cells transfected with the various forms of PKR (Fig. 3F), suggesting a translational and/or posttranslational function of the kinase in viral gene expression. We concluded that the catalytic activity of PKR is both necessary and sufficient, as judged from the function of PKRLS9, to inhibit NS protein synthesis.
Since NPTII protein synthesis from the subgenomic clone is HCV IRES dependent, we were interested in examining whether expression of this protein was affected by PKR (Fig. 3G). When protein extracts from Huh7 cells transiently expressing various forms of PKR and the subgenomic HCV DNA were subjected to immunoblot analysis, we found that, unlike that of the viral proteins, expression of NPTII (top panel) was resistant to the catalytically active forms of PKR (Flag-tagged wild-type PKR, lanes 2 to 4; Flag-PKRLS9, lanes 8 to 10). These results provided strong evidence for differential regulation of NS and NPTII protein expression by PKR.
Inhibition of viral protein synthesis by PKR is independent of eIF-2α phosphorylation.
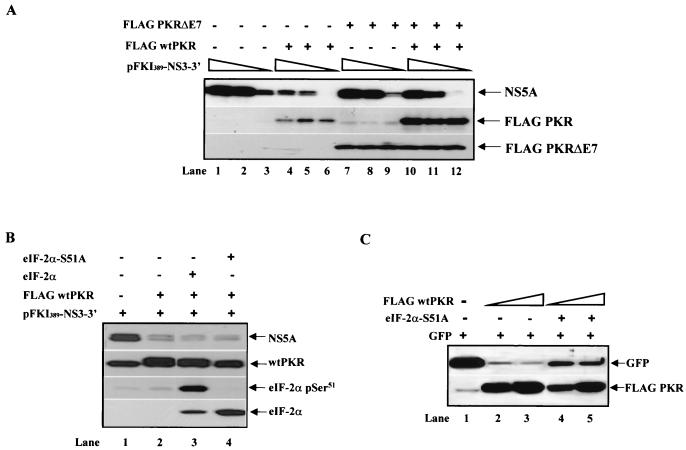
To better understand the molecular functions of PKR in NS protein synthesis, we next tested whether the presence of the dominant-negative PKRΔE7 (36) was capable of rescuing the inhibitory effects of PKR on NS5A protein expression (Fig. 4A). When various amounts of the subgenomic viral DNA were expressed in the absence (lanes 1 to 3) or presence of Flag-tagged wild-type PKR (lanes 4 to 6) or Flag-PKRΔE7 (lanes 7 to 9) or in the presence of both Flag-tagged wild-type PKR and Flag-tagged PKRΔE7 (lanes 10 to 12), we found that the inhibition of NS5A protein synthesis by wild-type PKR (top panel, lanes 4 to 6) was completely reversed by the coexpression of PKRΔE7 (lanes 10 to 12). The upregulation of endogenous PKR (middle panel, lanes 7 to 9) or exogenous wild-type PKR (lanes 10 to 12) is explained by the dominant-negative function of PKRΔE7. Specifically, we previously showed that ectopic expression of PKRΔE7 enhances the protein synthesis of endogenous PKR or transfected PKR by blocking endogenous eIF-2α phosphorylation (36).
FIG. 4.
Control of subgenomic HCV protein synthesis by PKR in Huh7 cells is independent of eIF-2α phosphorylation. (A) Reversal of PKR-mediated inhibition of NS5A protein synthesis by the dominant-negative PKRΔE7. Huh7 cells were treated with recombinant vaccinia virus/T7 virus to express transiently 1 μg (lanes 1, 4, 7, and 10), 0.5 μg (lanes 2, 5, 8, and 11), or 0.1 μg (lanes 3, 6, 9, and 12) of pFKI389-NS3-3′ DNA in the absence (lanes 1 to 3) or presence of 0.010 μg of Flag-tagged wild-type (wt) PKR cDNA (lanes 4 to 6), 0.1 μg of Flag-PKRΔE7 cDNA (lanes 7 to 9), or 0.010 μg of Flag-tagged wild-type PKR cDNA and 0.1 μg of Flag-PKRΔE7 cDNA (lanes 10 to 12). Expression of viral proteins was monitored by immunoblot analysis with anti-NS5A MAb (top panel), whereas expression levels of wild-type PKR (middle panel) and PKRΔE7 (bottom panel) were monitored by immunoblotting with anti-human PKR MAb (F9 clone) and anti-Flag MAb, respectively. Note the upregulation of endogenous PKR in Huh7 cells transfected with Flag-PKRΔE7 (middle panel, lanes 7 to 9), as reported earlier (36). The levels of endogenous human PKR in cells transfected only with the subgenomic viral DNA (middle panel, lanes 1 to 3) were detected after longer exposure (data not shown). (B) Suppression of PKR-mediated viral protein synthesis is not rescued by the presence of the dominant-negative eIF-2α S51A mutant. Huh7 cells were treated with recombinant vaccinia virus/T7 virus and 1 μg of pFKI389-NS3-3′ DNA (lanes 1 to 4), 0.01 μg of Flag-tagged wild-type PKR cDNA (lanes 2 to 4), 0.5 μg of wild-type eIF-2α cDNA (lane 3), or 0.5 μg of eIF-2α S51A cDNA (lane 4). Protein extracts (50 μg) were subjected to immunoblot analysis for detection of NS5A (top panel), PKR (second panel from the top), eIF-2α serine 51 phosphorylation (third panel from the top), or eIF-2α protein expression (bottom panel). Note that the mouse anti-eIF-2α MAb (bottom panel) does not cross-react with endogenous human eIF-2α (lanes 1 and 2). (C) Dominant-negative effects of the mouse eIF-2α S51A mutant on nonviral protein synthesis in Huh7 cells. With the vaccinia virus/T7 virus system, Huh7 cells were treated to express 0.1 μg of GFP DNA and 0.5 μg (lanes 2 and 4) or 0.9 μg (lanes 3 and 5) of Flag-tagged wild-type PKR cDNA in the absence (lanes 1 to 3) or presence (lanes 4 and 5) of 1 μg of eIF2α S51A cDNA. Protein extracts (50 μg) were subjected to immunoblot analysis with anti-GFP antibody (top panel) or anti-Flag MAb (bottom panel).
To obtain more direct evidence for a role of eIF-2α phosphorylation, we examined whether expression of the dominant-negative phosphorylation mutant Ser51-Ala (eIF-2α S51A [12]) was capable of rescuing the inhibitory effects of PKR on NS protein expression. To this end, Flag-tagged wild-type PKR cDNA and subgenomic HCV DNA were coexpressed in the presence of either mouse wild-type eIF-2α (Fig. 4B, lane 3) or the mouse eIF-2α S51A mutant (lane 4). Immunoblot analysis with anti-NS5A antibody revealed that the eIF-2α S51A mutant was unable to reverse PKR-mediated suppression of NS5A protein expression (top panel, compare lane 2 with lane 4). Immunoblot analysis with eIF-2α phosphoserine 51-specific antibodies showed the high levels of phosphorylation of exogenous wild-type eIF-2α by wild-type PKR (third panel from the top, lane 3) as well as the dominant-negative function of eIF-2α S51A over endogenous eIF-2α (compare lane 4 with lane 2).
Expression of the transfected eIF-2α was detected by immunoblot analysis with a monoclonal antibody that specifically recognizes the mouse but not the endogenous human protein (bottom panel, lanes 3 and 4). To find out whether the mouse eIF-2α S51A indeed functions as a dominant-negative in human cells, we assessed the expression of a nonviral gene in Huh7 cells in the presence of the eIF-2α mutant protein (Fig. 4C). It was previously shown that eIF-2α S51A improves translation of plasmid-derived mRNAs without affecting global protein synthesis (29). Based on this, we expressed GFP in Huh7 cells in the presence of wild-type PKR alone or wild-type PKR and eIF2A S51A cDNAs (Fig. 4C). We found that eIF-2α S51A was capable of relieving the translational repression of GFP by wild-type PKR (compare lanes 4 and 5 with lanes 2 and 3), demonstrating its dominant-negative function in our system.
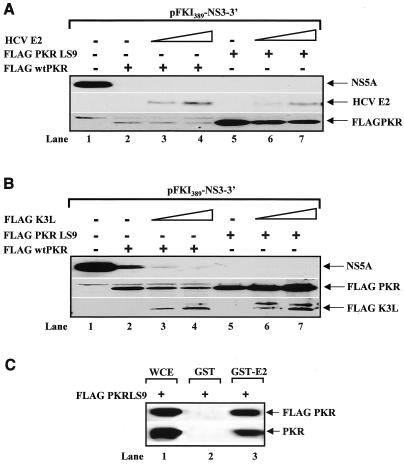
Then we tested whether the inhibitory effects of PKR were rescued by the HCV E2 (Fig. 5A) or the vaccinia virus K3L (Fig. 5B), since both proteins function as pseudosubstrate inhibitors of the kinase (8, 30, 57). We found that neither E2 (Fig. 5A, middle panel) nor K3L (Fig. 5B, bottom panel) was capable of blocking the inhibitory functions of either Flag-tagged wild-type PKR (Fig. 5A and B, top panels, compare lane 1 with lanes 2 to 4). Similar results were obtained with the catalytically active Flag-PKRLS9 (Fig. 5A and B, top panels, lanes 5 to 7). To find out whether the LS9 mutation had an effect on PKR interaction with E2 (57), we performed pulldown assays with glutathione S-transferase (GST)-E2 and Flag-PKRLS9 (Fig. 5C). We found that E2 interacted with PKRLS9 in vitro, suggesting that the lack of an effect in Fig. 5A was not due to the lack of an interaction between the two proteins. Together, these data supported the notion that suppression of HCV protein synthesis by PKR proceeds through a mechanism that does not involve eIF-2α phosphorylation.
FIG. 5.
Suppression of PKR-mediated viral protein synthesis is not rescued by expression of the pseudosubstrates E2 (A) and K3L (B) protein. Huh7 cells were treated with vaccinia virus/T7 virus to express 1 μg of pFKI389-NS3-3′ DNA (lanes 1 to 7), 0.05 μg of Flag-tagged wild-type (wt) PKR cDNA (lanes 2 to 4), 0.050 μg of Flag-PKRLS9 cDNA (lanes 5 to 7), 0.5 μg of E2 DNA (B, lanes 3 and 6), 0.95 μg of E2 DNA (B, lanes 4 and 7), 0.5 μg of Flag-K3L DNA (C, lanes 3 and 6), and 0.95 μg of Flag-K3L DNA (C, lanes 4 and 7), and 50 μg of protein extracts was used for immunoblotting with anti-NS5A MAb (C and D, top panel), anti-E2 MAb (C, middle panel), or anti-Flag MAb (C, bottom panel; D, middle and bottom panels). (C) E2 interacts with PKRLS9 in vitro. GST and GST-E2 proteins were expressed and purified as previously described (64). Equal amounts (1 μg) of GST fusion proteins were mixed with 500 μg of Huh7 protein extracts expressing Flag-PKRLS9. After pulldown with glutathione-Sepharose (64), the proteins were subjected to immunoblot analysis with anti-Flag antibody (top panel) or anti-human PKR MAb (clone E8). Lane 1, 50 μg of protein from Huh7 whole-cell extracts (WCE) used in the pulldown assays.
PKR does not significantly modulate NS protein stability.
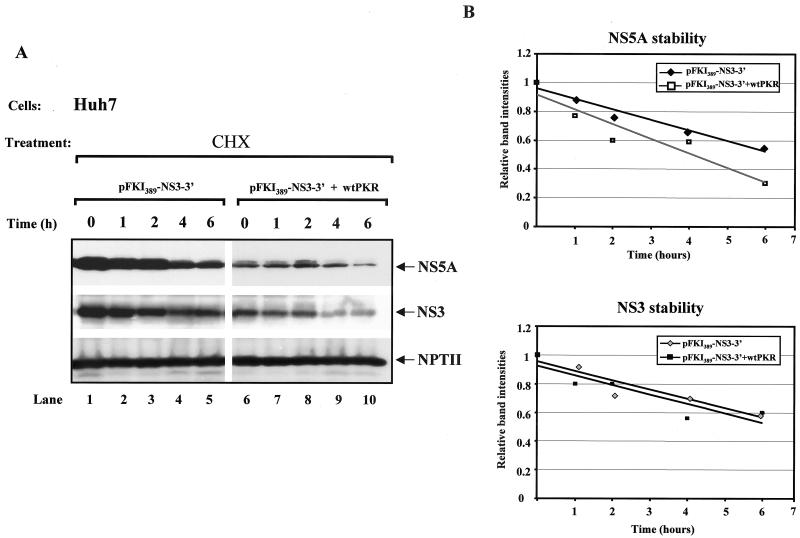
Although the above data argue for a translational role of PKR in NS protein synthesis, the possibility for a posttranslational role of the kinase in regulating protein stability was also examined. To this end, the subgenomic HCV DNA was transiently expressed in Huh7 cells in the absence or presence of Flag-tagged wild-type PKR, and protein stability was assessed by immunoblot analysis of protein extracts from untreated and cycloheximide-treated cells (Fig. 6). We found that both the NS5A (Fig. 6A, top panel) and NS3 (Fig. 6A, middle panel) proteins were susceptible to cycloheximide treatment (lanes 1 to 5). Although expression of Flag-tagged wild-type PKR reduced NS5A and NS3 protein expression (compare lane 6 with lane 1), it did not significantly change the rates of degradation of the viral proteins (compare lanes 6 to 10 with lanes 1 to 5).
FIG. 6.
Stability of NS proteins is not affected by wild-type (wt) PKR. (A) Huh7 cells were treated with recombinant vaccinia virus/T7 virus to express 1 μg of pFKI389-NS3-3′ DNA alone (lanes 1 to 5) or in the presence of 0.010 μg of Flag-tagged wild-type PKR cDNA (lanes 6 to 10). Sixteen hours after transfection, cells were treated with 50 μg of cycloheximide (CHX)/ml for the indicated times. Immunoblot analysis for NS5A (top panel), NS3 (middle panel), and NPTII (bottom panel) expression levels was performed with 50 μg of protein extracts. (B) The protein bands were quantified with NIH Image 1.54 software, and the average values from four separate experiments were plotted versus the time (hours) of cycloheximide treatment.
However, NPTII protein was resistant to the inhibitory effects of Flag-tagged wild-type PKR (compare lane 6 with lane 1) and cycloheximide treatment in either the absence or presence of Flag-tagged wild-type PKR (Fig. 6A, bottom panel, compare lanes 1 to 10 with lanes 1 to 5). The apparent long half-life of NPTII most likely accounts for its slow decrease in replicon cells in response to IFN-α (Fig. 1E). Quantification of the degradation rates of the viral proteins from four separate experiments showed a modest effect of PKR on NS5A stability (Fig. 6B, top panel), which, however, cannot account for the strong inhibitory effects of the kinase on NS5A expression (compare lanes 1 and 6). On the other hand, NS3 protein stability was unaffected by wild-type PKR (Fig. 6B). These data favor a translational role of PKR in NS protein synthesis.
Wild-type PKR induces HCV IRES-dependent translation.
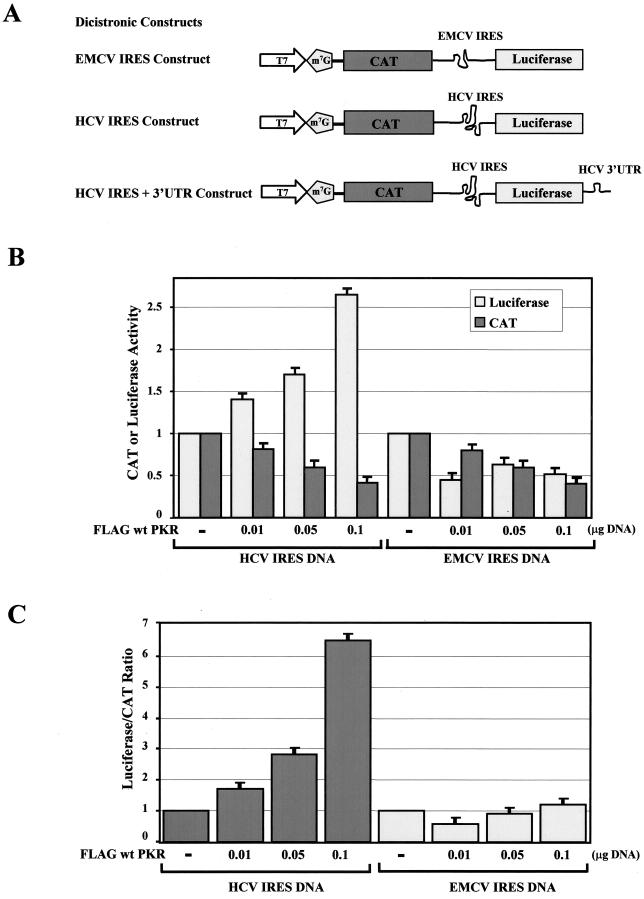
The presence of two different IRESs within the subgenomic HCV clone and the differential expression of genes under their control in the presence of active PKR prompted us to examine whether their activities are modulated by the kinase. To address this possibility, we used dicistronic constructs bearing either the HCV or EMCV IRES between the protein-coding regions of the bacterial CAT and firefly luciferase genes (54) (Fig. 7A). IRES-dependent translation was assessed in Huh7 cells with the vaccinia virus/T7 virus system because mRNAs produced by the T7 RNA polymerase are efficiently capped (16). Transient expression of the dicistronic constructs in Huh7 cells decreased cap-dependent translation of the CAT gene in the presence of increasing amounts of Flag-tagged wild-type PKR (Fig. 7B).
FIG. 7.
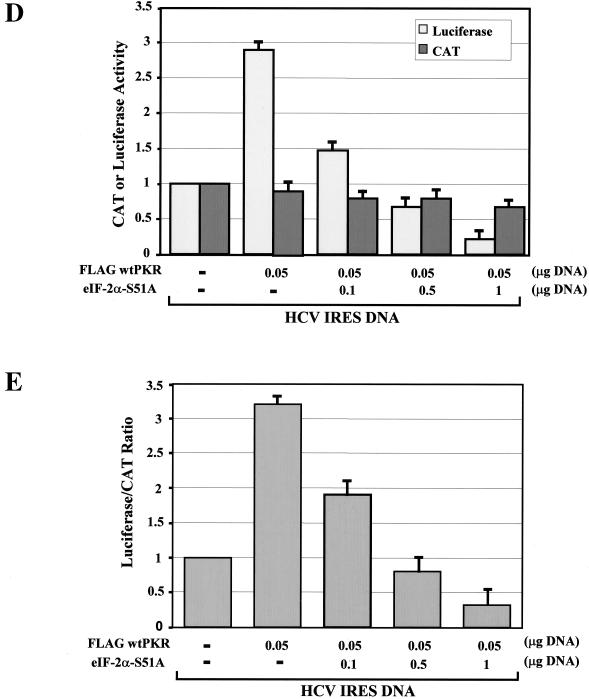
Wild-type PKR induces HCV IRES activity. (A) Schematic representations of the dicistronic IRES constructs used in this study. The EMCV or HCV IRES is positioned between the protein-coding regions for the CAT and luciferase genes, whereas the HCV 3′ UTR is located downstream of the luciferase gene. (B and C) Effects of wild-type PKR on HCV and EMCV IRES-dependent gene expression. With the vaccinia virus/T7 virus system, Huh7 cells transiently expressed 0.1 μg of HCV IRES or EMCV IRES dicistronic DNA alone (54) or in the presence of increasing amounts of Flag-tagged wild-type PKR cDNA (0.010 to 0.1 μg). Eighteen hours posttransfection, the actual enzymatic activities of CAT and luciferase were measured in normalized protein extracts. The actual levels of CAT and luciferase activity and the relative luciferase/CAT activity levels are shown in B and C, respectively. The values represent the averages of four separate experiments, each performed in triplicate. (D and E) Effect of eIF-2α S51A mutant on HCV IRES activity. Huh7 cells were treated with the vaccinia virus/T7 virus system to express 0.1 μg of HCV IRES DNA alone or in the presence of 0.050 μg of Flag-tagged wild-type PKR cDNA or in the presence of 0.050 μg of Flag-tagged wild-type PKR cDNA and increasing amounts of eIF-2α S51A cDNA (0.01 to 0.1 μg). The actual CAT and luciferase activity levels are shown in D, and the relative luciferase/CAT levels are shown in E. The values represent the averages of four separate experiments, each performed in triplicate.
Interestingly, translation of the luciferase gene from the HCV IRES was highly induced by the presence of Flag-tagged wild-type PKR (Fig. 7B). Contrary to this, EMCV IRES-driven translation was reduced when cells were transfected with the same amount of Flag-tagged wild-type PKR cDNA (Fig. 7B). When the luciferase activity was normalized to the CAT activity (Fig. 7C), we observed that HCV IRES-driven activity was induced up to sixfold by wild-type PKR, whereas EMCV IRES-driven activity remained unchanged. These findings provided strong evidence for differential regulation of these IRESs by wild-type PKR.
To investigate the role of eIF-2α phosphorylation in these events, we tested whether induction of HCV IRES activity by wild-type PKR was reversed by the expression of the eIF-2α S51A mutant. Transient expression of the HCV IRES dicistronic construct in the presence of Flag-tagged wild-type PKR and increasing amounts of eIF-2α S51A mutant resulted in the inhibition of PKR-induced IRES activity, which was proportional to the amount of transfected eIF-2α S51A cDNA (Fig. 7D and E). Taken together, these data suggested that PKR-mediated induction of HCV IRES is enhanced by eIF-2α phosphorylation.
Induction of HCV IRES requires the catalytic activity of PKR and is mitigated by the HCV 3′ UTR.
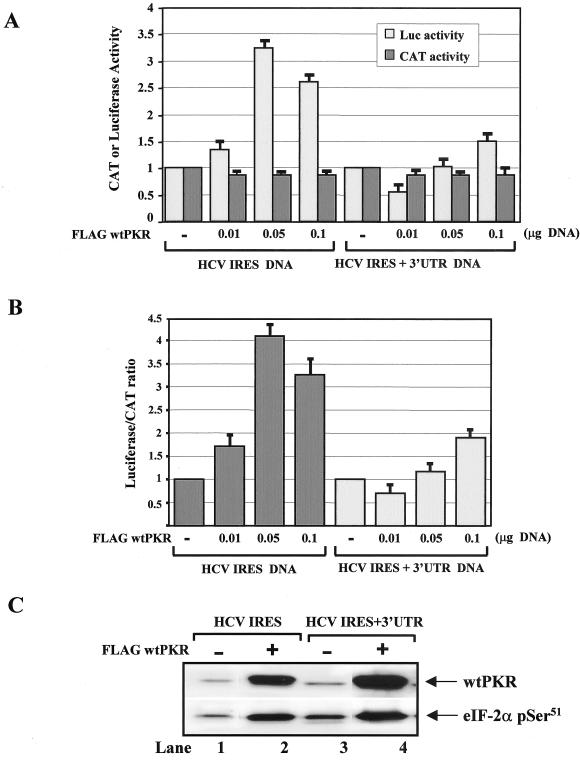
Since induction of HCV IRES activity by wild-type PKR was not seen in our experiments with the subgenomic clone (Fig. 3G), we hypothesized that the presence of other viral sequences may affect HCV IRES function. We thus tested whether the presence of the viral 3′ UTR, which was shown to modulate viral gene translation (25, 39), had an effect on the PKR-mediated induction of HCV IRES activity. To this end, Huh7 cells were treated with recombinant vaccinia virus/T7 virus to express HCV IRES dicistronic DNA that either lacks or contains the 3′ UTR in the presence of increasing amounts of Flag-tagged wild-type PKR cDNA (Fig. 8A).
FIG. 8.
Induction of HCV IRES activity by PKR requires catalytically active kinase and is controlled by the viral 3′ UTR. (A and B) Control of HCV IRES activity by the 3′ UTR. Huh7 cells were treated with recombinant vaccinia virus/T7 virus to express 0.1 μg of HCV-IRES dicistronic DNA that lacks or bears the 3′ UTR in the presence of increasing amounts (0.01 to 0.1 μg) of Flag-tagged wild-type PKR cDNA (A) or Flag-PKRLS9 cDNA (D). Eighteen hours later, CAT and luciferase activities were assessed (A and D), and the relative luciferase/CAT activity levels were quantified (B and E). The values represent the averages of four separate experiments, each performed in triplicate. (C) Protein extracts (50 μg) from cells expressing 0.1 μg of HCV IRES DNA alone (lane 1), 0.1 μg of HCV IRES DNA and 0.1 μg of Flag-tagged wild-type PKR cDNA (lane 2), 0.1 μg of HCV IRES and 3′ UTR DNA alone (lane 3), or 0.1 μg of HCV IRES and 3′ UTR DNA and 0.1 μg of Flag-tagged wild-type PKR cDNA (lane 4) were immunoblotted for PKR (top panel) and eIF-2α phosphorylation (bottom panel) levels with anti-human PKR MAb (F9 clone) and anti-eIF-2α phosphoserine 51 antibody, respectively.
We found that the kinetics of induction of HCV IRES activity by increasing amounts of wild-type PKR in this construct were different from those observed with the other HCV IRES construct shown in Fig. 7B. This can be explained by the differences in the backbone DNA of the two plasmids bearing the same dicistronic HCV IRES. Also, we noticed that inhibition of cap-dependent translation indicated by the CAT activity levels (Fig. 8A) was not as strong as with the HCV IRES construct in Fig. 7B. Since the second HCV IRES dicistronic construct contains the bovine growth hormone polyadenylation signal in the 5′ UTR, it is possible that the polyadenylated mRNAs interfere with cap-dependent translation in our system.
Interestingly, the presence of the viral 3′ UTR compromised the ability of wild-type PKR to induce HCV IRES-driven translation (Fig. 8A and B). In fact, a 10-fold-larger amount of Flag-tagged wild-type PKR cDNA was required to induce IRES activity in the presence of the 3′ UTR to equal the levels of IRES activity in the absence of the 3′ UTR (Fig. 8A and B). Immunoblot analysis showed that induction of eIF-2α phosphorylation by wild-type PKR was not diminished by the presence of the 3′ UTR (Fig. 8C, lower panel), suggesting that inhibition of IRES activity by the 3′ UTR may not involve eIF-2α phosphorylation.
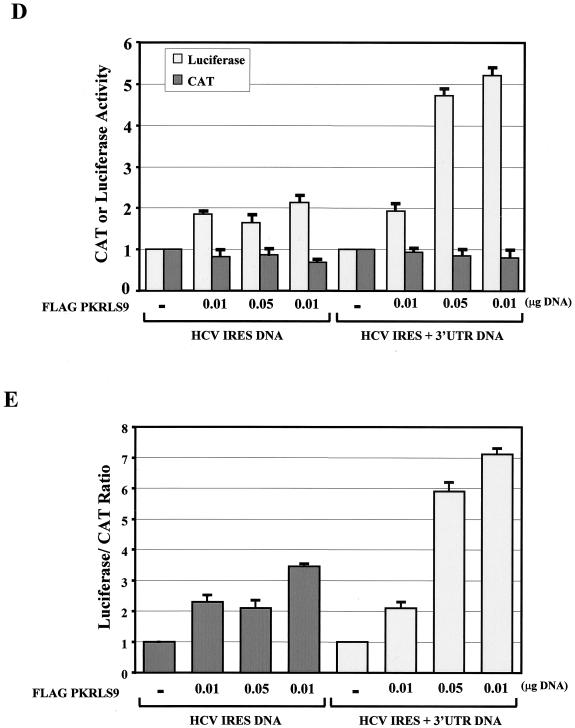
To get better insight into the molecular functions of IRES-dependent translation by PKR, we tested whether HCV IRES activity is induced by the catalytically active PKRLS9 and whether this function is controlled by the 3′ UTR. We found that Flag-PKRLS9 was able to induce HCV IRES activity from the dicistronic construct lacking the 3′ UTR (Fig. 8D and E), suggesting that the catalytic activity of PKR is both necessary and sufficient to mediate this stimulatory effect on IRES activity. Interestingly, the presence of the 3′ UTR did not suppress but rather enhanced the induction of HCV IRES activity by PKRLS9 (Fig. 8D and E). The data suggested that the inhibitory effects of the 3′ UTR on wild-type PKR require an intact dsRNA-binding domain of the kinase.
DISCUSSION
The primary scope of our study was to investigate the possible role of PKR and eIF-2α phosphorylation in the replication of the subgenomic HCV clone originally described by Lohmann and colleagues (38). With this prototype replicon, we found that expression of NS and PKR proteins and eIF-2α phosphorylation levels were variably regulated during the proliferation of Huh7 cells (Fig. 1C and D). In line with these findings, PKR activity was previously shown to be modulated in proliferating cells in a cell cycle-dependent manner (61, 66), whereas replication of an HCV subgenomic clone in Huh7 cells has been reported to be affected by cell density (23). Since our experiments were performed with unsynchronized Huh7 cells plated at low density, it is possible that eIF-2α phosphorylation levels are dependent on the plating efficiency and confluency of the cells.
We show that Huh7 cells contain PKR that is responsive to activation by autophosphorylation (Fig. 2A). However, eIF-2α phosphorylation levels 24 h after IFN-α treatment in both control and replicon cells was inversely proportional to PKR protein levels (Fig. 1C, lane 10; Fig. 1D, lanes 4 and 11), indicating the existence of PKR-independent pathways that target eIF-2α phosphorylation in proliferating Huh7 cells. Such pathways may involve the activities of PERK and/or GCN2 kinases, which have been demonstrated to play an important role in host protein synthesis by phosphorylating eIF-2α (11). However, our data do not exclude the possibility of action of a phosphatase that dephosphorylates eIF-2α, whose expression and/or activity is affected by cell proliferation and IFN-α treatment.
When replicon cells were treated with IFN-α, we observed a positive correlation between the inhibition of viral protein synthesis and upregulation of PKR (Fig. 1C and D). We also noticed that PKR protein levels were more highly induced by IFN-α in parental control cells than in replicon cells (Fig. 1D). Although it is not presently clear how the viral replicon regulates the induction of PKR by IFN-α, we hypothesize that activation of the Jak-Stat pathway and transcriptional induction of the pkr gene may be negatively regulated by the NS proteins, based on the previous observation that the HCV polyprotein can impair the Jak-Stat pathway (24). Interestingly, IFN-α treatment was accompanied by an overall induction of eIF-2α phosphorylation, which was higher in parental than in replicon Huh7 cells (Fig. 1D). Although these results implied a positive role of eIF-2α phosphorylation in the inhibition of NS protein synthesis in proliferating cells, in serum-starved replicon cells we found that suppression of NS protein expression did not require the induction of eIF-2α phosphorylation (Fig. 1F).
The experiments with replicon cells indicated an inverse correlation between PKR and NS protein expression, which could be explained either by the translation-inhibitory functions of PKR or by an ability of PKR to inhibit viral RNA replication. The former possibility was further addressed in transient-expression assays of various forms of PKR with the subgenomic HCV clone. These experiments demonstrated (i) the direct function of PKR in the suppression of NS protein synthesis and (ii) the essential role of the catalytic activity of PKR in this process (Fig. 2 and 3). Interestingly, NS protein expression was suppressed by PKRLS9 (Fig. 3D), which is defective in dsRNA binding but otherwise catalytically active (Fig. 3E). Our experiments with S. cerevisiae also showed that PKRLS9 was capable of inducing eIF-2α phosphorylation (data not shown), providing evidence for a distinct mode of activation of this PKR mutant that is independent of binding to dsRNA.
It was previously shown that in the inactive form, the N-terminal dsRNA-binding domain of PKR folds over the C-terminal kinase domain, keeping it in a “closed” conformation. Binding to dsRNA induces PKR homodimerization and exposes the kinase domain, resulting in activation by autophosphorylation (42). It is possible, then, that the LS9 mutation induces conformational changes that maintain PKR in an “opened” and constitutively active state. Although this interpretation still waits for the crystal structure of full-length PKR to be verified, our data clearly demonstrate that PKRLS9 is a catalytically active form of PKR capable of suppressing NS protein synthesis. Curiously, induction of eIF-2α phosphorylation by catalytically active PKR appears to be dispensable for the inhibition of NS protein synthesis from the subgenomic clone. That is, expression of the pseudosubstrate PKR inhibitor E2 (Fig. 5A) or K3L (Fig. 5B) or expression of the dominant-negative eIF-2α S51A mutant (Fig. 4B) was unable to rescue PKR-mediated inhibition of NS protein synthesis from the subgenomic clone.
Unlike that of NS proteins, expression of the NPTII protein was resistant to PKR activation (Fig. 3G). Since expression of NS proteins from the subgenomic clone is driven by the EMCV IRES and NPTII expression is driven by the HCV IRES, a plausible interpretation was that catalytically active forms of PKR differentially regulate the activities of these IRESs. We tested both hypotheses by assessing IRES-driven translation from dicistronic constructs containing either the HCV or EMCV IRES. We found that catalytically active PKR was capable of inducing the HCV IRES and inhibiting the EMCV IRES activity (Fig. 7B and C). Interestingly, induction of the HCV IRES by wild-type PKR was blocked by overexpression of the dominant-negative eIF-2α S51A mutant (Fig. 7D), suggesting that eIF-2α phosphorylation is implicated in this process.
Significantly, induction of HCV IRES activity by wild-type PKR was mitigated by the presence of the 3′ UTR (Fig. 8A), which is in line with a negative regulation of the HCV IRES by the 3′ UTR reported earlier (39). This result is consistent with the lack of induction of NPTII protein synthesis from the subgenomic clone by the expression of wild-type PKR (Fig. 3G). Most intriguing, however, was the finding that inhibition of PKR-mediated induction of HCV IRES by the 3′ UTR did not require reduction of eIF-2α phosphorylation (Fig. 8C), nor was the 3′ UTR able to block the induction of HCV IRES activity by PKRLS9 (Fig. 8D and E). These data suggested that the 3′ UTR probably functions downstream of eIF-2α phosphorylation and requires the dsRNA-binding properties of PKR.
One possible interpretation of our findings is that PKR induces the phosphorylation of an IRES trans-acting factor that positively regulates HCV IRES. In fact, the existence of an IRES trans-acting factor which mediates translation of the foot-and-mouth disease virus IRES has been demonstrated (45). Another interpretation is a possible competition between the 5′ cap and HCV IRES-dependent translation from dicistronic constructs for the recruitment of an initiation factor(s) that is utilized in both mechanisms. Recent biochemical and biological studies have shown the direct binding of the 40S ribosomal subunit at the site of the initiator AUG and the eIF3 through multiple and specific intermolecular contacts (7, 51, 52). Formation of the IRES/40S complex does not require additional translation initiation factors such as eIF1, eIF1A, eIF4A, eIF4B, eIF4E, and eIF4G (43, 52). A number of in vitro studies suggested that several proteins, including both conventional translational initiation factors such as eIF3 (7, 51) and noncanonical translation initiation factors such as La (2) and PTB (1, 3, 25), may stimulate HCV translation. Recently, eIF2Bγ and eIF2γ have been identified as cofactors in HCV IRES-mediated translation (33).
Our data show that the HCV IRES is more resistant to PKR-mediated translation inhibition than the EMCV IRES. The inhibition of EMCV IRES activity in dicistronic constructs by active PKR (Fig. 7B) is in agreement with a previous finding showing that induction of eIF-2α phosphorylation by endoplasmic reticulum stress negatively regulates EMCV IRES function (62). However, the inhibition of EMCV IRES function in the HCV subgenomic clone by active PKR is independent of eIF-2α phosphorylation (Fig. 4). Also, it is noteworthy that active PKR inhibits NS protein synthesis from the subgenomic clone (Fig. 2 and 3B) to a much higher degree than it inhibits translation of the luciferase gene from the dicistronic construct (Fig. 7B).
It is thus possible that the presence of viral RNA, such as the 3′ UTR, and/or the expression of the NS proteins amplifies the inhibitory effects of PKR on the EMCV IRES through a mechanism that does not require the induction of eIF-2α phosphorylation. It is also possible that the presence of the two IRESs within the subgenomic clone results in a competition for a translation factor(s) that is regulated by PKR in addition to eIF-2α. For example, a need for a translation factor that induces HCV IRES and inhibits EMCV IRES activity upon PKR activation could explain the functional differences of these IRESs when they are present together in the subgenomic clone and separately in the dicistronic constructs.
The physiological relevance of the control of HCV IRES activity by PKR in virus replication in vivo is not immediately clear because replicon cells represent an in vitro system. In the first instance, this result is inconsistent with the general notion of PKR as a negative regulator of viral protein synthesis and a mediator of the antiviral effects of IFN-α. Our hypothesis is that at the initial steps of the HCV life cycle, the presence of viral dsRNA or the core protein, which has recently been shown to function as an activator of the kinase (10), may induce the activity of PKR. Initially, activation of PKR may facilitate translation of the viral genes by enhancing IRES activity. At later stages of the viral life cycle, when sufficient amounts of the viral proteins have been produced, activation of PKR may be impaired in cells infected with viral quasispecies expressing NS5A and E2 proteins that are able to interact with and inhibit PKR (18, 57). Such a mechanism would be useful to the virus to maintain replication and bypass the destruction of infected cells by the prolonged activation of PKR (19).
Thus, in the case of HCV infection, the antiviral effects of PKR may be exerted at a level different from translation. For example, we show that the induction of PKR protein expression by IFN-α also coincides with the downregulation of viral RNA replication (Fig. 1) and that the viral 3′ UTR, which plays a crucial role in viral RNA replication (31, 65), functionally cross talks with PKR. These observations may provide a tentative, as yet unidentified link between the activation of PKR and inhibition of viral RNA replication.
Acknowledgments
We are grateful to R. Bartenschlager for pFKI389-NS3-3′ DNA and HCV replicon Huh7 cells, J. Dubuisson for anti-E2 antibodies, D. Taylor for GST-E2 and the E2 expression vector, K. F. Tsukiyama-Kohara for the pKIV vector, S. Pyronnet for the pcDNA3-CAT/Luc vector, A. Brasey (N. Sonenberg's lab) for rabbit polyclonal antibody to human eIF-2α, Boehringer Ingelheim Canada for anti-NS3 antibodies, K. Pantopoulos for critical reading of the manuscript and helpful suggestions, and members of our lab for helpful discussions.
This work was supported by a research grant from the Canadian Institutes for Health Research (CIHR) to A.E.K. and A.S. A.E.K. and M.L.L. are recipients of a CIHR scientist award and CIHR postdoctoral fellowship award, respectively.
REFERENCES
- 1.Ali, N., and A. Siddiqui. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 69:6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, N., and A. Siddiqui. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 94:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anwar, A., N. Ali, R. Tanveer, and A. Siddiqui. 2000. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 275:34231-34235. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R. F., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 5.Bisceglie, A. M., J. F. McHutchison, and C. M. Rice. 2002. New therapeutic strategies for hepatitis C. Hepatology 35:224-231. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Buratti, E. F., S. F. Tisminetzky, M. F. Zotti, and F. E. Baralle. 1998. Functional analysis of the interaction between HCV 5′ UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 26:3179-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, K., O. Elroy-Stein, B. Moss, and R. Jagus. 1993. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J. Biol. Chem. 268:12837-12842. [PubMed] [Google Scholar]
- 9.Chung, R. T., W. He, A. Saquib, A. M. Contreras, R. J. Xavier, A. Chawla, T. C. Wang, and E. V. Schmidt. 2001. Hepatitis C virus replication is directly inhibited by IFN-alpha in a full-length binary expression system. Proc. Natl. Acad. Sci. USA 98:9847-9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delhem, N. F., A. F. Sabile, R. F. Gajardo, P. F. Podevin, A. F. Abadie, M. A. Blaton, D. F. Kremsdorf, L. F. Beretta, and C. Brechot. 2001. Activation of the interferon-inducible protein kinase PKR by hepatocellular carcinoma derived-hepatitis C virus core protein. Oncogene 20:5836-5845. [DOI] [PubMed] [Google Scholar]
- 11.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. [DOI] [PubMed] [Google Scholar]
- 12.Donze, O., R. Jagus, A. E. Koromilas, J. W. Hershey, and N. Sonenberg. 1995. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 14:3828-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enomoto, N. F., I. F. Sakuma, Y. F. Asahina, M. F. Kurosaki, T. F. Murakami, C. F. Yamamoto, N. F. Izumi, F. F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enomoto, N. F., I. F. Sakuma, Y. F. Asahina, M. F. Kurosaki, T. F. Murakami, C. F. Yamamoto, Y. F. Ogura, N. F. Izumi, F. F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. [DOI] [PubMed] [Google Scholar]
- 15.Francois, C., G. Duverlie, D. Rebouillat, H. Khorsi, S. Castelain, H. E. Blum, A. Gatignol, C. Wychowski, D. Moradpour, and E. F. Meurs. 2000. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J. Virol. 74:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst, T. R., and B. Moss. 1989. Structure and stability of mRNA synthesized by vaccinia virus-encoded bacteriophage T7 RNA polymerase in mammalian cells. Importance of the 5′ untranslated leader. J. Mol. Biol. 206:333-348. [DOI] [PubMed] [Google Scholar]
- 17.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale, M., Jr., B. Kwieciszewski, M. Dossett, H. Nakao, and M. G. Katze. 1999. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J. Virol. 73:6506-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh, A. K., R. Steele, K. Meyer, R. Ray, and R. B. Ray. 1999. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J. Gen. Virol. 80:1179-1183. [DOI] [PubMed] [Google Scholar]
- 22.Green, S. R., and M. B. Mathews. 1992. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 6:2478-2490. [DOI] [PubMed] [Google Scholar]
- 23.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, T., S. M. Tahara, and M. M. Lai. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katze, M. G. 1995. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 3:75-78. [DOI] [PubMed] [Google Scholar]
- 27.Katze, M. G., M. Wambach, M. L. Wong, M. Garfinkel, E. Meurs, K. Chong, B. R. Williams, A. G. Hovanessian, and G. N. Barber. 1991. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated, 68,000-Mr protein kinase in a cell-free system. Mol. Cell. Biol. 11:5497-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman, R. J. 2000. The double-stranded RNA-activated protein kinase PKR, p. 503-527. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Kaufman, R. J., M. V. Davies, V. K. Pathak, and J. W. Hershey. 1989. The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol. Cell. Biol. 9:946-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawagishi-Kobayashi, M., J. B. Silverman, T. L. Ung, and T. E. Dever. 1997. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2α. Mol. Cell. Biol. 17:4146-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koromilas, A. E., S. Roy, G. N. Barber, M. G. Katze, and N. Sonenberg. 1992. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science 257:1685-1689. [DOI] [PubMed] [Google Scholar]
- 33.Kruger, M. F., C. F. Beger, Q. X. Li, P. J. Welch, R. F. Tritz, M. F. Leavitt, J. R. Barber, and F. Wong-Staal. 2000. Identification of eIF2Bγ and eIF2γ as cofactors of hepatitis C virus internal ribosome entry site-mediated translation using a functional genomics approach. Proc. Natl. Acad. Sci. USA 97:8566-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurosaki, M., N. Enomoto, T. Murakami, I. Sakuma, Y. Asahina, C. Yamamoto, T. Ikeda, S. Tozuka, N. Izumi, F. Marumo, and C. Sato. 1997. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology 25:750-753. [DOI] [PubMed] [Google Scholar]
- 35.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 36.Li, S., and A. E. Koromilas. 2001. Dominant negative function by an alternatively spliced form of the interferon-inducible protein kinase PKR. J. Biol. Chem. 276:13881-13890. [DOI] [PubMed] [Google Scholar]
- 37.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 39.Murakami, K. F., M. F. Abe, T. F. Kageyama, N. F. Kamoshita, and A. Nomoto. 2001. Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch. Virol. 146:729-741. [DOI] [PubMed] [Google Scholar]
- 40.Myung, J. F., N. F. Khalap, G. F. Kalkeri, R. F. Garry, and S. Dash. 2001. Inducible model to study negative strand RNA synthesis and assembly of hepatitis C virus from a full-length cDNA clone. J. Virol. Methods 94:55-67. [DOI] [PubMed] [Google Scholar]
- 41.Nagai, K. F., A. H. Wong, S. F. Li, W. N. Tam, A. R. Cuddihy, N. F. Sonenberg, M. B. Mathews, J. F. Hiscott, M. A. Wainberg, and A. E. Koromilas. 1997. Induction of CD4 expression and human immunodeficiency virus type 1 replication by mutants of the interferon-inducible protein kinase PKR. J. Virol. 71:1718-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nanduri, S., F. Rahman, B. R. Williams, and J. Qin. 2000. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 19:5567-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 46.Podevin, P., A. Sabile, R. Gajardo, N. Delhem, A. Abadie, P. Y. Lozach, L. Beretta, and C. Brechot. 2001. Expression of hepatitis C virus NS5A natural mutants in a hepatocytic cell line inhibits the antiviral effect of interferon in a PKR-independent manner. Hepatology 33:1503-1511. [DOI] [PubMed] [Google Scholar]
- 47.Polyak, S. J., D. M. Paschal, S. McArdle, M. J. Gale, Jr., D. Moradpour, and D. R. Gretch. 1999. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology 29:1262-1271. [DOI] [PubMed] [Google Scholar]
- 48.Pyronnet, S. F., L. F. Pradayrol, and N. Sonenberg. 2000. A cell cycle-dependent internal ribosome entry site. Mol. Cell 5:607-616. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Sarrazin, C., T. Berg, J. H. Lee, B. Ruster, B. Kronenberger, W. K. Roth, and S. Zeuzem. 2000. Mutations in the protein kinase-binding domain of the NS5A protein in patients infected with hepatitis C virus type 1a are associated with treatment response. J. Infect. Dis. 181:432-441. [DOI] [PubMed] [Google Scholar]
- 51.Sizova, D. V., V. G. Kolupaeva, T. V. Pestova, I. N. Shatsky, and C. U. Hellen. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 72:4775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spahn, C. M., J. S. Kieft, R. A. Grassucci, P. A. Penczek, K. Zhou, J. A. Doudna, and J. Frank. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959-1962. [DOI] [PubMed] [Google Scholar]
- 53.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 54.Svitkin, Y. V., A. F. Pause, A. F. Haghighat, S. F. Pyronnet, G. F. Witherell, G. J. Belsham, and N. Sonenberg. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 56.Taylor, D. R. 2001. Hepatitis C virus and interferon resistance: it's more than just PKR. Hepatology 33:1547-1549. [DOI] [PubMed] [Google Scholar]
- 57.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 58.Taylor, D. R., B. Tian, P. R. Romano, A. G. Hinnebusch, M. M. Lai, and M. B. Mathews. 2001. Hepatitis C virus envelope protein E2 does not inhibit PKR by simple competition with autophosphorylation sites in the RNA-binding domain. J. Virol. 75:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thibeault, D. F., R. F. Maurice, L. F. Pilote, D. F. Lamarre, and A. Pause. 2001. In vitro characterization of a purified NS2/3 protease variant of hepatitis C virus. J. Biol. Chem. 276:46678-46684. [DOI] [PubMed] [Google Scholar]
- 60.Tsukiyama-Kohara, K. F., N. F. Iizuka, M. F. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams, B. R. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18:6112-6120. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, J. E., T. V. Pestova, C. U. Hellen, and P. Sarnow. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511-520. [DOI] [PubMed] [Google Scholar]
- 63.Witherell, G. W., and P. Beineke. 2001. Statistical analysis of combined substitutions in nonstructural 5A region of hepatitis C virus and interferon response. J. Med. Virol. 63:8-16. [PubMed] [Google Scholar]
- 64.Wong, A. H., J. E. Durbin, S. Li, T. E. Dever, T. Decker, and A. E. Koromilas. 2001. Enhanced antiviral and antiproliferative properties of a STAT1 mutant unable to interact with the protein kinase PKR. J. Biol. Chem. 276:13727-13737. [DOI] [PubMed] [Google Scholar]
- 65.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zamanian-Daryoush, M., S. D. Der, and B. R. Williams. 1999. Cell cycle regulation of the double stranded RNA activated protein kinase PKR. Oncogene 18:315-326. [DOI] [PubMed] [Google Scholar]