Abstract
The RNA genome of the human T-cell leukemia virus type 1 (HTLV-1) codes for proteins involved in infectivity, replication, and transformation. We report in this study the characterization of a novel viral protein encoded by the complementary strand of the HTLV-1 RNA genome. This protein, designated HBZ (for HTLV-1 bZIP factor), contains a N-terminal transcriptional activation domain and a leucine zipper motif in its C terminus. We show here that HBZ is able to interact with the bZIP transcription factor CREB-2 (also called ATF-4), known to activate the HTLV-1 transcription by recruiting the viral trans-activator Tax on the Tax-responsive elements (TxREs). However, we demonstrate that the HBZ/CREB-2 heterodimers are no more able to bind to the TxRE and cyclic AMP response element sites. Taking these findings together, the functional inactivation of CREB-2 by HBZ is suggested to contribute to regulation of the HTLV-1 transcription. Moreover, the characterization of a minus-strand gene protein encoded by HTLV-1 has never been reported until now.
Human T-cell leukemia virus type 1 (HTLV-1), the etiologic agent of adult T-cell leukemia and HTLV-1-associated myelopathy/tropical spastic paraparesis, is a T-cell-tropic virus that belongs to the HTLV-bovine leukemia virus (HTLV-BLV) group of the retrovirus family. Other group members include HTLV-2, BLV, and the simian T-cell leukemia viruses. All retroviruses share similarity in their genome organization, with four genes encoding proteins necessary for the production of infectious virions, the order of these genes being invariably gag, pro, pol, env (from 5′ to 3′) (Fig. 1A). gag codes for internal structural proteins of the virion, pro codes for the viral protease, pol codes for reverse transcriptase, and env codes for envelope glycoproteins of the virion. The expression of the HTLV-1 genome is controlled by two regulatory genes, the rex and tax genes, located in the 3′ region of the viral genome. Tax is a potent transactivator of viral transcription (5, 30), and Rex acts at the posttranscriptional level by modulating the transport of the viral RNAs (14). Moreover, the HTLV-1 genome carries between the env gene and the 3′ long terminal repeat (LTR) other open reading frames encoding proteins with unknown functions. All these viral proteins are encoded by the viral plus-strand RNA.
FIG. 1.
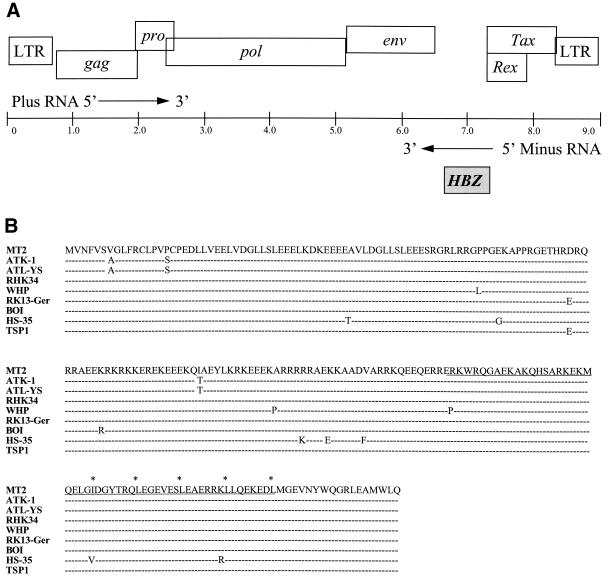
The HTLV-1 genome encodes the bZIP transcription factor HBZ. (A) Organization of the HTLV-1 genome. The HTLV-1 provirus genome (in kilobase pairs) is represented by a line. The viral genes encoded by the plus-strand RNA (white boxes) are shown above the genome line (only the genes whose functions have been clearly established are represented). The HBZ gene encoded by the minus-strand RNA is represented by a greyish box, below the line. (B) Comparison of the amino acid sequence of HBZ characterized from the HTLV-1-infected MT2 cell line with the HBZ protein from different HTLV-1 isolates. The amino acid sequence of MT2 HBZ is shown. The bZIP domain is underlined, and the leucine residues within the leucine zipper are indicated by asterisks. Only differences in the other HTLV-1 isolates are indicated, while homologies are represented by successive dashes. The different strains (with GenBank accession numbers in parentheses) are ATK-1 (J02029), ATL-YS (U19949), RHK34 (L03562), WHP (AF259264), RK13-Ger (AF042071), BOI (L36905), HS-35 (D13784), and TSP1 (M86840).
The trans-activation of the viral transcription by Tax operates through three 21-bp repeats (6, 28) located in the U3 region of the LTR. These conserved repeats, called Tax-responsive elements (TxREs), contain an imperfect cyclic AMP response element (CRE) (17) that is recognized by CREB (4, 37, 38) and CREM (29), two members of the activating transcription factor/CRE-binding protein (ATF/CREB) family. These factors are characterized by basic leucine zipper (bZIP) C-terminal structures required for DNA binding and protein dimerization. To activate transcription of the HTLV-1 genome, Tax interacts with CREB or CREM bound to the TxREs (4, 38) and then can recruit the transcriptional coactivator CREB-binding protein (CBP) (21). The recruitment of CBP to the HTLV-1 promoter could be involved in the acetylation of the core histone tails (7, 11).
Another member of the ATF/CREB family, CREB-2 (20), also known as ATF-4 (12) or TAXREB 67 (32), cooperates with the viral Tax protein to enhance the transcription of the HTLV-1 promoter (9, 27). We have shown that CREB-2 binds to the TxREs and that CREB-2 binding to each of the 21-bp motifs is enhanced by Tax (10). In this paper, we report that CREB-2 is able to interact with a second viral protein that had never been characterized until now. This novel viral protein is encoded by the minus-strand RNA that is transcribed by a functional promoter present in the antisense strand of the 1.8-kb 3′ terminus of the HTLV-1 proviral genome containing the 3′ LTR (22). As this protein contains a bZIP domain in its C-terminal region, we have named it HBZ (for HTLV-1 bZIP factor). We show here that HBZ contains a transcriptional activation domain and directly interacts with CREB-2, CREB-2 and HBZ associating via their bZIP domains. However, in spite of the presence of an activation domain, HBZ down-regulates the viral transcription. Indeed, HBZ can act as a negative regulator of CREB-2-dependent transcription by forming heterodimers that are unable to bind to the CRE and TxRE sites.
MATERIALS AND METHODS
Plasmids.
The complete HBZ coding region was amplified from cDNA prepared from MT2 cells and cloned into the plasmid pGEX-4T-2 (Pharmacia), and the HBZ DNA encoding the different truncated mutants was generated by PCR amplification on pGEX-HBZ. DNA amplified by PCR was then subcloned into different vectors: the bacterial expression vector pQE-31 (Qiagen), the eukaryotic vector pBind (Promega) containing the GAL4 DNA-binding (GAL4 DB) domain, the eukaryotic expression vector pCI-neo (Promega), and the plasmid pEGFP-C1 (Clontech). All the different plasmids used during this work and expressing CREB-2 (pGEX-CREB-2, pGEX-CREB-2263-351, pGEX-CREB-21-262,pQE-CREB-2, pLEXA-CREB-2263-351, pCI-CREB-2) Tax (pQE-Tax and pSG-Tax), and Tat (pBg312HIV-1Lai-Tat) have already been described elsewhere (8-10).
Screening for proteins that interact with the CREB-2 bZIP domain.
MT2 cDNA fused to the GAL4 activation domain of the pGAD10 vector (9) was screened using the CREB-2 bZIP domain as bait fused to the LEXA DNA-binding domain of the pBTM116 vector. The two-hybrid screen was performed as already described (9). Briefly, pLEXA-CREB-2 bZIP and the fusion cDNA library were cointroduced into the L40 Saccharomyces cerevisiae strain by the lithium acetate method (15). The L40 yeast strain possesses the His synthase gene (his3) and the lacZ gene under the control of LEXA binding sites. From approximately 9 × 106 clones screened, we selected robust colonies growing on agar medium lacking Trp, Leu, and His, for those that contained both types of plasmids (Leu+ and Trp+) and that also expressed interacting hybrid proteins (His+). Selected transformants were assayed for the expression of lacZ by the β-galactosidase filter assay as described in the Clontech protocol. Plasmid DNA of the clones that were strongly positive for β-galactosidase activity was extracted, analyzed by digestion with restriction enzymes, and sequenced.
Expression of HBZ tagged with GFP in COS7 cells.
To express HBZ and HBZΔbZIP with a green fluorescent protein (GFP) tag, the coding sequences of both proteins were subcloned into the vector pEGFP-C1. COS7 cells were transfected using the FuGENE 6-mediated transfection method (Roche) with 4 μg of expression vector. Cells were cultivated on the glass slides and then analyzed by fluorescence 48 h after transfection. Analysis of the green fluorescence was performed with a Leica DMR immunofluorescence microscope.
Transfections and luciferase assays.
CEM cells were transiently cotransfected according to the previously published procedure (31). Five micrograms of pACβ1 (β-galactosidase-containing reference plasmid) was included in each transfection for controlling of the transfection efficiency. The total amount of DNA in each transfection was the same, the balance being made up with empty plasmids. Cell extracts equalized for protein content were used for luciferase and β-galactosidase assays. For the assays with the GAL4-binding site promoter-reporter plasmid, the wild-type HBZ and the truncated mutants fused in frame with the GAL4 DB domain into the vector pBIND were cotransfected in CEM cells in the presence of the luciferase reporter plasmid pG5luc containing five GAL4 binding sites upstream of a minimal TATA box.
Protein expression and purification.
The bacterial expression vectors pQE containing either the HBZ bZIP domain, CREB-2, or Tax cDNA inserts were transformed into Escherichia coli M15. The N-terminal six-His-tagged proteins were purified as described by the manufacturer (Qiagen), dialyzed against binding buffer (see below) without bovine serum albumin (BSA), and kept at −80°C.
Streptavidin-biotin complex assay.
Biotinylated oligonucleotides corresponding to the HTLV-1 21-bp repeats TxRE III (5′-TCGACGTCCTCAGGCGTTGACGACAACCCCTCAC-3′) and somatostatin CRE (5′-GGTTCCTCCTTGGCTGACGTCAGAGAGAGA-3′) were annealed with their complementary oligonucleotides to form a double-stranded DNA. Biotinylated double-stranded DNA was incubated with bacterially produced proteins in 200 μl of binding buffer containing 50 mM Tris (pH 7.5), 500 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5 mM MgCl2, 0.1% Triton, 5% glycerol, and BSA (10 mg/ml) for 2 h at room temperature before addition of streptavidin beads (Pierce). After 1 h of incubation at 4°C, the beads were extensively washed with binding buffer without BSA. The proteins which remained bound to the beads were eluted in sodium dodecyl sulfate (SDS) loading buffer and analyzed by Western blotting.
Immunoprecipitation and Western blot assays.
Protein extracts were electrophoresed onto SDS-10% polyacrylamide gel (SDS-10% PAGE) and blotted to polyvinylidene difluoride membranes (Millipore). The blot was then incubated 1 h at room temperature with a blocking solution (Tris-buffered saline containing 5% milk) prior to addition of antiserum. After 2 h at 20°C, the blot was washed four times with Tris-buffered saline-0.2% Tween 20 and incubated for 1 h with either donkey anti-goat, goat anti-mouse, or goat anti-rabbit immunoglobulin-peroxidase conjugate. After three washes, the membrane was incubated with enhanced chemiluminescence reagent (Amersham). The membrane was then exposed for 0.5 to 5 min to Hyperfilms-ECL (Amersham). An immunoprecipitation assay was carried out as already described (3, 31).
The anti-CREB-2 polyclonal serum was purchased from Santa Cruz Biotechnology Inc., Santa Cruz, Calif., and the anti-Tax monoclonal antibody was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health; HTLV-1 Tax hybridoma 168A51-42 (Tab176) was obtained from B. Langton. The anti-HBZ serum was obtained by immunizing rabbits with purified six-His-tagged HBZ polypeptide corresponding to the bZIP domain of HBZ expressed from the bacterial expression vector pQE-31 (Qiagen). The N-terminal six-His-tagged protein was purified as described by the manufacturer.
RESULTS
The HTLV-1 genome encodes a bZIP transcription factor, HBZ.
CREB-2 is known to form heterodimers with other bZIP transcription factors such as C/EBPβ (26), Nrf2 (13), and CHOP (8). To identify other bZIP factors capable of interacting with CREB-2, the CREB-2 bZIP (amino acids 263 to 351) domain fused to the DNA-binding domain of LEXA was used as bait to screen, by the yeast two-hybrid approach, a cDNA library constructed from the HTLV-1-infected MT2 cell line by Gachon et al. (9). Of 9 × 106 transformants, several colonies positive for β-galactosidase activity were isolated from histidine-lacking plates. Two of these colonies contained the same cDNA coding for the C terminus (amino acids 160 to 209) of an unknown viral protein encoded by the complementary strand of the HTLV-1 RNA genome (Fig. 1A) of all the HTLV-1 isolates for which the sequences are accessible in the GenBank data base (Fig. 1B). Comparison of the amino acid sequence of this novel viral protein with databases revealed that the sequence has several characteristics of a bZIP transcription factor (Fig. 2). The C-terminal end contains one isoleucine and four leucines, each separated by six other amino acids, suggesting a leucine zipper structure (residues 164 to 192). Immediately upstream of this zipper sequence is an arginine-lysine-rich basic domain (residues 140 to 163), thought to be necessary for DNA binding by bZIP transcription factors.
FIG. 2.
Comparison of the amino acid sequences of the basic domains (at top; residues 140 to 164) and the leucine zipper structures (at bottom; residues 164 to 193) of HBZ and several other bZIP transcription factors. The conserved amino acids are indicated in boldface type.
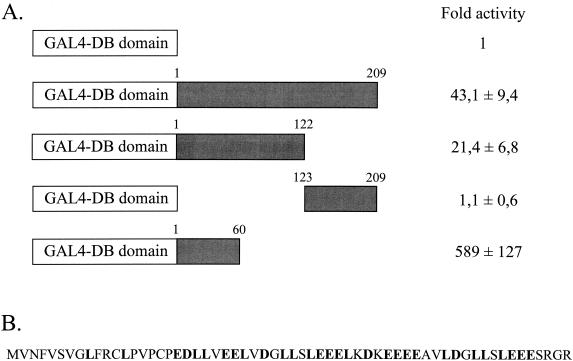
To confirm that HBZ could be a transcriptional factor, the yeast GAL4 DB domain was linked in frame to HBZ. GAL4-HBZ was assayed in CEM cells by using the luciferase reporter vector pG5luc that contains five GAL4-binding sites upstream of a minimal TATA box. We found that GAL4-HBZ stimulated the luciferase activity 43-fold (Fig. 3A). To map the transcriptional activation domain within HBZ, we fused the N-terminal (residues 1 to 122) or C-terminal (residues 123 to 209) regions to the GAL4 DB domain and measured luciferase activation in cotransfected CEM cells. The N terminus gave rise to a significant increase in reporter activity compared with the GAL4 DB domain alone, since the luciferase activity was overstimulated about 20-fold in the presence of the N-terminal domain of HBZ (Fig. 3A). When a shorter N-terminal domain of HBZ (residues 1 to 60) was fused to the GAL4 DB domain, the luciferase activity was overstimulated about 600-fold. On the other hand, the C terminus of HBZ was unable to activate the luciferase expression (Fig. 3A). This result shows that HBZ contains a potential transcriptional activation domain located in the N terminus (residues 1 to 60). As shown in Fig. 3B, the N-terminal domain of HBZ is a region rich in leucines (residues 9 to 53, 30% leucines) and acidic amino acids (residues 19 to 56, 46% acidic residues).
FIG. 3.
The N terminus of HBZ contains a potent transcriptional activation domain. (A) CEM cells were cotransfected with 5 μg of pACβ1 (β-galactosidase containing reference plasmid), 2 μg of the luciferase reporter vector pG5luc, and 2 μg of the eukaryotic vector pBIND expressing the GAL4 DB domain alone or fused to either the full-length HBZ product (209 amino acids long), the N-terminal region (residues 1 to 122 and 1 to 60), or the C-terminal domain (residues 123 to 209). Luciferase values were normalized for β-galactosidase activity and are expressed as increases relative to that of cells transfected with pSG-5 and pBIND only expressing the GAL4 DB domain. Values are presented as means ± standard deviations (n = 3). The GAL4-DB domain and the different regions of HBZ are represented by white and greyish boxes, respectively. (B) Amino acid sequence of the HBZ N terminus (residues 1 to 60). The N terminus is rich in acidic and leucine residues (indicated in boldface type in the sequence).
The bZIP family can be divided into three groups on the basis of binding site preference: the C/EBPs (23), the AP1 group of transcription factors (33), and the CREB/ATF family (25). To determine whether HBZ could belong to one of these groups, HBZ activity was tested on luciferase reporter constructs carrying synthetic promoters corresponding to either C/EBP, or AP-1, or CRE sites. However, HBZ was unable to activate significantly transcription from these different sites (data not shown), suggesting that HBZ could recognize unique DNA sequences that are distinct from the tested sites.
Expression of HBZ in vivo.
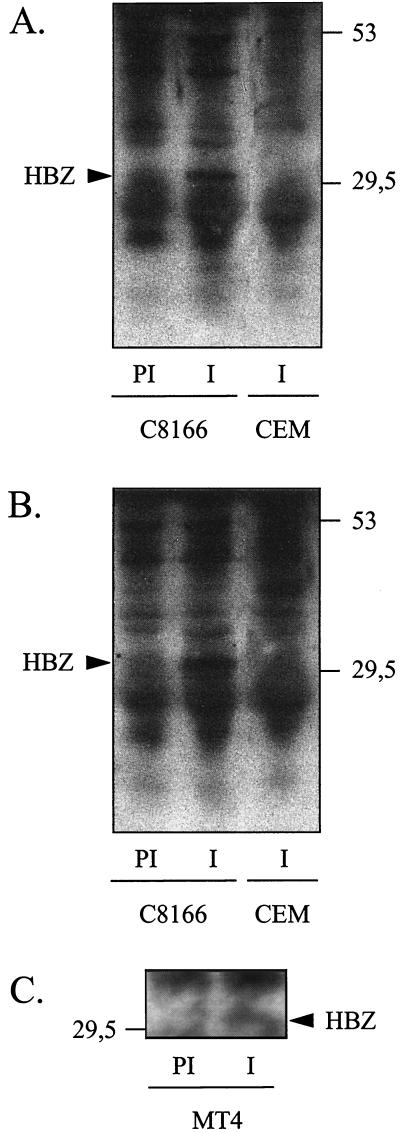
To raise polyclonal antibodies against HBZ, two rabbits were immunized with highly purified HBZ polypeptide corresponding to the last 87 amino acids of HBZ. We studied the expression of HBZ in vivo with both antisera by performing immunoprecipitation with anti-HBZ from extracts of an HTLV-1-infected C8166 cell line. A protein of 31 kDa, absent in extracts of uninfected cells, was detected with both antisera but not with the preimmune sera (Fig. 4A and B), the size of the detected protein being consistent with the size of recombinant HBZ expressed by transient transfection in eukaryotic cell lines (data not shown). In addition, by the same approach, we were able to detect HBZ also in MT4 cells that are infected by HTLV-1 too (Fig. 4C). These results clearly show that HBZ is expressed in T cells infected by HTLV-1. On the other hand, we were unable to detect HBZ by this approach in extracts from the MT2 cell line (data not shown), although the cDNA library screened by the two-hybrid method was constructed from MT2 mRNA. However, this inability to detect HBZ could be explained by a low level of HBZ expression in the MT2 cells. Differences in the level of viral protein expression between HTLV-1-infected cell lines have already been reported for Tax (16) due to the presence of defective proviruses. Thus, the profile of Tax in MT2 cells corresponds to a weakly expressed authentic 40-kDa Tax but also to a 69-kDa protein that is a fusion between the envelope and the Tax-coding sequence (16).
FIG. 4.
Detection of HBZ in vivo by immunoprecipitation. Proteins were immunoprecipitated from the lysate of either the HTLV-1-infected T-cell line C8166 or the uninfected T-cell line CEM using preimmune sera (PI) or the anti-HBZ sera (I) produced by two different rabbits (A and B). Precipitates were analyzed by SDS-PAGE and immunoblotting with anti-HBZ serum. The molecular mass markers (in kilodaltons) are indicated on the right of the figure. (C) The same approach was carried out from the lysate of the HTLV-1-infected MT4 cell line using preimmune serum (PI) or the anti-HBZ serum (I).
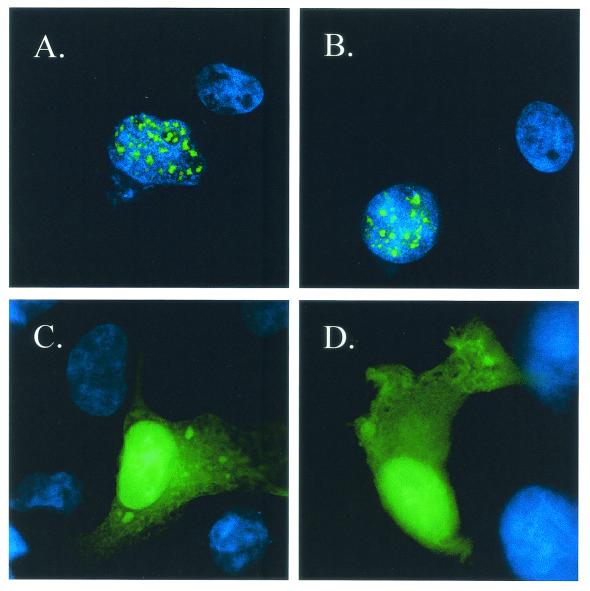
Next, we investigated the subcellular localization of HBZ in vivo. COS7 cells were transfected with vectors expressing HBZ tagged with GFP, fused to the N-terminal end of HBZ. As shown in Fig. 5, HBZ exhibits a granular distribution in the nucleus. To be sure that the position and the size of the GFP tag did not influence the HBZ localization, a smaller tag, containing the myc epitope, was fused to the C-terminal end of HBZ. In COS7 cells transfected with this construct, HBZ gives the same staining pattern as that observed with the GFP tag (data not shown). In addition, we fused to GFP a shorter form of HBZ (residues 1 to 122) from which the bZIP domain had been deleted. Indeed, the basic region of bZIP factors has been described to be involved in the targeting of the protein to the nucleus (2, 35). As shown in Fig. 5, this mutant exhibits a staining pattern identical to that of the wild-type GFP, with a diffuse distribution throughout the cytoplasm and the nucleus. Taken together, these results demonstrate that HBZ is a nuclear protein and confirm that HBZ belongs to the family of the bZIP transcription factors.
FIG. 5.
Immunofluorescence microscopy analysis of the subcellular localization of HBZ with an N-terminal GFP tag in vivo. COS7 cells were transfected with expression vectors encoding either GFP-HBZ (A and B), GFP (C), or GFP-HBZΔbZIP (D). Cells were cultivated on the glass slides, fixed, and stained with Hoechst solution, and then the green fluorescence was analyzed by immunofluorescence microscopy. The specificity of the immunofluorescent staining is indicated by the absence of signal in flanking untransfected cells. The blue fluorescence of the nuclei (Hoechst staining) is visualized by UV illumination.
HBZ interacts with CREB-2 in vitro and in vivo.
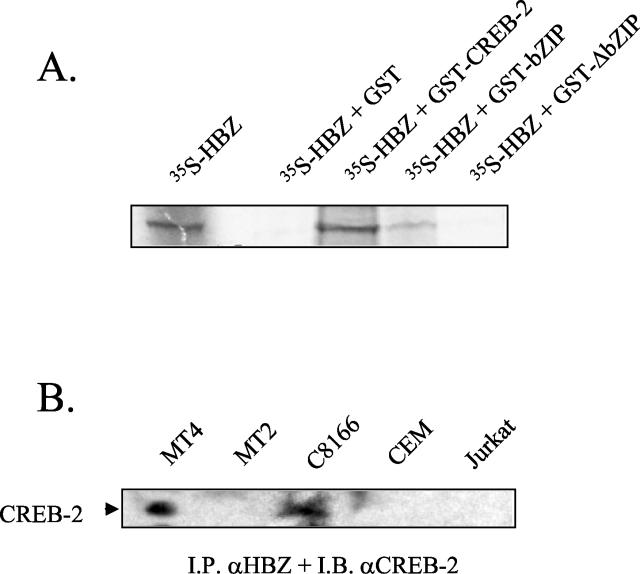
The physical interaction between HBZ and CREB-2 was first analyzed in vitro using recombinant proteins. A fusion protein of CREB-2 with glutathione S-transferase (GST) was produced in E. coli, and the binding to [35S]methionine-labeled HBZ that was produced in rabbit reticulocyte lysate was analyzed. As shown in Fig. 6A, [35S]HBZ bound to GST-CREB-2 but not to GST alone. In addition, [35S]HBZ bound to the CREB-2 bZIP domain fused to GST (Fig. 6A, lane GST-bZIP) but not to GST-CREB-2 from which the bZIP domain had been deleted (Fig. 6A, lane GST-ΔbZIP), confirming that CREB-2 interaction with HBZ is dependent on its bZIP domain. These results demonstrate that HBZ can interact with CREB-2 in vitro.
FIG. 6.
HBZ interacts with CREB-2. (A) In vitro binding assays of HBZ to CREB-2. Equal amounts of GST, GST-CREB-2, GST-CREB-2263-351 (lane GST-bZIP), and GST-CREB-21-262 (lane GST-ΔbZIP) immobilized on glutathione-Sepharose beads were incubated with [35S]HBZ, and bound proteins were analyzed by SDS-PAGE and autoradiography. The first lane corresponds to in vitro-translated [35S]HBZ. (B) Binding of HBZ to CREB-2 in vivo. Proteins from MT4, MT2, C8166, CEM, and Jurkat cell total lysate were immunoprecipitated with rabbit anti-HBZ (I.P. αHBZ), and immunoprecipitated proteins were analyzed by immunoblotting with goat anti-CREB-2 (I.B. αCREB-2).
To confirm that both endogenous HBZ and CREB-2 were also associated in vivo, immunoprecipitations of cell lysates of HTLV-1-infected (C8166, MT2, and MT4) and uninfected (Jurkat and CEM) cell lines were performed with anti-HBZ serum, and the immunoprecipitates were probed by Western blotting with a goat anti-CREB-2 serum. As expected, this approach revealed the presence of a complex between HBZ and CREB-2 in C8166 and MT4 extracts (Fig. 6B). On the other hand, such a complex was not detected in the Jurkat, CEM, and MT2 cell lines. Thus, this experiment demonstrates that endogenous HBZ and CREB-2 interact in C8166 and MT4 cell lines.
HBZ down-regulates the HTLV-1 transcription.
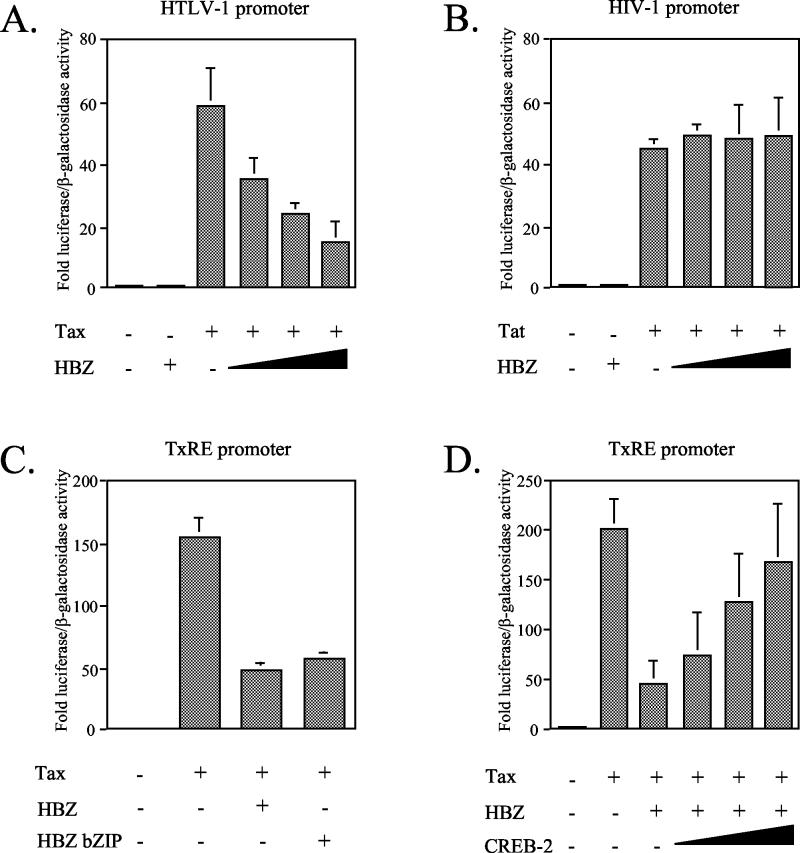
CREB-2 is known to function in association with Tax to trans-activate the HTLV-1 transcription (9, 27) by forming a stable complex CREB-2/Tax/CBP bound to the TxREs (10). For this reason, we analyzed the effects of HBZ on the viral transcription by cotransfecting CEM cells with a luciferase reporter construct carrying the HTLV-1 promoter and increasing amounts of pCI-HBZ in the presence of Tax expression vector pSG-Tax. As shown in Fig. 7A, Tax alone activated expression of the luciferase reporter gene by about 60-fold, but this stimulation was inhibited in the presence of HBZ (only a 15-fold stimulation). In order to be sure that this inhibition was not due to a nonspecific effect of HBZ on cellular transcription, HBZ was also tested in CEM cells with the HIV type 1 (HIV-1) promoter in the presence of the HIV-1 activator Tat. Under these conditions, HBZ did not down-regulate the HIV-1 transcription stimulated by Tat (Fig. 7B), confirming that the decrease in transactivation by HBZ from the HTLV-1 promoter in the presence of Tax was specific. Thus, HBZ is able to downregulate the HTLV-1 transcription.
FIG. 7.
HBZ down-regulates the HTLV-1 transcription by interacting with CREB-2. CEM cells were cotransfected with the following: (A) 2 μg of HTLV-1 LTR-luciferase, 1 μg of Tax expression vector pSG-Tax, and pCI-HBZ (0, 1, 3, or 9 μg) (the luciferase values are expressed as increases relative to that of cells transfected with pSG-5, pCI-neo, and HTLV-1 LTR-luciferase); (B) 2 μg of HIV-1 LTR-luciferase, 1 μg of the Tat expression vector pBg312HIV-1Lai-Tat, and pCI-HBZ (0, 1, 3, or 9 μg) (the luciferase values are expressed as increases relative to that of cells transfected with HIV-1 LTR-luciferase without Tat and HBZ); (C) 2 μg of HTLV-1 TxRE-luciferase, 1 μg of pSG-Tax, and 5 μg of pCI-neo expressing either HBZ or the HBZ bZIP domain (the luciferase values are expressed as increases relative to that of cells transfected with pSG-5, pCI-neo, and HTLV-1 TxRE-luciferase); (D) 2 μg of HTLV-1 TxRE-luciferase, 1 μg of pSG-Tax, 5 μg of pCI-HBZ, and pCI-CREB-2 (0, 1, 3, or 9 μg) (the luciferase values are expressed as fold increase relative to that of cells transfected with pSG-5, pCI-neo, and HTLV-1 TxRE-luciferase). For all the cotransfections, the total amount of DNA in each series of transfection was equal, the balance being made up with empty plasmids. and the luciferase values were normalized for β-galactosidase activity. Values represent means plus standard deviations (error bars) (n = 3).
To determine whether the negative effect of HBZ on the viral transcription might be mediated through an interaction with CREB-2, we performed two different luciferase reporter assays. We first analyzed by a transient-transfection assay performed in CEM cells the ability of the HBZ bZIP domain to inhibit the Tax transactivation of a synthetic promoter containing three tandem copies of the promoter-proximal TxRE. Effectively, the HBZ bZIP domain was able to down-regulate the transcription in the presence of Tax (Fig. 7C), confirming that an interaction between HBZ and endogenous CREB-2 could be involved in this negative effect. Next, we analyzed the effects of HBZ on the viral transcription by transfecting CEM cells with either HBZ alone or in the presence of increasing amounts of pCI-CREB-2. Interestingly, cotransfection of increasing amounts of pCI-CREB-2 resulted in a significant increase in luciferase activity (Fig. 7D). Lastly, we also checked that HBZ was unable to interact with Tax (data not shown). Taken together, our results suggest that HBZ is able to down-regulate the HTLV-1 transcription by interacting with CREB-2.
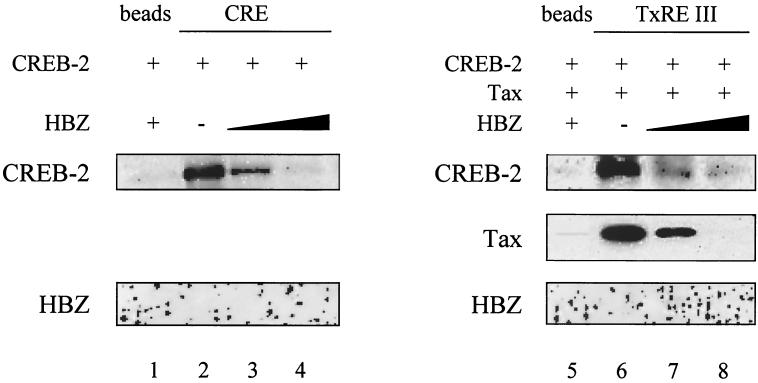
Recently, we have demonstrated that the cellular bZIP transcription factor CHOP was capable of inhibiting the HTLV-1 transcription (8), probably by forming heterodimers with CREB-2 that are unable to bind to the TxREs. In light of these results, we tested whether dimerization between HBZ and CREB-2 results in a heterodimer unable to bind to the cellular CRE consensus site. For this purpose, we used the streptavidin-biotin complex assay previously used by Gachon et al. for the characterization of the complexes formed among CREB-2, Tax, and TxREs (10). A double-stranded oligonucleotide corresponding to the somatostatin CRE was incubated with CREB-2 in the absence or in the presence of the bZIP domain of HBZ. CREB-2 alone bound to CRE (Fig. 8, lane 2), but in the presence of HBZ bZIP, CREB-2 interaction with CRE was inhibited (Fig. 8, lanes 3 and 4). We then asked whether HBZ also affects the binding of CREB-2 and Tax to the HTLV-1 TxRE III site (10). As shown in Fig. 8 (lanes 7 and 8), HBZ bZIP abolished the binding of CREB-2 and Tax to the TxRE site. In addition, by the same approach, we found that HBZ was unable to bind directly to the CRE and TxRE motifs (data not shown). In conclusion, our results demonstrate that HBZ inhibits CREB-2 from binding to CRE and the related TxRE site.
FIG. 8.
HBZ abolishes the binding of CREB-2 to the CRE and TxRE sites. Biotinylated oligonucleotides (100 ng) corresponding to the somatostatin CRE were incubated with 50 ng of CREB-2 in the absence (lane 2) or the presence (lanes 3 and 4) of the bZIP domain of HBZ (25 and 50 ng). The same experiment was performed with the HTLV-1 TxRE III, CREB-2, and Tax (50 ng) in the absence (lane 6) or the presence (lanes 7 and 8) of HBZ bZIP.
DISCUSSION
In this paper, we describe the identification of a novel viral bZIP factor, which we have named HBZ and which is encoded by the complementary strand of the HTLV-1 RNA genome. The HBZ gene is exactly located between the env and tax/rex genes of the HTLV-1 genome. Comparison of the HBZ amino acid sequence of different HTLV-1 isolates revealed that HBZ sequence is highly conserved (between 96 and 100% identity depending on the compared isolates), with a very high degree of conservation in the key domains such as the activation and bZIP domains. In addition, Larocca et al. have shown that the HBZ gene is transcribed from a promoter present in the 3′ terminus of the HTLV-1 provirus (22). By RNA blotting probed with single-stranded radiolabeled RNA, they characterized 2.5- and 2.9-kb minus-strand RNAs encompassing the HBZ gene. These RNAs accumulate in the HTLV-1-infected SLB1 cell line and are absent in uninfected cells (22). In this work, we have characterized the HBZ protein by using an immunological approach in two other HTLV-1-infected T-cell lines, C8166 and MT4. On the other hand, we were unable to detect HBZ protein in the MT2 cell line, probably due to a low expression of HBZ in these cells. Indeed, the abundance of HBZ-minus RNA in infected cells has been estimated to be 1/50 to 1/100 of the abundance of the Tax-plus RNA (22). Thus, taken together, these results clearly demonstrate that HBZ is a bona fide HTLV-1-encoded protein.
The HBZ gene is also present in the simian T-cell leukemia virus type 1 genome but does not exist in the genomes of HTLV-2 and BLV. Interestingly, HTLV-2 has not yet been proven to be the causative agent of any specific hematological disease, whereas BLV is involved in the development of B-cell lymphomas. This is the first time that the characterization of an HTLV-1 protein encoded by a minus-strand gene has been reported. An important aspect of our findings is to know whether such a minus-strand gene can also exist in the proviral genome of other retroviruses. In the case of the HIV-1 proviral genome, the existence of an antisense RNA encoding a protein with an apparent molecular mass of 19 kDa has been described previously (34), but the function of this protein has not been characterized until now. In the future, characterization of such hidden genes should allow us to better understand the molecular and cellular mechanisms involved in the development of the pathologies associated with the retroviruses.
We demonstrate here that HBZ can interact in vitro and in vivo with another bZIP protein, the cellular factor CREB-2, that has been described to function (9, 27) in association with Tax to trans-activate the transcription of the HTLV-1 genome. We show that, in the presence of HBZ, CREB-2 is no longer able to activate the transcription from the viral TxREs because the HBZ/CREB-2 heterodimer is unable to bind to the TxRE motif. Recently, we have demonstrated that the heterodimerization of CREB-2 with the cellular bZIP factor CHOP also down-regulates the HTLV-1 transcription (8). Thus, although CREB-2 is known to form heterodimers in vivo with other bZIP transcription factors, CREB-2 homodimerization, probably guided by Tax (1, 18, 36), seems to be necessary to activate the viral transcription. Consequently, CREB-2 heterodimerization represents a powerful means to regulate negatively the HTLV-1 transcription. However, CREB-2 is not the only bZIP factor cooperating with Tax to stimulate the HTLV-1 transcription, since CREB and CREM have also been suggested to be involved in the viral expression (4, 29, 37, 38). Preliminary data obtained by in vitro binding assays (G. Gaudray, unpublished results) suggest that HBZ is capable of interacting with CREB too. Of course, complementary approaches should be investigated further to clearly demonstrate whether HBZ could also inhibit CREB from binding to the HTLV-1 promoter. Moreover, our results suggest that balance between the production of Tax and HBZ could be involved in the regulation of the HTLV-1 transcription in the infected T cells. Production of Tax is necessary to promote the proliferation of cells infected by HTLV-1, but Tax is also responsible for a strong cytotoxic-T-lymphocyte response to HTLV-1-infected cells. By down-regulating the viral transcription, infected cells expressing HBZ could escape the cytotoxic-T-lymphocyte response.
HBZ obviously is not involved in only the down-regulation of the HTLV-1 transcription. Given the presence of an activation domain in its N-terminal domain, HBZ is likely to function as a trans-activator by binding enhancer motifs through homo- or heterodimer formation. In this perspective, recent results obtained in our laboratory by a yeast two-hybrid screening of the MT2 cDNA bank with HBZ show the existence of interactions between HBZ and other bZIP factors than CREB-2 (J. Basbous, unpublished data). From these observations, it is tempting to speculate that HBZ could mediate its trans-activation function through binding to promoters of cellular genes and disturb the normal expression of these genes. Moreover, Marek's disease virus, which is one of the most potent oncogenic avian herpesvirus, also encodes a bZIP factor, the MEQ protein (19). Marek's disease virus induces the rapid onset of T-cell lymphoma and a demyelinating disease, and MEQ has been suggested to be involved in the oncogenic process (24). Experiments are under way to further evaluate the possibility that HBZ could also be involved in the oncogenic process of the transformation of the HTLV-1-infected T-cells.
Acknowledgments
G.G. and F.G. contributed equally to this work.
This work was supported by institutional grants from the Centre National de la Recherche Scientifique (CNRS) and the Université Montpellier I (UM I) and grants to J.-M.M. from the Fondation de France and the Association pour la Recherche sur le Cancer (ARC 5933). G.G. and J.B. are supported by fellowships from the Ministère de l'Education Nationale, de la Recherche et de la Technologie (MENRT) and the Centre National de la Recherche Scientifique (Bourse Docteur Ingénieur du CNRS), respectively.
REFERENCES
- 1.Anderson, M. G., and W. S. Dynan. 1994. Quantitative studies of the effect of HTLV-I Tax protein on CREB protein-DNA binding. Nucleic Acids Res. 22:3194-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cibelli, G., S. Schoch, and G. Thiel. 1999. Nuclear targeting of cAMP response element binding protein 2 (CREB2). Eur. J. Cell Biol. 78:642-649. [DOI] [PubMed] [Google Scholar]
- 3.Coudronniere, N., J. Corbeil, V. Robert-Hebmann, J. M. Mesnard, and C. Devaux. 1998. The lck protein tyrosine kinase is not involved in antibody-mediated CD4 (CDR3-loop) signal transduction that inhibits HIV-1 transcription. Eur. J. Immunol. 28:1445-1457. [DOI] [PubMed] [Google Scholar]
- 4.Franklin, A. A., M. F. Kubik, M. N. Uittenbogaard, A. Brauweiler, P. Utaisincharoen, M. A. Matthews, W. S. Dynan, J. P. Hoeffler, and J. K. Nyborg. 1993. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 268:21225-21231. [PubMed] [Google Scholar]
- 5.Franklin, A. A., and J. K. Nyborg. 1995. Mechanisms of Tax regulation of human T cell leukemia virus type I gene expression. J. Biomed. Sci. 2:17-29. [DOI] [PubMed] [Google Scholar]
- 6.Fujisawa, J. I., M. Seiki, M. Sato, and M. Yoshida. 1986. A transcriptional enhancer sequence of HTLV-I is responsible for trans-activation mediated by p40x of HTLV-I. EMBO J. 5:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gachon, F., C. Devaux, and J. M. Mesnard. 2002. Activation of HTLV-I transcription in the presence of Tax is independent of the acetylation of CREB-2 (ATF-4). Virology 299:271-278. [DOI] [PubMed] [Google Scholar]
- 8.Gachon, F., G. Gaudray, S. Thebault, J. Basbous, A. J. Koffi, C. Devaux, and J. M. Mesnard. 2001. The cAMP response element binding protein-2 (CREB-2) can interact with the C/EBP-homologous protein (CHOP). FEBS Lett. 502:57-62. [DOI] [PubMed] [Google Scholar]
- 9.Gachon, F., A. Péléraux, S. Thébault, J. Dick, I. Lemasson, C. Devaux, and J. M. Mesnard. 1998. CREB-2, a cellular CRE-dependent transcription repressor, functions in association with Tax as an activator of the human T-cell leukemia virus type 1 promoter. J. Virol. 72:8332-8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gachon, F., S. Thebault, A. Peleraux, C. Devaux, and J. M. Mesnard. 2000. Molecular interactions involved in the transactivation of the human T-cell leukemia virus type 1 promoter mediated by Tax and CREB-2 (ATF-4). Mol. Cell. Biol. 20:3470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georges, S. A., W. L. Kraus, K. Luger, J. K. Nyborg, and P. J. Laybourn. 2002. p300-mediated Tax transactivation from recombinant chromatin: histone tail deletion mimics coactivator function. Mol. Cell. Biol. 22:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hai, T., F. Liu, W. J. Coukos, and M. R. Green. 1989. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 3:2083-2090. [DOI] [PubMed] [Google Scholar]
- 13.He, C. H., P. Gong, B. Hu, D. Stewart, M. E. Choi, A. M. K. Choi, and J. Alam. 2001. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. J. Biol. Chem. 276:20858-20865. [DOI] [PubMed] [Google Scholar]
- 14.Hidaka, M., J. Inoue, M. Yoshida, and M. Seiki. 1988. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 7:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, H., Y. Fukada, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeang, K.-T., D. Derse, M. Matocha, and O. Sharma. 1997. Expression status of Tax protein in human T-cell leukemia virus type 1-transformed MT4 cells: recall of MT4 cells distributed by NIH AIDS Research and Reference Reagent Program. J. Virol. 71:6277-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeang, K. T., I. Boros, J. Brady, M. Radanovich, and G. Khoury. 1988. Characterization of cellular factors that interact with the human T-cell leukemia virus type I p40x-responsive 21-base-pair sequence. J. Virol. 62:4499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, D.-Y., and K.-T. Jeang. 1997. HTLV-I Tax self-association in optimal trans-activation function. Nucleic Acids Res. 25:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, D., L. Lee, J.-L. Liu, H.-J. Kung, and J. K. Tillotson. 1992. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc. Natl. Acad. Sci. USA 89:4042-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karpinski, B. A., G. D. Morle, J. Huggenvik, M. D. Uhler, and J. M. Leiden. 1992. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc. Natl. Acad. Sci. USA 89:4820-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok, R. P. S., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H.-M. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the coactivator CBP. Nature 370:223-226. [DOI] [PubMed] [Google Scholar]
- 22.Larocca, D., L. A. Chao, M. H. Seto, and T. K. Brunck. 1989. Human T-cell leukemia virus minus strand transcription in infected T-cells. Biochem. Biophys. Res. Commun. 163:1006-1013. [DOI] [PubMed] [Google Scholar]
- 23.Lekstrom-Himes, J., and K. G. Xanthopoulos. 1998. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 273:28545-28548. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J.-L., Y. Ye, L. Lee, and H.-J. Kung. 1998. Transforming potential of the herpesvirus oncoprotein MEQ: morphological transformation, serum-independent growth, and inhibition of apoptosis. J. Virol. 72:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montminy, M. 1997. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 56:807-822. [DOI] [PubMed] [Google Scholar]
- 26.Podust, L. M., A. M. Krezel, and Y. Kim. 2001. Crystal structure of the CCAAT box/enhancer-binding protein β activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA. J. Biol. Chem. 276:505-513. [DOI] [PubMed] [Google Scholar]
- 27.Reddy, T. R., H. Tang, X. Li, and F. Wong-Staal. 1997. Functional interaction of the HTLV-1 transactivator Tax with activating transcription factor-4 (ATF4). Oncogene 14:2785-2792. [DOI] [PubMed] [Google Scholar]
- 28.Shimotohno, K., M. Takano, T. Teruuchi, and M. Miwa. 1986. Requirement of multiple copies of a 21 nucleotide sequence in the U3 region of human T-cell leukemia virus type I and type II long terminal repeats for trans-acting activation of transcription. Proc. Natl. Acad. Sci. USA 83:8112-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, T., J. I. Fujisawa, M. Toita, and M. Yoshida. 1993. The trans-activator Tax of human T-cell leukemia virus type I (HTLV-I) interacts with cAMP-responsive element (CRE) binding and CRE modulator proteins that bind to the 21-base-pair enhancer of HTLV-I. Proc. Natl. Acad. Sci. USA 90:610-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thébault, S., F. Gachon, G. Gaudray, and J. M. Mesnard. 2001. Regulation of human T-cell leukemia virus type I genome transcription by the viral Tax protein. Recent Res. Dev. Virol. 3:151-164. [Google Scholar]
- 31.Thébault, S., F. Gachon, I. Lemasson, C. Devaux, and J. M. Mesnard. 2000. Molecular cloning of a novel human I-mfa domain-containing protein that differently regulates HTLV-I and HIV-1 expression. J. Biol. Chem. 275:4848-4857. [DOI] [PubMed] [Google Scholar]
- 32.Tsujimoto, A., H. Nyunoya, T. Morita, T. Sato, and K. Shimotohno. 1991. Isolation of cDNAs for DNA-binding proteins which specifically bind to a tax-responsive enhancer element in the long terminal repeat of human T-cell leukemia virus type I. J. Virol. 65:1420-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Dam, H., and M. Castellazzi. 2001. Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene 20:2453-2464. [DOI] [PubMed] [Google Scholar]
- 34.Vanhee-Brossollet, C., H. Thoreau, N. Serpente, L. D'Auriol, J. P. Levy, and C. Vaquero. 1995. A natural antisense RNA derived from the HIV-1 env gene encodes a protein which is recognized by circulating antibodies of HIV+ individuals. Virology 206:196-202. [DOI] [PubMed] [Google Scholar]
- 35.Waeber, G., and J. F. Habener. 1991. Nuclear translocation and DNA recognition signals colocalized within the bZIP domain of cyclic adenosine 3′,5′-monophosphate response element-binding protein CREB. Mol. Endocrinol. 5:1431-1438. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, S., and M. R. Green. 1993. HTLV-I Tax protein stimulation of DNA binding of bZIP proteins by enhancing dimerization. Science 262:395-399. [DOI] [PubMed] [Google Scholar]
- 37.Yin, J. Y., and R. B. Gaynor. 1996. Complex formation between CREB and Tax enhances the binding affinity of CREB for the human T-cell leukemia virus type 1 21-base-pair repeats. Mol. Cell. Biol. 16:3156-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, L. J., and C.-Z. Giam. 1992. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc. Natl. Acad. Sci. USA 89:7070-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]