Abstract
Kinetics and voltage dependence of inactivation of a prokaryotic voltage-gated sodium channel (NaChBac) were investigated in an effort to understand its molecular mechanism. NaChBac inactivation kinetics show strong, bell-shaped voltage dependence with characteristic time constants ranging from ∼50 ms at depolarized voltages to a maximum of ∼100 s at the inactivation midpoint. Activation and inactivation parameters for four different covalently linked tandem dimer or tandem tetramer constructs were indistinguishable from those of the wild-type channel. Point mutations in the outer part of the pore revealed an important influence of the S195 residue on the process of inactivation. For two mutants (S195D and S195E), the maximal and minimal rates of inactivation observed were increased by ∼2.5-fold, and the midpoint of the steady-state inactivation curve was shifted ∼20 mV in the hyperpolarizing direction, compared to the wild-type channel. Our data suggest that pore vestibule structure is an important determinant of NaChBac inactivation, whereas the inactivation mechanism is independent of the number of free cytoplasmic N- and C-termini in the functional channel. In these respects, NaChBac inactivation resembles C-type or slow inactivation modes observed in other voltage-gated K and Na channels.
INTRODUCTION
One of the common properties of voltage-gated channels is their ability to inactivate in response to membrane depolarization. Two general mechanisms of inactivation have been characterized. The first mechanism is usually typified by relatively fast inactivation during a sustained depolarization, effected at the molecular level by a “ball and chain” (1–5) or “hinged-lid” mechanism (6). Molecular mechanisms of this type of inactivation, called N-type inactivation, have been studied in detail in K and Na channels. The groups responsible for this type of inactivation are localized at the cytoplasmic side of the channel. For example, in Shaker K-channel, the N-terminal region of the channel plays the role of the “ball”, which occludes the inner part of the pore after channel activation and thus closes the channel (7). A similar mechanism has been described for Na channels. In this case channel block is probably provided by the III–IV interdomain linker latched by a triplet of hydrophobic residues (IFM motif) to form a hinged lid (8).
A second type of inactivation process, usually with significantly slower kinetics, has been observed in a number of channels (9–11). This type of inactivation is called C-type or slow inactivation, with yet a third inactivation process (P-type) also recognized in potassium channels (12). Although the detailed mechanisms of slow inactivation processes are incompletely understood, it is believed that rearrangement of the pore, rather than motion of a cytoplasmic region of the channel, is likely to play a central role (13–15). The importance of better understanding of slow inactivation in sodium voltage-gated channels is evident since its alteration is linked to such clinical disorders as periodic paralysis, myotonia, idiopathic ventricular fibrillation, and long-QT syndrome, as recently reviewed by Vilin and Ruben (16).
The prokaryotic voltage-gated sodium channel (NaChBac) shows homology with eukaryotic voltage-gated channels though its sequence shows closer similarity with calcium channels than with sodium channels (17). NaChBac activates in response to membrane depolarization and then enters an inactivated state (18). It is likely that activation in NaChBac occurs through similar mechanisms as in voltage-gated eukaryotic channels, based on the fact that the structure of NaChBac contains a highly charged, S4 transmembrane (TM) segment in a six-TM-segment domain (Fig. 1), which is homologous to the voltage sensor of eukaryotic channels (19). Nonetheless, the mechanism of NaChBac inactivation is not well understood. There are no interdomain linkers in the homotetrameric structure, and neither is there an N-terminal region resembling a “ball and chain” inactivation gate in wild type NaChBac. These features hint that cytoplasmic parts of the channel may not be important for inactivation. Thus, the suggestion arose that inactivation of NaChBac occurs through a mechanism that involves rearrangement of its pore region, similar to C-type or slow inactivation observed in eukaryotic channels (20).
FIGURE 1.
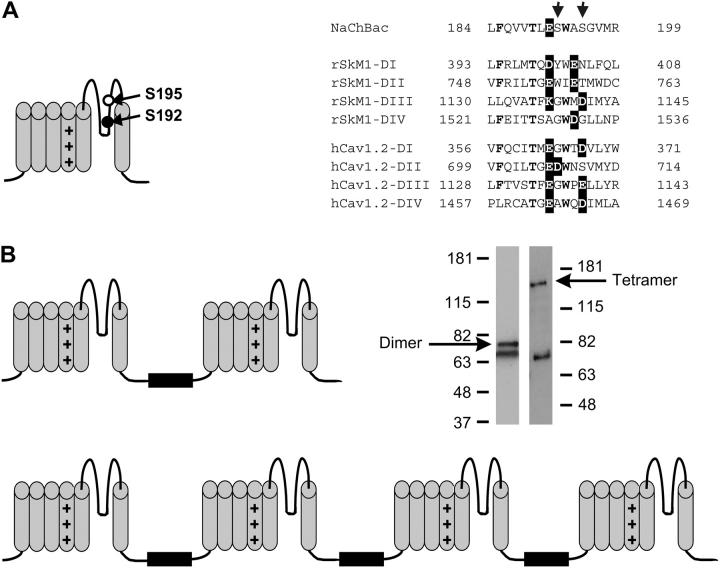
Likely transmembrane topologies of NaChBac point mutants and tandem constructs. (A) Point mutations of wild-type NaChBac were made at position S192 or S195. A total of four point mutants were constructed. Serine at position 192 or 195 was substituted by aspartate (S192D or S195D) or glutamate (S192E or 195E). The pore-region sequence alignment shows NaChBac, with eukaryotic channels rSkM1 (NaV1.4), and hCaV2.1 for comparison. The arrows indicate positions 192 and 195 in the NaChBac sequence. (B) Tandem dimers and tetramers were used for functional studies. Interdomain linkers between WT NaChBac subunits were either poly-glycine (Gly20) or an extended linker with an additional 42-amino acid segment containing the 6H2 epitope (see Methods). Western blots were used to confirm the expression of full-length dimerBL and tetramerBL.
We have tested this hypothesis by creating tandem constructs, with modified N-terminal regions in the resulting linked homologous domains, and channels carrying mutations in a selectivity filter of the pore region. We have identified a residue, located in the outer pore region which has an important influence on NaChBac inactivation. Overall, our data suggest that inactivation of NaChBac may be similar to C-type inactivation found in eukaryotic channels.
METHODS
Mutagenesis
Single amino acid substitutions in the bacterial Na channel were generated essentially as described previously (21), using overlapping polymerase chain reaction (PCR) amplification with oligonucleotide primers (Qiagen Operon, Alameda, CA) containing the sequence for the desired amino acid substitutions, followed by subcloning into pTracer (Invitrogen, Carlsbad, CA).
Tandem oligomers (dimers, trimers, and tetramers) of the NaBacCh, with polyglycine linkers, were prepared by engineering unique restriction sites into the bacterial channel template using high-fidelity PCR amplification (ProofStart, Qiagen). Schematics of the constructs are shown in Fig. 1. The C-terminal oligonucleotides (Qiagen Operon) of each subunit included a polyglycine spacer (20 glycine residues) and had the translation termination codon removed, resulting in an open reading frame. After appropriate restriction endonuclease digestion, the resulting domains—I (Ecor1-Xba1), II (Ecor1-Bamh1), and III (Ecor1-Spe1)—were isolated from agarose gels using the QiaQuick Gel Extraction Kit (Qiagen), and were cloned into the appropriate sites in pBluescript SK (Stratagene, La Jolla, CA) before assembly into pTracer. All clones were completely sequenced to ensure that no unwanted PCR-induced mutations had been introduced.
To test the effect of extending the linkers, and to incorporate a convenient antibody site, we subsequently added a 42-amino acid segment between the unique restriction sites demarking the subunit (domain) boundaries. This segment corresponds to an extracellular loop of the bovine NCX1 protein (22), and includes the 6H2 epitope, antibodies to which are commercially available from Chemicon International (MAB1590, Temecula, CA). This lengthened the spacers between each pair of wild-type (WT) subunits to 62 amino acids from 20 (poly-Gly). The original pTracer NaBacCh clone (kindly provided by Dr. David Clapham) was used as domain IV of the chimeric tetramer. As above, subunits were cloned stepwise into the “base vector” from the C-terminus backward, resulting in three chimeric clones (i.e., III/IV, II/III/IV, and I/II/III/IV). These subunits were also generated by PCR amplification, cloned into the unique sites separating the subunits, checked for orientation, and sequenced as described above for the polyglycine tandem constructs. DNA for transfection of tsA201 or HEK293 cells was prepared using the Endo-Free Maxi Kit (Qiagen). In the following text, tandem constructs incorporating the extended interdomain linkers, including the 6H2 epitope, are referred to as dimerBL, trimerBL, and tetramerBL.
Protein assay by Western blotting and Sypro ruby staining
Plasmids pcDNA4/HisMaxA and pcDNA4/HisMaxC were obtained from Invitrogen Canada (Burlington, Ontario). The His-Select nickel cartridge and imidazole were purchased from Sigma-Adrich (St. Louis, MO). Mouse anti-Na+/Ca++ exchanger (NCX-1) Monoclonal antibody was from Chemicon.
DimerBL and TetramerBL NaChBac cDNA were subcloned in-frame into, respectively, the pcDNA4/HisMaxA and pcDNA4/HisMaxC mammalian expression vectors. Hek293 cells were transfected with DimerBL NaChBac DNA or TetramerBL NaChBac DNA using Polyfect (Qiagen) and grown for a further 36 h. Cells were harvested, resuspended in EDTA-free RIPA buffer (1% NP-40, 0.5% DOC, 0.14M NaCl, 25 mM Tris, pH 7.5, 0.1 mM PMSF, and 1X EDTA-free complete protease inhibitor cocktail (Roche, Germany) and were then shaken for 15 min at room temperature. After centrifuging the lysed cells for 15 min at 15,000 × g to pellet the cellular debris. the supernatant was collected and loaded onto a His-Select Cartridge, which had been prewashed with deionized water and equilibration buffer (50 mM NaH2PO4, pH 8.0, 0.3M NaCl, 10 mM imidazole). The supernatant flowed out under gravity, and the cartridge was then washed three times with each of the following: first, buffer without detergent (50 mM NaH2PO4, pH 8.0, 0.3 M NaCl, 10 mM imidazole), then with buffer with a mild detergent (50 mM NaH2PO4, pH 8.0, 0.3 M NaCl, 10 mM imidazole, 0.2% TWEEN 20), and, finally, a wash buffer with a stronger detergent (50 mM NaH2PO4, pH 8.0, 0.3 M NaCl, 10 mM imidazole, 0.2% CTAB). For both fusion proteins, after these nine washing steps, the eluate contained no detectable protein as indicated by the ultraviolet absorbance (A280). The fusion protein was then eluted from the cartridge with a buffer containing 50 mM NaH2PO4, pH 8.0, 0.3 M NaCl, 250 mM imidazole.
After the affinity chromatographic purification, the concentration of each fusion protein was determined using the bicinchoninic acid method (Pierce, Rockford, IL) and run over 4–12% sodium dodecyl sulfate NUPAGE gels (Invitrogen). The proteins were detected by Western blotting with the NCX1 monoclonal antibody, which binds to the BL linker insert (Fig. 1 B). A nonspecific band at ∼70 kDa was identified by control blots using nontransfected cells and mock transfections using a plasmid lacking any NaChBac insert.
Electrophysiology
Mammalian cells (tSA201) were transiently transfected, using either calcium phosphate precipitation or Polyfect transfection reagent (Qiagen), with a plasmid carrying the gene encoding WT NaChBac or a related construct. These cells were voltage-clamped at room temperature using the whole-cell variant of the patch-clamp technique with an Axopatch 200B (Axon Instruments, Union City, CA) or List EPC-7 (HEKA Elektronik, Lambrecht/Pfalz, Germany) amplifier. Recording pipettes were pulled from Corning 8161 (Potash-Rubium-Lead) glass with a softening temperature of 600°C and dielectric constant of 8.3 and had a resistance of 0.8–2 MΩ. Recordings were made 1–3 days after transfection for WT and point mutant channels and 2–5 days after transfection for tandem dimer and tetramer constructs. Values of the maximal conductance (Gmax, as defined in the following section) of the transfected cells used ranged from ∼4 nS to ∼120 nS. The external bathing solution contained 140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, and 10 mM HEPES (pH 7.4); internal (pipet) solution contained 130 mM CsF, 10 mM EGTA, 10 mM NaCl, and 10 mM HEPES (pH 7.4). In a few experiments designed to examine the Ca permeability of NaChBac pore mutants, the external solution (20 Ca) contained 20 mM CaCl2, 1 mM MgCl2, 65 mM CsCl, 40 mM tetraethylammonium Cl, 10 mM HEPES titrated to pH 7.2 with tetraethylammonium hydroxide. The 20 Ca external solution was used in conjunction with the normal internal solution.
Data analysis
Data were analyzed by using Clampfit and SigmaPlot software. The steady-state inactivation curves were fitted by the equation: I/Imax = 1/(1 + exp((Vh-i − V)/b)), where Vh-i is the potential of half-maximal inactivation and b is a slope factor (mV/e-fold).
Current-voltage curves were fitted using Ohm's law/Boltzmann expression: I = (V − Vrev)Gmax(1/(1 + exp((Vh-a − V)zF/RT))), where I is the peak current for the test potential V, Vrev is the reversal potential, Gmax is the maximal conductance, z is the apparent valence of the gating charge, and Vh-a is the half maximal activation potential.
For summary data used to compare WT and mutated channels, time constants for recovery and inactivation were obtained by single-exponential fits, as follows. Time dependence of channel inactivation was fitted with I = I0 + a[exp(−bt)], and channel recovery from inactivation was fitted with I = I0 + a[1 − exp(−bt)]. In the case of the most negative voltages (−120 to −140 mV) shown in Fig. 3 E, a two-exponential gave slightly better fits to the data.
FIGURE 3.
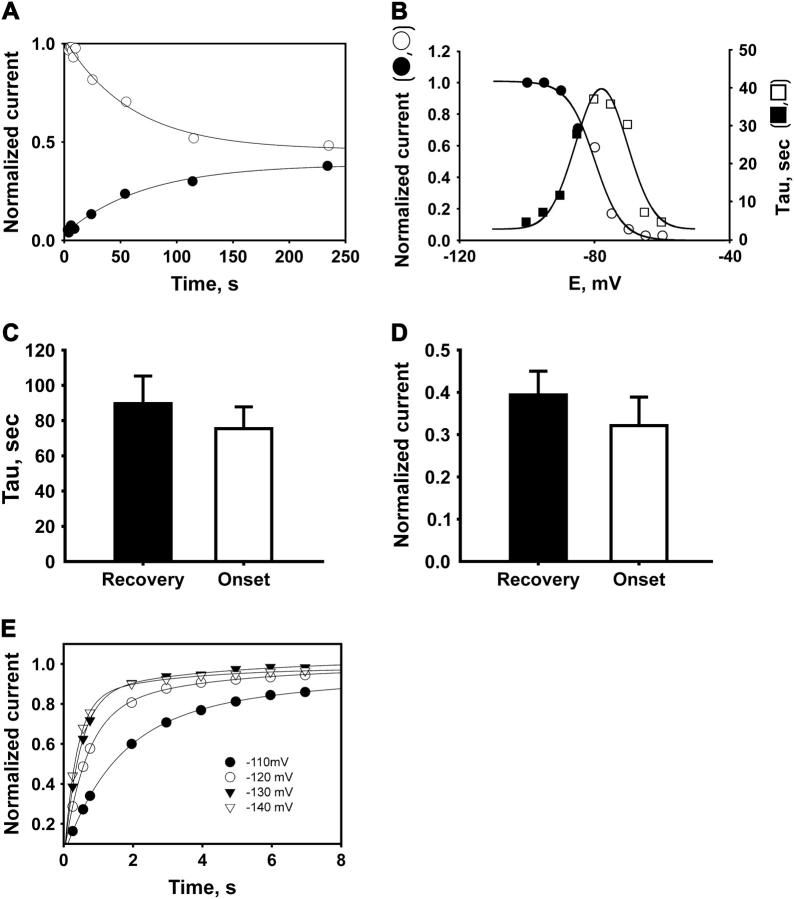
Inactivation and recovery for WT NaChBac channels. Neither time constants for approach to the steady state nor steady-state current levels depend on initial conditions. Kinetics of approach to the steady state were determined starting with either 100% inactivation or 100% availability. These measurements were done at the voltages where only a part of the channels inactivate in the steady state. (A) Kinetics of recovery and inactivation measured at −80 mV. (•) Time course of channel recovery (τ = 71); (○) time course of inactivation (τ = 59). Lines represent single-exponential fits. (B) Time constants and normalized current (fraction of noninactivated channels) determined from recovery (solid symbols) or inactivation (open symbols) as in Fig. 2. Plot of normalized, steady-state current levels was fitted with I/Imax = 1/(1 + exp((Vh-i − V)/b)), with Vh-i = −79 mV and b = 4 mV/e-fold. Plot for time constants was fitted with τ = τVh/[exp((Vh − V) × b) + exp((V − Vh) × a)], with Vh = −75 mV. (C and D) Averaged values of recovery/inactivation time constants and steady-state currents, respectively, at −80 mV from 15 independent experiments. Error bars show the SEM. Parameters measured from the recovery process were not significantly different from those obtained from inactivation experiments. (E) WT channel recovery from inactivation at hyperpolarized voltages. A single-exponential fit is shown for −110 mV (τ = 1.7 ms). For the range −120 to −140 mV, use of a two-exponential fit, with a minority component showing slower recovery, gave a slightly improved fit. Time constants and relative amplitudes, expressed as a percentage, for these traces were: −120 mV, 0.6 s (77%) and 3 s (23%); −130 mV, 0.5 s (81%) and 5 s (19%); and −140 mV, 0.4 s (85%) and 3 s (19%).
All summary data are presented as mean ± SEM (n), where n is the number of determinations. Statistical significance was evaluated using the unpaired Student's t-test.
RESULTS
Kinetics of channel recovery/inactivation does not depend on the initial state of the channel
Kinetics of NaChBac inactivation and recovery from inactivation were measured by using two-pulse or three-pulse voltage-clamp protocols (Fig. 2, A and B). To measure inactivation rates all channels were first transferred into their resting state by application of a −140 mV holding potential for 20 s, after which a conditioning pulse of varying duration was applied (Fig. 2 A). The fraction of noninactivated channels was estimated as a ratio between peak current induced by application of −10 mV test pulse after the conditioning pulse, and peak control current measured in the absence of a conditioning pulse. The values obtained were plotted as a function of conditioning-pulse duration and fitted with a single-exponential fit to determine kinetic parameters and the fraction of noninactivated channels at steady state (Fig. 2 C). In the more negative range of voltages, the kinetics of recovery from inactivation were measured. To do this, before the conditioning (recovery) pulse, all channels were inactivated by 1-s pulse to −10 mV and the rate of recovery from inactivation was measured (Fig. 2, B and D).
FIGURE 2.
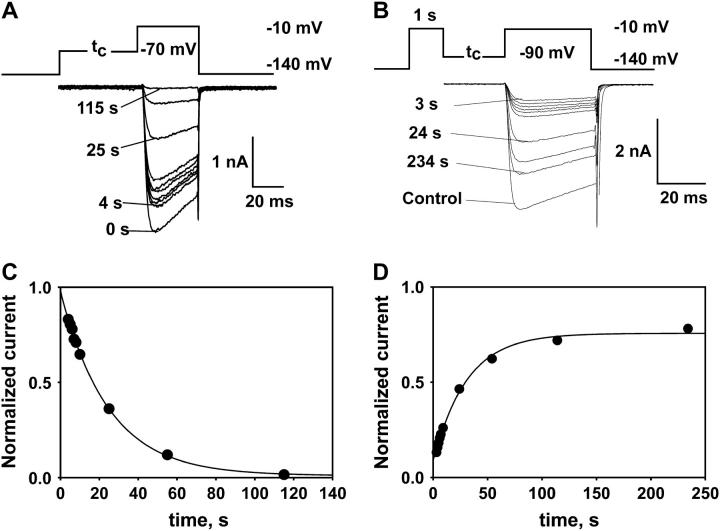
Two-pulse protocols were used to measure the kinetics of transition of the channel to its steady state from either the inactivated or the resting state. (A) Records used to determine kinetics of inactivation. Conditioning pulses of different durations were applied to the channel in its resting state. The fraction of noninactivated channels was estimated by measuring the peak current induced by short membrane depolarization to −10 mV. (B) Records used to determine kinetics of recovery. Channels were inactivated by membrane depolarization to −10 mV before the conditioning pulse. The degree of recovery from inactivation was determined as a ratio of the peak current, for the control test pulse from −140 mV to −10 mV, to the peak current induced by depolarization to −10 mV after a conditioning pulse. (C) Time course of inactivation at −70 mV (τ = 25 s) from records in A. (D) Time course of recovery from inactivation at −90 mV (τ = 31 s), from records in B. For further analyses, steady-state current levels after inactivation or recovery were determined from single-exponential fits, as shown in the examples here.
At intermediate voltages, where not all channels recovered or inactivated in the steady state, it was possible to measure both inactivation and recovery rates by applying different protocols to the same cell. An example of such measurements at −80 mV is shown in Fig. 3 A. At −80 mV, ∼50% of all channels are inactivated at the steady state. Thus, it was possible to obtain reliable single-exponential fits to both recovery and inactivation time courses. Plots in Fig. 3, C and D, summarize average of fitting parameters measured at −80 mV in five to eight different experiments. At any given conditioning potential, values of time constants and the steady-state fraction of “available” channels were indistinguishable after recovery and inactivation. This confirms that the kinetics of reaching the steady state do not depend on the initial conditions and can be measured by either inactivation or recovery protocols. Fig. 3 B shows typical experiments, where recovery-inactivation parameters were measured by using different protocols at different voltages (solid symbols, recovery protocol; open symbols, inactivation protocol). In most experiments, we monitored kinetics by the most convenient and accurate protocol, based on the fraction of recovered/inactivated channels in the steady state.
Voltage dependence of inactivation of WT NaChBac
The steady-state inactivation curve of the WT NaChBac is shown on Fig. 4 A (solid circles). The half-inactivation voltage, Vh-i of inactivation, obtained from a Boltzmann fit is equal to −84 ± 3.1 mV. The time constant for inactivation and recovery from inactivation (τi/r) was determined from single-exponential fits of the plots obtained from two-pulse protocols. τi/r shows a strong, bell-shaped, voltage dependence with the peak of 100 s, at ∼−85 mV (Fig. 4 C, solid circles), which corresponds to the midpoint of steady-state inactivation curve. At the more negative voltages, the time dependence of channel recovery from inactivation was best fitted with a two-exponential function. At −140 mV, slow (τ = 3 s) and fast (τ = 0.4 s) time components of recovery were observed. At depolarized voltages (>−50 mV), inactivation time constants were measured from single-exponential fits to current traces induced by pulses to the test voltage from the −140-mV holding potential (Fig. 4 D, solid circles). In this voltage range, inactivation is ∼1000× faster than at −85 mV.
FIGURE 4.
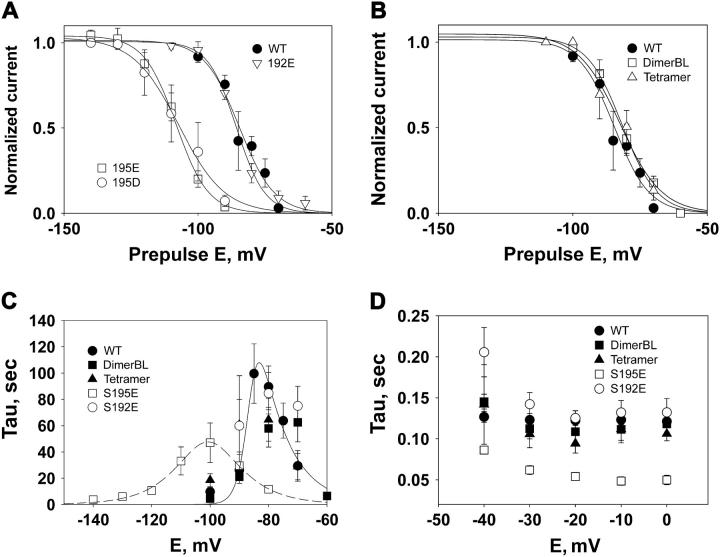
Voltage dependence of steady-state inactivation, and of the kinetics of recovery/inactivation for WT and mutant channels. (A) Steady-state inactivation of wild-type and point mutant channels. Data points were obtained from fits to recovery or inactivation time courses (see Fig. 2, C and D), or from measurements of peak current induced by −10 mV test pulse applied after a 4 min conditioning pulse at the indicated voltage. Vh for the WT channel was −84 mV. Steady-state inactivation voltage dependence of the 192E mutant was similar to WT (for S195E, Vh = −85 mV), but the half-inactivation voltage was shifted to more negative values for S195E and S195D mutants (Vh values: S195E, −106 mV; S195D, −107 mV). (B) Voltage dependence of steady-state inactivation for tetramer and dimerBL channels (Vh values: tetramer, −82 mV; dimerBL, −78 mV). (C) Voltage dependence of recovery-inactivation time constants. Lines show the result of fits to data for WT (solid line) and S195E channels (dashed line) by the function τ = τVh/[exp((Vh − V) × b) + exp((V − Vh) × a)]. The values of parameter Vh (voltage at the peak) obtained from fits were −85 mV for the WT and −101 mV for the S195E. (D) Fast-inactivation time constants measured by fitting current traces elicited by step pulses from holding potential at −140 mV.
Expression of pore mutants and tandem oligomers
Maximal conductances for S192E, S195D, and S195E mutants were similar to those for the wild-type. Neither the A194D nor A194E point mutants, nor the tandem trimer transfected alone, showed any measurable current, and currents for S192D were insufficiently reliable for detailed analysis. Current amplitudes in the cells transfected with poly-Gly tetramer and dimer constructs were ∼3 times lower than in the case of the wild-type. For the dimerBL and tetramerBL constructs, currents were ∼2-fold higher than for the comparable poly-Gly tandem construct. Expression of full-length dimeric and tetrameric constructs was confirmed by Western blot using antibodies against BL linker (Fig. 1 B).
Changes in inactivation voltage dependence associated with mutations in the pore region
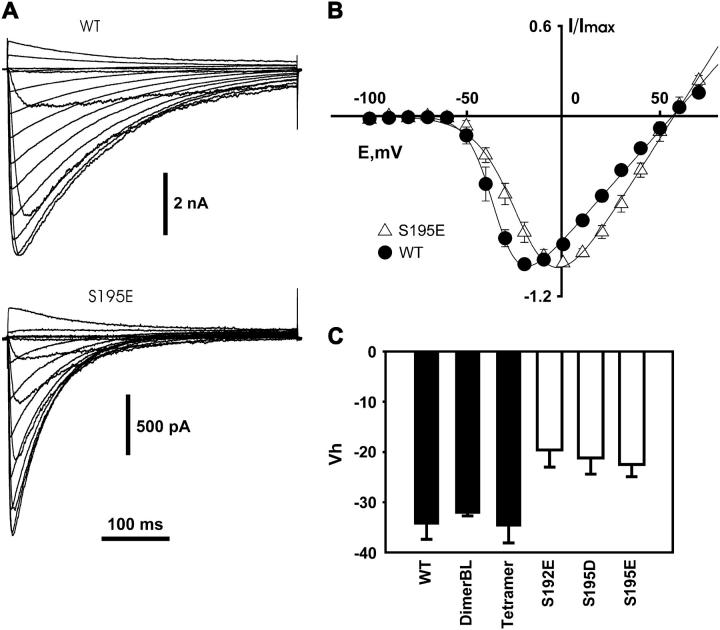
The steady-state inactivation curves and voltage dependence of inactivation kinetics were measured in the mutants where an uncharged residue in the pore region (S195) was replaced by a negatively charged residue (S195E or S195D). In these cases, the steady-state inactivation curves were shifted ∼20 mV in the hyperpolarizing direction compared to the WT channel (Fig. 4 A and Table 1). The midpoints of steady-state inactivation curves were −106 ± 0.6 mV for S195E and −107 ± 1.5 mV for S195D. A similar shift was observed for the voltage dependence of recovery and inactivation time constants (Fig. 4 C). In addition, overall kinetics for these mutants became faster than kinetics of WT NaChBac. This effect was observed at both strongly hyperpolarized and strongly depolarized voltages. At depolarized voltages, the inactivation kinetics of the mutants studied were 1.75–2.5 times faster than for WT (Fig. 4 D). A similar relationship was observed between the slowest time constants for pore mutants and wild-type, estimated as the peak values of the plots shown in Fig. 4 C. By contrast with the S195 mutants, in the case of S192E, the midpoint of the steady-state inactivation curve (Vh-I = −85 ± 0.9 mV) (Fig. 4 A) and inactivation kinetics were not significantly different from the WT channel (Fig. 4, C and D). Based on sequence alignment, position 192 is immediately C-terminal to the “inner ring” location that is conventionally taken to define the selectivity filter in eukaryotic Na and Ca channels.
TABLE 1.
Inactivation parameters for NaChBac and selected mutants
| Construct | Inactivation midpoint, Vh-i (mV) | Slope factor (mV/e-fold) |
|---|---|---|
| WT | −84 ± 3.1 | 6 ± 2 |
| DimerBL | −78 ± 1.3 | 6.5 ± 0.9 |
| Tetramer | −82 ± 2.6 | 7 ± 2 |
| S195E | −106 ± 0.6 | 6.1 ± 0.5 |
| S195D | −107 ± 1.5 | 8 ± 1 |
| S192E | −85 ± 0.9 | 5.3 ± 0.7 |
Inactivation parameters shown here were derived from Boltzmann fits to steady-state inactivation curves (Fig. 4, A and B), which were obtained by averaging values of normalized current at each voltage (from five to eight separate experiments at each voltage). Values of the normalized, steady-state currents were obtained from single-exponential fits to recovery/inactivation time courses, as in Fig. 3 A.
Cytoplasmic linkers between NaChBac subunits do not change voltage dependence or kinetics of inactivation
We measured inactivation kinetics for tandem constructs that had polypeptide linkers connecting adjacent 6TM domains (see Methods). The linkers consisted either of a Gly20 segment (tetramer), or Gly20 plus a 42-amino acid segment (22). The extended linker was used in the dimerBL and tetramerBL constructs (see Methods). Vh-i for steady-state inactivation (Fig. 4 B) and inactivation kinetics (Fig. 4, C and D) of both types of constructs were similar to the wild-type (−78 ± 1.3 mV for the dimerBL and −82 ± 2.6 mV for the tetramer, see also Table 1), suggesting that modifications of the terminal region of NaChBac did not significantly change its inactivation characteristics. Less extensive experiments on the Gly20-linked dimer, and the tetramerBL showed similar behavior (data not shown). In contrast to the dimer and tetramer, transfection with the trimer constructs formed by three linked NaChBac domains did not produce measurable currents. This is consistent with the idea that active channels assembled only from four monomers, two dimers, or one tetramer, making exactly four domains in a functional unit.
Cells cotransfected with DNA for trimer and wild-type monomer showed small, detectable currents, but it was unclear whether the functional channels were composed of four monomers, one monomer co-assembled with one trimer, or some other possibility. Relatively low levels of current were observed when wild-type and trimer plasmids were cotransfected, and for this reason we did not study them in detail. Nonetheless, the lack of detectable current observed after trimer-only transfections suggests that the assembly of aberrant channels, in which, for example, only the first protomer of a tandem subunit contributed to a functioning channel, is a very low-probability event, at most. Although we cannot rigorously exclude the complicating specter of “spare” protomers in a channel (23–26), the simplest explanation of our functional data, together with the Western blot verification of dimer and tetramer expression, is that functional channels are made up of exactly four monomers or homologous domains, assembled into a fourfold symmetric unit. Given the expression of full-length dimer and tetramer protein, and the near-WT functional properties of dimers and tetramers, we speculate that the absence of substantial functional expression of trimers reflects faulty assembly, but the reason for this is not clear.
Activation gating of NaChBac and its mutants
The half-activation voltage for WT and mutant channels was measured from the families of currents induced by stepping from the holding potential of −140 mV to the range of voltages from −100 mV to 70 mV at 10-mV intervals. Representative currents, recorded from WT channels and the S195E mutant are shown in Fig. 5 A. Data from seven experiments for the WT and S195E mutants were normalized to maximal current, averaged and plotted against voltage (Fig. 5 B). The midpoint of activation for the S195E mutant (Vh-a = −22 ± 2.5 mV) was shifted 13 mV more positive than for the WT NaChBac (Vh-a = −35 ± 2.2 mV). Fig. 5 C represents the summarized results of the measurements of Vh-a for WT NaChBac and its mutants. Plotted values represent the mean of fitted parameters measured from separate experiments. The activation midpoints (Vh-a) for the S195E; S195D, and S192E mutants were significantly shifted in the positive direction compared to the WT (p < 0.05), whereas the difference between Vh-a values for the dimerBL (−32 ± 0.5 mV), tetramer (−35 ± 3.4 mV), and WT (−35 ± 2.2 mV) was not significant. Activation shifts seen for the pore mutants are in the opposite direction to those for inactivation. Also, unlike the case of inactivation, a shift is seen for S192E, as well as the S195E and S195D mutants (Fig. 5 C and Table 2). Thus, the inactivation process is not rigidly linked to voltage-dependent activation.
FIGURE 5.
Voltage dependence of activation of wild-type and mutant channels. (A) Representative current traces of wild-type and S195E mutant channels obtained by stepping the voltage from −140 mV to a range of voltages from −100 mV to 70 mV, in 10-mV increments. (B) Averaged peak I/V curves for WT and the S195E mutant (five and seven experiments, respectively). Currents were normalized to their maximal value. Lines are results of a fit with I = (V − Vrev)Gmax(1/(1 + exp((Vh-a − V)zF/RT))), where Vh-a = −34 mV for WT and Vh-a = −19 mV for the S195E mutant. (C) Vh of activation of WT and different mutants. Values of Vh were obtained by averaging parameters obtained from different experiments (5–12 experiments for each group).
TABLE 2.
Activation parameters for NaChBac and selected mutants
| Construct | Activation midpoint, Vh-a (mV) | Apparent valence, z |
|---|---|---|
| WT | −35 ± 2.2 (7) | 4.8 ± 0.6 (7) |
| DimerBL | −32 ± 0.5 (4) | 4.8 ± 0.7 (4) |
| Tetramer | −35 ± 3.4 (6) | 3.9 ± 0.6 (5) |
| 195E | −22 ± 2.5 (7) | 2.4 ± 0.1 (7) |
| 195D | −21 ± 3.2 (5) | 2.1 ± 0.1 (4) |
| 192E | −15 ± 3.4 (3) | 3.1 ± 1.3 (3) |
Activation parameters were obtained from separate Ohm's Law/Boltzmann fits of the current-voltage curves for individual experiments, as described in Methods. Indicated values are the mean ± SEM, with the number of experiments indicated as (n).
Cadmium and inactivation kinetics of NaChBac
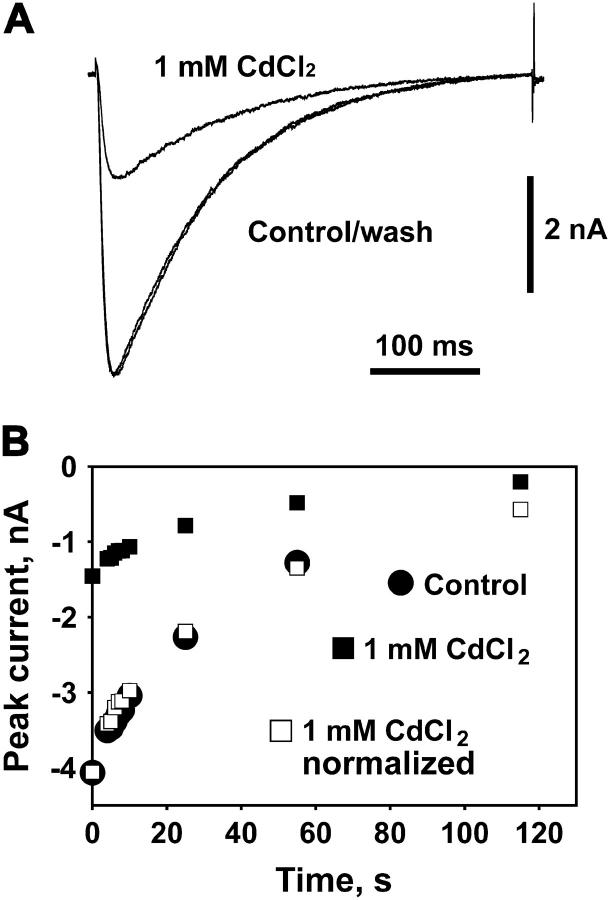
As previously shown (27), cadmium blocks NaChBac, when applied on the extracellular side. We sought to determine whether this pore-blocking ion affects the kinetics of the apparently pore-based mechanism of NaChBac inactivation. For mammalian sodium channels, slow or ultra-slow inactivation can be modulated either by external sodium (28) or by a pore-blocking conotoxin (29). In our records, the form of the NaChBac current transient was not visibly changed by addition of cadmium, consistent with binding/dissociation that equilibrates very rapidly compared with the timescale of the conductance changes. Kinetics of inactivation of the wild-type NaChBac were measured by using both single- and two-pulse protocols. Fig. 6 A shows current traces recorded in control and in the presence of 1 mM cadmium. Kinetics of current decay at −10mV in the presence of cadmium was unchanged compared to control. Similarly, we observed no detectable change of inactivation kinetics at −60 mV, measured by a two-pulse protocol (Fig. 6 B). Effectiveness of cadmium block of S195E mutant (IC50 = 1.3 ± 0.3 mM, n = 6) was similar to the WT NaChBac (IC50 = 1.5 ± 0.2 mM, n = 8). Summarizing the kinetics of current decay at −10 mV, inactivation time constants were τ = 149 ± 16 ms for WT controls, and τ = 150 ± 30 ms in the presence of Cd (n = 6). For S195E, the values were τ = 66 ± 4 ms for controls, and τ = 59 ± 7 ms in the presence of Cd (n = 4). Both the absence of a cadmium effect on inactivation kinetics, and the lack of effect of the mutation S195E on cadmium block suggest that the site of cadmium block is distinct from the pore residue(s) that determine inactivation rates.
FIGURE 6.
Kinetics of inactivation of the wild-type channel are not changed in the presence of the externally applied blocker cadmium. (A) Representative current traces for control and 1 mM Cd2+. Currents were induced by step pulse from −140 mV to −10mV. Time constants of inactivation were determined from single-exponential fits of decaying current. (B) Kinetics of channel inactivation at −60 mV, measured with a two-pulse protocol (see Fig. 2) for control and in the presence of 1 mM Cd2+. The ratio between peak current in control and in the presence of Cd2+ at 0 s was used to normalize currents in the presence of Cd2+ (□). The normalized currents superpose on the control data, showing that the time courses in the presence and absence of Cd2+ are indistinguishable.
DISCUSSION
This study is a first attempt to identify the possible molecular bases of inactivation of the bacterial voltage-gated sodium channel, NaChBac. We have found that inactivation characteristics changed when point mutations are introduced at one location in the pore region of the channel – substitution of the neutral serine at position 195 with a negatively charged aspartate or glutamate residue shifted the midpoint of steady-state inactivation curve to more negative voltages and increased the speed of the channel inactivation. In contrast, a similar substitution for S192 did not significantly change inactivation parameters. We also found that modification of terminal regions of the channel monomers by linking channel subunits into tandem constructs had no effect on inactivation. On this basis, we conclude that inactivation of NaChBac involves participation of its pore region, and thus may be similar to the mechanisms of C-type inactivation of eukaryotic voltage-gated channels.
Inactivation of WT NaChBac
Firstly, we measured inactivation parameters of wild-type NaChBac. We found that the kinetics of NaChBac inactivation are strongly dependent on voltage. The half-inactivation voltage, which we measured in the steady state, was more negative than that reported in a previous study (30). This difference appears to arise from the need to use longer pulses to ensure that the channels reach a steady state over the whole voltage range of interest. Conditioning pulses longer than 100 s were required for the voltages near the midpoint of the inactivation curve (−85 mV). We routinely used a conditioning duration of 240 s. Inactivation time constants showed a bell-shaped voltage dependence, which is typical of voltage-gated channels described by the Hodgkin-Huxley model (31), but is generally consistent with models in which forward and backward rate constants show oppositely directed voltage dependence. Overall, NaChBac gating is significantly slower than for eukaryotic voltage-gated channels. The time to peak current for NaChBac, during a depolarization to −10 mV, was ∼20 ms, whereas in most eukaryotic sodium voltage-gated channels it is <1 ms (32–35).
The striking difference between inactivation time constants of NaChBac and eukaryotic sodium channels offers a hint about the underlying mechanism. In the case of NaChBac, the time constant of channel inactivation decreases on the order of 1000-fold, from a maximum of ∼100 s to 100 ms at more depolarized voltages. By contrast, τh for squid axon changes only ∼10-fold over a comparable range, e.g., ∼9 ms to 1 ms in Fig. 2.17 of Hille's book (36). Hence, kinetic analysis alone suggests that the mechanism of inactivation gating in NaChBac might differ from that of “fast” inactivation in eukaryotic voltage-gated channels. However, the longer inactivation time constants for NaChBac are very close to the values for “ultraslow inactivation” time constants measured in rat skeletal muscle channels (37,38).
Inactivation mechanisms in eukaryotic channels and NaChBac
The sequence of NaChBac contains a charged S4 segment, which is conserved in all types of voltage-gated channels and is most likely responsible for NaChBac activation by depolarization (39). In contrast, NaChBac does not have any region which can be obviously identified as a fast inactivation gate. However, the region responsible for inactivation does vary among the broader family of voltage-gated eukaryotic channels. By using mutagenesis, we tried to identify the regions of NaChBac that might be involved in its inactivation.
Based on the fact that NaChBac has many similarities to eukaryotic voltage-gated channels, including sequence homology and electrophysiological properties, we hypothesized that inactivation of NaChBac might occur through some mechanism in common with eukaryotic channels. Experimental data obtained from studying eukaryotic sodium and potassium channels suggest the existence of at least two distinct mechanisms of channel inactivation. N-type inactivation is often characterized by faster kinetics and can be eliminated by deletion of the N-terminal region (for certain potassium channels), or mutation of the IFM motif of the interdomain linker (for sodium channels) (40). Also, the kinetics of N-type inactivation of potassium channels can be modified by changing the number of functional N-terminals in the assembled channel (41). In that case, the kinetics of inactivation of nonconcatenated tetramers became slower, compared to wild-type channels, as the number of N-terminal inactivation gates per channel was reduced by coexpression of an N-terminal deletion mutant with WT Shaker channels. Using tandem-linked dimers and tetramers, we did not detect any significant difference between inactivation of wild-type channel and the tandem concatemers. Hence, we conclude that, in contrast to N-type inactivation in eukaryotic channels, NaChBac inactivation does not involve either N- or C-terminal cytoplasmic regions of the WT monomer.
Slow inactivation in sodium channels may involve a mechanism similar to that of C-type inactivation in potassium channels (42). C-type inactivation usually has slower kinetics than N-type and is sensitive to the mutations in the pore region. Similarly, in sodium channels, mutations that change the charge of groups located at the outer side of the selectivity filter change slow-inactivation kinetics (43–46). For example, it was shown that, for the rat skeletal muscle sodium channel, substitution of a negatively charged residue (aspartate) for the alanine at position 1529 of domain IV in the selectivity filter favors ultraslow inactivation (47). We have found that change-changing substitutions at position 195 of the outer pore loop of NaChBac lead to changes in inactivation kinetics and voltage dependence. Negative shifts of inactivation associated with the S195D/E mutations (Fig. 4 A) are not secondary to shifts in the voltage dependence of activation, which, for these mutants, occurs at more positive voltages (Fig. 5, B and C). The mutation S192E, which presumably lies further into the pore from the outer mouth of the channel, did not change inactivation compared to the wild-type channel, but does result in a positive shift in activation. Overall, our data are consistent with an NaChBac inactivation mechanism that resembles the ultraslow inactivation in skeletal-muscle sodium channels.
Cadmium, a pore blocker, did not affect inactivation kinetics
It has been shown that cations which enter voltage-gated channels from the extracellular side can affect kinetics of inactivation, presumably by interactions with the pore region (48,49). We found that externally applied cadmium, in concentrations sufficient to block >50% of NaChBac current, did not change inactivation kinetics. This seemingly surprising result suggests to us that the cadmium binding site is distinct from the region participating in channel inactivation. This idea is supported by the fact that effectiveness of cadmium block of pore mutant, S195E, is the same as for the WT channel, whereas inactivation kinetics and voltage dependence for this mutant differ from those of the WT.
NaChBac as a model channel
The homology between a number of bacterial and eukaryotic channels is well-established. Bacterial channels have been successfully used for description of a number of fundamental structural and functional properties believed to be similar to those of eukaryotic channels (50,51). NaChBac is an important model channel because it is relatively simple, having relatively small monomeric units that presumably assemble into a homotetramer to form the functional channel. At the same time, it possesses properties common to all three families of eukaryotic voltage-gated channels: it has only one domain, as do potassium channels; it is sodium-selective; and it is sensitive to the blockers of eukaryotic Ca channels. When first described, the gene which encodes NaChBac was proposed to be a gene for a voltage-gated Ca channel (CaBac 1.1) with only one 6TM domain (52). This conclusion was very intriguing, because in eukaryotic organisms voltage-gated Ca channels are tetramers. In 2001, this channel was expressed in eukaryotic CHO cells and characterized electrophysiologically by patch-clamp (53). As predicted from sequence homology, this channel was voltage-gated, but surprisingly, it turned out to be sodium selective. Based on its selectivity, the channel was renamed NaChBac. However, in the same study, it was shown that NaChBac is sensitive to certain drugs that block eukaryotic voltage-gated calcium channels.
NaChBac has been used successfully to investigate the molecular basis of channel selectivity between sodium and calcium. It becomes permeable to calcium when additional negatively charged amino acids are introduced into the pore region by point mutagenesis (54). In a series of preliminary experiments, we observed reversal potentials consistent with increased permeability to calcium for channels containing the substitution S195E (only the D substitution at this position was studied by Yue et al. (54)), or additional negative charges. With 20 Ca external solution (see Methods), mean values of Vrev were −30mV for WT (n = 3); +17 mV for WT coexpressed with S195E (n = 2), and Vrev = +27 to +35 mV (one experiment each for S195E expressed alone, and for coexpression with A194D/S195E or A194D/S195D). Our data thus confirm the published observations that Ca permeability increases with an increasing number of negative charge residues in the lining of the outer pore.
NaChBac has also been used as a model channel for study of activation kinetics. Measurements of ionic (55) and gating currents (56) allowed the development of kinetic models for NaChBac activation. Novel fluorescence studies performed using a mammalian cell expression system suggest that NaChBac has a voltage sensor similar to that of the voltage-gated bacterial potassium channel, KVAP, but that both of these prokaryotic channels differ in some functional details from Shaker (57). Our finding, that NaChBac inactivation has many parallels with slow, or ultraslow, inactivation in eukaryotic voltage-gated channels, supports the possibility that NaChBac could be a valuable tool in the study of molecular mechanisms of these processes. It also should be noted that since the discovery and initial description of NaChBac, several other voltage-gated sodium channels in bacteria have been identified (58) and roles for them in motility, chemotaxis, and pH homeostasis have been proposed (59). Thus, electrophysiological studies of NaChBac may also illuminate the possible roles played by ionic currents in the life of bacteria.
CONCLUSION
NaChBac appears to inactivate by a mechanism similar to C-type inactivation of eukaryotic channels. It seems likely that, during evolution, N-type inactivation appeared later than other key elements of voltage-gated channels, such as the voltage sensor and the basic 6TM structure.
Acknowledgments
We thank Drs. David Clapham and Dejian Ren for providing the original NaChBac clone, Dr. Paul Schnetkamp for DNA encoding the part of NCX1 that was used in the extended interdomain linker, and Dr. Harry Fozzard for comments on a draft of the manuscript.
This work was supported by operating grants from the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Alberta, NWT, and Nunavut. R.J.F. is a Heritage Medical Scientist of the Alberta Heritage Foundation for Medical Research.
References
- 1.Armstrong, C. M., F. Bezanilla, and E. Rojas. 1973. Destruction of sodium conductance inactivation in squid axons perfused with pronase. J. Gen. Physiol. 62:375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, C. M., and F. Bezanilla. 1977. Inactivation of the sodium channel. II. Gating current experiments. J. Gen. Physiol. 70:567–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezanilla, F., and C. M. Armstrong. 1977. Inactivation of the sodium channel. I. Sodium current experiments. J. Gen. Physiol. 70:549–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshi, T., W. N. Zagotta, and R. W. Aldrich. 1990. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 250:533–538. [DOI] [PubMed] [Google Scholar]
- 5.Hoshi, T., W. N. Zagotta, and R. W. Aldrich. 1991. Two types of inactivation in Shaker K+ channels: effects of alterations in the carboxy-terminal region. Neuron. 7:547–556. [DOI] [PubMed] [Google Scholar]
- 6.Vassilev, P. M., T. Scheuer, and W. A. Catterall. 1988. Identification of an intracellular peptide segment involved in sodium channel inactivation. Science. 241:1658–1661. [DOI] [PubMed] [Google Scholar]
- 7.Hoshi, T., W. N. Zagotta, and R. W. Aldrich. 1990. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 250:533–538. [DOI] [PubMed] [Google Scholar]
- 8.Vassilev, P. M., T. Scheuer, and W. A. Catterall. 1988. Identification of an intracellular peptide segment involved in sodium channel inactivation. Science. 241:1658–1661. [DOI] [PubMed] [Google Scholar]
- 9.Balser, J. R., H. B. Nuss, N. Chiamvimonvat, M. T. Perez-Garcia, E. Marban, and G. F. Tomaselli. 1996. External pore residue mediates slow inactivation in μ1 rat skeletal muscle sodium channels. J. Physiol. 494:431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Townsend, C., and R. Horn. 1997. Effect of alkali metal cations on slow inactivation of cardiac Na+ channels. J. Gen. Physiol. 110:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Barneo, J., T. Hoshi, S. H. Heinemann, and R. W. Aldrich. 1993. Effects of external cations and mutations in the pore region on C-type inactivation of Shaker potassium channels. Receptors Channels. 1:61–71. [PubMed] [Google Scholar]
- 12.Loots, E., and E. Y. Isacoff. 1998. Protein rearrangements underlying slow inactivation of the Shaker K+ channel. J. Gen. Physiol. 112:377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogielska, E. M., W. N. Zagotta, T. Hoshi, S. H. Heinemann, J. Haab, and R. W. Aldrich. 1995. Cooperative subunit interactions in C-type inactivation of K channels. Biophys. J. 69:2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong, B. H., G. F. Tomaselli, and J. R. Balser. 2000. A structural rearrangement in the sodium channel pore linked to slow inactivation and use dependence. J. Gen. Physiol. 116:653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loots, E., and E. Y. Isacoff. 2000. Molecular coupling of S4 to a K+ channel's slow inactivation gate. J. Gen. Physiol. 116:623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilin, Y. Y., and P. C. Ruben. 2001. Slow inactivation in voltage-gated sodium channels: molecular substrates and contributions to channelopathies. Cell Biochem. Biophys. 35:171–190. [DOI] [PubMed] [Google Scholar]
- 17.Durell, S. R., and H. R. Guy. 2001. A putative prokaryote voltage-gated Ca2+ channel with only one 6TM motif per subunit. Biochem. Biophys. Res. Commun. 281:741–746. [DOI] [PubMed] [Google Scholar]
- 18.Ren, D., B. Navarro, H. Xu, L. Yue, Q. Shi, and D. E. Clapham. 2001. A prokaryotic voltage-gated sodium channel. Science. 294:2372–2375. [DOI] [PubMed] [Google Scholar]
- 19.Durell, S. R., and H. R. Guy. 2001. A putative prokaryote voltage-gated Ca2+ channel with only one 6TM motif per subunit. Biochem. Biophys. Res. Commun. 281:741–746. [DOI] [PubMed] [Google Scholar]
- 20.Catterall, W. A. 2001. Physiology. A one-domain voltage-gated sodium channel in bacteria. Science. 294:2306–2308. [DOI] [PubMed] [Google Scholar]
- 21.Light, P. E., C. Bladen, R. J. Winkfein, M. P. Walsh, and R. J. French. 2000. Molecular basis of protein kinase C-induced activation of ATP-sensitive potassium channels. Proc. Natl. Acad. Sci. USA. 97:9058–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, K., P. J. Bauer, T. G. Kinjo, R. T. Szerencsei, W. Bonigk, R. J. Winkfein, and P. P. Schnetkamp. 2003. Assembly of retinal rod or cone Na+/Ca2+-K+ exchanger oligomers with cGMP-gated channel subunits as probed with heterologously expressed cDNAs. Biochemistry. 42:4593–4600. [DOI] [PubMed] [Google Scholar]
- 23.Heginbotham, L., and R. MacKinnon. 1992. The aromatic binding site for tetraethylammonium ion on potassium channels. Neuron. 8:483–491. [DOI] [PubMed] [Google Scholar]
- 24.McCormack, K., L. Lin, L. E. Iverson, M. A. Tanouye, and F. J. Sigworth. 1992. Tandem linkage of Shaker K+ channel subunits does not ensure the stoichiometry of expressed channels. Biophys. J. 63:1406–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, Y., Y. Yan, and F. J. Sigworth. 1997. How does the W434F mutation block current in Shaker potassium channels? J. Gen. Physiol. 109:779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurst, R. S., R. A. North, and J. P. Adelman. 1995. Potassium channel assembly from concatenated subunits: effects of proline substitutions in S4 segments. Receptors Channels. 3:263–272. [PubMed] [Google Scholar]
- 27.Ren, D., B. Navarro, H. Xu, L. Yue, Q. Shi, and D. E. Clapham. 2001. A prokaryotic voltage-gated sodium channel. Science. 294:2372–2375. [DOI] [PubMed] [Google Scholar]
- 28.Townsend, C., and R. Horn. 1997. Effect of alkali metal cations on slow inactivation of cardiac Na+ channels. J. Gen. Physiol. 110:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todt, H., S. C. Dudley, Jr., J. W. Kyle, R. J. French, and H. A. Fozzard. 1999. Ultra-slow inactivation in μ1 Na+ channels is produced by a structural rearrangement of the outer vestibule. Biophys. J. 76:1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren, D., B. Navarro, H. Xu, L. Yue, Q. Shi, and D. E. Clapham. 2001. A prokaryotic voltage-gated sodium channel. Science. 294:2372–2375. [DOI] [PubMed] [Google Scholar]
- 31.Hodgkin, A. L., and A. F. Huxley. 1952. Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J. Physiol. 116:449–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell, D. T., and B. Hille. 1976. Kinetic and pharmacological properties of the sodium channel of frog skeletal muscle. J. Gen. Physiol. 67:309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armstrong, C. M., F. Bezanilla, and E. Rojas. 1973. Destruction of sodium conductance inactivation in squid axons perfused with pronase. J. Gen. Physiol. 62:375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo, M. V., and P. Shrager. 1981. Block and inactivation of sodium channels in nerve by amino acid derivatives. I. Dependence on voltage and sodium concentration. Biophys. J. 35:31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavlov, E., C. Bladen, C. Diao, and R. J. French. 2005. Bacterial Na channels: progenitors, progeny, or parallel evolution? In Bacterial Ion Channels and Eukaryotic Homologues. A. Kubalski and B. Martinac, editors. American Society for Microbiology, Washington, D.C. 191–207.
- 36.Hille, B. 2001. Ion Channels of Excitable Membranes. Sinauer Associates Sunderland, MA.
- 37.Todt, H., S. C. Dudley, Jr., J. W. Kyle, R. J. French, and H. A. Fozzard. 1999. Ultra-slow inactivation in μ1 Na+ channels is produced by a structural rearrangement of the outer vestibule. Biophys. J. 76:1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hilber, K., W. Sandtner, O. Kudlacek, I. W. Glaaser, E. Weisz, J. W. Kyle, R. J. French, H. A. Fozzard, S. C. Dudley, and H. Todt. 2001. The selectivity filter of the voltage-gated sodium channel is involved in channel activation. J. Biol. Chem. 276:27831–27839. [DOI] [PubMed] [Google Scholar]
- 39.Durell, S. R., and H. R. Guy. 2001. A putative prokaryote voltage-gated Ca2+ channel with only one 6TM motif per subunit. Biochem. Biophys. Res. Commun. 281:741–746. [DOI] [PubMed] [Google Scholar]
- 40.Vassilev, P. M., T. Scheuer, and W. A. Catterall. 1988. Identification of an intracellular peptide segment involved in sodium channel inactivation. Science. 241:1658–1661. [DOI] [PubMed] [Google Scholar]
- 41.MacKinnon, R., R. W. Aldrich, and A. W. Lee. 1993. Functional stoichiometry of Shaker potassium channel inactivation. Science. 262:757–759. [DOI] [PubMed] [Google Scholar]
- 42.Liu, Y., M. E. Jurman, and G. Yellen. 1996. Dynamic rearrangement of the outer mouth of a K+ channel during gating. Neuron. 16:859–867. [DOI] [PubMed] [Google Scholar]
- 43.Xiong, W., R. A. Li, Y. Tian, and G. F. Tomaselli. 2003. Molecular motions of the outer ring of charge of the sodium channel: do they couple to slow inactivation? J. Gen. Physiol. 122:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kambouris, N. G., L. A. Hastings, S. Stepanovic, E. Marban, G. F. Tomaselli, and J. R. Balser. 1998. Mechanistic link between lidocaine block and inactivation probed by outer pore mutations in the rat μι1 skeletal muscle sodium channel. J. Physiol. 512:693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomaselli, G. F., N. Chiamvimonvat, H. B. Nuss, J. R. Balser, M. T. Perez-Garcia, R. H. Xu, D. W. Orias, P. H. Backx, and E. Marban. 1995. A mutation in the pore of the sodium channel alters gating. Biophys. J. 68:1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balser, J. R., H. B. Nuss, N. Chiamvimonvat, M. T. Perez-Garcia, E. Marban, and G. F. Tomaselli. 1996. External pore residue mediates slow inactivation in μ1 rat skeletal muscle sodium channels. J. Physiol. 494:431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilber, K., W. Sandtner, O. Kudlacek, I. W. Glaaser, E. Weisz, J. W. Kyle, R. J. French, H. A. Fozzard, S. C. Dudley, and H. Todt. 2001. The selectivity filter of the voltage-gated sodium channel is involved in channel activation. J. Biol. Chem. 276:27831–27839. [DOI] [PubMed] [Google Scholar]
- 48.Townsend, C., and R. Horn. 1997. Effect of alkali metal cations on slow inactivation of cardiac Na+ channels. J. Gen. Physiol. 110:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardo, L. A., S. H. Heinemann, H. Terlau, U. Ludewig, C. Lorra, O. Pongs, and W. Stuhmer. 1992. Extracellular K+ specifically modulates a rat brain K+ channel. Proc. Natl. Acad. Sci. USA. 89:2466–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doyle, D. A., C. J. Morais, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- 51.MacKinnon, R., S. L. Cohen, A. Kuo, A. Lee, and B. T. Chait. 1998. Structural conservation in prokaryotic and eukaryotic potassium channels. Science. 280:106–109. [DOI] [PubMed] [Google Scholar]
- 52.Durell, S. R., and H. R. Guy. 2001. A putative prokaryote voltage-gated Ca2+ channel with only one 6TM motif per subunit. Biochem. Biophys. Res. Commun. 281:741–746. [DOI] [PubMed] [Google Scholar]
- 53.Ren, D., B. Navarro, H. Xu, L. Yue, Q. Shi, and D. E. Clapham. 2001. A prokaryotic voltage-gated sodium channel. Science. 294:2372–2375. [DOI] [PubMed] [Google Scholar]
- 54.Yue, L., B. Navarro, D. Ren, A. Ramos, and D. E. Clapham. 2002. The cation selectivity filter of the bacterial sodium channel, NaChBac. J. Gen. Physiol. 120:845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, Y., V. Yarov-Yarovoy, T. Scheuer, and W. A. Catterall. 2004. A gating hinge in Na+ channels; a molecular switch for electrical signaling. Neuron. 41:859–865. [DOI] [PubMed] [Google Scholar]
- 56.Kuzmenkin, A., F. Bezanilla, and A. M. Correa. 2004. Gating of the bacterial sodium channel, NaChBac: Voltage-dependent charge movement and gating currents. J. Gen. Physiol. 124:349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blunck, R., D. M. Starace, A. M. Correa, and F. Bezanilla. 2004. Detecting rearrangements of Shaker and NaChBac in real-time with fluorescence spectroscopy in patch-clamped mammalian cells. Biophys. J. 86:3966–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koishi, R., H. Xu, D. Ren, B. Navarro, B. W. Spiller, Q. Shi, and D. E. Clapham. 2004. A superfamily of voltage-gated sodium channels in bacteria. J. Biol. Chem. 279:9532–9538. [DOI] [PubMed] [Google Scholar]
- 59.Ito, M., H. Xu, A. A. Guffanti, Y. Wei, L. Zvi, D. E. Clapham, and T. A. Krulwich. 2004. The voltage-gated Na+ channel NaVBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus. Proc. Natl. Acad. Sci. USA. 101:10566–10571. [DOI] [PMC free article] [PubMed] [Google Scholar]