Abstract
Latency-associated nuclear antigen 1 (LANA1) of Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) persistently maintains a plasmid containing the KSHV latent origin of replication (oriP) as a closed circular episome in dividing cells. In this study, we investigated the involvement of chromosome binding activity of LANA1 in persistent episome maintenance. Deletion of the N-terminal 22 amino acids of LANA1 (ΔN-LANA) inhibited the interaction with mitotic chromosomes in a human cell line, and the mutant concomitantly lost activity for the long-term episome maintenance of a plasmid containing viral oriP in a human B-cell line. However, a chimera of ΔN-LANA with histone H1, a cellular chromosome component protein, rescued the association with mitotic chromosomes as well as the long-term episome maintenance of the oriP-containing plasmid. Our results suggest that tethering of KSHV episomes to mitotic chromosomes by LANA1 is crucial in mediating the long-term maintenance of viral episomes in dividing cells.
Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) is a gamma 2 herpesvirus and is associated with the development of Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease (5, 6, 10, 17, 21, 24). KSHV can establish long-term persistent infections in tumor cells and lymphoma-derived cell lines. In such cells, the double-stranded KSHV DNA genome can persist as multiple copies of closed circular episomes (5, 9), like the genome of its close relative, Epstein-Barr virus (EBV).
Two viral components can mediate the long-term episome maintenance of KSHV in infected cells. One is latency-associated nuclear antigen 1 (LANA1), encoded by open reading frame 73, and the other is a cis-acting DNA sequence (latent origin of replication [oriP]) in the 5′ end of the KSHV genome (terminal repeat [TR] sequence) (1, 2, 7). LANA1 persistently maintains a plasmid containing multiple TRs as an episome in a human B-cell line (1, 2).
Previous studies showed that LANA1 was directly bound to the KSHV TR sequence (2, 8, 11). Furthermore, an imperfect 20-bp palindrome in the TR sequence exhibited binding activity for LANA1, and a plasmid containing three TRs was persistently maintained as an episome in a human cell line (2, 11). Domain map analysis revealed that the C-terminal region of LANA1 bound to this 20-bp palindrome in the TR sequence (11). Since this region contains the LANA1 dimerization domain (22), it seems likely that LANA1 binds to the TR sequence as a dimer, like EBNA1, the episome maintenance protein of EBV.
About 40 to 80 copies of episomes are stably maintained in KSHV-infected lymphoma cells in vivo (1, 4). The constant KSHV copy number in dividing cells indicates that KSHV has an efficient system for segregating viral episomes into two daughter cells in every cell division. In the present study, we examined the involvement of the chromosome interaction activity of LANA1 in long-term episome maintenance. A 22-amino-acid deletion from the N terminus of LANA1 (ΔN-LANA) inhibited both the interaction with mitotic chromosomes and the long-term episome maintenance of a KSHV oriP-based plasmid. Moreover, these two activities were rescued by fusion of LANA1 with histone H1, a cellular chromosome component protein. Thus, our results support the hypothesis that LANA1 tethers viral episomes to mitotic chromosomes, a process which is essential for the proper segregation of viral DNAs in dividing cells.
MATERIALS AND METHODS
Cell lines.
BJAB is an EBV-negative Burkitt lymphoma cell line. The cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum (FCS), penicillin (500 U/ml), and streptomycin (500 μg/ml). The human cervical carcinoma cell line HeLa was cultured in Dulbecco's modified Eagle's medium supplemented with 10% FCS, penicillin (500 U/ml), streptomycin (500 μg/ml), and 2 mM l-glutamine.
Plasmids.
The plasmid pSG5, with the simian virus 40 early promoter, was used as the expression plasmid for LANA1 and its derivatives. The plasmid pSGLANA contains wild-type full-length LANA1 (1) and was kindly provided by Kenneth Kaye (Department of Medicine, Harvard Medical School). The plasmid pSGLANA also contains the viral FLIP and viral cyclin genes. To eliminate accidental expression of the viral FLIP and viral cyclin genes, we removed these genes from pSGLANA by cutting with NheI and BglII, blunt ending, and religating with the addition of a BglII linker. The plasmid obtained, pSGWT-LANA, was used as the expression plasmid for wild-type LANA1. The plasmid pSGΔN-LANA was constructed by PCR with primers 5′-GAATTCACCATGTGTAGGAAACGAAACAGG-3′ (underlined nucleotides indicate the EcoRI site, which was used for cloning into pSG5) and 5′-ACTCGAGCTAGCTGCAGTTATGTCATTTCCTGTGGAGAGTC-3′. The amplified ΔN-LANA fragment was introduced into pSGWT-LANA cut with EcoRI and BamHI. To construct a chimeric gene, H2B-ΔNLANA, the entire histone H2B gene without a termination codon was amplified by PCR from the plasmid pCMV-HA-HistoneH2B. The ΔN-LANA fragment was also amplified from pSGWT-LANA by PCR. The amplified H2B fragment and a mutant LANA1 fragment were introduced into pSGWT-LANA cut with EcoRI and BamHI.
The PCR primers used were 5′-CGAATTCCCACCATGTACCCATACGATGTT-3′ (underlined nucleotides indicate the EcoRI site) and 5′-ACGCGTCTTGGAGCTGGTGTACTTGGTGAC-3′ (underlined nucleotides indicate the MluI site) for histone H2B and 5′-ACGCGTTGTAGGAAACGAAACAGGTCTCCG-3′ (underlined nucleotides indicate the MluI site) and 5′-ACTCGAGCTAGCTGCAGTTATGTCATTTCCTGTGGAGAGTC-3′ for a LANA1 mutant.
An H1-ΔNLANA expression plasmid was constructed by replacing the EcoRI-MluI fragment of the H2B gene in the H2B-ΔNLANA expression plasmid with the BspEI-EcoRI fragment containing the full-length histone H1 gene in the pEGFP-HistoneH1 plasmid (kindly provided by S. C. Hung, National University of Singapore) by blunt-end ligation. The recombinant plasmids were confirmed by DNA sequencing. The plasmid Z6-13 contains the first ∼13 kb of the KSHV genome derived from BC-1 cells cloned into SuperCos, which carries a neomycin resistance gene regulated by the simian virus 40 promoter (Stratagene, La Jolla, Calif.) (1, 20). The plasmid sCOS was constructed by removing all of the KSHV sequence from Z6-13 by cutting with EcoRI followed by self-ligation and was used as a control for Z6-13. The pMik-HygB plasmid contains a hygromycin resistance gene and was used as the selectable marker plasmid for cotransfection with the LANA1 expression plasmids into BJAB cells.
Immunofluorescence analysis.
HeLa cells (15 × 103 cells/well) were cultured on eight-well chamber slides (Nalgen Nunc International, Naperville, Ill.) overnight and transfected with the LANA1 expression plasmids (0.086 μg) by using PolyFect (Qiagen Inc., Valencia, Calif.). At 48 h after transfection, the cells on the slides were fixed with 1% paraformaldehyde containing 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 5 min at room temperature. After being washed with PBS, the cells were further incubated with serum from a Kaposi's sarcoma patient with a high antibody titer against LANA1 (kindly provided by Fransis Kasolo, University Teaching Hospital, Lusaka, Zambia) for 60 min at 37oC. After being washed with PBS, the cells were incubated with a fluorescein-conjugated goat anti-human antibody (Cappel, Costa Mesa, Calif.) and with 0.5 μg of propidium iodide (PI)/ml in PBS for 60 min at 37oC. After being washed with PBS, the cells were visualized by using a laser scanning confocal microscope (MRC-1024; Bio-Rad Laboratories, Hercules, Calif.) and LaserSharp software (Bio-Rad Laboratories).
Visualization of LANA1 association with chromosomes.
To visualize the interaction of LANA1 with mitotic chromosomes, HeLa cells on eight-well chamber slides were transfected with the LANA1 expression plasmids. At 36 h after transfection, the cells were treated with 0.6 μg of Colcemid/ml for 6 h, washed twice with PBS, and allowed to swell in a hypotonic A buffer (1% trisodium citrate, 0.5 mM CaCl2, 0.5 mM MgCl2). The cells on the slides were then fixed with 1% paraformaldehyde containing 0.1% Triton X-100 in PBS for 5 min at room temperature. After three washes with PBS, the cells were stained with serum from a Kaposi's sarcoma patient. The following steps used to visualize LANA1 and chromosomes were identical to those described above.
To visualize the interaction of LANA1 with individual chromosomes in metaphase spreads, HeLa cells (4 × 105 cells/well) cultured on six-well culture plates were transfected with the LANA1 expression plasmids (3 μg). At 36 h after transfection, the cells were treated with 0.5 μg of Colcemid/ml for 4 h at 37oC. The cells were harvested with 0.05% trypsin and 0.53 mM EDTA and then collected by centrifugation at 260 × g for 5 min. The collected cells were suspended in a hypotonic B buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 5 mM MgCl2), dropped immediately onto slides, and air dried for 10 min. The cells on the slides were then fixed with 1% paraformaldehyde containing 0.1% Triton X-100 in 25% PBS for 5 min at room temperature. The cells were washed with PBS and stained with serum from a Kaposi's sarcoma patient. The following steps used to visualize LANA1 and chromosomes were identical to those described above.
Western blotting.
BJAB cells (106) were collected, washed twice with PBS, and directly lysed in 100 μl of sample buffer (50 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 850 mM β-mercaptoethanol, 10% glycerol). The lysates (10 μl) were heated at 95°C for 5 min and then size separated under reducing conditions by electrophoresis on an SDS-6% polyacrylamide gel. Separated proteins were electronically transferred to a polyvinylidene difluoride membrane by using a wet method. The membrane was incubated with Block Ace (Dainippon Seiyaku, Osaka, Japan) for 1 h at room temperature and further incubated overnight with anti-LANA antibody (Applied Biosystems). After being washed with 10 mM Tris-HCl (pH 8.0)-150 mM NaCl-0.05% Tween 20, the membrane was further incubated with anti-rat immunoglobulins conjugated with horseradish peroxidase (Bio-Rad Laboratories, Hercules, Calif.). Proteins recognized by the antibodies in the membrane were visualized by using an enhanced chemiluminescence Western blotting detection system (Amersham Pharmacia Biotech, Piscataway, N.J.).
Establishment of BJAB/LANA1 clones.
To establish BJAB cell lines expressing LANA1 (BJAB/LANA1 cells), BJAB cells (5 × 106) were washed twice with PBS, suspended in RPMI 1640 containing linearized pSGLANA (25 μg) and pMik-HygB (1 μg) in an electroporation cuvette, and pulsed with an electroporator (Gene Pulser; Bio-Rad Laboratories) at 200 V and 975 μF. The cells were then cultured in RPMI 1640 supplemented with FCS for 48 h. After the addition of hygromycin B (0.4 mg/ml), the cells were seeded on 96-well plates and further cultured for 2 weeks. Hygromycin-resistant clones were screened for the expression of LANA1 protein by Western blotting.
Episome maintenance assay.
BJAB/LANA1 cells (5 × 106) were transfected with 25 μg of Z6-13 or sCOS by the electroporation method described above. At 48 h after transfection, the cells were cultured in the presence of G418 (0.75 mg/ml) for a minimum of 3 weeks. Episomal DNA was extracted from the resistant cells (5 × 106) by the alkaline lysis method. Briefly, the cells were lysed with 200 μl of a solution containing 50 mM glucose, 25 mM Tris-HCl (pH 8.0), and 10 mM EDTA; 200 μl of a solution containing 200 mM NaOH-1% SDS; and 200 μl of 6 M potassium acetate. Lysates were centrifuged (20,400 × g for 15 min at 4°C), and the supernatant was treated with a mixture of phenol and chloroform. The extracted plasmid was precipitated with 70% ethanol, and the precipitate was dissolved in 10 μl of distilled H2O. Then, 1 μl of the plasmid solution was introduced into Escherichia coli DH10B competent bacteria by electroporation. The diluted bacterial culture was plated on an ampicillin plate and cultured overnight at 37°C, and the number of colonies was counted. Plasmids propagated in E. coli were extracted by the alkaline lysis method as described above.
RESULTS
The N-terminal 22 amino acids of LANA1 are essential for its interaction with mitotic chromosomes.
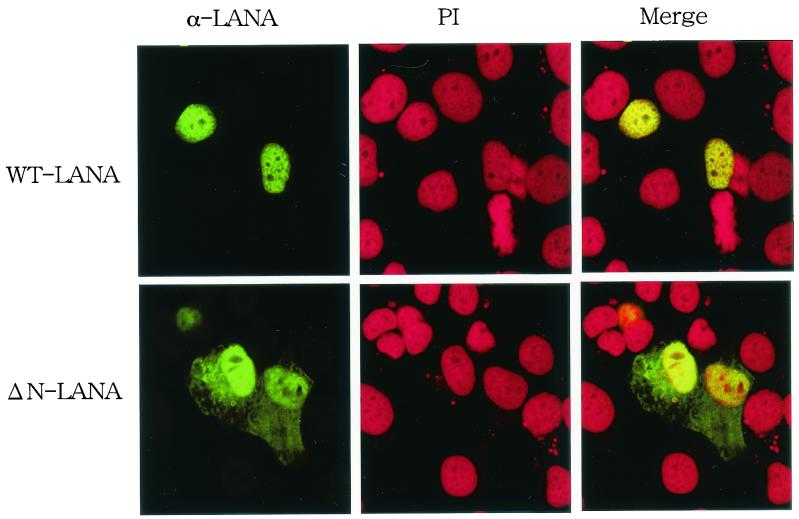
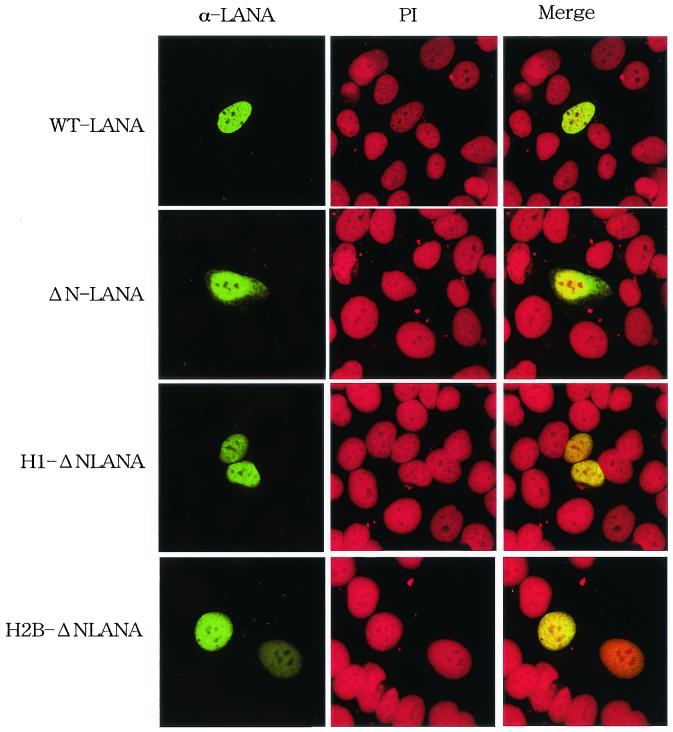
The N-terminal 32 amino acids of LANA1 fused with green fluorescent protein were localized at mitotic chromosomes, and deletion of this region inhibited the interaction of LANA1 with mitotic chromosomes (18). To examine the role of this chromosome binding site (CBS) of LANA1 in episome maintenance activity, we initially constructed a plasmid encoding the ΔN-LANA mutant (Fig. 1). The wild-type and mutant constructs were then transiently transfected into a human cervical carcinoma cell line, HeLa. Serum from a Kaposi's sarcoma patient with a high antibody titer against LANA1 detected the accumulation of wild-type LANA1 (green) in the nucleus (red) (Fig. 2). On the other hand, ΔN-LANA was detected both in the nucleus and in the cytoplasm, and the signal was stronger in the nucleus than in the cytoplasm.
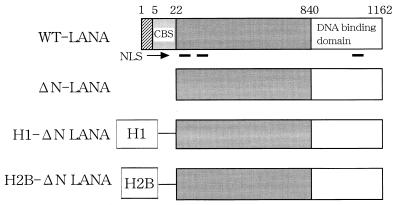
FIG. 1.
Structures of LANA1 and mutants. Numbers of amino acids of LANA1, ΔN-LANA, histone H1, and histone H2B and locations of the CBS, nuclear localization signal (NLS), and DNA binding domain of LANA1 are indicated. WT, wild type.
FIG. 2.
Localization of LANA1 and ΔN-LANA in HeLa cells. HeLa cells on glass slides were transfected with the indicated plasmids and cultured for 48 h. The cells were fixed with 1% paraformaldehyde containing 0.1% Triton X-100. Fixed cells were then stained with serum from a Kaposi's sarcoma patient (α-LANA) (green) and PI (red) and examined by laser scanning confocal microscopy (magnification, ×588). WT, wild type.
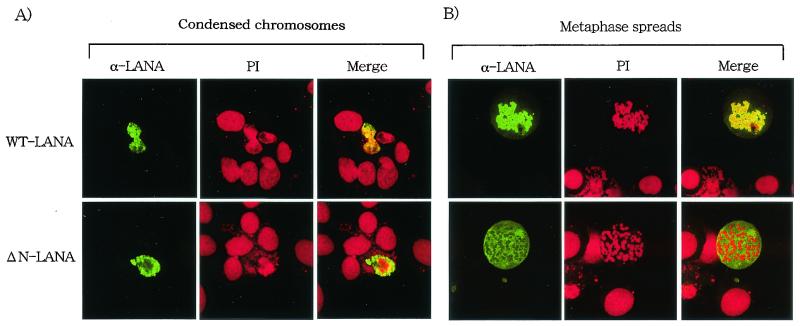
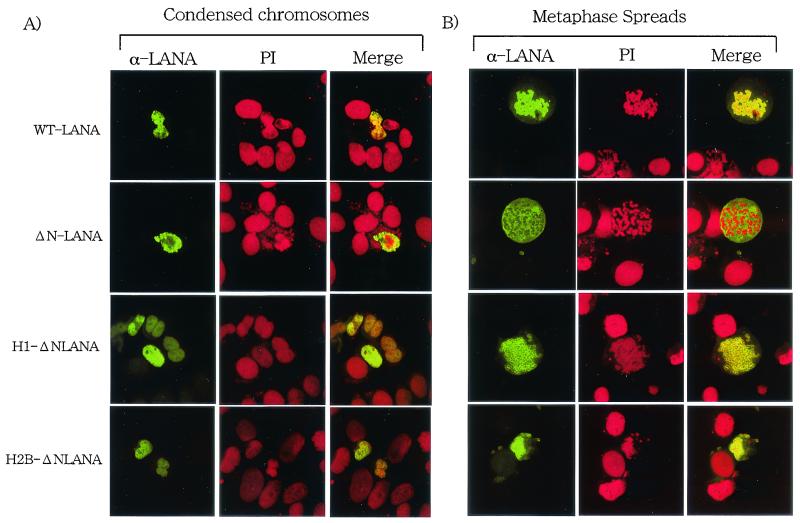
To study the chromosome binding activity of LANA1, HeLa cells transfected with the plasmids were treated with Colcemid and then further treated with a hypotonic buffer. Colcemid treatment increases the number of cells in the mitotic phase, and the hypotonic buffer causes the cells to swell, facilitating easier detection of the association with chromosomes in treated cells. In Colcemid-treated cells, LANA1 (green) was colocalized with mitotic chromosomes that were counterstained with PI (red) (Fig. 3A). On the other hand, much less ΔN-LANA was associated with chromosomes. Next, Colcemid-treated HeLa cells were harvested with trypsin. The harvested cells were further treated with the hypotonic buffer and were dropped immediately onto slides. This procedure disperses individual chromosomes on the slides (Fig. 3B). Again, wild-type LANA1 was mostly colocalized with individual chromosomes in metaphase spreads, and the signal was detected throughout individual spread chromosomes. In contrast, ΔN-LANA was mostly detected separately from mitotic chromosomes. These results indicated that the N-terminal 22 amino acids of LANA1 are essential for the tight association with mitotic chromosomes in human cells.
FIG. 3.
Association of LANA1 with mitotic chromosomes in HeLa cells. HeLa cells on glass slides were transfected with the indicated plasmids and cultured for 24 h. (A) The cells were treated with Colcemid, further treated with a hypotonic buffer, and fixed with 1% paraformaldehyde containing 0.1% Triton X-100. Fixed cells were then stained with serum from a Kaposi's sarcoma patient (α-LANA) (green) and PI (red) and examined by laser scanning confocal microscopy (magnification, ×828). WT, wild type. (B) Metaphase spreads of HeLa cells transfected with the indicated plasmids were prepared as described in Materials and Methods. The metaphase spreads were stained with serum from a Kaposi's sarcoma patient (α-LANA) (green) and PI (red) and examined bylaser scanning confocal microscopy (magnification, ×828).
The N-terminal CBS of LANA1 is essential for persistent episome maintenance.
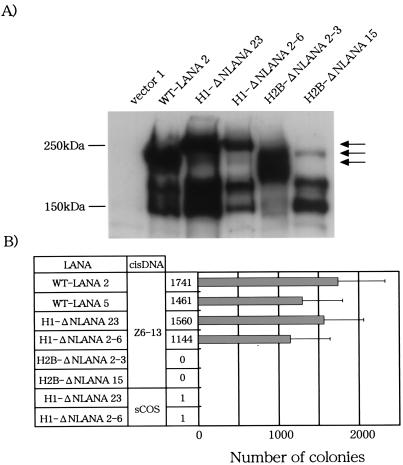
We next examined the plasmid maintenance activity of ΔN-LANA. Expression plasmids encoding ΔN-LANA and wild-type LANA1 were transfected into a human B-cell line (BJAB) together with the hygromycin resistance gene, and the cells were seeded in 96-well plates and cultured in the presence of hygromycin B. Resistant clones were examined for the expression of LANA1 by Western blot analysis (Fig. 4A). Anti-LANA antibody detected an approximately 220-kDa product in two BJAB clones transfected with LANA1 and ΔN-LANA, while the specific product was undetectable in BJAB clones transfected with the control plasmid (Fig. 4A). The antibody also detected several smaller products ranging from 130 to 190 kDa. The same antibody detected a similar 220-kDa product and several of the smaller ones in a KSHV-infected cell line (BCBL-1) (data not shown). These smaller products were likely to be produced by proteolytic cleavage, as discussed previously (18).
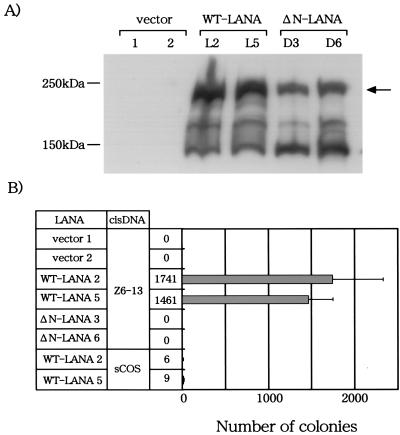
FIG. 4.
Long-term episome maintenance activity of LANA1 in BJAB cells. (A) Cell lysates were prepared from BJAB cells transfected with the indicated plasmids, and the amount of LANA1 protein in each lysate was measured by Western blot analysis with an anti-LANA antibody. The arrow indicates bands corresponding to the expected sizes of full-length LANA1 or ΔN-LANA proteins. WT, wild type. (B) BLAB/LANA1 and BJAB/ΔN-LANA cells were transfected with Z6-13 or sCOS by electroporation and then selected against G418 for at least 3 weeks. Extrachromosomal DNA was extracted from G418-resistant cells by the alkaline lysis method and introduced into competent bacteria. The numbers of ampicillin-resistant colonies were counted (actual numbers are listed to left of bar graph). Data represent the mean and standard deviation of at least three independent experiments.
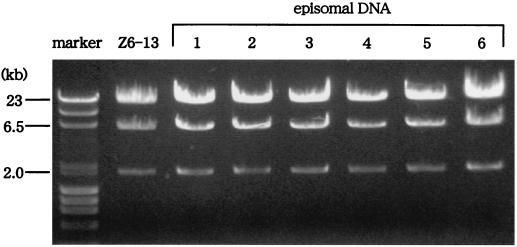
The BJAB clones expressing LANA1 or ΔN-LANA were then transfected with the Z6-13 plasmid containing the KSHV oriP or the sCOS control plasmid by the electroporation method, and the cells were selected against G418 resistance conferred by Z6-13 for 3 weeks. Episomal DNA was extracted from BJAB cells cultured in the presence of G418 for 3 weeks, and the plasmids were transfected into competent bacteria. The extracts from two independent BJAB/LANA1 clones transfected with Z6-13 induced approximately 1,500 bacterial colonies, whereas those from BJAB/vector clones did not produce bacterial colonies (Fig. 4B). Thus, LANA1 seems to maintain the Z6-13 plasmid in a human B-cell line in the long term. The plasmid maintenance activity of LANA1 was specific to Z6-13, since sCOS (which did not encode the KSHV sequence) was not well maintained in BJAB/LANA1 clones. Restriction enzyme digestion confirmed that all six plasmids recovered from BJAB/LANA1 clones transfected with Z6-13 were indeed Z6-13 (Fig. 5). These results were consistent with previous findings that LANA1 can maintain the KSHV oriP-based plasmid as an episome in BJAB cells (1). In contrast, the G418-selected BJAB cell line expressing ΔN-LANA (BJAB/ΔN-LANA cells) could not retain Z6-13 as an episome, although ΔN-LANA contains the Z6-13 binding domain (Fig. 1) (11). These results indicated that the N-terminal CBS of LANA1 is essential for the long-term maintenance of the KSHV oriP-based plasmid as an episome in BJAB cells.
FIG. 5.
The Z6-13 plasmid was maintained in BJAB/LANA1 clones. Plasmids were extracted from bacterial colonies originating from BJAB/LANA1/Z6-13 cells. The extracted plasmids and the Z6-13 plasmid were cut with EcoRV and electrophoresed on a 0.7% agarose gel, and the separated plasmid fragments were stained with 0.5 μg of ethidium bromide/ml. The markers are λ DNA fragments cut with HindIII and φX174 DNA fragments cut with HaeIII.
A chimera of LANA1 with histone H1 but not histone H2B mediates long-term episome maintenance.
The above results suggested that the interaction of LANA1 with mitotic chromosomes is essential for long-term episome maintenance. If this hypothesis were correct, then the episome maintenance activity of ΔN-LANA would be rescued by fusion with a chromosome binding protein. To evaluate this hypothesis, we generated a chimera of ΔN-LANA with either histone H1 or histone H2B, each of which is a well-known chromosome component (Fig. 1). Immunofluorescence analysis showed that these chimeric proteins predominantly accumulated in the nucleus of HeLa cells (Fig. 6) and were tightly associated with mitotic chromosomes, like wild-type LANA1 (Fig. 7).
FIG. 6.
Localization of chimeric LANA1 proteins in HeLa cells. HeLa cells on glass slides were transfected with the indicated plasmids and cultured for 36 h. The cells were fixed with 1% paraformaldehyde containing 0.1% Triton X-100. Fixed cells were then stained with serum from a Kaposi's sarcoma patient (α-LANA) (green) and PI (red) and examined by laser scanning confocal microscopy (magnification, ×732). WT, wild type.
FIG. 7.
Association of chimeric LANA1 proteins with mitotic chromosomes in HeLa cells. HeLa cells on glass slides were transfected with the indicated plasmids and cultured for 36 h. (A) The cells were treated with Colcemid, further treated with a hypotonic buffer, and fixed with 1% paraformaldehyde containing 0.1% Triton X-100. Fixed cells were then stained with serum from a Kaposi's sarcoma patient (α-LANA) (green) and PI (red) and examined by laser scanning confocal microscopy (magnification, ×888). WT, wild type. (B) Metaphase spreads of HeLa cells transfected with the indicated plasmids were prepared by a dropping method as described in Materials and Methods. The metaphase spreads were stained with serum from a Kaposi's sarcoma patient (α-LANA) (green) and PI (red) and examined by laser scanning confocal microscopy (magnification, ×888).
We next transfected the chimeric constructs into BJAB cells, and cells expressing the chimeric proteins were selected by hygromycin resistance. Two clones expressed a protein of the size expected for a LANA1 fusion with histone H1 (H1-ΔNLANA), and two clones expressed a protein of the size expected for a LANA1 fusion with H2B (H2B-ΔNLANA). These BJAB clones were then transfected with Z6-13 or sCOS and selected against G418. Episomal DNA was extracted from BJAB cells selected against G418 for 3 weeks, and the number of episomes was determined by a bacterial colony assay. Samples from two BJAB clones expressing H1-ΔNLANA reproducibly produced bacterial colonies, and the numbers were equivalent to those obtained with wild-type LANA1 (Fig. 8). Like that of wild-type LANA1, the episome maintenance activity of H1-ΔNLANA was specific to Z6-13, since sCOS was not rescued from BJAB clones expressing H1-ΔNLANA. These results supported the hypothesis that the association of LANA1 with mitotic chromosomes is essential for the long-term maintenance of the oriP-based plasmid in dividing cells. In contrast to the LANA1 chimera with histone H1, the LANA1 chimera with H2B could not maintain Z6-13 episomes, measurable as bacterial colonies, in BJAB cells, although the reason for the difference between the two chimeric proteins is unknown.
FIG. 8.
Episome maintenance activity of chimeric LANA1 proteins. (A) Cell lysates were prepared from BJAB cells transfected with the indicated plasmids, and the amount of LANA1 protein in each lysate was measured by Western blot analysis with an anti-LANA antibody. Arrows indicate bands corresponding to the expected sizes of full-length LANA1, H1-ΔNLANA, or H2B-ΔNLANA proteins. WT, wild type. (B) BJAB cells expressing H1-ΔLANA and BJAB cells expressing H2B-ΔLANA were transfected with Z6-13 or sCOS by the electroporation method and then selected against G418 for at least 3 weeks. Extrachromosomal DNA was extracted from G418-resistant cells by the alkaline lysis method and introduced into competent bacteria. The numbers of ampicillin-resistant colonies were counted (actual numbers are listed to left of bar graph). Data represent the mean and standard deviation of at least three independent experiments.
DISCUSSION
Several viruses, including herpesvirus, persistently infect host cells as extrachromosomal double-stranded episomal DNA. These episomes replicate only once per cell cycle. About 50 to 100 copies of episomes are stably maintained in virus-infected cells. To constantly maintain such low-copy-number episomes in host cells, almost equal numbers of viral episomes should segregate into the two daughter cells every cell division. Therefore, these viruses should have a skillful system for segregating viral episomes in dividing cells.
In the present study, we showed that deletion of the N-terminal 22 amino acids (CBS) of LANA1 severely reduced the interaction with mitotic chromosomes and inhibited the long-term episome maintenance of the Z6-13 plasmid containing the KSHV oriP in a human B-cell line (Fig. 2 to 4). Moreover, a cellular chromosome component protein, histone H1, rescued both the chromosome interaction activity of the mutant (ΔN-LANA) and its long-term episome maintenance activity (Fig. 6 to 8). These results indicated that the interaction of LANA1 with mitotic chromosomes is essential for the long-term episome maintenance of the KSHV oriP-based plasmid. Several reports have demonstrated that LANA1 tethers viral episomes to mitotic chromosomes (1, 7). Considered together with these earlier findings, our results support the hypothesis that LANA1 tethers viral episomes to mitotic chromosomes, an activity which is essential for the appropriate segregation of viral episomes into new daughter cells during cell division (1, 7).
Unlike histone H1, histone H2B fused with ΔN-LANA did not support the long-term episome maintenance of the KSHV oriP-based plasmid in BJAB cells, although the chimeric protein was detected in association with mitotic chromosomes. H1 is a linker histone, while H2B is a core histone. Thus, the proper topology of LANA1 within mitotic chromosomes may be required for this function. Alternatively, histone H1 but not histoneH2B may have additional functions required for long-term episome maintenance by LANA1, since histone H1 was shown to interact with LANA1 in vitro as well as in vivo (7).
EBV EBNA1 and bovine papillomavirus E2 maintain the viral genome as an episome in host cells, and these proteins are also associated with mitotic chromosomes (3). Mutation of the E2 CBS inhibited episome maintenance activity (15). Like LANA1 mutants, EBNA1 mutants that lost the ability to interact with chromosomes were inactive for episome maintenance, but these activities of EBNA1 were rescued by fusion with histone H1 (12). Thus, these DNA viruses probably utilize similar strategies to efficiently segregate viral episomes in infected cells, an activity which is essential for long-term persistent infections in human cells.
EBP2 is a cellular factor that interacts with the EBNA1 CBS; it was isolated by a yeast two-hybrid assay (23). A subsequent study indicated that EBP2 mediates the interaction of EBNA1 with mitotic chromosomes and that EBP2 is an essential factor for the episome maintenance activity of EBNA1. For instance, EBNA1 does not support the long-term retention of the EBV oriP-based plasmid in yeast cells, but it can do so in the presence of EBP2 (13). A cellular protein may also interact with the LANA1 CBS to recruit mitotic chromosomes. It is, however, unlikely that EBP2 is the protein recruiting mitotic chromosomes for LANA1, since LANA1 and EBNA1 localize at different sites in mitotic chromosomes (25).
LANA1 interacts with several cellular proteins, such as histone H1, ATF4/CREB2, RING3, and SAP30 (7, 14, 16, 19). Among them, histone H1 is a candidate for mediating the interaction of LANA1 with mitotic chromosomes (7). The N-terminal CBS (5 to 22 amino acids) of LANA1 contains a cluster of basic amino acids. Thus, this region of LANA1 may not be the interaction surface for histone H1, since histone H1 is also a basic protein. On the basis of these observations, further analysis is necessary to elucidate precisely how LANA1 interacts with mitotic chromosomes.
We have used a new method to measure the episome maintenance activity of LANA1. The numbers of plasmids (Z6-13) that were maintained in BJAB/LANA1 cells were estimated as bacterial colonies. At least two methods are available to analyze the episome maintenance activity of viral proteins, the Gardella assay and the Southern blot assay (1, 2). The bacterial transfer assay is easier and more quantitative than the Gardella assay and is therefore a good alternative for measuring the episome maintenance activity of viral proteins, including LANA1.
Acknowledgments
H.S. and M.F. contributed equally to this study.
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas and for Scientific Research of Japan.
We thank Mary Ballestas, Kenneth Kaye, and Patrick Moore for pSGLANA and Z6-13 plasmids and Fransis Kasolo for serum from a Kaposi's sarcoma patient with a high antibody titer against LANA1. We also thank Ryoko Fujita for excellent technical assistance.
REFERENCES
- 1.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through a cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calos, M. P. 1998. Stability without a centromere. Proc. Natl. Acad. Sci. USA 95:4084-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 9.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupin, N., T. L. Diss, P. Kellam, M. Tulliez, M. Q. Du, D. Sicard, R. A. Weiss, P. G. Isaacson, and C. Boshoff. 2000. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood 95:1406-1412. [PubMed] [Google Scholar]
- 11.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 17.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 18.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultz, T. F., Y. Chang, and P. S. Moore. 1998. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8), p. 87-134. In D. J. McCance (ed.), Human tumor viruses. ASM Press, Washington, D.C.
- 22.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 25.Szekely, L., C. Kiss, K. Mattsson, E. Kashuba, K. Pokrovskaja, A. Juhasz, P. Holmvall, and G. Klein. 1999. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J. Gen. Virol. 80:2889-2900. [DOI] [PubMed] [Google Scholar]