Abstract
The phosphoprotein (P protein) of respiratory syncytial virus (RSV) is a key component of the viral RNA-dependent RNA polymerase complex. The protein is constitutively phosphorylated at the two clusters of serine residues (116, 117, and 119 [116/117/119] and 232 and 237 [232/237]). To examine the role of phosphorylation of the RSV P protein in virus replication, these five serine residues were altered to eliminate their phosphorylation potential, and the mutant proteins were analyzed for their functions with a minigenome assay. The reporter gene expression was reduced by 20% when all five phosphorylation sites were eliminated. Mutants with knockout mutations at two phosphorylation sites (S232A/S237A [PP2]) and at five phosphorylation sites (S116L/S117R/S119L/S232A/S237A [PP5]) were introduced into the infectious RSV A2 strain. Immunoprecipitation of 33Pi-labeled infected cells showed that P protein phosphorylation was reduced by 80% for rA2-PP2 and 95% for rA2-PP5. The interaction between the nucleocapsid (N) protein and P protein was reduced in rA2-PP2- and rA2-PP5-infected cells by 30 and 60%, respectively. Although the two recombinant viruses replicated well in Vero cells, rA2-PP2 and, to a greater extent, rA2-PP5, replicated poorly in HEp-2 cells. Virus budding from the infected HEp-2 cells was affected by dephosphorylation of P protein, because the majority of rA2-PP5 remained cell associated. In addition, rA2-PP5 was also more attenuated than rA2-PP2 in replication in the respiratory tracts of mice and cotton rats. Thus, our data suggest that although the major phosphorylation sites of RSV P protein are dispensable for virus replication in vitro, phosphorylation of P protein is required for efficient virus replication in vitro and in vivo.
The phosphoprotein (P protein) of human respiratory syncytial virus (RSV), a prototype Pneumovirus of the family Paramyxoviridae, is an essential component of the viral RNA polymerase, along with the large polymerase (L) and nucleocapsid (N) proteins (12, 35). Interaction of the RSV P protein with the N and L proteins promotes the formation of a transcriptase complex that is essential for viral RNA transcription and replication (10, 19, 20). Although L protein is the catalytic RNA polymerase, P protein is essential for transcription and replication of viral RNA (7, 14). In addition to the N, P, and L proteins, several viral proteins are required for RSV RNA synthesis. The antitermination function of M2-1 is essential for processive RNA synthesis and suppression of transcription termination in intergenic regions (6, 13). M2-2 has been postulated to have a role in regulating the switch between viral RNA transcription and replication processes (3, 17).
The P protein of RSV subgroup A2 is 241 amino acids in length, which is much shorter than the P proteins of other paramyxoviruses (5, 21), and forms homotetramers (1), similar to the Sendai virus P protein (29, 30). The interaction of the N and P proteins enables proper folding of N protein and enables N protein to encapsidate viral RNA during RNA replication (4, 15, 23). By analogy with the other paramyxovirus P proteins, the P protein of RSV likely acts as a cofactor that serves both to stabilize the L protein and to place the polymerase complex on the N:RNA template (14).
RSV P protein is constitutively phosphorylated within the virion core as well as in infected cells. Phosphorylation is mediated by the cellular casein kinase II (8, 33) on two clusters of serines: 116, 117, and 119 in the central region and 232 and 237 in the C-terminal region (26, 27, 31, 33). Approximately 80% of P protein phosphorylation is localized to Ser-232, and the remaining 20% is distributed among Ser-116, -117, -119, and -237. The role of phosphorylation is not well defined. Bacterially expressed, nonphosphorylated P protein cannot form tetramers (1) to support transcription in an in vitro system (2). Phosphorylation of bacterially expressed P protein, however, restores its ability to support transcription, suggesting that the phosphorylated P protein is required to convert the newly initiated polymerase into a stable complex (2). In contrast to these observations, inhibition of phosphorylation in RSV-infected cells did not abolish viral transcription or replication (2, 32). The bulk of P protein phosphorylation is also not required for RNA synthesis in an RSV minigenome system, although absolute levels of RNA may have been reduced (31). In addition, substitutions of S-232 or S-237 by alanine retained its ability to interact with N protein, as shown by the formation of the inclusion bodies in the cotransfected cells (10) and reduction of phosphorylation by phosphorylation inhibitors did not impact tetramer formation of P protein (1).
To determine the role of the RSV P protein phosphorylation in virus replication cycles, the five serine residues in P protein were altered to eliminate their phosphorylation potential. The functions of the resulting P protein mutants were analyzed with an RSV minigenome system. Two recombinant RSVs that had mutations of the serines at two (233/237) or five (116/117/119/232/237) phosphorylation sites were obtained by a reverse genetic technique described previously (6, 18). The effects of P protein phosphorylation on the synthesis of viral RNA and virus replication in cell culture, as well as in the respiratory tracts of mice and cotton rats, were studied.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Monolayer cultures of HEp-2 and Vero cells (obtained from American Type Culture Collection) were maintained in minimal essential medium (MEM) containing 5% fetal bovine serum (FBS). Recombinant RSV A2 (rA2) was recovered from an antigenomic cDNA derived from an RSV A2 strain, pRSVC4G (18), and grown in Vero cells. The modified vaccinia virus Ankara strain expressing bacteriophage T7 RNA polymerase, MVA-T7 (34), was provided by Bernard Moss and grown in CEK cells. Polyclonal anti-RSVA2 antibodies were obtained from Biogenesis (Sandown, N.H.). Monoclonal anti-RSV P protein antibodies 1P, 02/021P, and 76P were gifts from Jose A. Melero.
Functional analysis of P protein mutants by RSV minigenome replication assay.
The plasmids expressing RSV N, P, and L proteins under the control of the T7 promoter (in the pCITE vector) were described previously (18). The RSV minigenome, pRSVCAT, encodes a negative-sense chloramphenicol acetyltransferase (CAT) gene under the control of the T7 promoter (22). pRSVCAT/EGFP was constructed by inserting an enhanced green fluorescent protein (EGFP) gene, which was flanked by the RSV gene start and gene end sequence downstream of the CAT gene, into pRSVCAT. Phosphorylation mutations were engineered in the P gene by using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The major phosphorylation mutations engineered in P protein are indicated in Fig. 1.
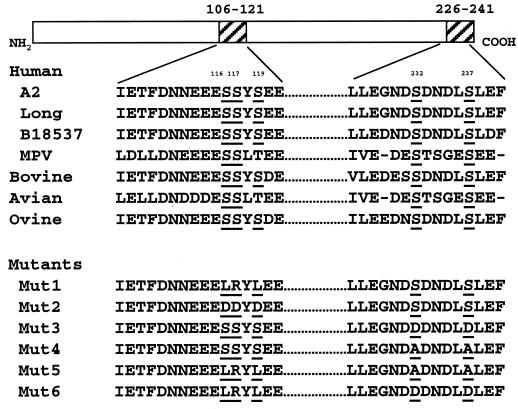
FIG. 1.
Sequence comparison of the P proteins in the central region (nt 106 to 121) and in the C-terminal region (nt 226 to 241) among pneumoviruses. The serine residues in these regions are underlined. RSV-A2, human RSV subgroup A2 strain; Long, human RSV subgroup A long strain; B18537, human RSV subgroup B strain 18537; MPV, human metapneumovirus; Bovine, bovine RSV; Avian, avian pneumovirus; Ovine, ovine RSV. Mutations introduced in each P protein mutant are indicated.
The effect of the P protein phosphorylation mutations on RSV replication was assayed with an RSV CAT minigenome system. HEp-2 cells in 12-well plates were infected with MVA-T7 at a multiplicity of infection (MOI) of 5 for 1 h, followed by transfection with 0.2 μg of pRSV-CAT or pRSVCAT/EGFP together with 0.2 μg of plasmid pN, 0.1 μg of pL, and 0.2 μg of wild-type pP or mutant pP in triplicates. The amount of CAT protein expressed in pRSVCAT- or pRSVCAT/EGFP-transfected cells was determined by an enzyme-linked immunosorbent assay (ELISA) (Roche Molecular Biochemicals). The expression of the genomic RNA and CAT mRNA in the transfected cells was examined by Northern blotting with a digoxigenin (DIG)-labeled negative-sense CAT riboprobe.
Recovery of recombinant RSV.
Two phosphorylation mutations containing two serine site substitutions (SSSAA [PP2]) or five serine site substitutions (LRLAA [PP5]) were introduced into rA2 (18). Mutations were initially introduced into the P gene in an RSV cDNA subclone, pRSV-(A/S), which contains the RSV A2 sequences from nucleotide (nt) 2128 (AvrII) to nt 4485 (SacI), by the QuikChange Site-Directed Mutagenesis kit (Stratagene). The AvrII-SacI fragment carrying the introduced mutations was then inserted into the full-length RSV A2 antigenomic cDNA clone, pRSVC4G (18). pRSVC4G contains the C-to-G change at the fourth position of the leader region in the antigenomic sense. Two recombinant viruses were recovered from the transfected HEp-2 cells by the method described previously (18) and designated as rA2-PP2 (SSSAA) and rA2-PP5 (LRLAA). The recovered viruses were plaque purified and amplified in Vero cells. Virus titer was determined by plaque assay on Vero cells, and the plaques were enumerated after immunostaining with a polyclonal anti-RSV A2 serum (Biogenesis). The presence of each mutation in the recombinant viruses was confirmed by sequence analysis of the P gene cDNA amplified by reverse transcription-PCR (RT-PCR) with viral genomic RNA as template.
Replication of rA2-PP2 and rA2-PP5 in HEp-2 and Vero cells.
The plaque formation efficiency of each mutant was examined in HEp-2 and Vero cells. Cell monolayers in six-well plates were infected with 10-fold serially diluted virus and incubated under an overlay consisting of L15 medium containing 2% FBS and 1% methylcellulose for 6 days at 35°C. The plaques were visualized and enumerated after immunostaining with a polyclonal anti-RSV A2 serum.
The growth kinetics of rA2-PP2 and rA2-PP5 in comparison with those of rA2 were studied in both HEp-2 and Vero cells. Cells in six-well plates were infected with rA2, rA2-PP2, or rA2-PP5 at an MOI of 1.0 or 0.01. After 1 h of adsorption at room temperature, the infected cells were washed three times with phosphate-buffered saline (PBS), overlaid with 3 ml of Opti-MEM I (Invitrogen), and incubated at 35°C. At 24-h intervals, 200 μl of culture supernatant was collected and stored at −80°C in the presence of SPG (0.2 M sucrose, 3.8 mM KH2 PO4, 7.2 mM K2HPO4, 5.4 mM monosodium glutamate) prior to virus titration. After each aliquot was removed, an equal amount of fresh medium was added to the cells. The virus titer was determined by plaque assay on Vero cells at 35°C.
Virus release analyses were performed with HEp-2 and Vero cells. Cells in six-well plates were infected with rA2, rA2-PP2, or rA2-PP5 at an MOI of 1.0. At each time point, the culture supernatants were collected, and then the cell monolayers were washed twice with PBS and scraped in 1 ml of OptiMEM I. Viruses associated with the infected cells were released by a one-time freeze-thaw. Infectious virus present in the culture medium or in the infected cells was titrated by plaque assay on Vero cells.
Replication of rA2-PP2 and rA2-PP5 in mice and cotton rats.
Virus replication in vivo was determined in respiratory pathogen-free BALB/c mice and cotton rats (Sigmodon hispidus) obtained from Harlan. Mice or cotton rats in groups of eight were inoculated intranasally under light methoxyflurane anesthesia with 0.1 ml of inoculum containing 106 PFU of virus per animal. Four days postinoculation, the animals were sacrificed by CO2 asphyxiation, and the lung tissues were harvested. The tissues were homogenized in OptiMEM I (Invitrogen), and the virus titer per gram of lung tissue was determined by plaque assay on Vero cells.
Metabolic labeling of viral proteins in infected cells.
To examine phosphorylation of P protein in virus-infected cells, Vero cells were infected with rA2, rA2-PP2, or rA2-PP5 at an MOI of 1.0 in duplicate. After incubation at 35°C for 16 h, the cells were incubated for 30 min in Dulbecco's MEM (DMEM) lacking either cysteine and methionine or phosphate. One set of samples was then incubated with [35S]Cys and [35S]Met (Amersham Biosciences) at 100 μCi/ml, and the other set was incubated with 33Pi (ICN) at 100 μCi/ml for 4 h. The radiolabeled proteins were extracted by lysis of the cell monolayers in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodcyl sulfate).
Immunoprecipitation and Western blotting.
The radiolabeled polypeptides were immunoprecipitated either by polyclonal goat anti-RSV A2 antibodies or by a mixture of anti-P protein monoclonal antibodies (1P/021P/76P) at 4°C overnight. The antibody-protein complex was precipitated by the addition of 30 μl of protein G-agarose beads (Invitrogen), incubated at 4°C for 1 h, and washed three times with RIPA buffer containing 300 mM NaCl. The immunoprecipitated polypeptides were electrophoresed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 15% polyacrylamide) and detected by autoradiography. The N and P proteins detected on the autoradiographs were quantified by densitometry with a Molecular Dynamics densitometer by using ImageQuant 5.0 for Windows NT (Molecular Dynamics). For Western blotting, Vero cells were infected with each virus at an MOI of 1.0, and the cells were lysed in protein lysis buffer at 48 h postinfection. Detection of viral proteins in the blot by polyclonal anti-RSV antibody was performed as described by Lu et al. (22).
Northern blotting analysis of viral RNA synthesis.
To examine RSV RNA expression, Vero or HEp-2 cells were infected with rA2, rA2-PP2, or rA2-PP5 at an MOI of 1.0. The total cellular RNA was prepared at 48 h postinfection with an RNeasy RNA extraction kit (Qiagene). Equal amounts of total RNA were separated on 1.2% agarose gels containing formaldehyde and transferred to nylon membranes (Amersham Pharmacia Biotech) with a Turboblotter apparatus (Schleicher & Schuell). The membranes were hybridized with RSV gene-specific riboprobes labeled with DIG. The positive-sense F gene probe was used to detect viral genomic RNA, and the negative-sense P gene was used to detect viral mRNA. Hybridization of the membranes with riboprobes was performed at 65°C. Signals from the hybridized probes were detected by using a DIG-Luminescent Detection kit (Roche Molecular Biochemicals) and visualized by exposure to BioMax film (Kodak).
RESULTS
Generation of P protein phosphorylation mutants.
The five phosphorylation sites in P protein at serines 116, 117, and 119 (central region) and 232 and 237 (C-terminal region) are well conserved in the pneumoviruses (Fig. 1). To examine the role of P protein phosphorylation in virus replication, the serine residues in these two clusters were mutagenized to remove their phosphorylation potential. The three serines in the central region were replaced with leucine, arginine, and leucine, respectively (Mut1 [LRLSS]), or aspartic acid to mimic the negative charges of the phosphate groups (Mut2 [DDDSS]). The two serines in the C-terminal region were changed to either aspartic acid (Mut3 [SSSDD]) or alanine (Mut4 [SSSAA]). In addition, all five serines were changed to LRLAA (Mut5) or LRLDD (Mut6) to eliminate all of the major P protein phosphorylation sites. The positions of the substituted residues in each mutant are summarized in Fig. 1.
In vitro functions of phosphorylation-defective P protein.
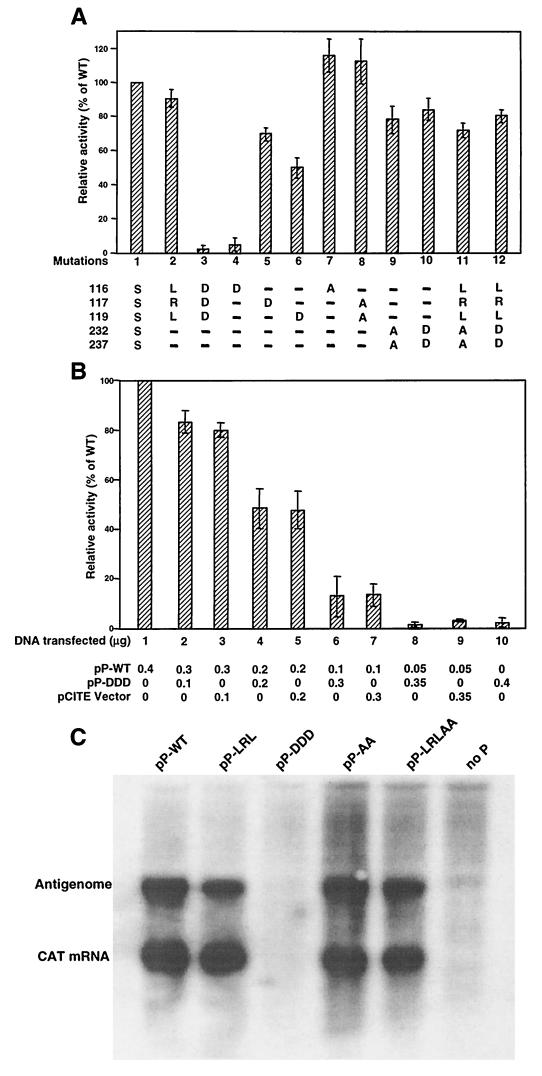
The functions of the altered P protein were evaluated in the RSV CAT minigenome assay. HEp-2 cells were transfected with pRSVCAT along with pL, pN, and wild-type or mutant pP, and expression of the CAT gene was measured. The function of each P protein mutant was calculated as its relative activity compared to that of wild-type P protein. As shown in Fig. 2A, substitution of the three central serines by LRL (lane 2) had little effect on protein function, but substitution of these three residues by aspartic acid (DDD, lane 3) almost completely abolished the protein's function. To evaluate each position independently, three single aspartic acid substitutions were made. As shown in Fig. 2A, S116D was not functional (lane 4), and the other two mutants (S117D, lane 5; S119D, lane 6) remained functional, albeit at a reduced level. However, substitution of Ser-116 or Ser-117/119 by alanine had no effect on P protein function in the minigenome assay. These observations indicated that the serines at 116/117/119 were not required for P protein function and that the aspartic acid residues might have a structural impact on the P protein. Mutation of the P protein at the C-terminal phosphorylation sites, 232/237, substituting alanine (lane 9) or aspartic acid (lane 10), reduced the P protein function by approximately 10 to 20% (Fig. 2A). A slightly reduced level of reporter gene activity was detected in cells expressing mutant P protein that had all five serines removed (LRLAA, lane 11; LRLDD, lane 12). All of the P protein mutants expressed a level of P protein comparable to that of the wild type in these assays as determined by Western blotting (data not shown). Therefore, the minigenome assay indicated that removal of all five phosphorylation sites from RSV P protein did not have a significant impact on protein function in vitro. The difference in the protein activity among these P protein mutants could be due to the reduction of P protein phosphorylation or due to an alteration of P protein structure caused by substitutions of the phosphorylation sites.
FIG. 2.
Functional analysis of RSV P protein phosphorylation mutants. (A) MVA-T7-infected HEp-2 cells were transfected with pRSVCAT together with pN, pL, and wild-type (WT) or mutant pP plasmids. The CAT reporter gene activities were determined by CAT-ELISA and expressed as the percentage of that of wild-type P protein. Error bars represent the standard deviation of three replicate experiments. The serine substitution mutations are shown at the bottom of the graph. (B) Cells were cotransfected with wild-type pP in decreasing amounts together with increasing amounts of pP-DDD or pCITE vector, and the reporter gene activity was expressed as a percentage of that of wild-type P protein. (C) Northern blot analysis of transcription and replication of the RSVCAT/EGFP minigenome in cells expressing the indicated mutant P protein. The CAT mRNA and antigenomic RNA are indicated.
Since Mut3 (DDDSS) almost completely abolished the P protein function, it was thus interesting to know if this mutant would exhibit any dominant-negative effect on the function of wild-type P protein. Plasmid pP-DDD was cotransfected with the wild-type P protein plasmid pP-wt in different ratios together with 0.4 μg of pN and 0.2 μg of pL to determine if this mutant would interfere with wild-type P protein function in the minigenome assay (Fig. 2B). The T7 expression vector (pCITE) was used as a control. The levels of reporter gene expression decreased in correlation with the decreased amount of wild-type pP, which was most likely due to suboptimal ratio among the N, P, and L proteins. However, pP-DDD reduced the reporter gene expression at a level similar to that of the pCITE vector control. Thus, it appeared Mut3 did not have any dominant-negative effect on wild-type P protein function.
Transcription and replication of the pRSVCAT/EGFP minigenome in cells expressing several P protein mutants were analyzed by Northern blotting analysis. pRSVCAT/EGFP was used in Northern blotting in order to better distinguish mRNA from antigenome or read-through RNA. The CAT mRNA and antigenomic RNA were not detected in cells expressing pP-DDD (Fig. 2C), confirming that this mutant P protein was not able to form functional polymerase. For pP-LRL, pP-AA, and pP-LRLAA, which were functional by the pRSVCAT minigenome assay, both CAT mRNA and antigenomic RNA were detected. However, it appeared that the amount of the antigenomic RNA was slightly lower for the P protein mutants containing substitutions of LRL residues.
Replication of recombinant viruses rA2-PP2 and rA2-PP5 in cell culture.
To examine the effect of P protein phosphorylation mutations on virus replication, two mutants were introduced into the RSV A2 antigenomic cDNA clone: one with mutations at the two C-terminal serines (SSSAA [PP2]) and the other with mutations at five serines (LRLAA [PP5]). Both recombinant viruses were obtained from the transfected cDNA and designated rA2-PP2 and rA2-PP5, respectively. Each virus was amplified in Vero cells, and both the released and cell-associated viruses were collected. rA2-PP2 and rA2-PP5 had titers of approximately 2 × 107 PFU/ml in Vero cells, a level comparable to that of wild-type rA2.
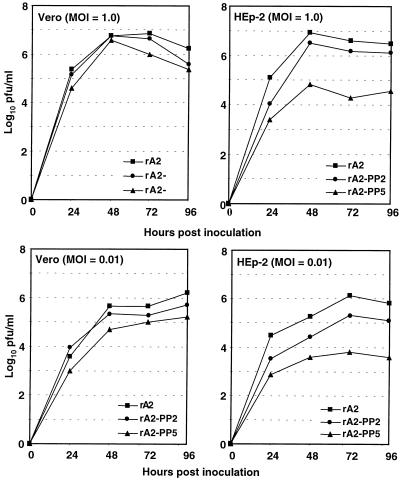
The single-cycle (MOI = 1.0) and multicycle (MOI = 0.01) replication kinetics of rA2-PP2 and rA2-PP5 released into the culture medium were compared to that of rA2 in both HEp-2 and Vero cells (Fig. 3). In Vero cells, both mutants reached peak titers slightly lower than that of wild-type rA2. In HEp-2 cells, however, rA2-PP2 and, to a greater extent, rA2-PP5 reached peak titers much lower than that of wild-type rA2. At an MOI of 1.0, the peak titer of rA2-PP2 was only slightly reduced (0.4 log10), but rA2-PP5 had a peak titer reduction of 2.1 log10. At an MOI of 0.01, the reductions in their peak titers were even greater: 0.8 log10 for rA2-PP2 and 2.3 log10 for rA2-PP5 (Fig. 3).
FIG. 3.
Growth kinetics of rA2-PP2 and rA2-PP5 in Vero and HEp-2 cells. Vero or HEp-2 cells were infected with virus at an MOI of 1.0 or 0.01 and incubated at 35°C. Aliquots of culture supernatant (200 μl) were harvested at 24-h intervals for 96 h. The virus titers are an average of two experiments.
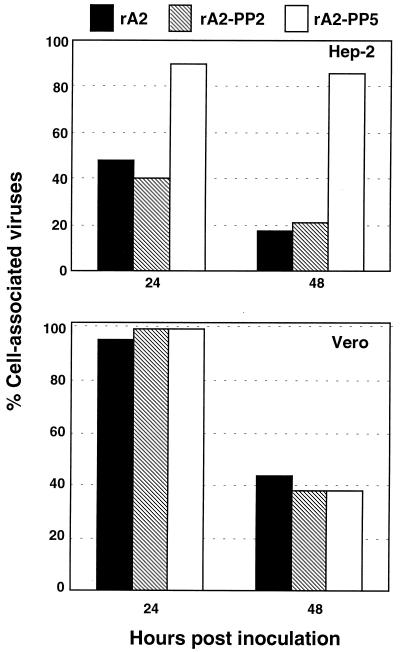
To investigate whether rA2-PP5 was inefficiently released from infected HEp-2 cells compared to Vero cells, HEp-2 or Vero cells were infected with rA2, rA2-PP2, or rA2-PP5, and the amount of virus released into the culture medium supernatant or associated with the cells was monitored (Fig. 4). In HEp-2 cells, at 24 h postinfection, less than 50% of rA2 and rA2-PP2 was associated with the cells. In contrast, approximately 90% of rA2-PP5 was associated with the cells. The percentages of cell-associated viruses for both rA2 and rA2-PP5 at 48 h postinfection were decreased to around 20%. However, about 85% of rA2-PP5 remained cell associated (Fig. 4, upper panel). In contrast to the result obtained from the infected HEp-2 cells, rA2, rA2-PP2, and rA2-PP5 had a similar level of virus associated with the infected Vero cells. The majority of the viruses were cell associated at 24 h postinfection, and about 40% of the viruses remained cell associated at 48 h postinfection (Fig. 4, lower panel). These data suggested that dephosphorylation of P protein affected virus release from the infected HEp-2 cells, but not from the infected Vero cells.
FIG. 4.
Cell association of rA2-PP5 in HEp-2 cells. HEp-2 or Vero cells were infected with rA2, rA2-PP2, or rA2-PP5 at an MOI of 1.0, and at 24 and 48 h postinfection, the amounts of virus released in the culture media or in the infected cells were determined by plaque assay. The percentages of virus that remained associated with the cells are represented.
Phosphorylation of P protein in rA2-PP2- and rA2-PP5-infected cells.
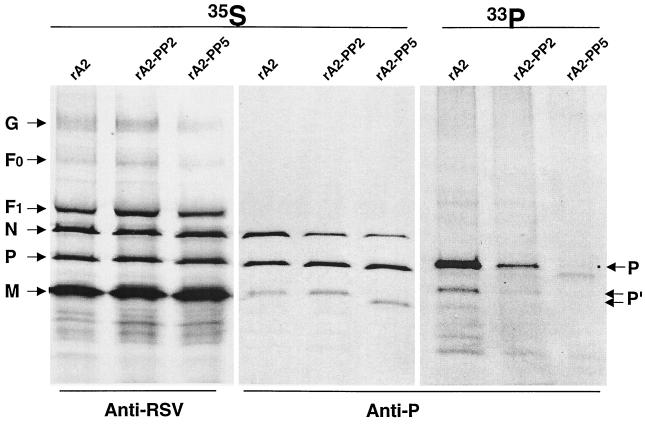
To examine the level of phosphorylation of P protein in infected cells, Vero cells were infected with rA2, rA2-PP2, or rA2-PP5 and labeled with 33Pi or [35S]Met and [35S]Cys, respectively. The polypeptides were immunoprecipitated with anti-RSV polyclonal antibody or a mixture of anti-P protein monoclonal antibodies (Fig. 5). The level of P protein expressed in rA2-PP2- and rA2-PP5-infected cells was comparable to that of wild-type rA2, as shown by immunoprecipitation of 35S-labeled infected cells. It appeared that the migration pattern of the mature form of P protein was not significantly changed by the P protein phosphorylation status. In addition to the major P protein species that migrated at approximately 35 kDa, a faster-migrating protein band was also detected by anti-P protein antibodies, and the band of rA2-PP5 migrated even faster. Phosphorylation of P protein was reduced by about 80% for rA2-PP2 and 95% for rA2-PP5 compared to that of rA2. Only a trace amount of P protein labeled with 33Pi was detected in rA2-PP5-infected cells.
FIG. 5.
Immunoprecipitation of RSV-infected proteins infected with wild-type or phosphorylation mutants. Vero cells were infected with rA2, rA2-PP2, or rA2-PP5 at an MOI of 1.0 and incubated at 35°C. At 18 h of postinfection, proteins were radiolabeled with [35S]Cys and [35S]Met (100 μCi/ml) in DMEM deficient in cysteine and methionine or 33Pi (100 μCi/ml) in DMEM deficient in phosphate for 4 h, immunoprecipitated either by anti-RSV polyclonal or by anti-P protein monoclonal antibodies, separated by SDS-PAGE (15% polyacrylamide), and autoradiographed. P indicates the mature form of the P protein, and P′ represents the immature form of P protein.
Anti-P monoclonal antibodies also immunoprecipitated the N protein in addition to P protein because of the specific N-P protein interaction in the infected cells. As shown in Fig. 5, The N protein immunoprecipitated by anti-P antibodies was reduced in rA2-PP2- and rA2-PP5-infected cells. The reduction of N protein was greater in rA2-PP5-infected cells (60%) than in rA2-PP2-infected cells (30%). Both rA2-PP2 and rA2-PP5 had an N/P protein ratio similar to that of wild-type rA2 when precipitated by anti-RSV antibodies. Thus, removal of the potential phosphorylation sites in P protein affected the interactions between the N and P proteins.
Viral RNA and protein synthesis in rA2-PP2- and rA2-PP5-infected cells.
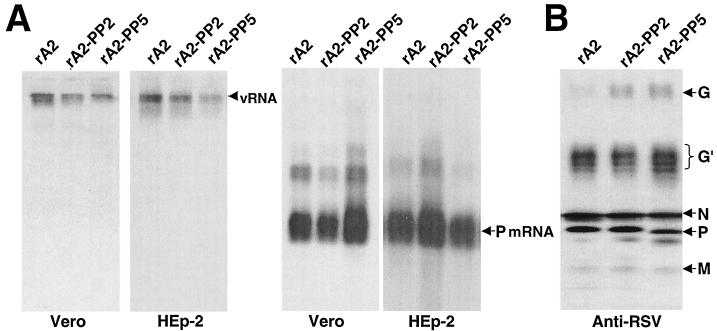
Synthesis of viral RNA and protein in rA2-PP2- and rA2-PP5-infected cells was evaluated by Northern and Western blotting analyses. Vero or HEp-2 cells were infected with wild-type rA2, rA2-PP2, and rA2-PP5 at an MOI of 1.0, and viral RNA was extracted 48 h postinfection. As shown in Fig. 6A, in the infected Vero cells, genomic RNA synthesis was slightly reduced for rA2-PP2 and more reduced for rA2-PP5. However, the P mRNA level was not reduced in rA2-PP5-infected cells. Instead, a slightly increased amount of mRNA was detected in rA2-PP5-infected cells. In the infected HEp-2 cells, rA2-PP5 also had a reduced ratio of genomic RNA to mRNA. Interestingly, the change in the genomic RNA/mRNA ratio was consistently observed throughout the course of infection only when an MOI of 1.0 was used. To examine whether viral protein synthesis was also increased in rA2-PP5-infected Vero cells, Western blotting was performed (Fig. 6B). Except for the slightly increased G protein synthesis, the levels of N, P, and M proteins were not increased in rA2-PP5-infected cells. Thus, the increased mRNA produced in rA2-PP5-infected cells did not result in a concomitant increase in protein expression.
FIG. 6.
Synthesis of viral RNA and proteins in infected cells. (A) Vero or HEp-2 cells were infected with rA2, rA2-PP2, or rA2-PP5 at an MOI of 1.0. At 48 h postinfection, total intracellular RNA was extracted for Northern blotting to detect viral genomic RNA or P mRNA. (B) Protein synthesis in the infected Vero cells was examined by Western blotting with anti-RSV polyclonal antibodies. Viral proteins are indicated on the left. G′ represents the partially glycosylated forms of G protein.
Genetic stability of the P protein phosphorylation mutations.
To examine the genetic stability of the P protein phosphorylation mutations, rA2-PP2 and rA2-PP5 were passaged in Vero and HEp-2 cells in duplicate for five consecutive times. Consistent with the virus release experiment (Fig. 4), infection took longer with each increased passage in HEp-2 cells for rA2-PP5, and a reduced number of virus progeny were released from the infected cells. Viral RNA was extracted from the infected cell culture supernatant at the 5th passage, and the P protein gene cDNA was obtained by RT-PCR and sequenced. All of the introduced mutations were maintained throughout the passages for both rA2-PP2 and rA2-PP5.
Replication of rA2-PP2 and rA2-PP5 in mice and cotton rats.
Replication of rA2-PP2 and rA2-PP5 in the lower respiratory tracts of mice and cotton rats was examined (Table 1). Consistent with its growth kinetics in cell culture, rA2-PP5 was more attenuated in replication in the lower respiratory tracts of mice and cotton rats. The replication of rA2-PP2 and rA2-PP5 was reduced by 1.84 and 3.06 log10, respectively, in the lungs of mice and by 1.81 and 3.11 log10, respectively, in the lungs of cotton rats.
TABLE 1.
Replication of recombinant RSV in mice and cotton rats
| Virus | Virus titer in lungs (mean log10 PFU/g ± SE)a
|
|
|---|---|---|
| Mice | Cotton rats | |
| rA2 | 4.64 ± 0.08 | 4.72 ± 0.08 |
| rA2-PP2 | 2.80 ± 0.29 | 2.91 ± 0.29 |
| rA2-PP5 | 1.58 ± 1.06 | 1.61 ± 0.80 |
Groups of eight Balb/c mice or cotton rats were inoculated with 106 PFU of virus intranasally under light anesthesia on day 0 and sacrificed on day 4. Virus titers per gram of lung tissue were determined by plaque assay.
DISCUSSION
Many viruses have evolved mechanisms to use cellular machinery to modify viral proteins to regulate their functions. Like its counterparts in other paramyxoviruses, RSV P protein is the major phosphorylated protein. Phosphorylation of RSV P protein is mediated by cellular casine kinase II (8, 24) at the two clusters of serines: the central region at residues 116, 117, and 119 and the C-terminal region at residues 232 and 237 (2, 26, 27, 33). These phosphorylation sites are well conserved in the P protein of pneumoviruses, but their roles in the virus life cycle have not been well understood. Although a total of 16 serine residues are present in RSV P protein, removal of the five major phosphorylation sites did not appear to activate alternative phosphorylation sites, and only a trace amount of phosphorylated P protein was detected in rA2-PP5-infected cells. This study thus provides direct evidence to demonstrate that the bulk of the P protein phosphorylation is not essential for RSV replication.
P protein phosphorylation adds a negative charge to the polypeptide via the phosphate group. It has been shown previously that removal of the phosphate group from Ser-232 of P protein halted transcription elongation in vitro, but substitution of Ser-232 by aspartic acid restored transcription activity to 50% of that of wild-type P protein (8). In this paper, replacement of both residues at positions 232 and 237 with alanine had no significant impact on RNA transcription and replication. Similar results have been reported for vesicular stomatitis virus (9) and Marburg virus (25). Thus, in addition to changing the five serines at positions 116, 117, and 119 and 232 and 237 to LRL and AA, respectively, these two clusters of serines were also changed to aspartic acid to mimic the negative charges. The functions of the phosphorylation mutants were analyzed by a minigenome replicon assay as described by Lu et al. (22). A similar level of activity was observed for P protein with S232D/S237D or S232A/S237A changes. Surprisingly, substitutions of the three serines at 116, 117, and 119 by aspartic acid completely abolished P protein function, and the single S116D change had the most dramatic effect. Substitutions of the same residues by LRL had only a slight effect on P protein function. Since the central phosphorylation region is highly charged (Fig. 1), introduction of an additional aspartic acid at Ser-116 may have disturbed the structure of P protein and thus resulted in loss of the protein function.
Coimmunoprecipitation analysis indicated that interactions of the N and P proteins were reduced by dephosphorylation of P protein. About 70% N-P protein interaction was observed for rA2-PP2, from which the two major phosphorylation sites had been removed, and only 40% N-P protein interaction remained for rA2-PP5, from which all five phosphorylation sites had been removed. This observation is consistent with a previous report in which alteration of S-232 and S-237 reduced the ability of P protein to interact with N protein by about 50% in a two-hybrid system (28). Since addition of the newly synthesized P protein to the N protein RNA template is critical for RNA replication (4), a reduction in N-P protein interaction may affect the proper folding of N protein and thus specific encapsidation of RNA by N protein, which could result in reduced virus replication. Recently, it was discovered that phosphorylation of Marburg virus VP30, also a component of the viral RNA polymerase complex, is critical for its interaction with the NP inclusion bodies (25). The interaction of VP30 with NP inclusions is likely mediated through the negative charges of the phosphate group, because substitution of serine residues 40 and 42 by aspartic acid restored the interaction.
The C-terminal region of RSV P protein is the major determinant for its interaction with the N protein (10). Recently, we and the other groups have reported that other regions in P protein are also involved in its interaction with N protein (20, 22). This report also demonstrated that phosphorylation of RSV P protein was important for N-P protein interaction. It remains to be determined whether reduction of the N-P protein interaction for the phosphorylation-deficient P protein is the result of a change in the protein conformation or a disruption of the direct N-P protein interaction through the phosphate groups.
It was observed that the level of mRNA synthesis was slightly elevated in rA2-PP5-infected Vero cells, but not in HEp-2 cells, whereas genomic RNA synthesis was slightly reduced in the infected Vero and HEp-2 cells, suggesting P protein phosphorylation may be involved in regulating viral RNA transcription and replication. The reduced RNA synthesis in rA2-PP5-infected HEp-2 cells was probably due to its less efficient replication. The minigenome analysis suggested that a slightly lower antigenome/mRNA ratio correlated with the LRL change. Therefore, it remains to be determined if the phenotype observed for rA2-PP5 is due to its lack of P protein phosphorylation instead of the structural change caused by these mutations. Further studies are needed to determine if P protein phosphorylation plays a role in regulating viral RNA synthesis. An in vitro study with VSV P protein has suggested that phosphorylation of the carboxyl-terminal domain II residues in P protein of VSV is required for optimal replication, but not for transcription (16).
Since infectious virus rA2-PP5 replicated efficiently in Vero cells, it is presumed that RSV P protein oligomerization was not affected by P protein phosphorylation. P protein phosphorylation and oligomerization have been studied in vitro for other negative-strand RNA viruses. It has been shown that phosphorylation of VSV P protein was required for P protein oligomerization, which is critical for protein function (9). In contrast, rabies virus P protein phosphorylation was not required for its oligomerization (11). Asenjo and Villanueva (1) reported that RSV P protein with the five phosphorylation sites removed formed tetramers in the transiently expressed HEp-2 cells, but not in bacterial cells (1). Thus, it is suggested that unphosphorylated P protein in infected HEp-2 cells could have a transitory phosphorylation that does not take place in bacteria. Barik et al. (2) have reported that the bacterially expressed P protein with S-232 and S-237 replaced by alanine was inactive in vitro (2). Although our experiment indicated that only a trace amount (<5%) of P protein was phosphorylated in rA2-PP5-infected cells, it is not known whether this amount of phosphorylation is required for protein oligomerization.
Interestingly, removal of the major phosphorylation sites from P protein significantly reduced virus budding from rA2-PP5-infected cells, and the majority of viruses remained cell associated. rA2-PP5 was also not able to undergo extensive in vitro passage and was highly attenuated in mice and cotton rats, demonstrating a critical role of phosphorylation for RSV replication in vivo. Future work will be needed to understand the mechanism of how P protein phosphorylation affects RSV budding and replication.
Acknowledgments
We thank the animal facility of Medimmune Vaccines for assistance with the mice and cotton rat experiments; the tissue culture facility for supplying cells; Xing Cheng, Helen Zhou, and HyunJung Park for technical assistance; and George Kemble and Richard Spaete for discussions and critical review of the manuscript. We are grateful to Jose Melero for the gift of the anti-P monoclonal antibodies.
This work was supported in part by NIH SBIR grants (2R44A145267-01/02).
REFERENCES
- 1.Asenjo, A., and N. Villanueva. 2000. Regulated but not constitutive human respiratory syncytial virus (HRSV) P protein phosphorylation is essential for oligomerization. FEBS Lett. 467:279-284. [DOI] [PubMed] [Google Scholar]
- 2.Barik, S., T. McLean, and L. C. Dupuy. 1995. Phosphorylation of Ser232 directly regulates the transcriptional activity of the P protein of human respiratory syncytial virus: phosphorylation of Ser237 may play an accessory role. Virology 213:405-412. [DOI] [PubMed] [Google Scholar]
- 3.Bermingham, A., and P. L. Collins. 1999. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. USA 96:11259-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, M. C., S. Smallwood, and S. A. Moyer. 1999. Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J. Virol. 73:6474-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 6.Collins, P. L., M. G. Hill, E. Camargo, H. Grosfeld, R. M. Chanock, and B. R. Murphy. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 92:11563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran, J., R. Boeck, and D. Kolakofsky. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 10:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupuy, L. C., S. Dobson, V. Bitko, and S. Barik. 1999. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the viral polymerase; phosphorylation by casein kinase 1 occurs mainly at Ser215 and is without effect. J. Virol. 73:8384-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, Y., and J. Lenard. 1995. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J. 14:1240-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Barreno, B., T. Delgado, and J. A. Melero. 1996. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J. Virol. 70:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gigant, B., F. Iseni, Y. Gaudin, M. Knossow, and D. Blondel. 2000. Neither phosphorylation nor the amino-terminal part of rabies virus phosphoprotein is required for its oligomerization. J. Gen. Virol. 81:1757-1761. [DOI] [PubMed] [Google Scholar]
- 12.Grosfeld, H., M. G. Hill, and P. L. Collins. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy, R. W., and G. W. Wertz. 2000. The Cys3-His1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 74:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber, M., R. Cattaneo, P. Spielhofer, C. Orvell, E. Norrby, M. Messerli, J. C. Perriard, and M. A. Billeter. 1991. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 185:299-308. [DOI] [PubMed] [Google Scholar]
- 16.Hwang, L. N., N. Englund, T. Das, A. K. Banerjee, and A. K. Pattnaik. 1999. Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J. Virol. 73:5613-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, H., X. Cheng, H. Z. Y. Zhou, S. Li, and A. Seddiqui. 2000. Respiratory syncytial virus that lacks open reading frame 2 of the M2 gene (M2-2) has altered growth characteristics and is attenuated in rodents. J. Virol. 74:74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, H., D. Clarke, H. Z.-Y. Zhou, X. Cheng, K. Coelingh, M. Bryant, and S. Li. 1998. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology 251:206-214. [DOI] [PubMed] [Google Scholar]
- 19.Khattar, S. K., A. S. Yunus, P. L. Collins, and S. K. Samal. 2001. Deletion and substitution analysis defines regions and residues within the phosphoprotein of bovine respiratory syncytial virus that affect transcription, RNA replication, and interaction with the nucleoprotein. Virology 285:253-269. [DOI] [PubMed] [Google Scholar]
- 20.Khattar, S. K., A. S. Yunus, and S. K. Samal. 2001. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P and P-L interactions using a minigenome system. J. Gen. Virol. 82:775-779. [DOI] [PubMed] [Google Scholar]
- 21.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, respiratory syncytial virus, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 22.Lu, B., R. Brazas, C.-H. Ma, T. Kristoff, X. Cheng, and H. Jin. 2002. Identification of temperature-sensitive mutations in the phosphoprotein of respiratory syncytial virus that are likely involved in its interaction with the nucleoprotein. J. Virol. 76:2871-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masters, P. S., and A. K. Banerjee. 1988. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 62:2658-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazumder, B., and S. Barik. 1994. Requirement of casein kinase II-mediated phosphorylation for the transcriptional activity of human respiratory syncytial viral phosphoprotein P: transdominant negative phenotype of phosphorylation-defective P mutants. Virology 205:104-111. [DOI] [PubMed] [Google Scholar]
- 25.Modrof, J., C. Moritz, L. Kolesnikova, T. Konakova, B. Hartlieb, A. Randolf, E. Muhlberger, and S. Becker. 2001. Phosphorylation of Marburg virus VP30 at serines 40 and 42 is critical for its interaction with NP inclusions. Virology 287:171-182. [DOI] [PubMed] [Google Scholar]
- 26.Navarro, J., C. Lopez-Otin, and N. Villanueva. 1991. Location of phosphorylated residues in human respiratory syncytial virus phosphoprotein. J. Gen. Virol. 72:1455-1459. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Seco, M. P., J. Navarro, R. Martinez, and N. Villanueva. 1995. C-terminal phosphorylation of human respiratory syncytial virus P protein occurs mainly at serine residue 232. J. Gen. Virol. 76:425-430. [DOI] [PubMed] [Google Scholar]
- 28.Slack, M. S., and A. J. Easton. 1998. Characterization of the interaction of the human respiratory syncytial virus phosphoprotein and nucleocapsid protein using the two-hybrid system. Virus Res. 55:167-176. [DOI] [PubMed] [Google Scholar]
- 29.Tarbouriech, N., J. Curran, C. Ebel, R. W. Ruigrok, and W. P. Burmeister. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99-109. [DOI] [PubMed] [Google Scholar]
- 30.Tarbouriech, N., J. Curran, R. W. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7:777-781. [DOI] [PubMed] [Google Scholar]
- 31.Villanueva, N., R. Hardy, A. Asenjo, Q. Yu, and G. Wertz. 2000. The bulk of the phosphorylation of human respiratory syncytial virus phosphoprotein is not essential but modulates viral RNA transcription and replication. J. Gen. Virol. 81:129-133. [DOI] [PubMed] [Google Scholar]
- 32.Villanueva, N., J. Navarro, and E. Cubero. 1991. Antiviral effects of xanthate D609 on the human respiratory syncytial virus growth cycle. Virology 181:101-108. [DOI] [PubMed] [Google Scholar]
- 33.Villanueva, N., J. Navarro, E. Mendez, and I. Garcia-Albert. 1994. Identification of a protein kinase involved in the phosphorylation of the C-terminal region of human respiratory syncytial virus P protein. J. Gen. Virol. 75:555-565. [DOI] [PubMed] [Google Scholar]
- 34.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]
- 35.Yu, Q., R. W. Hardy, and G. W. Wertz. 1995. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol. 69:2412-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]