Abstract
RNA-dependent RNA polymerases (RdRps) that initiate RNA synthesis by a de novo mechanism should specifically recognize the template initiation nucleotide, T1, and the substrate initiation nucleotide, the NTPi. The RdRps from hepatitis C virus (HCV), bovine viral diarrhea virus (BVDV), and GB virus-B all can initiate RNA synthesis by a de novo mechanism. We used RNAs and GTP analogs, respectively, to examine the use of the T1 nucleotide and the initiation nucleotide (NTPi) during de novo initiation of RNA synthesis. The effects of the metal ions Mg2+ and Mn2+ on initiation were also analyzed. All three viral RdRps require correct base pairing between the T1 and NTPi for efficient RNA synthesis. However, each RdRp had some distinct tolerances for modifications in the T1 and NTPi. For example, the HCV RdRp preferred an NTPi lacking one or more phosphates regardless of whether Mn2+ was present or absent, while the BVDV RdRp efficiently used GDP and GMP for initiation of RNA synthesis only in the presence of Mn2+. These and other results indicate that although the three RdRps share a common mechanism of de novo initiation, each has distinct preferences.
RNA viruses initiate RNA synthesis by either of two major mechanisms: de novo synthesis (where the primer is 1 nucleotide [nt]) or primer-dependent synthesis (where synthesis is initiated with an oligonucleotide or a protein covalently linked to nucleotides). De novo initiation is used by numerous RNA viruses, including those with genomes of positive, negative, and ambisense RNA (17). Examples include the double-stranded RNA viruses such as φ6 (7) and rotavirus (8), negative-strand RNA viruses such as vesicular stomatitis virus, positive-stranded alphavirus-like viruses, and members of the Flaviviridae (1, 22, 26, 43).
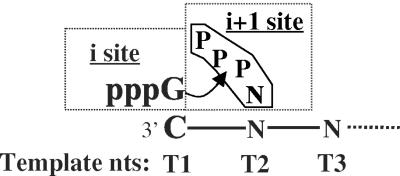
At its essence, de novo initiation is a simple process involving the active site of the polymerase, the initiation nucleotide (the NTPi) that provides the 3′-hydroxyl for nucleotidyl transfer to a second nucleoside triphosphate (NTP), and a template initiation site (the T1) (Fig. 1A). Additional structures in the polymerase or conformational changes may contribute to the specificity of this process. The NTPi binds to the i site in the polymerase and is base paired to T1 (12). The second NTP binds to the i + 1 site and is paired with T2. After the synthesis of the first phosphodiester bond, either the polymerase or the template translocates and the i + 1 site is used to incorporate subsequent nucleotides. The initiated RNA can then be coupled to other processes, such as the addition of the 5′ cap (35).
FIG. 1.
Nomenclature and RdRps used in this study. (A) Schematic of a polymerase active site responsible for de novo initiation. The initiation nucleotide(pppG) is base paired to the 3′-most C of the template RNA. The second nucleotide that binds to the i + 1 site is also represented (PPPN). The arrow shows the reaction leading to the formation of a phosphodiester bond. Template nucleotides are named sequentially, starting with T1.
General requirements for de novo initiation of viral RNA synthesis include a higher Km for the NTPi than for other NTPs (14, 22, 43). Having a special site and Km for the NTPi in comparison to other NTPs indicates that initiation is subject to additional regulations and/or specificity in recognition (reference 34 and reference therein). Specific recognition of the NTPi during de novo initiation has been preliminarily demonstrated for a number of viral RNA-dependent RNA polymerases (RdRps), including the ones from influenza virus, bovine viral diarrhea virus (BVDV), and brome mosaic virus (BMV) (14, 18).
In this work, we seek to better understand the mechanism of de novo initiation of RNA synthesis by RdRps by examining the requirements for the T1 nucleotide and the NTPi. We find that recombinant flaviviral RdRps and replicase complexes of plant-infecting RNA viruses specifically recognize both nucleotides. Despite the specific recognition, however, each enzyme has distinguishable preferences for the NTPi and the T1 nucleotide.
MATERIALS AND METHODS
Materials.
Nucleotides, nucleotide analogs, and dinucleotide NTPi analogs were from Sigma, Inc. (St. Louis, Mo.). The nucleotides were dissolved and neutralized with NaOH to a pH between 7 and 7.5. All RNAs and oligonucleotides longer than 2 nt were synthesized chemically by Dharmacon, Inc. (Boulder, Colo.) and deprotected according to the manufacturer's protocol, and a single band of the expected size was purified from a 7.5 M urea-polyacrylamide gel. The RNA was eluted and quantified as described by Ranjith-Kumar et al. (33). DNA oligonucleotide dLE19 was from MWG, Inc. (High Point, N.C.).
Plant viral RNA replicases.
BMV replicase was enriched from infected barley according to the protocol of Sun et al. (40). Cucumber mosaic virus (CMV) replicase was prepared from infected tobacco leaves as described by Sivakumaran et al. (38). Cowpea chlorotic mottle virus (CCMV) replicase complex was extracted from infected Chenopodium quinoa leaves as described by Adkins and Kao (2). Replicase activity assays were carried out in a 40-μl reaction mixture containing final concentrations of 20 mM sodium glutamate at pH 8.2, 12 mM dithiothreitol, 4 mM MgCl2, 0.5% (vol/vol) Triton X-100, 500 μM GTP, 200 μM ATP, 200 μM UTP, and 250 nM [α-32P]CTP (400 Ci/mmol; 10 μCi/ml; Amersham). Some reactions were amended with MnCl2. The template was used at a concentration of 125 nM. Following incubation for 60 min at 25°C, the reaction was extracted with phenol-chloroform (1:1, vol/vol), adjusted to a 0.4 M final concentration of ammonium acetate, and precipitated with ethanol (6:1, vol/vol) and 10 μg of glycogen. Loading buffer (45% [vol/vol] deionized formamide, 1.5% [vol/vol] glycerol, 0.04% [wt/vol] bromophenol blue, and 0.04% [wt/vol] xylene cyanol) was used to dissolve the pellet. The samples were heated at 90°C for 3 min prior to electrophoresis on denaturing (7.5 M urea) gels of 10 to 20% polyacrylamide. Gels were wrapped in plastic and exposed to film at −80°C. Product bands were quantified using a PhosphorImager (Molecular Dynamics). Within each experiment, results were normalized to those of wild-type control run within that experiment.
Recombinant NS5B proteins and activity assays.
The NS5B proteins from BVDV, hepatitis C virus (HCV), and GB virus-B (GBV) were expressed with C-terminal truncations of 23, 21, and 23 amino acids to increase the solubility of the proteins. This change does not affect de novo initiation of RNA synthesis. In addition, six histidines encoded by the pET21 plasmid were added to the C termini of these proteins. Escherichia coli strain BL21(DE3)LysS harboring the plasmids were grown at 30°C in standard Luria-Bertani medium supplemented with final ampicillin and chloramphenicol concentrations of 50 and 34 μg/ml, respectively, until the culture reached an optical density at 600 nm of 1.0. The culture temperature was then lowered to 25°C and expression was induced for 4 h with 1 mM isopropyl-thiogalactoside. Cells were harvested after centrifugation at 5,000 × g for 10 min. The preparation of the lysate was essentially as described by Behrens et al. (4), and the proteins were enriched sequentially through columns of Talon nickel and poly(U) RNA resins (Pharmacia Inc.). The N termini of the expressed proteins were sequenced to confirm the correct translation of each protein, and the masses of the proteins were determined by mass spectrometry. NS5B was quantified by a Bradford colorimetric assay using bovine serum albumin as a concentration standard.
Standard RdRp assays consisted of 2.5 pmol of template (unless stated otherwise) with 100 ng of NS5B in 20 μl of reaction in buffer A. MnCl2 was added to some reactions, as will be specified. The samples were processed as described for the plant viral RNA replicases. Where standard deviations are shown, the results were replicated at least thrice. Km determinations were performed as mentioned by Ranjith-Kumar et al. (32) by measuring the initial reaction rates at different substrate concentrations.
RESULTS
Recognition of the initiation cytidylate.
To study and compare the mechanism of de novo initiation by recombinant Flaviviridae RdRps, the NS5B proteins of HCV, GBV, and BVDV were expressed and purified as described in Materials and Methods. The final preparations used for characterization contained only the recombinant RdRps in Coomassie brilliant blue-stained gels (see Fig. 1 of the accompanying manuscript by Ranjith-Kumar et al. [32]). N-terminal sequencing of the proteins and mass spectrometry confirmed that the proteins were expressed correctly. However, the N-terminal methionine is absent in the final proteins, presumably due to processing by E. coli.
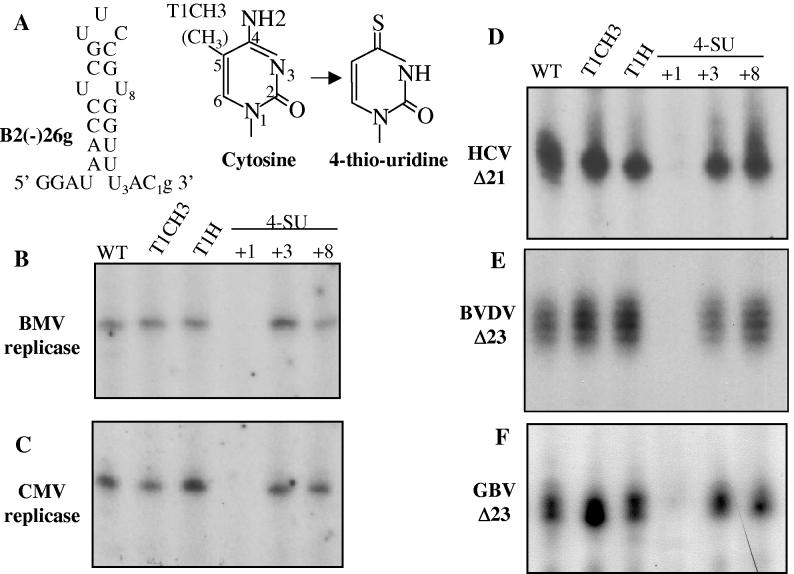
A predicted characteristic of the de novo initiation of viral RNA synthesis is that the T1 nucleotide has special requirements in comparison to other template nucleotides. To establish that all three recombinant viral RdRps have special recognition of the T1 cytidylate in the absence of other RdRp-associated proteins, we used a chemically synthesized RNA named B2(-)26g as a prototype for chemical modification (Fig. 2A). B2(-)26g is the core promoter for genomic RNA2 plus-strand initiation of BMV (39), and it is known to be recognized by the CMV replicase (38). It has a guanylate 3′ of the initiation cytidylate, a requirement for initiation by the BMV and CMV replicases (13, 38). The 3′ guanylate could negatively affect RNA synthesis by recombinant RdRps compared to RNAs where the initiation cytidylate is the 3′-most nucleotide (15). All RNAs tested in this series have a 3′ guanylate to facilitate comparisons.
FIG. 2.
Specific recognition of the initiation cytidylate by plant viral replicases and by recombinant viral RdRps. (A) Sequence and RNA secondary structure of the template B2(-)26g and its chemically synthesized variants that contain specific modifications. The modified nucleotides are numbered according to their position as template nucleotides. The secondary structure of B2(-)26g was established by nuclear magnetic resonance analysis (39). The two bases denote a normal cytosine and a 4SU. The methyl group attached to the C-5 position in T1CH3 is shown in parentheses. (B) RNA synthesis by the BMV replicase using either B2(-)26g or the modified variants of B2(-)26g listed on top of the autoradiogram. (C) RNA synthesis by the CMV replicase. (D to F) RNA synthesis by the HCV Δ21, the BVDV Δ23, and the GBV Δ23 RdRps using the templates listed above the autoradiogram.
Several versions of B2(-)26g containing nucleotide analogs were synthesized to examine the requirements for the initiation cytidylate (Fig. 2A). An RNA with a methyl group added to the non-hydrogen-bonding C-5 position of the T1 cytidylate was named T1CH3. B2(-)26g with a deoxyribose at only the T1 position was named T1H. Lastly, three RNAs were made with the base analog 4-thio-uridine (4SU) at the T1, T3, or T8 positions. A 4SU differs from a cytidylate by having a thio instead of an NH2 group at the C-4 position (Fig. 2A), thus causing a loss of the double bond between C-4 and the N-5 of guanylate, which should significantly affect the base pairing with the NTPi and the template. When these five modified RNAs were tested in comparison to prototype B2(-)26g, RNA synthesis from each varied slightly between the three RdRps (Fig. 2D to F) and also between the two RNA replicases (Fig. 2B and C). Most notable is that the BVDV RdRp has a propensity to add nontemplated nucleotides to the 3′ terminus of the nascent RNA, resulting in a ladder of bands (Fig. 2E). This activity is present without Mn2+ but is especially prominent in the presence of Mn2+ (33; also see below). However, all five polymerases had poor synthesis with the template containing a 4SU at the T1 position, but not at the T3 and T8 positions, indicating a special requirement for the T1 cytidine (Fig. 2B to F). Also, the changes to the T1 ribose in T1H had no detrimental effect on any of the polymerases tested. These results are in agreement with and extend the previous observations of Kim et al. (18) and demonstrate that the specific recognition of the initiation cytidylate is a property of the recombinant RdRp, even in the absence of the other subunits of the replicase complex.
To further probe recognition in the de novo initiation complex, we focused on the recombinant RdRps. LE19 contains the 3′-terminal 19 nt of minus-strand BVDV RNA and has a cytidylate as the 3′-most nucleotide (Fig. 3A). It does not direct RNA synthesis by the plant viral RNA replicases (C. C. Kao, unpublished results) but has been characterized for de novo initiation of RNA synthesis by the BVDV and the HCV RdRps (18, 33). LE19 is useful for studies of de novo initiation in that it contains only one cytidylate at the T1 position. Hence, GTP is used only for initiation and not during elongation.
FIG. 3.
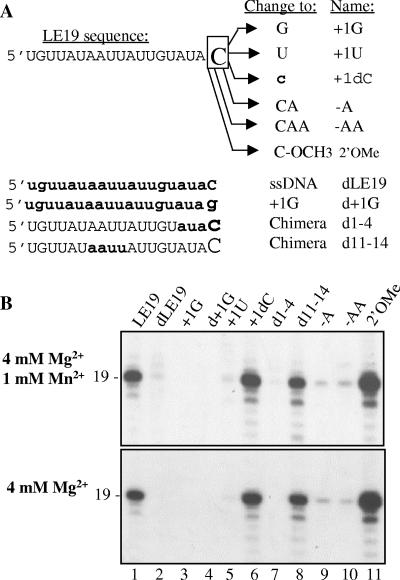
RNA synthesis by RdRps using LE19 and LE19 with modified nucleotides. (A) Sequence of chemically synthesized LE19. The modifications are focused on the initiation cytidylate (boxed C). Changes in LE19 and the names of the variants of LE19 are indicated in the two columns to the right. Deoxyribonucleotides, where present, are shown with lowercase boldface letters, while ribonucleotides are shown in capital letters. (B) Autoradiogram of the RNA products of the GBV RdRp. The size of the 19-nt RNA was determined by comparison to a ladder of RNA markers routinely used in the laboratory. The divalent metals used in the reactions are listed to the left of the autoradiograms.
Derivatives of LE19 were synthesized (Fig. 3A) and assayed for RNA synthesis by all three recombinant RdRps. The reactions were either with 4 mM Mg2+ or with 4 mM Mg2+ and 1 mM Mn2+. Only the autoradiograph for results with the GBV RdRp is shown (Fig. 3B), while quantitative results from all three RdRps are compiled in Table 1.
TABLE 1.
RNA synthesis by recombinant RdRps with different LE19 modificationsa
| Template | Mean % RNA synthesis ± SD
|
|||||
|---|---|---|---|---|---|---|
| HCV Δ21
|
BVDV Δ23
|
GBV Δ23
|
||||
| +Mn2+ | −Mn2+ | +Mn2+ | −Mn2+ | +Mn2+ | −Mn2+ | |
| RNA | ||||||
| LE19 | 100 | 100 | 100 | 100 | 100 | 100 |
| +1G | 1 ± 1 | 0 | 4 ± 1 | 0 | 0 | 0 |
| +1U | 16 ± 1 | 0 | 15 ± 1 | 0 | 7 ± 0 | 3 ± 0 |
| −A | 99 ± 0 | 246 ± 64 | 46 ± 9 | 3 ± 0 | 10 ± 1 | 10 ± 1 |
| −AA | 9 ± 5 | 7 ± 2 | 5 ± 1 | 0 | 14 ± 1 | 9 ± 2 |
| DNA | ||||||
| dLE19 | 93 ± 5 | 6 ± 1 | 131 ± 7 | 11 ± 8 | 5 ± 1 | 0 |
| d+1G | 17 ± 1 | 1 ± 1 | 0 | 0 | 1 ± 0 | 0 |
| Chimera | ||||||
| +1dC | 212 ± 14 | 371 ± 50 | 137 ± 9 | 93 ± 13 | 173 ± 3 | 127 ± 16 |
| 2′OMe | 115 ± 39 | 149 ± 52 | 126 ± 0 | 41 ± 0 | 325 ± 4 | 714 ± 138 |
| d1-4 | 83 ± 0 | 33 ± 7 | 86 ± 7 | 4 ± 5 | 3 ± 0 | 1 ± 0 |
| d11-14 | 73 ± 5 | 50 ± 27 | 131 ± 2 | 68 ± 2 | 83 ± 5 | 51 ± 6 |
All reactions contained 4 mM Mg2+. Where indicated (+Mn2+) Mn2+ was added to a final concentration of 1 mM. −Mn2+, no Mn2+ added.
The GBV RdRp initiated RNA synthesis using LE19 (Fig. 3B, lane 1), generating a 19-nt product. It also produced a 19-nt RNA from LE19P, which has a puromycin added to the 3′ terminus of LE19 (Ranjith-Kumar, data not shown). The puromycin prevents primer extension, thus demonstrating that RNA synthesis is initiated by a de novo mechanism. RNA synthesis by the GBV RdRp can take place without Mn2+, contradicting the previous claim of Zhong et al. that the GBV RdRp is dependent on Mn2+ for RNA synthesis (48). Changing the T1 cytidylate to a guanylate in template +1G abolished RNA synthesis, although a uridylate in RNA +1U resulted in 7% of the synthesis seen from LE19 when Mn2+ was present (Fig. 3B, lane 5; Table 2). The GBV RdRp required the initiation cytidylate in a position-dependent manner; the addition of one or two adenylates to the 3′ terminus of LE19 in RNAs named −A and −AA decreased RNA synthesis significantly (Fig. 3B, lanes 9 and 10). We note, however, that the products synthesized from −A and −AA were 19 nt, indicating that the cytidylate remained as the initiation nucleotide.
TABLE 2.
RNA synthesis by recombinant RdRps using different GTP analogsa
| Analog | Mean % RNA synthesis ± SD
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Recombinant RdRps
|
Replicases
|
||||||||
| HCV Δ21
|
BVDV Δ23
|
GBV Δ23
|
BMV | CMV (+Mn2+) | CCMV | ||||
| +Mn2+ | −Mn2+ | +Mn2+ | −Mn2+ | +Mn2+ | −Mn2+ | ||||
| Ribose | |||||||||
| GTP | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| dGTP | 71 ± 8 | 0 ± 0 | 252 ± 3 | 1 ± 0 | 98 ± 1 | 0 ± 0 | 60 ± 2 | 73 ± 4 | 65 ± 15 |
| Phosphate | |||||||||
| GDP | 348 ± 29 | 117 ± 1 | 209 ± 40 | 4 ± 1 | 149 ± 2 | 50 ± 7 | 7 ± 0 | 69 ± 6 | 57 ± 7 |
| GMP | 535 ± 75 | 277 ± 30 | 234 ± 6 | 21 ± 2 | 154 ± 3 | 153 ± 8 | 4 ± 0 | 55 ± 5 | 50 ± 8 |
| Guanosine | 79 ± 12 | 0 ± 0 | 11 ± 3 | 0 ± 0 | 40 ± 6 | 2 ± 1 | NTb | NT | NT |
| GTP-γ-S | 85 ± 3 | 189 ± 16 | 116 ± 12 | 72 ± 2 | 43 ± 2 | 62 ± 5 | 120 ± 11 | 76 ± 6 | 36 ± 4 |
| GDP-β-S | 501 ± 50 | 173 ± 3 | 257 ± 23 | 9 ± 1 | 80 ± 9 | 27 ± 1 | 26 ± 0 | 78 ± 10 | 61 ± 4 |
| βγ-imido GTP | 118 ± 7 | 844 ± 90 | 394 ± 13 | 217 ± 26 | 136 ± 4 | 109 ± 4 | 50 ± 1 | 84 ± 8 | 69 ± 3 |
| Base | |||||||||
| XTP | 6 ± 3 | 2 ± 1 | 1 ± 1 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 | 2 ± 3 | 1 ± 1 |
| ITP | 44 ± 8 | 1 ± 0.4 | 25 ± 1 | 1 ± 0 | 23 ± 1 | 1 ± 0 | 13 ± 2 | 73 ± 7 | 82 ± 8 |
| N7-meGTP | 87 ± 5 | 9 ± 4 | 61 ± 1 | 4 ± 0 | 128 ± 19 | 2 ± 0 | 12 ± 1 | 18 ± 3 | 19 ± 5 |
All reactions contained 4 mM Mg2+. Where indicated Mn2+ (+c), Mn2+ was added to a final concentration of 1 mM. LE19 is the template used for all of the RNA synthesis reactions using recombinant RdRps. The plant viral replicases used template −20/13, the subgenomic promoter for BMV. −Mn2+, no Mn2+ added.
NT, not tested.
The GBV RdRp was unable to efficiently synthesize RNA from a wholly deoxyribose version of LE19, dLE19 (Fig. 3B, lane 2), although the HCV and BVDV RdRps and plant viral RNA replicases can use single-stranded DNAs to initiate RNA synthesis (36, 39, 42). To examine which portion of the template is required to be a ribonucleotide(s) to allow synthesis by the GBV RdRp, templates +1dC and 2′OMe were tested. These two nucleic acids have the ribose C2′ moiety of the T1 cytidylate modified to be, respectively, a deoxyribose and an O-methoxy group. Both templates directed RNA synthesis at a higher level than LE19 (Fig. 3B, lanes 6 and 11), indicating that the T1 position can be a deoxyribonucleotide, in agreement with the observations in Fig. 2E. However, a change of positions T2 to T4 to deoxyribonucleotides abolished RNA synthesis, while a change of positions T11 to T14 did not significantly affect synthesis (Fig. 3B, lanes 7 and 8). Therefore, ribonucleotides near the initiation cytidylate are preferred for RNA synthesis by the GBV RdRp.
In contrast to the GBV RdRp, the HCV and BVDV RdRps appear to use a wider range of templates containing modified nucleotides for RNA synthesis. However, previous observations with the BVDV and HCV RdRps were made with templates with different sequences, thus making comparison of the results difficult. Therefore, requirements of the BVDV and HCV RdRps were tested with the templates derived from LE19 (Fig. 3A). RNA synthesis by each RdRp is expressed relative to that from LE19 (Table 1).
All three RdRps efficiently use LE19 as a template for de novo initiation in the presence of both Mg2+ and Mn2+. Template +1G decreased RNA synthesis significantly regardless of whether the reactions have Mg2+ as the only divalent metal or are supplemented with Mn2+. However, Mn2+ had a number of significant effects. For example, +1U directed the BVDV and HCV RdRps to produce RNAs at 15 and 16%, respectively, only in the presence of Mn2+ (Table 1, +1U), indicating that Mn2+ relaxed the recognition of the T1 nucleotide. However, synthesis cannot initiate efficiently from template purine nucleotides even in the presence of Mn2+.
Mn2+ also had different effects on the three RdRps. RNA synthesis from template −A by the HCV RdRp did not depend on the presence of Mn2+ (Table 1). In contrast, RNA synthesis by the BVDV RdRp was reduced to half when Mn2+ was present and synthesis was at background level in its absence. This effect was seen with several RNAs, indicating that higher nontemplated nucleotide addition in the presence of Mn2+ is an inherent property of the BVDV RdRp (Kao, unpublished results). Synthesis by the GBV RdRp from −A and −AA was at 10% of that from LE19 whether Mn2+ was present or not (Table 1). Consistent with previous reports (15, 18), the HCV and BVDV RdRps could initiate RNA synthesis from the single-stranded DNA, dLE19, in the presence of Mn2+ (Table 1). The 3′-terminal cytidylate in dLE19 is required since a change of that nucleotide to a deoxyguanylate in template d + 1G significantly decreased RNA synthesis (Table 1). These differences indicate that the three RdRps may have distinct requirements during some phase of RNA synthesis and that Mn2+ can be a contributing factor to template recognition.
Use of the NTPi.
Brassenelli et al. (6) had demonstrated that the HCV RdRp has a low-affinity GTP binding site in addition to the GTP-specific NTPi site. Mutation of the key residues in this low-affinity GTP binding site did not affect de novo initiation with GTP or several of the GTP analogs tested, including GMP and dGTP (Ranjith-Kumar, unpublished data). Furthermore, treatment with RNase T1, which cleaves 3′ of a guanylate, was used to demonstrate that GTP was used as the NTPi in our assay (33). These results indicate that LE19 and GTP analogs could be used to examine the requirements for the NTPi, as described below.
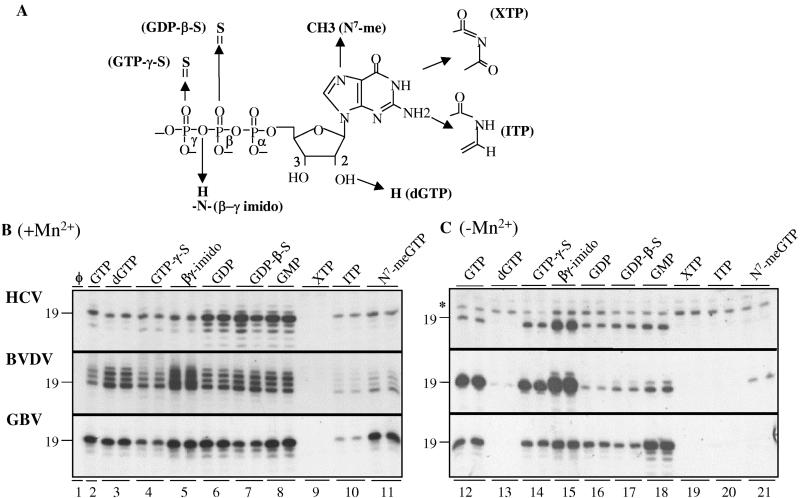
The GTP analogs used to functionally probe the requirement for the NTPi can be divided into groups that are altered in the phosphates, ribose, or base (Fig. 4A). Analogs with phosphate modifications are represented by GDP, GMP, guanosine 5′-O-3′-thiotriphosphate (GTP-γ-S), guanosine 5′-O-2′thio-diphosphate (GDP-β-S), and βγ-imido GTP. dGTP was the one ribose modification that was tested. Base modifications include XTP, ITP, and N7-methyl GTP (N7-meGTP). Qualitative results from representative experiments are shown in Fig. 4B and C, and quantitative ones from several experiments are in Table 2. A reaction lacking GTP or GTP analogs had no detectable RNA synthesis even though ATP and UTP were present at 100 μM, indicating specificity for GTP (Fig, 4B, lane 1). GTP at 200 μM was able to support RNA synthesis of an expected 19-nt RNA (Fig. 4B, lanes 2, and 4C, lane 12). The amount of product initiated with GTP differed significantly with the three RdRps in the absence of Mn2+, with the HCV RdRp producing severalfold less product compared to reactions with the BVDV and GBV RdRps (Fig. 4C, lanes 12, all three panels). This difference is not due to differing amounts of active enzyme in the three RdRp preparations since RNA syntheses by the three RdRps can be more comparable in the presence of Mn2+ and with some of the GTP analogs (compare the three panels in Fig. 4B and 4C). Instead, this result suggests that the HCV RdRp is more dependent on Mn2+ for de novo initiation than the RdRps from BVDV and GBV. As previously noted, the BVDV RdRp has increased terminal nucleotide addition to the nascent RNA in the presence of Mn2+ (Fig. 2E; also compare Fig. 4B and C, middle panels). Also, a band at 21 nt is seen with the HCV RdRp in the absence of Mn2+ (Fig. 4C, asterisk) and also in the absence of GTP (Fig. 5C, top panel, lane 1), arising from nontemplated nucleotide addition to LE19 (33).
FIG. 4.
Use of GTP analogs in the de novo initiation of RNA synthesis by three RdRps. (A) Chemical structure of GTP and the moieties that are affected in the GTP analogs. Only the portions of GTP that are affected are shown, and the names of the analogs are in parentheses. (B) RNA synthesized by the three RdRps in the presence of 1 mM Mn2+ and 4 mM Mg2+. Except for the positive and negative controls, all GTP analogs were tested in duplicate, independent reactions. The quantitation of these and other results are presented in Table 1. Positions of the 19-nt RNA expected of accurate de novo initiation and termination are indicated to the left of the autoradiogram. (C) RNA synthesized by the three RdRps in the presence of only Mg2+ (4 mM). The asterisk identifies a 21-nt band in RNA synthesis reactions with the HCV RdRp that likely arose by the addition of nontemplated nucleotides to the 3′ terminus of LE19 or the nascent RNA.
FIG. 5.
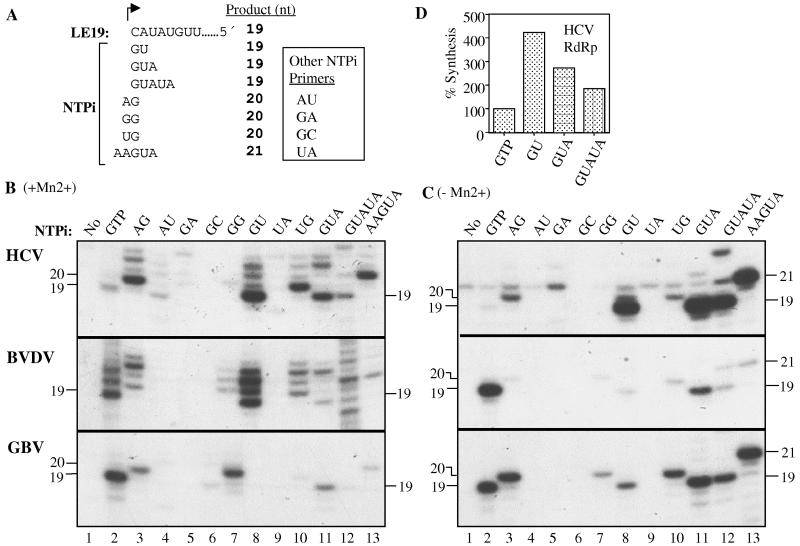
Use of oligonucleotide NTPi primers by the recombinant RdRps. (A) Schematic of potential base pairing between the NTPi analog and the template LE19. Should base pairing between the primer and LE19 take place, the expected length of the RdRp product would be the number of nucleotides indicated in the column to the right. The products may also have nontemplated nucleotides at their 3′ terminus; hence, a ladder of bands may be observed. Four oligonucleotides that are not complementary to LE19 were also tested: AU, GA, GC, and UA. (B) Products of RNA synthesis in the presence of both 4 mM Mg2+and 1 mM Mn2+ by the HCV, BVDV and GBV RdRps. (C) Products of RNA synthesis by the GBV RdRp in the presence of only 4 mM Mg2+. (D) Quantification of the effect of NTPi length on the accumulation of HCV RdRp products.
In reactions with GTP analogs substituted for GTP, several similarities and differences in RNA syntheses among the three RdRps were seen in an Mn2+-dependent manner. For example, dGTP initiated synthesis by all three RdRps only in the presence of Mn2+ (compare Fig. 4B, lane 3, and 4C, lane 13). Also, initiation with ITP and N7-meGTP by all three RdRps improved in the presence of Mn2+ (Table 2). Together with the results from modified templates (Table 1), Mn2+ affects multiple aspects of de novo initiation. The fact that the three RdRps responded differently with NTPi analogs in a metal-dependent manner suggests that divalent metals may act on RdRp structure.
Requirements for the phosphates of the NTPi were examined. The presence of Mg2+ and Mn2+ allowed all three RdRps to use analogs lacking one or more phosphates to synthesize products in the following relative amounts (from most to least): GMP, GDP, GTP, guanosine (Table 2). However, relative to the synthesis with GTP, the HCV, BVDV, and GBV RdRps had, respectively, 5.4-, 2.3-, and 1.5-fold the RNA synthesis seen with GMP. With the nucleoside guanosine, the HCV, BVDV, and GBV RdRps had syntheses of 79, 11, and 40% of that seen with GTP (Table 2). In the presence of only Mg2+, the preferred order of the same analogs differed among the three RdRps. The HCV and GBV RdRps still preferred GMP for initiation, while the BVDV RdRp strongly preferred GTP and had relatively low levels of synthesis when the NTPi lacked one or more phosphates. Furthermore, without Mn2+, guanosine was not used by any of the three RdRps.
We used GTP-γ-S, GDP-β-S, and βγ-imido GTP to further compare the effects of modifications in the phosphate portion of the GTP on RNA synthesis. The sulfur that replaces the oxygen in the phosphate in GTP-γ-S and GDP-β-S should delocalize the net negative charge of the phosphates. Compared to results with GDP, GDP-β-S increased RNA synthesis by the HCV RdRp both in the presence and absence of Mn2+ (Table 2), while the BVDV RdRp required Mn2+ to initiate with GDP-β-S. With its three phosphates, GTP-γ-S directed RNA synthesis by the HCV and BVDV RdRps similar to the levels seen with GTP. The GBV RdRp had a moderate reduction of RNA synthesis with both GDP-β-S and GTP-γ-S either in the presence or absence of Mn2+ (Fig. 4B, lanes 4 and 7; Fig. 4C, lanes 14 and 17). With βγ-imido GTP, all three RdRps increased initiation both in the presence and absence of Mn2+. These results indicate that one or more phosphates in the NTPi can be dispensable for initiation in the presence of Mn2+, but the requirements differ for each RdRp. The HCV RdRp generally prefers to initiate RNA synthesis with less negatively charged NTPi, while the BVDV RdRp prefers to use NTPi with three phosphates unless Mn2+ is present—in which case, the BVDV RdRp will no longer require the γ-phosphate. The GBV RdRp has an intermediate requirement for the phosphates compared to the other two RdRps.
Base modifications in the NTPi were examined next. A methyl modification at the non-hydrogen-bonding N-7 position of guanine affected RNA synthesis in the absence of Mn2+ (Table 2). For the H-bonding face of guanine, moieties in XTP and ITP will alter hydrogen bond formation with the T1 cytidylate (Fig. 4A). The absence of the C2 amino and the N1 imino group in XTP should abolish normal hydrogen bonds between the G:C base pair. Not surprisingly, XTP was not used for RNA synthesis by any of the three RdRps, even in the presence of Mn2+. The absence of the C2 amino group in ITP should cause the loss of one H-bond between a G:C base pair. In the absence of Mn2+, RNA synthesis by all three RdRps was at background levels. In the presence of Mn2+, however, ITP was tolerated better by the three RdRps, with synthesis being from 23 to 44% of that with GTP (Table 2). These results indicate that base pairing is an important feature of the de novo initiation complex.
The concentration of several GTP analogs for which RNA synthesis is at half of the maximum velocity, the Km, was determined. Although the Km cannot provide details regarding the affinity of the specific analog-RdRp interaction, it may help explain the differences in accumulation of the final RNA products when different NTPi analogs are used. The presence of Mn2+ decreased the Kms for GTP by 28-, 7-, and 34-fold for the GBV, BVDV, and HCV RdRps, respectively (Table 3). All analogs tested had similar or higher Kms for RNA synthesis in comparison to GTP. The slightly higher Km in comparison to GTP indicates that although these analogs resulted in increased initiation of RNA synthesis, it is not due to more-efficient use of the analogs. Perhaps GDP or GMP allows more rounds of initiation or faster polymerase escape from initiation. Information on pre-steady-state kinetics of RNA synthesis by RdRp will be necessary to obtain additional details in the mechanism of RNA-dependent RNA synthesis.
TABLE 3.
Effect of GTP analogs on the Km of RNA synthesisa
| Analog | Mean Km ± SD (μM)
|
|||||
|---|---|---|---|---|---|---|
| HCV Δ21
|
BVDV Δ23
|
GBV Δ23
|
||||
| +Mn2+ | −Mn2+ | +Mn2+ | −Mn2+ | +Mn2+ | −Mn2+ | |
| GTP | 3 ± 0.3 | 103 ± 12 | 24 ± 5 | 168 ± 16 | 1 ± 0.2 | 28 ± 5 |
| dGTP | 48 ± 11 | 87 ± 29 | 103 ± 28 | |||
| GDP | 16 ± 3 | 54 ± 3 | ||||
| GMP | 9 ± 2 | 434 ± 70 | 31 ± 6 | 512 ± 44 | ||
All reactions contained 4 mM Mg2+. Where indicated (+Mn2+), Mn2+ was added to a final concentration of 1 mM. LE19 was the template used for the Km determinations. −Mn2+, no Mn2+ added.
NTPi use by three plant viral replicases.
We seek to determine whether closely related plant viral replicases have specific preferences in the use of NTPi analogs. RNA −20/13, which contains a 20-nt nontemplated subgenomic core promoter and a 13-nt template sequence, was used for this analysis (37). Similar to LE19, −20/13 contains only one cytidylate in the template at the T1 position, thus allowing the examination of the GTP analogs as substitutes for the NTPi. RNA −20/13 is recognized by the replicases from BMV, CMV, and the CCMV (2; M. Chen and C. C. Kao, unpublished results).
The three plant viral replicases used the GTP analogs as the NTPis to different extents (Table 2). With the exception of GTP-γ-S in initiation by the BMV replicase, all three replicases preferred GTP as the NTPi. This is in contrast to the results seen with the recombinant RdRps. Consistent with the results from recombinant RdRps, XTP resulted in the lowest level of RNA synthesis, between 1 to 2% of that seen with GTP (Table 2). Therefore, base pairing between T1 and the NTPi is a key requirement for de novo initiation by recombinant RdRps and plant viral replicases.
Use of oligonucleotide NTPi analogs.
Oligonucleotides of five bases or fewer could replace GTP for de novo initiation by RNA replicases (10, 14). The BMV replicase requires specific positioning and recognition of the oligonucleotide NTPi primers (14). The HCV RdRp used some dinucleotide primers to initiate RNA synthesis, but these were not used to probe the requirements of the NTPi site (15, 47). Hence, we examined the use of oligonucleotide NTPi primers in de novo initiation more systematically. Oligonucleotide NTPi primers may be complementary to one or more of the bases in LE19 (Fig. 5A) and are named by their base sequence in the 5′-to-3′ orientation. All primers used contain a 5′ hydroxyl instead of one or more phosphates.
Oligonucleotide GU is complementary to the T1 and T2 nucleotides in LE19 and should initiate the synthesis of a 19-nt RNA. In the presence of Mn2+, GU substituted for GTP in initiation by the HCV RdRp, synthesizing the 19-nt product at 420% of the level seen with GTP (Fig. 5B, lane 8, top panel). The BVDV was able to use GU to initiate synthesis only when Mn2+ was in the reaction (middle panels of Fig. 5B and C, lanes 8). Unexpectedly, the GBV RdRp used GU and other oligonucleotide NTPi primers inefficiently in the presence of Mn2+ (Fig. 5B, lane 8, bottom panel). For GBV RdRp use of the oligonucleotide NTPi analogs improved in the absence of Mn2+ (compare Fig. 5B and C, bottom panels), suggesting that Mn2+ may participate in the exclusion of NTPi primers that form extensive base pairing with the template. These observations further reinforce the differences in the initiation complex between closely related RdRps.
Dinucleotides AU, GA, GC, and UA that were not complementary to T1 and T2 yielded less than 8% of the products yielded by GU (Fig. 5B, lanes 4, 5, 6, and 9), indicating that base pairing to T1 and T2 of the template allows more-efficient de novo initiation. AU and UA were unable to direct initiation by the BVDV and GBV RdRps in the presence or absence of Mn2+ even though they could base pair with several internal sites in the template. For example, UA could form Watson-Crick base pairs with T2 and T3 to produce an 18-nt RNA (Fig. 5A) but did not. Therefore, for efficient RNA synthesis base pairing must include the T1 nucleotide. The HCV RdRp did inefficiently initiate RNA synthesis with AU in the presence of Mn2+ (less than 8% of the amount in comparison to reactions with GU; Fig. 5B, top panel, lane 4). The AU-initiated RNA was 19 nt long, indicating that mispairing occurred with the T1 and T2 nucleotides during initiation by the HCV RdRp.
GUA and GUAUA were used to examine the effects of increasing the length of base pairing between the NTPi analog and the template (Fig. 5D). For RNA synthesis by the HCV RdRp in the presence of Mn2+, GUA and GUAUA produced RNA at 64 and 44%, respectively, relative to that in the reaction initiated with GU (Fig. 5D). Therefore, while base pairing between the NTPi and T1 is required, more-stable base pairing does not necessarily translate into increased synthesis.
Shifting of the oligonucleotide NTPi analogs relative to T1.
Although oligonucleotides AG, GG, and UG are not complementary to T1 and T2 of LE19 (Fig. 5A), one or more of them initiated synthesis by each of the three RdRps (Fig. 5B and C, lanes 3, 7, and 10, all three panels). Notably, the GBV RdRp, which generally did not efficiently initiate synthesis with complementary primers GU and GUA, produced more abundant products with AG and GG (Fig. 5B, lanes 3 and 7, bottom panel). Products of all three RdRps were initiated with AG, GG, and/or UG and were at least 20 nt, not 19 nt, suggesting that the 3′-most nucleotide of the dimer base paired with T1 while the 5′-most nucleotide protruded over the 3′ terminus of the template (Fig. 5A). We shall refer to this phenomenon as “primer shift.” No products were observed in reactions lacking the NTPi analogs, indicating that the 20-nt RNA did not result from terminal nucleotide addition (Fig. 5B, lane 1). To test the hypothesis that the NTPi did shift relative to T1, we used AAGUA, where the three underlined nucleotides could base pair with T1 to T3 of LE19. If AAGUA performed the primer shift, a 21-nt product would be expected. Indeed, all three RdRps produced RNAs that are minimally 21 nt, with the longer RNAs having nontemplated nucleotides added at the end of RNA synthesis (Fig. 5B, lane 13, all three panels). Primers GUA and GUAUA, which could base pair starting from the T1 nucleotide of LE19, produced RNAs that are minimally 19 nt in length in the absence of Mn2+ (Fig. 5C). Furthermore, the HCV and GBV RdRps initiated synthesis with AAGUA at levels comparable to that initiated from GUA, indicating that the protruding two 5′ nucleotides did not negatively affect RNA synthesis. These results show that the catalytic pocket within the RdRp can accommodate NTPi primers that have at least two 5′ nucleotides that are not base paired to the template.
ATP could not initiate RNA synthesis efficiently even if the T1 nucleotide in the template was a uridylate (Table 1). We examined whether more-extensive base pairing between the NTPi primers and the template can overcome the innate specificity for the NTPi and T1. These reactions used RNA +1U, which is identical to LE19 except for the uridylate at the T1 position (Fig. 3A), and primers AU and GU, which are, respectively, complementary to T1 and T2 of +1U and LE19. Only the RdRps from HCV and BVDV were assayed, since synthesis by the GBV RdRp was minimal with GU and LE19 and a comparison with +1U would not be productive (Fig. 5B, lane 8). In the presence of Mn2+ and GU, the HCV and BVDV RdRps produced a 19-nt RNA from +1U at less than 19% of that produced by reactions with LE19 (Table 4). With AU and LE19, RNA synthesis was at 11 and 3% for the HCV and BVDV RdRps. The HCV and BVDV RdRps produced RNA from primer AU and +1U at 32 and 14%, respectively. The fact that synthesis is not restored to the level seen with GU indicates that specificity for the NTPi and T1 nucleotide cannot be overcome simply by more-stable base pairing of the two components.
TABLE 4.
Effects of the T1 nucleotide on RNA synthesis by dinucleotide NTPi analogs
| NTPi | HCV Δ21
|
BVDV Δ23
|
||
|---|---|---|---|---|
| LE19 | +1U | LE19 | +1U | |
| GU | 100 | 19 | 100 | 12 |
| AU | 11 | 32 | 3 | 14 |
DISCUSSION
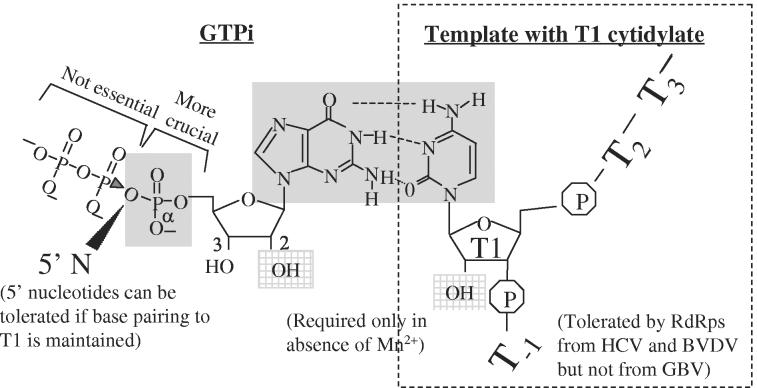
While proteins in the flaviviral replicase could influence recognition of the T1 and NTPi, their recognition is directly mediated by the RdRp. In characterizing the requirements for de novo initiation in vitro, we found that Mn2+ can affect substrate recognition by inducing a rearrangement in the RdRps (32). Furthermore, a comparison of the recognition of the T1 and the NTPi by three Flaviviridae RdRps revealed specificity for a cytidylate as the T1 nucleotide and GTP as the NTPi (summarized in Fig. 6). This specificity can be modulated by changes in the stability of the NTPi and template interactions but is highly preferred by the RdRps. Despite the specificities, however, significant differences exist in how the three recombinant RdRps recognize the template and NTPi. These functional studies should complement results from emerging structural studies (5, 6, 20, 45).
FIG. 6.
Summary of features in the NTPi and the template required for de novo initiation of RNA synthesis by the flaviviral RdRps. The features in shaded boxes are crucial for RNA synthesis, while the ribose 2′ hydroxyls in hatched boxes are required only in the absence of Mn2+. Oligonucleotide primers having 1 nt or more 5′ of the NTPi (5′ N) that can be tolerated for de novo initiation could be attached to the alpha phosphate of the NTPi as shown by the shaded triangle.
NTPi recognition.
The HCV RdRp has been reported to specifically require GTP for RNA synthesis (21). A low-affinity GTP binding site was identified in crystals of the HCV RdRp lacking the C-terminal 55 amino acids (6). The site is about 30 Å from the catalytic pocket and is formed by four amino acids of the thumb domain and two from the N-terminal finger domain. The low-affinity GTP binding site (estimated Kd of 250 to 450 μM) is specific for GTP and has been proposed to play a role in regulating the oligomeric state of the HCV RdRp, perhaps in a manner similar to the poliovirus 3Dpol (27, 45).
The existence of this specific secondary GTP binding site raises the possibility that GTP and GTP analogs used in this study could act through this ancillary site rather than as the NTPi. Several results indicate that we are probing the requirements of the NTPi site. (i) There is no RNA synthesis in the absence of GTP even though ATP could have served as the NTPi in the templates with a +1 uridylate. (ii) The RNAs initiated with GTP or the NTPi analogs have a 5′ guanylate, as detected by RNase T1 digestion. (iii) The low-affinity GTP binding site is specific for rGTP (6), while RNA synthesis could initiate with dGTP when Mn2+ was present (Table 2). (iv) Oligonucleotide NTPi primers that could perform a primer shift generated products of the expected lengths, indicating that they are used in polymerization. (v) Lastly, several mutations in the low-affinity GTP site exhibited properties similar to those of the wild type with different GTP analogs (Ranjith-Kumar et al., unpublished data). All of these results indicate that the low-affinity GTP binding site and the NTPi site that recognizes GTP have different biochemical properties and distinct functions.
The three RdRps and the three plant viral replicases accepted GTP and GTP analogs to various degrees (Table 2 and Fig. 4 and 5). In general, the replicases have a more limited use of the NTPi analogs, indicating either that the RdRps within these replicases are innately more specific or that the association with other replicase subunits confers additional levels of specificity to de novo initiation. Consistent with the recognition of different RNA core promoters, the BMV replicase appears to retain the highest level of specificity for the substrates needed for de novo initiation. Of the three flaviviral RdRps, the one from HCV was the most capable of accepting a range of NTPi analogs, perhaps indicating a lesser inherent specificity for the initiation process.
The structure of the HCV RdRp in complex with NTPs shows that there are several contacts with the phosphates of the NTPi (6). The contacts were not completely resolved but have been shown to be residues that are C-terminal to the GDD motif that coordinates metal ions. The residues should bond to the GTP through its phosphates, with the nearby metals shielding the negative charges of the phosphates. These interactions likely change significantly in the presence of the template, since we have functionally determined that specific base pairing between the NTPi and the template is the key criterion for initiation.
For the HCV RdRp, the lack of one or two phosphates in the NTPi analogs increased RNA synthesis and guanosine was able to initiate synthesis at 79% compared to GTP (Table 2). Therefore, the contacts between the RdRp and the triphosphate portion of the NTPi are not required for RNA synthesis, at least in vitro (Fig. 6). Other DNA-dependent RNA polymerases (DdRps), such as the T7 RNA polymerase, could initiate synthesis with guanosine (23, 24), and DdRps and RdRps can use dinucleotide NTPi analogs that also lack 5′ phosphates (3, 14). With the T7 RNA polymerase, the Km for the NTPi was reported to be unaffected by the presence of the phosphates (23), while we observed a three- to fivefold increase in the Km or GDP and GMP (Table 3). Also, differences in RNA synthesis by the three RdRps predict a different set of interactions with the triphosphate portions of the NTPi. Perhaps the differences could be partially accounted for by the amino acids within the respective catalytic pockets of the three RdRps. These predictions will be tested by determination of the structures of flaviviral RdRps in complex with initiation-competent templates and NTPs.
At least two nucleotides in the NTPi analog can protrude 5′ of the T1 nucleotide of the template. This observation can be used to probe the exit site of the nascent RNA in the RdRp ternary complex. In addition, primer shift during initiation of RNA synthesis by the flaviviral RdRps resembles a step in a process observed in minus-strand RNA viruses, named prime-and-realign (9, 19). In essence, prime-and-realign allows repair of mutations in the T1 nucleotide by initiating synthesis of an oligonucleotide nascent RNA using a template nucleotide several positions downstream (toward the 5′ end) of the normal T1 nucleotide. The nascent oligonucleotide RNA is then realigned relative to the 3′ end of the template before the polymerase resumes RNA synthesis. This process then generates RNA with a repaired initiation nucleotide. The three recombinant RdRps use oligonucleotide NTPi analogs that are staggered to different extents with respect to the template T1. The HCV RdRp can accept these staggered primers as the NTPi at almost the same level as a dimeric oligonucleotide that is correctly base paired to T1 and T2 (Fig. 5B). Interestingly, the BVDV RdRp had decreased initiation from the staggered NTPi analogs, while the GBV RdRp generally did not prefer oligonucleotide primers in the presence of Mn2+. Despite differences in the use of the NTPi primers, all three RdRps require that the 3′-most nucleotides of the primer retain base pairing with at least the T1 nucleotide.
Effect of manganese.
Polymerases are metalloproteins that use divalent metals for coordinating the substrates and products of the polymerization reaction. Mn2+ has been known to increase the utilization of various modified substrates by DdRps (30, 41). We have observed similar effects on the recognition of the NTPi and the T1 nucleotide by RdRps, indicating a common theme in polymerization in the two classes of RNA polymerases. Our data are also consistent with the idea that Mn2+ shields negative charges of the NTPi. However, we also observed that Mn2+ prevented some use of oligonucleotide NTPi primers by the GBV RdRp. It is likely that the myriad effects mediated by Mn2+ are a result of both direct interaction with the substrates for polymerization and conformational changes in the RdRp itself (32). Perhaps Mn2+ induced the rearrangement of some portions of the GBV active site to prevent extensively base-paired NTPi and template from being extended. An Mn2+-induced rearrangement was not obvious in the crystal structure of the C-terminally deleted version of the HCV RdRp (6). However, we have results consistent with the C-terminal residues of the HCV Δ21 protein being functionally involved in a conformational change that leads to altered modes of RNA synthesis (32). We note that the crystal structure of the HCV RdRp in complex with GTP is lacking 55 C-terminal residues (6).
In DdRps, a two-magnesium ion model is responsible for catalysis (12). A similar model is generally accepted for RdRps. Mn2+ is not thought to play a physiologically relevant role in RNA-dependent RNA synthesis, in part due to the intracellular Mn2+ concentration being in the low-micromolar range while free magnesium is at approximately 0.5 mM (31, 46). Mg2+ therefore has a greater chance to occupy the RdRp active site. However, the structures of RdRps known to date have revealed that Mn2+ can specifically bind in the catalytic pocket (6, 7, 25), and NTP occupancy at the i + 1 site was claimed to be better in the presence of Mn2+ (6). Since viral RNA replication takes place by means of a population of viral replication enzymes, it is possible that a subset of RdRp molecules would contain Mn2+ in their active site. These RdRps could not only initiate synthesis more efficiently but also increase the diversity of the viral replication progenies due to error-prone RNA synthesis (29). It is also possible that the identity of the metals within an RdRp's active site may change during different stages of RNA synthesis. Other enzymes do require Mn2+ for activity, giving precedence for selective sequestration of Mn2+ (e.g., see references 28 and 44). While de novo initiation is now widely accepted as a common process for the flaviviral RdRps (1, 16, 22), this work demonstrates that specific requirements for de novo initiation can differ between viral RdRps and allows comparison with the determinants of specificity for DdRps (11). The innate specificity will likely reflect differences in the structures of the RdRps and may provide ways for viruses to distinguish their RNA synthesis from those of other viruses present in the same cell. The observed differences could also be exploited in the design of drugs specific to each RdRp.
Acknowledgments
We thank the IU Cereal Killers for helpful discussions.
We thank the USDA and the NSF for funding the Kao laboratory. C. Kao acknowledges a fellowship from the Linda and Jack Gill Foundation.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation, but not the elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, S., and C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. [DOI] [PubMed] [Google Scholar]
- 3.Adkins, S., S. Stawicki, G. Faurote, R. Siegel, and C. Kao. 1998. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens S.-E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, and R. DeFrancesco. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, F. A. Rey, and R. DeFrancesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bramford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., and J. T. Patton. 2000. De novo synthesis of minus-strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA 6:1455-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcin, D., M. Lezzi, M. Dobbs, R. M. Elliott, C. Schmaljohn, C. Y. Kang, and D. Kolakofsky. 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol. 69:5754-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda, A., K. Mizumoto, and A. Ishihama. 1986. RNA polymerase of influenza virus: dinucleotide-primed initiation of transcription at specific positions on viral RNA. J. Biol. Chem. 261:5987-5991. [PubMed] [Google Scholar]
- 11.Huang, Y., A. Beaudry, J. McSwiggen, and R. Sousa. 1997. Determinants of ribose specificity in RNA polymerization: effects of Mn2+ and deoxynucleoside monophosphate incorporation into transcripts. Biochemistry 36:13718-13728. [DOI] [PubMed] [Google Scholar]
- 12.Joyce, C. M., and T. A. Steitz. 1995. Polymerase structure and function: variations on a theme? J. Bacteriol. 177:6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao, C. 2002. Lessons learned from the core RNA promoters of Brome mosaic virus and Cucumber mosaic virus. Mol. Plant Pathol. 3:55-62. [DOI] [PubMed] [Google Scholar]
- 14.Kao, C., and J. H. Sun. 1996. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J. Virol. 70:6826-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant Flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 17.Kao, C. C., D. Ecker, and P. Singh. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:252-260. [DOI] [PubMed] [Google Scholar]
- 18.Kim, M.-J., W. Zhong, Z. Hong, and C. C. Kao. 2000. Template nucleotide moieties for de novo initiation of RNA synthesis by a recombinant RNA-dependent RNA polymerase. J. Virol. 74:10312-10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo, L., R. Fearns, and P. L. Collins. 1997. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J. Virol. 71:4944-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesberg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 22.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, C. T., and J. E. Coleman. 1989. T7 RNA polymerase does not interact with the 5′-phosphate of the initiating nucleotide. Biochemistry 28:2760-2762. [DOI] [PubMed] [Google Scholar]
- 24.Muller, D. K., C. T. Martin, and J. E. Coleman. 1989. T7 RNA polymerase interacts with its promoter from one side of the DNA helix. Biochemistry 28:3306-3313. [DOI] [PubMed] [Google Scholar]
- 25.Ng, K., M., Cherney, A., López-Vázquez, A., Machín, J. Alonso, F., Parra, and M. James. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277:1381-1387. [DOI] [PubMed] [Google Scholar]
- 26.Oh, J.-W., G.-T. Sheu, and M. M. C. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 27.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 28.Pei, Y., K. Lehman, L. Tian, and S. Shuman. 2000. Characterization of Candida albicans RNA triphosphatase and mutational analysis of its active site. Nucleic Acids Res. 28:1885-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pezo, V., and S. Wain-Hobson. 1997. Hypermutagenic in vitro transcription employing biased NTP pools and manganese cations. Gene 186:67-72. [DOI] [PubMed] [Google Scholar]
- 30.Pinto, D., M. T. Sarocchi-Landousy, and W. Guschlbacher. 1979. 2′-deoxy-2′fluorouridine-5′-triphosphates: a possible substrate for E. coli RNA polymerase. Nucleic Acids Res. 6:1041-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quamme, G. A., L.-J. Dai, and S. Rabkin. 1993. Dynamics of intracellular free Mg2+ changes in a vascular smooth muscle cell line. Am. J. Physiol. 265:H281-H288. [DOI] [PubMed] [Google Scholar]
- 32.Ranjith-Kumar, C. T., Y.-C. Kim, L. Gutshall, C. Silverman, S. Khandekar, R. T. Sarisky, and C. C. Kao. 2002. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 76:12513-12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, R. Maley, R. Sarisky, and C. C. Kao. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 75:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy, P. S., and D. Chatterji. 1994. Evidence for a pyrimidine nucleotide-specific initiation site (the i site) on Escherichia coli RNA polymerase. Eur. J. Biochem. 225:737-745. [DOI] [PubMed] [Google Scholar]
- 35.Shuman, S., and B. Schwer. 1995. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol. 17:405-410. [DOI] [PubMed] [Google Scholar]
- 36.Siegel, R., L. Bellon, L. Beigelman, and C. Kao. 1999. Use of RNA, DNA and chimeric templates revealed that a viral RNA-dependent RNA polymerase can bridge the gap from the RNA to the DNA world. J. Virol. 73:6424-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegel, R., S. Adkins, and C. Kao. 1997. Sequence-specific recognition of an RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sivakumaran, K., Y. Bao, M. Roossnick, and C. Kao. 2000. Recognition of the core RNA promoter for minus-strand RNA synthesis by the brome mosaic virus and Cucumber mosaic virus RNA replicases. J. Virol. 74:10323-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivakumaran, K., C. H. Kim, R. Tayon, and C. Kao. 1999. RNA sequence and structural determinants for the recognition and efficiency of RNA synthesis by a viral RNA replicase. J. Mol. Biol. 294:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun, J. H., S. Adkins, G. Faurote, and C. Kao. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 226:1-12. [DOI] [PubMed] [Google Scholar]
- 41.Tabor, S., and C. Richardson. 1989. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. USA 86:4076-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tayon, Jr. R., M.-J. Kim, and C. Kao. 2001. Nucleotides near the 5′-terminus of the template contributes to the completion of RNA synthesis by viral RNA replicases. Nucleic Acids Res. 29:3583-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Testa, D., and A. K. Banerjee. 1979. Initiation of RNA synthesis in vitro by vesicular stomatitis virus. Role of ATP. J. Biol. Chem. 254:2053-2058. [PubMed] [Google Scholar]
- 44.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parson, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, R., and K. Ellis. 1989. In vivo measurement of total body magnesium and manganese in rats. Am. J. Physiol. 257:R1136-R1140. [DOI] [PubMed] [Google Scholar]
- 47.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, A. Gosh, C. Cameron, J. Lau, and Z. Hong. 2000. Template-primer requirements and single-nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong, W., P. Ingravallo, J. Wright-Minogue, A. S. Uss, A. Skelton, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. RNA-dependent RNA polymerase activity encoded by GB virus-B nonstructural protein 5B. J. Viral Hepat. 7:335-342. [DOI] [PubMed] [Google Scholar]