Abstract
Constitutive levels of production of STAT-1 were reduced by 10 h postinfection (p.i.) and significantly lost by 24 h p.i. in FL cells acutely infected with mumps virus (MuV). This result was consistent with that observed in previous studies and experiments with cells persistently infected with MuV (FLMT cells). There was a marked decrease in the amount of STAT-1 in cells expressing MuV accessory protein V (MuV-V). Furthermore, single amino acid substitutions in the Cys-rich region of V protein (Vc189a, Vc207a, and Vc214a) showed that each cysteine residue plays an important role in the decrease in STAT-1 production, but substitution of a histidine residue at amino acid position 203 had no effect. These events and the resultant suppression of the alpha interferon (IFN-α) response were confirmed by a luciferase reporter gene assay with five tandem repeats of the IFN-α-stimulated response element as an enhancer element of the firely luciferase gene. STAT-1 production was restored and detectable in FLMT cells treated with a proteosome inhibitor, such as MG132 or lactacystin. In the presence of MG132, ubiquitination of STAT-1 and the interaction of MuV-V with STAT-1 were demonstrated in FLMT cells by immunoprecipitation with anti-STAT-1 antibody. The same results for the interaction and ubiquitination were obtained in experiments with an expression vector for a C-terminal deletion mutant of STAT-1. The truncated STAT-1 molecules were degraded in the presence of MuV-V. Therefore, the C-terminal region (transcriptional activation and Src homology 2 domains) of STAT-1 is not necessary for its degradation caused by MuV-V. Our data suggest that MuV-V promotes ubiquitination and degradation of STAT-1.
Interferon (IFN)-induced antiviral activity is mediated by IFN-inducible proteins, such as 2′,5′-oligoadenylate synthetase (2-5AS), double-stranded RNA-activated protein kinase, and MxA protein. The expression of these proteins is induced by IFN upon activation of the intracellular IFN signal transduction pathway, which consists of Janus protein kinases (JAKs) and proteins in the family of signal tranducers and activators of transcription (STAT). Therefore, inactivation of antiviral proteins or dysfunction of the IFN signal transduction pathway results in the suppression of IFN-induced antiviral activity. It has been reported that many viruses have the ability to inhibit the antiviral function of IFN for the purpose of their replication (4, 5, 6).
Viruses belonging to the Paramyxoviridae family block the IFN signal transduction pathway in order to circumvent host cellular defense activity. Mumps virus (MuV) and human parainfluenza virus type 2 (hPIV2) reduced the constitutive production of STAT-1 and STAT-2, respectively (3, 7, 15, 20, 21, 22, 28, 30). These proteins are components of the transcription factor IFN-stimulated gene factor 3 (ISGF3) or gamma IFN (IFN-γ)-activated factor (GAF). The ISGF3 complex is formed from STAT-1α, STAT-2, and IFN regulatory factor-9 (IRF-9), and the GAF complex is a homodimer of STAT-1α. ISGF3 and GAF bind to the IFN-responsive promoter elements alpha IFN (IFN-α)-stimulated response element (ISRE) and IFN-γ-activated sequence (GAS), respectively. Didcock et al. reported that simian virus 5 (SV5) belongs to the same genus as MuV and induced the proteosome-mediated degradation of STAT-1 (1). The proteosome inhibitor MG132 prevented SV5-induced STAT-1 degradation. Moreover, our recent studies also noted that MG132 partly inhibited the degradation of STAT-1 in FL cells persistently infected with MuV (FLMT cells) and that basal levels of STAT-1 were detectable in the presence of the reagent (15). Therefore, it is reasonable to consider that the mechanism(s) of reduction of STAT-1 induced by MuV infection is the same as that found in SV5 infection. However, the detailed mechanism of STAT-1 degradation is not clearly understood, although it has been shown that SV5 protein V (SV5-V)-induced STAT-1 reduction seems to be mediated by a proteosome-mediated degradation pathway. These results are supported by one finding only from an experiment with the proteosome inhibitor MG132.
It is important to clarify whether the MuV protein V (MuV-V)-induced decrease in STAT-1 production is mediated through the ubiquitination and proteosome degradation pathways. Kim and Maniatis reported that the carboxy-terminal region of STAT-1 is necessary for the degradation of its activated or phosphorylation form (13). In this study, we sought to evaluate the role of the C-terminal region of STAT-1 in its degradation caused by MuV-V and to examine the function of the active or essential site of MuV-V (seven cysteine residues highly conserved among different paramyxoviruses) in the interaction between MuV-V and STAT-1.
MATERIALS AND METHODS
Cell cultures and virus infection.
Human aminion cells (FL cells), FLMT cells (28), and 293T cells were cultured with RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. FL cells were infected with MuV strain Torii at a multiplicity of infection (MOI) of 1.0 for various times.
Antibodies and reagents.
Rabbit polyclonal antibodies against STAT-1 p84/p91 (E-23) and Jak-2 (M-126) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Rabbit antibodies against Jak-1 and Tyk-2 were obtained from Upstate Biotechnology (Lake Placid, N.Y.). Antibody to STAT-1 phosphorylated at Tyr-701 [anti-STAT1pTyr(701) antibody], antibody to Jak-1 phosphorylated at Tyr-1022 and Tyr-1023 [anti-Jak1pYpY(1022/1023) antibody], and anti-Jak2pYpY(1007/1008) antibody were obtained from BioSource International (Camarillo, Calif.). Anti-Tyk2pYpY(1054/1055) antibody and antiactin antibody (C-4) were obtained from New England Biolabs (Cambridge, Mass.) and Chemicon (Temecula, Calif.), respectively. Antimultiubiquitin monoclonal antibody (clone FK2) was purchased from Medical and Biological Laboratories (Nagoya, Japan). Antiphosphotyrosine antibody PY20 and anti-STAT-2 antibody were obtained from Transduction Laboratories (San Diego, Calif.). Rabbit antibody against the C-terminal region of MuV-V was provided by A. Kato and K. Takeuchi (National Institute of Infectious Diseases, Tokyo, Japan).
MG132 and clasto-lactacystin β-lactone were purchased from Peptide Institute (Osaka, Japan) and Medical and Biological Laboratories, respectively.
IFN-α and IFN-γ were purchased from Serotec (Oxford, United Kingdom) and Genzyme (Cambridge, Mass.), respectively. Both were used at a final concentration of 1,000 IU/ml.
Western blotting.
After treatment of cells with IFN or a proteosome inhibitor, the cells were lysed in lysis buffer (20 mM HEPES-NaOH [pH 7.5], 150 mM NaCl, 1% Triton X-100, complete protease inhibitor, 20 μM MG132), and the cell lysates obtained were analyzed by a Western blot technique with appropriate antibodies as described previously (3, 28, 29).
Substitution of single amino acids in V protein and transient expression.
The expression vector for MuV-V, pTM-V, was described previously (15). A substitution of a cysteine or a histidine residue with an alanine residue in V protein (c189a, h203a, c207a, and c214a) was introduced by using a QuickChange site-directed mutagenesis kit (Stratagene) and the pTM-V vector, and the mutation was confirmed by DNA sequencing. A monolayer of 293T cells grown to 50% confluence on polylysine-coated six-well plates was transfected with 5 μg of vector DNA (pTM-V, pTM-Vc189a, pTM-Vh203a, pTM-Vc207a, or pTM-Vc214a) and Superfect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. After cultivation for 40 h, the transfectants were lysed in lysis buffer, and the cell lysates were solubilized by boiling with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer (62.5 mM Tris-HCl [pH 6.8], 5% 2-mercaptoethanol, 2% SDS, 0.005% bromphenol blue, 10% glycerol) and analyzed by a Western blot technique.
Construction of deletion mutants of STAT-1.
For preparation of deletion mutants of STAT-1, DNA fragments were amplified by PCR with appropriate primer sets for the target STAT-1 DNA (pCI-neo-Flag-STAT-1; provided by K. Nagawa, School of Pharmaceutical Sciences, Hokkaido University). The pT-FlagST1C684 (amino acid sequence of STAT-1, 1 to 684) and pT-FlagST1C577 (amino acid sequence of STAT-1, 1 to 577) expression vectors were constructed with the pTarget vector and the amplified PCR product by using primer set FtagF/ST1CR684 (CTACCATGGACTACAAGGACG and CTCTGGTGCTTCCTATGGCCTG) or FtagF/ST1CR577 (CTACCATGGACTACAAGGACG and AGCCCATGATTCACCCATCATTC). pTSTAT-1 was constructed with pTarget vector and cDNA of STAT-1.
FL and FLMT cells grown on 10-cm dishes to approximately 50 to 60% confluence were transfected with pT-STAT-1, pCI-neo-Flag-STAT-1, pT-FlagST1C684, or pT-FlagST1C577 by using Superfect. After cultivation for 24 h, MG132 was added at a final concentration of 20 μM, and the mixture was further incubated for 6 h. The cells were harvested and lysed in lysis buffer. The supernatants obtained by centrifugation were incubated with an anti-Flag M2 agarose affinity gel (Sigma, St. Louis, Mo.) for 2 h at 4 C, washed with MG132-free lysis buffer, and extracted with SDS-PAGE sample buffer for analysis by a Western blot technique with anti-Flag M2 monoclonal antibody (Sigma).
Reporter gene assay with luciferase.
Activation of the IFN-α signal transduction pathway was determined by a luciferase reporter assay with a transiently transfected reporter plasmid (pISRE-Luc) containing the enhancer element IFN-α-stimulated response element as described previously (29). Briefly, 293T cells were grown on a 24-well plate to approximately 40% confluence the day before transfection. Reporter plasmids (0.5 μg), 0.05 μg of pRL-TK reference plasmid vector (Promega, Madison, Wis.), and 0.75 μg of expression vector (pTM-V, pTM-Vc189a, pTM-Vh203a, pTM-Vc207a,or pTM-Vc214a) were mixed and transfected into 293T cells by using 5 μl of Superfect. After cultivation for 24 h, IFN-α was added, and the mixture was further incubated for 6 h. Luciferase activity in the cell lysates was measured by using a dual-luciferase reporter assay system (Promega). The experiments were performed in triplicate. Calculation of reporter activity and expression of the results were carried out as described previously (29).
Immunoprecipitation analysis.
Immunoprecipitation analysis with anti-STAT-1 antibody was carried out as described previously (29). Briefly, after solubilization of cellular protein with lysis buffer, the cell lysates and 5 μg of antibody were mixed and incubated at 4 C for 1 h. The resultant immunocomplex was recovered with protein G-Sepharose 4B (Amersham Pharmacia, Uppsala, Sweden). The materials binding to the resin were solubilized and applied on a polyacrylamide gel, followed by Western blot analysis. Anti-Flag M2 agarose affinity gel and anti-Flag M2 monoclonal antibody were used for immunoprecipitation experiments with Flag- STAT-1.
RT-PCT.
Total RNAs were isolated from cells treated with IFN by using Isogene (Nippon Gene, Toyama, Japan) in accordance with the manufacturer's protocol. RNAs then were treated with DNase to digest the contaminated DNA. To examine the levels of induction of the 40-kDa 2-5AS and IRF-1 transcripts by treatment with IFN-α and IFN-γ, respectively, reverse transcription (RT)-PCR analysis was performed by using appropriate primer sets as described previously (3, 28).
RESULTS
Decrease in STAT-1 production and phosphorylation of JAKs in MuV-infected FL cells.
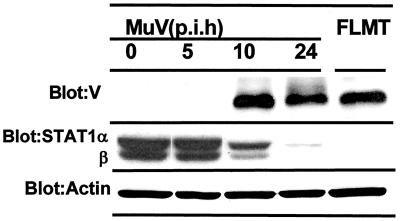
Evidence showing a decrease in the basal expression of STAT-1 and dysfunction of the responses to IFN-α and IFN-γ in FLMT cells has been reported (15, 28). However, it is still unknown whether the decrease in STAT-1 production is found in acute infection with MuV. To clarify the association between the expression of MuV-V and the decrease in STAT-1 production, FL cells were infected with MuV at an MOI of 1.0 and analyzed for STAT-1 and MuV-V by Western blotting at various times after infection (Fig. 1). Constitutive levels of STAT-1 were significantly reduced to about 40% the control level (no infection) at 10 h postinfection (p.i.) in acute infection, and large amounts of STAT-1 were lost within 24 h. The disappearance of STAT-1 correlated with the production of MuV-V, which was faint at 5 h p.i. and notable at 10 h p.i. These findings are consistent with those reported for FLMT cells. The results obtained for FLMT cells are also shown in Fig. 1.
FIG. 1.
Decrease in STAT-1 production caused by MuV infection. Cell lysates from FL cells infected with MuV for various times at an MOI of 1.0 and FLMT cells (persistent infection) were analyzed by SDS-PAGE and Western blotting. p.i.h, hours postinfection.
On the other hand, the phosphorylation of tyrosine kinases (Jak-1, Jak-2, and Tyk-2) is essential for activation of the IFN-α and IFN-γ signaling pathway. Therefore, the lack of kinase activity causes disruption of the IFN signal transduction pathway. Although there is evidence for suppression of kinase activity by infection with Sendai virus (14), measles virus (S. Yokota et al., Virology, in press), and herpes simplex virus type 1 (29), no other types of viral infection capable of suppressing kinase activity have been identified. It is important to investigate the activation of the IFN receptor-associated tyrosine kinases, Jak-1, Jak-2, and Tyk-2, in cells infected with MuV, which belongs to the same family (Paramyxoviridae) as Sendai virus and measles virus.
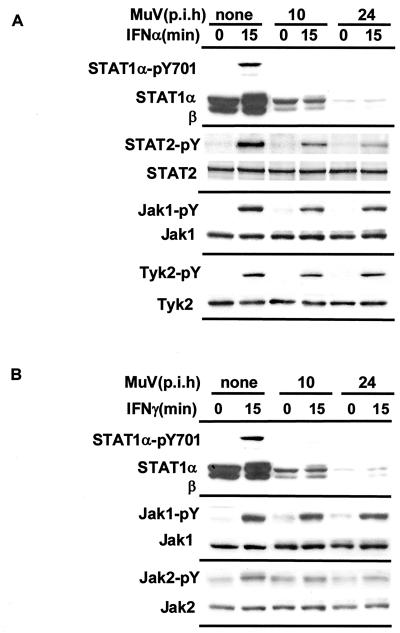
We examined the levels of phosphorylation of the kinases. Cells were treated with 1,000 IU of IFN-α (Fig. 2A) or IFN-γ (Fig. 2B) per ml for 15 min at various times after infection with MuV. As shown in Fig. 2A, the phosphorylation of Jak-1 and Tyk-2 was not suppressed at all compared with that of the control. These activated kinases first phosphorylate STAT-2 and then phosphorylate STAT-1. To evaluate the active functions of these kinases, the levels of phosphorylation of STAT-2 were examined in cells infected with MuV. Phosphorylation was slightly suppressed at 10 h p.i. and significantly suppressed at 24 h p.i. (Fig. 2A). These results were rather unexpected, because the phosphorylation of Jak-1 and Tyk-2 was not inhibited at all by MuV. The reason is still unknown. However, we suggest that the reduction in STAT-2 phosphorylation was dependent on the decrease in STAT-1 production caused by MuV infection. In other words, STAT-1 is needed for the phosphorylation of STAT-2.
FIG. 2.
Phosphorylation of the JAK family after infection with MuV. FL cells were infected with MuV for various times at an MOI of 1.0. For each time, the cells were treated with 1,000 IU of IFN-α (A) or IFN-γ (B) per ml for 15 min and then lysed. The lysates were analyzed by SDS-PAGE and Western blotting. For the detection of phosphorylated STAT-2, cell lysates were immunoprecipitated with anti-STAT-2 antibody and then analyzed by Western blotting with antiphosphotyrosine antibody. p.i.h, hours postinfection.
In the IFN-γ signaling pathway, the binding of IFN-γ to its receptor leads to the transphosphorylation and activation of Jak-1 and Jak-2. The phosphorylation of Jak-1 was not inhibited at 10 or 24 h p.i.. For Jak-2, phosphorylation was slightly suppressed in a manner dependent on the course of infection. The phosphorylation of Jak-2 occurs first and is needed for the subsequent activation of Jak-1. The phosphorylation of Jak-1 in MuV-infected cells was comparable to that in uninfected cells in spite of the slight suppression of Jak-2 phosphorylation and activation. We therefore suggest that the reduced level of Jak-2 phosphorylation is sufficient to activate Jak-1. The suppressive effect of MuV on JAK family phosphorylation is not seen in acute infection. From what we have shown above, it is evident that blocking of IFN signaling by MuV is due mainly to a decrease in STAT-1α production and partly to suppression of STAT-2 activation.
Role of the Cys-rich region of MuV-V in the decrease in STAT-1 production.
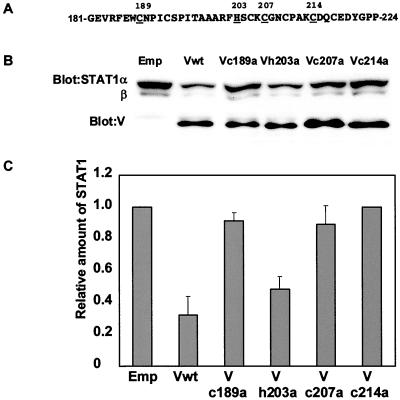
Recently, Kubota et al. reported that the Cys-rich region of MuV-V is involved in the suppression of the IFN-α and IFN-γ signal transduction pathway through a decrease in STAT-1 production (15). The Cys-rich region, which is a highly conserved sequence among viruses of the Paramyxoviridae family, contains seven cysteine residues and acts as a zinc binding domain (10, 19, 24). Lin et al. noted that each cysteine residue in SV5-V plays a key role in binding to damage-specific DNA binding protein (DDB) 1 (DDB1) (16). They also demonstrated that MuV-V interacts with DDB1. However, it is still not clear what region or domain in MuV-V is associated with the loss of STAT-1. To elucidate the active region, four mutant V proteins (Vc189a, Vh203a, Vc207a, and Vc214a), which have single amino acid substitutions (Fig. 3A), were expressed in 293T cells; the basal levels of STAT-1 were examined by immunoblotting (Fig. 3B and C). A reduction of STAT-1 production was seen in 293T cells expressing wild-type MuV-V but not with the introduction of an empty vector. The expression of Vh203a reduced STAT-1 production to the same level as that seen in cells expressing wild-type MuV-V. However, all three mutant proteins containing substitutions of the cysteine residues produced no decrease in STAT-1 production. Considering these results together, we propose that each cysteine residue in the Cys-rich region, but not histidine, is implicated in both the interaction of V protein with DDB1 and the association between V protein and STAT-1.
FIG. 3.
Protein levels for STAT-1 in 293T cells expressing wild-type or mutant V protein. (A) Amino acid positions (numbers) and presence (underlining) of cysteine and histidine residues in the Cys-rich region of V protein. (B) Cell lysates from 293T cells transfected with mutants of V protein with single amino acid substitutions were analyzed by Western blotting. Emp, empty vector; Vwt, wild-type MuV-V. (C) Densitometric analysis of STAT-1 bands was performed with a scanner (Epson GT-9500) and ScnImage softwear (Scion Corp.). Error bars indicate standard deviations of triplicate analysis.
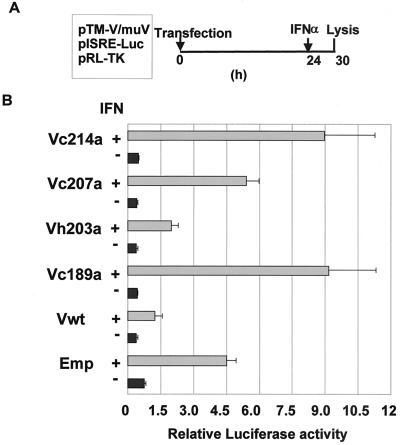
As STAT-1 is an essential component of transcriptional factors ISGF3 and GAF, depletion of the component results in the suppression of induction of the IFN response through the disruption of IFN signaling. Therefore, we next performed a luciferase reporter assay to confirm that there was no reduction of STAT-1 production in 293T cells expressing mutant MuV-V. 293T cells were transfected with reporter and expression plasmids and treated with IFN-α under the protocol indicated in Fig. 4A. Figure 4B shows enhancement and suppression of IFN-α-inducible gene induction by the expression of mutant MuV-V. In 293T cells transfected with an empty vector, firefly luciferase activity was increased about sixfold by treatment with IFN-α compared to that seen in non-IFN-treated control cells. Similar to the results found for transfection with the empty vector, the expression of mutant MuV-V with no STAT-1-decreasing activity (Vc189a, Vc207a, and Vc214a) did not result in any significant suppression of luciferase activity and enhanced the enzyme activity about seven- to ninefold in the presence of IFN treatment, although the expression of wild-type MuV-V or another mutant MuV-V (Vh207a) contributed to the suppression of IFN-induced luciferase induction. These results indicate an association of two events, the loss of STAT-1 and the suppression of IFN-inducible gene expression. Substitution of cysteine residues with alanine in the Cys-rich region inhibited the ability of MuV-V to reduce STAT-1 production. Therefore, we propose that cysteine residues in the Cys-rich region are involved in the loss of STAT-1 and the subsequent disruption of IFN signaling.
FIG. 4.
Effects of V proteins on induction of IFN-inducible genes. (A) Protocol of experiment. 293T cells were transfected with a reporter plasmid (pISRE-Luc), an expression vector, and pRL-TK as an internal reference for transfection efficiency. After incubation for 24 h, the transiently expressed transfectants were treated with IFN-α for 6 h, and then the cells were lysed. (B) Luciferase activities in cell lysates. The results are expressed as activity relative to that obtained in a control experiment without IFN. Vwt, wild-type MuV-V; Emp, empty vector. Error bars indicate standard deviations. Data are averages of three determinations.
Proteosome-mediated degradation of STAT-1 by MuV-V.
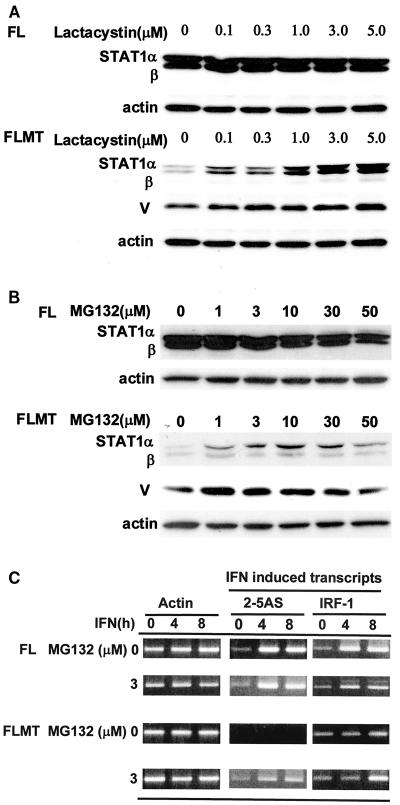
The mechanism(s) for the selective loss of STAT-1 caused by MuV-V is not clearly understood. However, there is much evidence to support the involvement of SV5-V or hPIV2 V protein (hPVI2-V) in the proteosome-mediated degradation of STAT-1 or STAT-2 (1, 30). MuV, like these two viruses, belongs to the Rublavirus genus in the Paramyxoviridae family. It is therefore expected that the depletion of STAT-1 by MuV-V is regulated by ubiquitination and the proteosome degradation system. The ability of a proteosome inhibitor to restore the levels of STAT-1 following its loss was examined in FLMT cells. FLMT cells were treated with lactacystin (Fig. 5A) or MG132 (Fig. 5B) at various concentrations for 6 h. A small amount of STAT-1 was detectable in the absence of the inhibitors. However, the effect of MuV-V on STAT-1 disappearance was blocked by the inhibitors in a dose-dependent manner. Maximum restoration of STAT-1 production was observed with 3.0 or 5.0 μM lactacystin and 10 μM MG132. To test whether the recovery of STAT-1α was associated with the restoration of the IFN response in FLMT cells, the levels of IFN-α-induced 2-5AS and IFN-γ-induced IRF-1 transcripts were examined in total RNAs from FL and FLMT cells treated with MG132. Both transcripts were significantly induced in MG132-treated FLMT cells, although the induction was of a lower level than in MG132-treated FL cells (Fig. 5C). The IFN response in FLMT cells treated with a proteosome inhibitor was restored in a manner dependent on the recovery of STAT-1α. This restoration was not found in FLMT cells treated with E64d (a cysteine protease inhibitor that inhibits calpain and lysosomal proteases). Furthermore, interestingly, the levels of MuV-V also increased after treatment with these drugs, indicating that MuV-V levels are, at least in part, regulated by the proteosome degradation system.
FIG. 5.
Effects of proteosome inhibitors on basal STAT-1 production. FL or FLMT cells were treated with the indicated concentrations of lactacystin (A) or MG132 (B) for 6 h, and then the cells were harvested and lysed to analyze basal levels of STAT-1 production by Western blotting with anti-STAT-1 antibody. (C) Restoration of IFN signaling in FLMT cells treated with MG132. After treatment without or with 3 μM MG132 for 6 h, cells were treated with 1,000 IU of IFN-α or IFN-γ per ml for 4 and 8 h. Levels of IFN-induced transcripts were analyzed by RT-PCR.
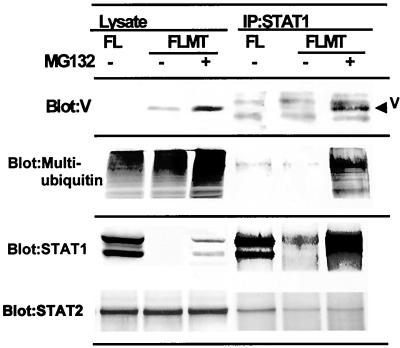
Multiubiquitination of STAT-1 and interaction between MuV-V and STAT-1.
To examine the multiubiquitination of STAT-1, extracts from FLMT cells treated or not treated with MG132 were subjected to immunoprecipitation analysis with anti-STAT-1 antibody; the resultant precipitates were analyzed by immunoblotting with anti-MuV-V (Fig. 6, upper panel), antimultiubiquitin (Fig. 6, middle panel), anti-STAT-1 (Fig. 6, lower panel), and anti-STAT-2 (Fig. 6, lower panel) antibodies. In the presence of MG132, anti-STAT-1 antibody coimmunoprecipitated multiubiquitinated STAT-1 and MuV-V. However, they were undetectable in FL and FLMT cells in the absence of the drug. These results strongly suggest that STAT-1 is multiubiquitinated before its degradation by the 26S proteosome and furthermore that an interaction between MuV-V and STAT-1 may occur.
FIG. 6.
Multiubiquitination of STAT-1 and interaction between MuV-V and STAT-1. FL or FLMT cells were treated (+) or not treated (−) with 20 μM MG132 for 6 h and then lysed in lysis buffer. The resulting cell lysates were immunoprecipitated (IP) with anti-STAT-1 antibody. The precipitates were subjected to immunoblotting with antimultiubiquitin, anti-MuV-V, or anti-STAT-2 antibody.
On the other hand, Parisien et al. reported that SV-5- STAT-1- STAT-2 complex formation is thought to play a key role in the selective degradation of STAT-1 (22). To elucidate the involvement of STAT-2 in the degradation complex containing MuV-V, coimmunoprecipitation of STAT-2 from cell lysates was examined by using anti-STAT-1 antibody in the presence of MG132. As shown in Fig. 6, lower panel, the coimmunoprecipitation of STAT-2 supported the hypothesis proposed by Parisien et al. (22).
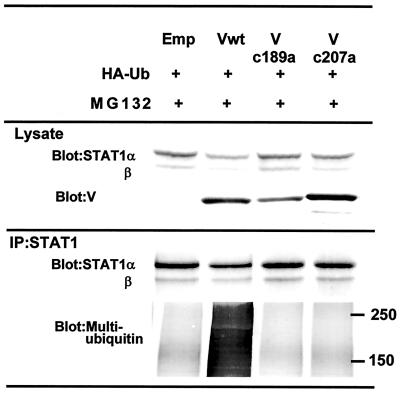
The effect of mutant MuV-V on the ubiquitination of STAT-1 was investigated in 293T cells transfected with expression vectors for HA-tagged ubiquitin and mutant MuV-V in the presence of MG132 (Fig. 7). Lysates from 293T cells expressing wild-type MuV-V or mutant MuV-V (Vc189a and Vc207a) were immunoprecipitated with anti-STAT-1 antibody. The precipitates were probed by Western blot analysis with antimultiubiquitin antibody (Fig. 7, lower panel). Multiubiquitinated STAT-1 was found in cells expressing wild-type MuV-V but not in cells expressing mutant MuV-V. In addition, we were not able to detect an interaction between STAT-1 and mutant V proteins (Vc189a and Vc207a) in this coimmunoprecipitation experiment (data not shown). Mutant MuV-V, which was not associated with a decrease in STAT-1 production, could not promote the ubiquitination of STAT-1 because of poor binding activity. Therefore, we suggest that cysteine residues in the Cys-rich region play an important role in the binding of MuV-V to STAT-1. The evidence presented here suggests that the multiubiquitination of STAT-1 is dependent on the presence of wild-type or active MuV-V and is probably mediated through an interaction of the latter with STAT-1.
FIG. 7.
Association of mutant MuV-V with ubiquitination of STAT-1. 293T cells were transfected with pCI-neo-HA-Ubiquitin (HA-Ub) and an expression vector. After cultivation for 19 h, the transfectants were treated with 20 μM MG132 for 5 h. The cells were lysed and immunoprecipitated (IP) with anti-STAT-1 antibody to analyze for multiubiquitination of STAT-1. Emp, empty vector; Vwt, wild-type MuV-V. Numbers at right indicate kilodaltons.
The C-terminal region of STAT-1 is not necessary for its ubiquitination.
Kim and Maniatis reported that activated or phosphorylated STAT-1 is degraded by ubiquitin-dependent proteolysis via the 26S proteosome (13). The phosphorylation of Tyr-701 and Ser-727 in STAT-1 is a prerequisite for its ubiquitination. These phosphorylation sites are located in the transcriptional activation domain of the C-terminal region. Therefore, it is important to investigate whether the C-terminal region of STAT-1 is also necessary for its degradation by MuV-V.
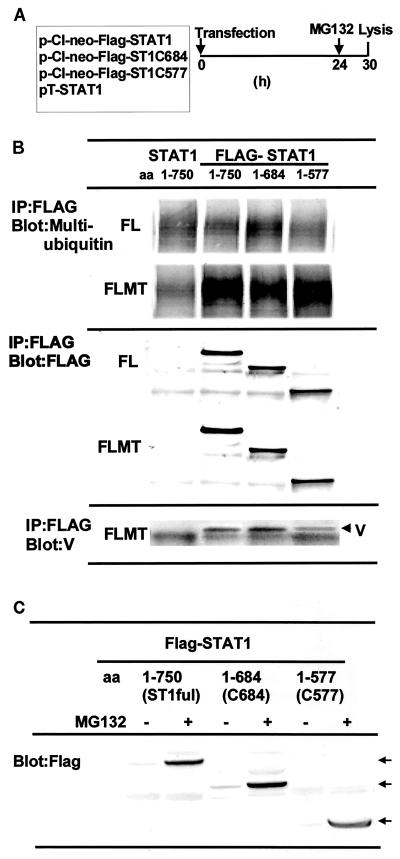
We constructed expression vectors for Flag-tagged full-length STAT-1 (pCI-neo-Flag-STAT-1) and two Flag-tagged C-terminal deletion mutants (pT-FlagST1C684 and pT-FlagST1C577). The deletion mutants lack the transcriptional activation domain (pT-FlagST1C684) or both the transcriptional activation domain and the Src homology 2 domain (pT-FlagST1C577). FL and FLMT cells were transfected with these expression vectors and treated with MG132 as indicated in Fig. 8A. After immunoprecipitation of cell lysates on an anti-Flag M2 agarose affinity gel, the precipitates were analyzed by Western blotting with antimultiubiquitin antibody or anti-MuV-V antibody (Fig. 8B). Multiubiquitination of the two mutant STAT-1 proteins was comparable to that seen with the expression of full-length STAT-1 in FLMT cells (Fig. 8B, upper panel). The middle panel of Fig. 8B indicates expression of Flag-STAT-1. Furthermore, anti-Flag antibody (M2; Sigma) coimmunoprecipitated MuV-V from lysates of FLMT cells expressing full-length STAT-1 or the two deletion mutants (Fig. 8B, lower panel). Moreover, the expression of MuV-V promoted not only the ubiquitination of C-terminally truncated STAT-1 but also its degradation in the absence of MG132 (Fig. 8C). On the basis of these observations, we propose that the N-terminal region of STAT-1 is needed for its degradation or ubiquitination and interaction with MuV-V.
FIG. 8.
Ubiquitination and degradation of C-terminally truncated STAT-1. (A) Protocol of experiment. FL or FLMT cells were transfected with expression vectors for mutant STAT-1 and cultured for 24 h. The transfectants were treated with MG132 for 6 h. The cells were lysed, and the resulting cell lysates were immunoprecipitated on an anti-Flag (M2; Sigma) agarose affinity gel. (B) Immunoprecipitates (IP) from the cell lysates were analyzed for multiubiquitination of the deletion mutant and coimmunoprecipitation of MuV-V with mutant STAT-1 by Western blotting with anti-MuV-V and antimultiubiquitin antibodies. aa, amino acids. (C) Degradation of C-terminally truncated STAT-1. Cell lysates from FLMT cells expressing C-terminally truncated STAT-1 were analyzed by Western blotting with anti-STAT-1 antibody in the presence (+) or absence (−) of MG132. ST1ful is full-length STAT-1. Arrows indicate full-length and C-terminally truncated STAT-1.
DISCUSSION
The genome of viruses of the Paramyxoviridae family contains six or seven genes that code for eight proteins, with the exception of the Pneumovirus genus. The P gene codes for two (P and V; Rublavirus genus) or three (P, V, and C; Morbillivirus genus and Respirovirus genus) proteins. These accessory V and C proteins are thought to be associated with virus pathogenesis, virus replication, and modulation of the cell cycle (2, 8, 10, 11, 12, 18, 23). Recently, it was reported that an antiviral function of IFN is inhibited by the V or C protein through disruption of the IFN signal transduction pathway, as determined by several strategies, such as (i) suppression of the phosphorylation of STATs and JAKs (Sendai virus C protein) and (ii) reduction of the constitutive or basal levels of STATs (SV5-V, hPIV2-V, and MuV-V) (1, 3, 7, 14, 15, 20, 21, 28, 30). SV5-V and MuV-V are associated with the blocking of IFN signaling through a decrease in the production of STAT-1, which is an essential component of transcriptional factors ISGF3 and GAF. MuV infection and production of V protein do not induce the suppression of JAK phosphorylation during the early stage of infection. However, the suppression of STAT-2 phosphorylation was demonstrated in acute infection with MuV. Therefore, we propose that the dysfunction of IFN signaling caused by MuV infection results mainly from the decrease in STAT-1 production and partly from the suppression of STAT-2 phosphorylation. It is still unknown what mechanism is involved in the suppression of STAT-2 phosphorylation. As STAT-2 generally associates with STAT-1, we cannot exclude the possibility that the suppression may result from a decrease in STAT-1 production.
Didcock et al. reported that SV5-V induces the proteosome-mediated degradation of STAT-1 (1). We also demonstrated that the V protein or the Cys-rich region in the V protein contributes to the induction of STAT-1 disappearance, probably via regulation by the ubiquitin-proteosome pathway (15). The V protein contains a unique carboxy-terminal domain (Cys-rich region). Furthermore, the position and presence of seven cysteine residues in the Cys-rich region are highly conserved among different paramyxoviruses.
SV5-V, MuV-V, hPIV2-V, and measles virus V protein can interact with DDB1 (17). Furthermore, the association of SV5-V with nucleocapsids or RNA has been demonstated (16). The highly conserved seven cysteine residues in the Cys-rich region play a critical role in binding specifically to zinc and DDB1 (10, 17, 19, 24). On the other hand, the amino-terminal domain of V protein is necessary for interactions with soluble NP and RNA (16, 25).
Kubota et al. reported that the Cys-rich region in the C terminus of MuV-V is associated with blocking of the IFN signal transduction pathway through a decrease in STAT-1 production (15). Therefore, it is important to investigate the role of the seven cysteine residues in the Cys-rich region of MuV-V in the loss of STAT-1. The possible and critical role of the cysteine residues in STAT-1 disappearance was confirmed by Western blot and luciferase reporter assays with several mutants of MuV-V which have single amino acid substitutions of the cysteine residues at positions 189, 207, and 214. Recently, it was found that the interaction of MuV-V with RACK1 occurs independently of the cysteine residues in the Cys-rich region, although the Cys-rich region is sufficient for the interaction with RACK1 (15a). These results suggested that the binding of MuV-V to STAT-1 or RACK1 is variable.
As MuV-V induces a reduction of constitutive or basal levels of STAT-1, multiubiquitination of STAT-1 is undetectable in MuV-infected cells. However, restoration of STAT-1 production was seen in FLMT cells treated with proteosome inhibitor MG132. After treatment with MG132, immunoprecipitates obtained from FLMT cell lysates with anti-STAT-1 antibody were subjected to Western blot analysis with anti-MuV-V or antimultiubiquitin antibody. Multiubiquitination of STAT-1 was found to be dependent on the presence of V protein. Furthermore, the most striking interaction between MuV-V and STAT-1 was obtained in this experiment, but not in the absence of MG132. In addition to these results, MuV-V mutants (Vc189a and Vc207a) interacted poorly with STAT-1. We suggest that the cysteine residues in the Cys-rich region of MuV-V are required for the ubiquitination of STAT-1, probably because they bind with STAT-1. Therefore, V protein was shown to promote the multiubiquitination of STAT-1.
It is obvious that the amounts of phosphorylated STAT-1 in IFN-γ-treated cells are controlled by the ubiquitin-dependent proteolysis of STAT-1 (13). Phosphorylation sites for Tyr-701 and Ser-727 residues in STAT-1 are located in the transcriptional activation domain of the carboxy-terminal region. Phosphorylation of STAT-1, in other words, the C-terminal region is necessary for STAT-1 ubiquitination. However, the mechanism underlying the MuV-V-induced ubiquitination of unphosphorylated STAT-1 has not yet been elucidated. To determine whether the C-terminal region of unphosphorylated STAT-1 is involved in ubiquitination and proteosome-mediated degradation, we constructed two deletion mutants lacking the transcriptional activation domain (pT-FlagST1C684) or the transcriptional activation and Src homology 2 domains (pT-FlagST1C577). In contrast to the ubiquitination of phosphorylated STAT-1, our experiments on the ubiquitination and stability of the C-terminal deletion mutants indicated that neither domain conferred multiubiquitination on STAT-1. The interaction between MuV-V and the deletion mutant of STAT-1 was clearly demonstrated in FLMT cells. Therefore, we suggest that the degradation signal and the binding site for MuV-V may be located in the N-terminal region of unphosphorylated STAT-1. The N terminus of STAT-1, containing two functional domains, the N-terminal and coiled-coil domains, has been implicated in protein-protein interactions (9, 22, 27). Therefore, we postulate that these domains function as a region of interaction with MuV-V. These results strongly suggest that the association of MuV-V and STAT-1, at least in part, promotes the ubiquitination of STAT-1. However, we do not rule out the possibility that MuV-V functions at other steps or in other pathways to cause the disappearance of STAT-1.
Recent studies on selective STAT degradation induced by paramyxoviruses revealed that SV5-V does not induce the degradation of endogenous STAT-1 in the absence of STAT-2, whereas this degradation is achieved by the interaction of SV5-V with STAT-1 and STAT-2 (22). These authors also discussed the association of the complex formed by SV5-V, STAT-1, and STAT-2 with STAT-1 degradation. Therefore, it is important to elucidate whether MuV-V-mediated-degradation of STAT-1 also requires the presence of STAT-2. Analysis of the coimmunoprecipitation of STAT-2 in FLMT cells (Fig. 6, lower panel) suggested that the formation of a functional MuV-V- STAT-1- STAT-2 complex plays a key role in the degradation of STAT-1 by MuV-V.
In the ubiquitin-proteosome pathway, E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase) enzymes are required for the multiubiquitination of target proteins (26). Identification of these enzymes (especially E3), which catalyze the sequential reaction that produces multiubiquitin chains via isopeptide bonds, is necessary for a further understanding of the proteosome-mediated degradation of STAT-1 caused by MuV-V.
REFERENCES
- 1.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT-1 for proteosome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escoffier, C., S. Manie, S. Vincent, C. P. Muller, M. Billeter, and D. Gerlier. 1999. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J. Virol. 73:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujii, N., N. Yokosawa, and S. Shirakawa. 1999. Suppression of interferon response gene expression in cells persistently infected with mumps virus, and restoration from its suppression by treatment with ribavirin. Virus Res. 65:175-185. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 5.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signaling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 6.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2001. Paramyxovirus accessory proteins as interferon antagonists. Microbiol. Immunol. 45:787-800. [DOI] [PubMed] [Google Scholar]
- 7.Hariya, Y., N. Yokosawa, N. Yonekura, G. Kohama, and N. Fujii. 2000. Mumps virus can suppress the effective augmentation of HPC-induced apoptosis by IFN-gamma through disruption of IFN signaling in U937 cells. Microbiol. Immunol. 44:537-541. [DOI] [PubMed] [Google Scholar]
- 8.Hasan, M. K., A. Kato, M. Muranaka, R. Yamaguchi, Y. Sakai, I. Hatano, M. Tashiro, and Y. Nagai. 2000. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74:5619-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath, C. M., G. R. Stark, I. M. Kerr, and J. E. Darnell. 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 16:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, C., K. Kiyotani, Y. Fujii, N. Fukuhara, A. Kato, Y. Nagai, T. Yoshida, and T. Sakaguchi. 2000. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J. Virol. 74:7834-7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, T. Shioda, and Y. Nagai. 1997. Importance of the cysteine-rich carboxy-terminal half of V protein for Sendai virus pathogenesis. J. Virol. 71:7266-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, T. K., and T. Maniatis. 1996. Regulation of interferon-γ-activated STAT1 by the ubiquitin-proteasome pathway. Science 273:1717-1719. [DOI] [PubMed] [Google Scholar]
- 14.Komatsu, T., K. Takauchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-rich region of mumps virus structural V protein correlates with block of interferon α and γ signal transduction pathway through decrease of STAT-1α. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 15a.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2002. Association of mumps virus V protein with RACK1 results in dissociation of STAT-1 from the alpha interferon receptor complex. J. Virol. 76:12676-12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, G. Y., R. G. Paterson, and R. A. Lamb. 1997. The RNA binding region of the paramyxovirus SV5 V and P proteins. Virology 238:460-469. [DOI] [PubMed] [Google Scholar]
- 17.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 18.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liston, P., and D. J. Briedis. 1994. Measles virus V protein binds zinc. Virology 198:399-404. [DOI] [PubMed] [Google Scholar]
- 20.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 22.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson, J. B., D. Thomas, H. Lewicki, M. A. Billeter, and M. B. Oldstone. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80-89. [DOI] [PubMed] [Google Scholar]
- 24.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 25.Randall, R. E., and A. Bermingham. 1996. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology 224:121-129. [DOI] [PubMed] [Google Scholar]
- 26.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Mol. Cell. Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 27.Xu, X. A., Y. L. Sun, and T. Hoey. 1996. Cooperative DNA binding and sequence selective recognition conferred by the STAT amino terminal domain. Science 273:794-797. [DOI] [PubMed] [Google Scholar]
- 28.Yokosawa, N., T. Kubota, and N. Fujii. 1998. Poor induction of interferon induced 2′,5′-oligoadenylate synthetase (2-5AS) in cells persistently infected with mumps virus is caused by decrease of STAT-1alpha. Arch. Virol. 143:1985-1992. [DOI] [PubMed] [Google Scholar]
- 29.Yokota, S., N. Yokosawa, T. Kubota, T. Suzutani, I. Yoshida, S. Miura, K. Jimbow, and N. Fujii. 2001. Herpes simplex virus type 1 suppresses the interferon signaling pathway by inhibiting phophorylation of STATs and Janus kinases during an early infection stage. Virology 286:119-124. [DOI] [PubMed] [Google Scholar]
- 30.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]