Abstract
We measured actin turnover in lamellipodia and lamellae of migrating cells, using quantitative Fluorescent Speckle Microscopy. Lamellae disassembled at low rates from the front to the back. However, the dominant feature in their turnover was a spatially random pattern of periodic polymerization and depolymerization moving with the retrograde flow. Power spectra contained frequencies between 0.5 and 1 cycle/min. The spectra remained unchanged when applying Latrunculin A and Jasplakinolide in low doses, except that additional frequencies occurred beyond 1 cycle/min. Whereas Latrunculin did not change the rate of mean disassembly, Jasplakinolide halted it completely, indicating that the steady state and the dynamics of actin turnover are differentially affected by pharmacological agents. Lamellipodia assembled in recurring bursts at the leading edge and disassembled ∼2.5 μm behind. Events of polymerization correlated spatially and temporally with transient formation of Arp2/3 clusters. In lamellae, Arp2/3 accumulation and polymerization correlated only spatially, suggesting an Arp2/3-independent mechanism for filament nucleation. To acquire these data we had to enhance the resolution of quantitative Fluorescent Speckle Microscopy to the submicron level. Several algorithmic advances in speckle image processing are described enabling the analysis of kinetic and kinematic activities of polymer networks at the level of single speckles.
INTRODUCTION
Many cell functions rely on cytoskeleton plasticity, i.e., the cytoskeletal polymer structures continuously assemble, disassemble, and reassemble at the timescale of the function they support. One of the most striking examples of cytoskeleton turnover can be observed in migrating cells. A highly polarized, dendritic network of filamentous actin (F-actin) polymerizes next to the plasma membrane of the leading edge, supposedly generating the forces that push the cell boundary forward (1). Replenishment of the pool of monomeric actin (G-actin) for polymerization occurs by depolymerization of the network in regions more distant to the cell edge.
In migrating cells, F-actin turnover is accompanied by retrograde flow of the network away from the leading edge (2–5). It is powered either by forces counteracting polymerization at the leading edge (6,7), or by network contraction mediated by myosin motor activity (7,8). In many cell types, retrograde flow is observed only in the first few microns from the cell edge. Further back, retrograde flow of the front network and anterograde flow of the cortical F-actin network underneath the cell body merge in a convergence zone (9), rich in myosin II (10).
The degree of F-actin plasticity has remained one of the main unknowns of cell migration mechanics. In this study, we focus on the plasticity between the leading edge and the convergence zone. Previously, we discovered that in this region the actin cytoskeleton consists of two spatially overlapping arrays with distinct assembly dynamics (11). At the leading edge, a thin band of polymerization is juxtaposed to a band of depolymerization. We referred to this fast treadmilling array as the lamellipodium. The spatial separation of assembly and disassembly agreed with the picture conveyed by the dendritic nucleation model (12), which proposes Arp2/3-mediated monomer association at the front, and ADF/cofilin-mediated monomer dissociation at the base of the network. As expected, blocking either assembly or disassembly (11), or disrupting Arp2/3 function perturbed the formation of a lamellipodium, surprisingly without inhibition of cell protrusion (13). In unperturbed cells, the space between lamellipodium and convergence zone was filled by a lamella, where regions of assembly and disassembly were randomly distributed. Peaks of assembly alternated with peaks of disassembly, similar to cortical F-actin in nonmigrating cells (14). In cells without a lamellipodium, the lamella reached all the way to the leading edge and the pattern of turnover did not deviate from its randomness, even in phases of pronounced protrusion (11,13). We concluded that lamellipodium and lamella together form a highly plastic bipartite F-actin structure that mediates cell protrusion in a not yet fully understood manner.
In this study we focus on differences between network assembly and disassembly in lamella and lamellipodium and seek answers to the following questions:
Are repeated cycles of lamella assembly and disassembly temporally organized, i.e., do they obey a specific periodicity?
If this is the case, will the time span between assembly and disassembly in random patterns be equivalent to the time it takes to polymerize and depolymerize the lamellipodium?
Are the mechanisms for lamellipodium and lamella assembly the same?
What is the response of the lamella to small molecule inhibitors of F-actin assembly and disassembly, which selectively remove the lamellipodium?
To examine these questions we utilized quantitative Fluorescent Speckle Microscopy (qFSM) (15). In FSM, incorporation of a low concentration of fluorescently labeled actin monomers into the F-actin network and subsequent imaging with high NA optics results in a random pattern of intensity maxima and minima. A speckle is defined as a local intensity maximum, significantly brighter than its surroundings. Changes in speckle intensity and speckle movements are in a stochastic sense related to network turnover and polymer flow. In a previous article, we presented a model for how statistical analysis of the intensity changes in the time-points of speckle appearance (birth) and disappearance (death) can be converted into temporal and spatial maps of F-actin turnover (14). We defined the rate of intensity changes in these incidences as a single-speckle kinetic score, which is, on average, proportional to the number of labeled monomers exchanged with the network in this location. By appropriate spatial and/or temporal filtering of the scores from thousands of speckles, maps of the momentary state of F-actin turnover could be resolved for each frame of the movie. Evidently, this analysis requires a robust tracking of speckle trajectories from initiation to termination. Therefore, we had initially tested the model on contact-inhibited newt lung epithelial cells, where F-actin flow was stalled and conventional single-particle tracking was sufficient to follow random speckle motion associated with thermal fluctuations and local network contraction (14). Speckle tracking in migrating cells with retrograde flow had turned out to be more challenging. We had proposed graph-theoretical approaches to link corresponding speckles between consecutive frames (16), with a performance sufficient to map out polymer flow, but not to reconstruct complete speckle trajectories from birth to death. Instead, we had exploited conservation laws of speckle field intensity to produce low-resolution maps of network turnover that could be correlated to network translocation (7).
For this study, we had to extend the resolution to the submicron level and use single speckle analysis (14) to measure polymer turnover. Besides higher resolution, the evaluation of birth and death events of single speckles adds robustness to the mapping of F-actin turnover. In both events, intensity changes are dominated by local association or dissociation of labeled subunits whereas contributions from bleaching, focus drift, or specimen movements are less significant. By contrast, the evaluation of the conservation equation includes large areas in between speckles, which do not contain information on monomer exchange but where imaging artifacts may be the main cause of signal variation.
In the following section we describe the methodological improvements necessary to process single speckle birth and death events in migrating cells. We present several methods of validation to indicate that single speckle tracking can be performed with high fidelity despite the high density and the low contrast and stability of the signal. We then utilize this ability to elucidate the characteristics and mechanisms of F-actin assembly in lamellipodium and lamella networks.
MATERIALS AND METHODS
Cell culture and preparation for FSM imaging
PtK1 cells were cultured in Ham's F12 medium containing 25 mM HEPES (Sigma-Aldrich, St. Louis, MO) and 10% FBS (Gibco BRL, Carlsbad, CA) at 37°C, 5% CO2. Primary cultures of newt lung epithelial cells were maintained in half-strength L-15 medium in 10% FCS and antibiotics at room temperature. X-rhodamine-conjugated skeletal muscle actin was prepared as described previously (17) and microinjected into cells at 0.5–1 mg/ml. After injection, the cells were incubated for ∼2 h at 37°C or at room temperature to allow fluorescent actin to incorporate into the cytoskeleton. PtK1 cells grew in small islands, where cells at the border of the island established a polarized morphology with the characteristic motile machinery of a protruding edge. However, because of strong contacts at the trailing edge to neighboring cells in the island they remain nearly stationary, with a movement of the leading edge at least one-order-of-magnitude slower than F-actin flow analyzed in this article. Newt cells grew in epithelial sheets that extended from the explant of lung tissue during the wound healing response in culture. Cells at the border of the sheet established the same polarized morphology as PtK1 cells. Cells in the center of the sheet, which were completely surrounded by neighboring cells, lost their polarity, and turned off retrograde flow. These cells are referred to as nonmigrating, contact-inhibited cells (18).
Plasmid cDNA (100 μg/ml) encoding the GFP-p34 component of the Arp2/3 complex was microinjected together with X-rhodamine actin into the nuclei of PtK1 cells. Cells were incubated at room temperature for 6–8 h to allow GFP-p34 expression.
Time-lapse spinning disk confocal FSM
Time-lapse FSM was performed using the spinning disk confocal microscope system on an inverted microscope (TE300 Quantum, Nikon, Melville, NY) using a 100× 1.4 NA Plan-Apochromatic DIC objective lens (19). Images with an average exposure time of 500–1500 ms were collected at 5- or 10-s intervals on an Orca 2 camera (Hamamatsu, Bridgewater, NJ) containing a 1280 × 1024 array of 6.7 × 6.7 μm pixels in progressive scan interline transfer configuration operated in the slow-scan (1.25 mHz) 14 bit-depth mode, where the noise is typically 3–5 electrons (root-mean-square).
Image analysis
Raw TIFF image sequences were transferred from MetaMorph (Universal Imaging, Downingtown, PA) to our qFSM software. This program has been developed under MATLAB (The MathWorks, Natick, MA) with computationally expensive modules written in C/C++ for faster execution. Algorithms for speckle extraction and tracking are described either in Ponti et al. (14) or in the following section. Graphical representations of the results have been programmed under MATLAB, using MATLAB's standard graphics functionality.
ANALYSIS OF FLUORESCENT SPECKLES
In this section, we describe the algorithms added to the previously described qFSM package (14). It includes two new modules: 1), a method for iterative speckle extraction (ISE), which prevents loss of speckles in crowded image regions, i.e., when speckle signals partially fuse; and 2), an iterative method for general nearest-neighbor (GNN) tracking of speckles, which improves the assignment of speckle correspondences in consecutive frames by adaptive propagation of speckle movements.
Iterative speckle extraction
Speckle fusions induced by low-pass filtering and local maximum detection
In a noise-free image, speckles could be detected as diffraction-limited local maxima, representing clusters of network-bound fluorophores. To identify speckles in a real image among the much more numerous local maxima associated with noise, we first filter the raw image with the optical point-spread function. This retains features within the bandpass of the microscope while suppressing noise components with higher spatial frequencies. Subsequently, we test the intensity differences between local maxima and surrounding local minima against a calibrated model of camera noise to exclude maxima associated with low-frequency noise components (14).
Because of the low-pass filtering and the finite resolution of the discrete local maximum operator, speckle pairs that are separated in the raw image by a distance close to the diffraction limit will not be distinguished by this procedure (see Supplementary Materials and Methods). Also, noise-induced variations of the raw signal are convolved into the filtered image and may obscure local maxima associated with true speckles. Therefore, two proximate speckles which are resolved as separate in time-point t − 1 can become a single detected maximum in t, but may later be resolved again, e.g., in t + 1. The statistics of speckle births and deaths will, for this example, be contaminated by one false death in t − 1 and one false birth in t + 1. To alleviate this problem, we designed a new speckle detection scheme, which largely recovers the local maxima lost due to low-pass filtering.
Recovery of higher-order speckles to resolve fused speckles
Speckles extracted by the detection scheme described in Ponti et al. (14) are termed primary speckles. The goal of our new approach was to recover higher-order speckles that were fused with primary or any lower-order speckles. The optimal method to achieve this is by fitting mixture models of an a priori unknown number of point-spread functions to the raw signal (20). However, for FSM with potentially thousands of higher-order speckles, the computational expense for this approach is prohibitive.
Instead, we implemented a fast approximation to mixture model fitting. The algorithm starts with the detection of primary speckles. Then, the noise-free signals of all primary speckles are subtracted from the low-pass-filtered raw image (see Fig. S1 in Supplementary Materials and Methods). The difference image is subjected to renewed speckle extraction by local maximum detection and statistical significance testing to detect previously masked, secondary speckles. The procedure is repeated until the fraction of additionally extracted speckles falls below a user-defined threshold (see below). Some of the higher-order speckles are artifacts generated by the simple image subtraction. They violate the generalized Sparrow criterion and are therefore excluded from the analysis, as thoroughly explained in the online Supplementary Materials and Methods.
Besides the generalized Sparrow criterion, newly extracted speckles have to pass the significance test against noise (14). The combination of the two tests guarantees rapid convergence of the algorithm. We found that in nearly all data sets the relative amount of speckles added in iteration 4 and higher is <0.5% (21). Therefore, we usually stop the search after three iterations, although the software also supports iteration control via a minimum threshold for the percentage of new speckles.
Enhancement of tracking and identification of birth and death events by ISE
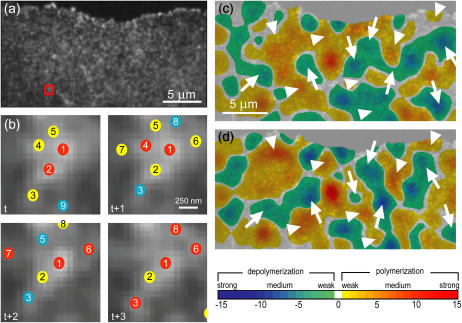
Fig. 1, a and b, demonstrates how ISE recovers multiple speckles despite temporary fusion and how it reduces the number of false birth and death events. From a cluster of partially fused speckles, as the one indicated by the small red box in Fig. 1 a, the original speckle extraction module (14) would only recover primary speckles (red in Fig. 1 b). In contrast, two additional iterations of ISE recover several new higher-order speckles (second-order in yellow, third-order in cyan). Since the analysis of the F-actin turnover relies on complete speckle trajectories from birth to death, it is obvious that the kinetic analysis performed on primary speckles only reports several falsely scored kinetic events. For instance, the disappearance of speckle 2 from frame t to t + 1 would be considered as a speckle death and, dependent on the statistical significance of foreground and background intensity changes, would be mistakenly scored as a polymerization or depolymerization event.
FIGURE 1.
Iterative speckle extraction and effects on mapping of F-actin turnover. (a) Raw FSM image. (b) Speckle extraction in four consecutive time-points. (Red, first-order speckles; yellow, second-order speckles; and cyan, third-order speckles.) Numbers identify the speckles in different time-points. F-actin turnover mapped from primary speckles only (c) and speckles up to order-three (d). The color representation of polymerization and depolymerization agrees with illustrations in Ponti et al. (14), allowing better comparison with previous studies. Arrowheads and arrows highlight regions where the additional information generated by high-order speckles yields significant shifts in the location and magnitude of polymerization and depolymerization peaks, respectively.
Full recovery of all speckles not only eliminates false birth and death events, but also avoids false links in trajectories. For example, considering only the primary speckles any tracking algorithm will associate speckle 1 to speckle 1, and speckle 2 to speckle 4 between frame t and t + 1. Continuation of tracking will likely lead to a link of speckle 7 in t + 2 to speckle 3 in t + 3, and so forth.
Fig. 1, c and d, compare maps of cortical F-actin turnover in a contact-inhibited cell generated based on primary speckles only (Fig. 1 c) and speckles up to order-three (Fig. 1 d). Although the key properties of periodic assembly and disassembly of the network (14) did not change (see Supplementary Materials and Methods, Movie 1), the panels indicate significant shifts in the locations and magnitudes of peak polymerization (arrowheads) and depolymerization (arrows). These shifts are critical when FSM is used for mapping turnover at the micron scale.
General nearest-neighbor tracking with motion propagation
Previously, we used local nearest-neighbor assignment to identify corresponding speckles in consecutive frames (14). This was possible because the F-actin cortex of nonmigrating cells was overall stationary. The random displacements of speckles between frames had a magnitude 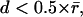 where
where 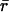 denotes the average distance between two neighboring speckles. In this regime, particle association via shortest distance identification provides the optimal tracking solution (22). Occasionally, the fluctuations were increased so that the displacements for a few speckles fell into the regime
denotes the average distance between two neighboring speckles. In this regime, particle association via shortest distance identification provides the optimal tracking solution (22). Occasionally, the fluctuations were increased so that the displacements for a few speckles fell into the regime 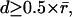 where topological conflicts cause two speckles in time-point t to compete for the same nearest neighbor in time-point t + 1. We resolved such conflicts with a set of heuristic rules. With directed retrograde F-actin flow in migrating cells, most speckle displacements fall into the regime
where topological conflicts cause two speckles in time-point t to compete for the same nearest neighbor in time-point t + 1. We resolved such conflicts with a set of heuristic rules. With directed retrograde F-actin flow in migrating cells, most speckle displacements fall into the regime 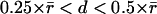 or beyond. In this case a local nearest-neighbor search no longer has an unambiguous solution and must be replaced by rigorous global nearest-neighbor (NN) association according to
or beyond. In this case a local nearest-neighbor search no longer has an unambiguous solution and must be replaced by rigorous global nearest-neighbor (NN) association according to
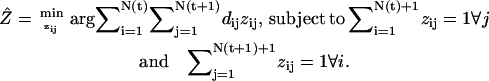 |
(1) |
The matrix Z describes the unknown assignment matrix, whose elements zij = 1 if speckle i in t corresponds to speckle j in t + 1, and zij = 0 otherwise. The matrix has dimensions N(t) + 1 × N(t + 1) + 1 with N(t) and N(t + 1) denoting the speckle numbers extracted in t and t + 1, respectively. Nonzero elements zN(t)+1,j in the last row of the matrix refer to birth events, i.e., there is no speckle present in t that links to speckle j in t + 1. Analogously, nonzero elements in zi,N(t+1)+1 refer to death events. Equation 1 formulates a standard problem of combinatorial optimization, where elements of Z are determined so that the overall sum of speckle displacements is minimized. Speckles with a birth or death incidence between the two time-points have an auxiliary displacement value dN(t)+1,j = di,N(t+1)+1 > dmax associated, where dmax defines the maximal speckle displacement between two consecutive time-points. This is the only parameter required to run the tracking algorithm. It can easily be determined by coarse assessment of the F-actin flow speed prior to precise tracking. Innumerable solutions to Eq. 1 have been proposed (23). For this article we implemented a modified version of the self-organized map neural-network algorithm (24,25).
Obviously, if the majority of speckle displacements are 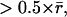 the application of Eq. 1 will no longer be appropriate. In this case the tracking scheme requires support from a motion model that predicts the displacement of each speckle between t and t + 1. Data association according to Eq. 1 is executed on truncated displacements
the application of Eq. 1 will no longer be appropriate. In this case the tracking scheme requires support from a motion model that predicts the displacement of each speckle between t and t + 1. Data association according to Eq. 1 is executed on truncated displacements 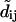 between predicted and observed speckle position in t + 1 (GNN tracking; (22)).
between predicted and observed speckle position in t + 1 (GNN tracking; (22)).
To derive a motion model we exploited the fact that in F-actin networks, speckles probe the superposition of a coherent flow component, i.e., the velocities of speckles are correlated over a certain distance, and an uncorrelated, random component associated with the sources of positional fluctuations described above. Given a first assignment 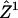 from time-point t to t + 1 based on global NN tracking we extracted the coherent component of speckle flow ui = u(xi) by filtering the speckle displacements
from time-point t to t + 1 based on global NN tracking we extracted the coherent component of speckle flow ui = u(xi) by filtering the speckle displacements
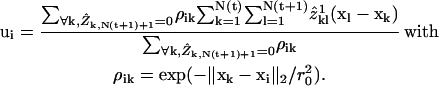 |
(2) |
The correlation length r0 of the filter kernel was chosen as a function of the local flow incoherence (see below). Using the flow field thus found, truncated displacements 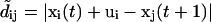 were calculated for all speckle pairs and the assignment matrix updated by a renewed solution of Eq. 1. The iteration was stopped if there was no change in the assignment matrix. In practice, the scheme proved to converge very quickly and a single iteration was sufficient. This means that many of the initially calculated global NN links provided accurate estimates of speckle flow, and motion propagation was necessary only in a few places of the network with high flow speeds (see Supplementary Materials and Methods).
were calculated for all speckle pairs and the assignment matrix updated by a renewed solution of Eq. 1. The iteration was stopped if there was no change in the assignment matrix. In practice, the scheme proved to converge very quickly and a single iteration was sufficient. This means that many of the initially calculated global NN links provided accurate estimates of speckle flow, and motion propagation was necessary only in a few places of the network with high flow speeds (see Supplementary Materials and Methods).
The same motion prediction was also applied to the closing of gaps in speckle trajectories. In Ponti et al. (14), we proposed to link trajectories terminating in frame t − 1 to trajectories initiated in t + 1, if a statistically insignificant, weak local intensity maximum fell into the intersection of two circular search regions of radius dmax, centered at the positions of termination and initiation in t − 1 and t + 1, respectively. With motion propagation, the criterion is modified to the search of a weak local intensity maximum in t inside the intersection of two circles, the first centered at 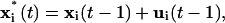 the second at
the second at 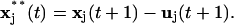
Locally adaptive flow filtering
The correlation length r0 in Eq. 2 has to be set as a compromise between preservation of details in the flow field and adequate averaging of random displacements. F-actin networks in migrating cells contain pronounced flow gradients, e.g., in regions of strong contraction or at the transition from lamellipodium to lamella. In both cases the flow filtering has to be decorrelated to preserve the local flow structure. We achieved this by adaptive attenuation of
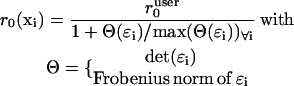 |
(3) |
applied to the strain tensor ɛi = 1/2(∇ui + ∇ui)T. High values Θ(ɛi) indicate significant deformations that deviate from the normal behavior of an F-actin network. Accordingly, the user-defined parameter 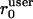 should reflect the correlation length of the mechanically coupled motion of two points in a normal F-actin network. We set
should reflect the correlation length of the mechanically coupled motion of two points in a normal F-actin network. We set 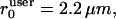 motivated by 1/r-decay distances measured in two-point microrheology of F-actin networks (unpublished data). Although the algorithm was implemented as an iterative scheme, i.e., the filtered network flow could again be employed to update the strain tensor, the flow maps did not significantly change after the first iteration. We thus stopped adaptive filtering after one round of strain estimation and adaptive flow filtering.
motivated by 1/r-decay distances measured in two-point microrheology of F-actin networks (unpublished data). Although the algorithm was implemented as an iterative scheme, i.e., the filtered network flow could again be employed to update the strain tensor, the flow maps did not significantly change after the first iteration. We thus stopped adaptive filtering after one round of strain estimation and adaptive flow filtering.
Probing F-actin retrograde flow in migrating cells by single-speckle tracking
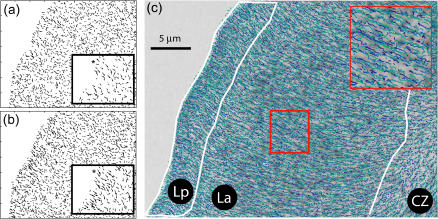
Fig. 2 gives an example of how GNN tracking enhances the quality of speckle trajectories. The merits of GNN are most obvious in regions where high rates of birth and death events and faster flow (in the regime 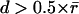 ) result in a large number of topological conflicts, as in the region close to the leading edge of the cell. We compare the GNN solution with global nearest-neighbor tracking (Fig. 2 a), which contains too few links and a broad distribution of motion directions (some even pointing toward the cell edge), indicating that, in many cases, the rate of retrograde flow exceeds the limit
) result in a large number of topological conflicts, as in the region close to the leading edge of the cell. We compare the GNN solution with global nearest-neighbor tracking (Fig. 2 a), which contains too few links and a broad distribution of motion directions (some even pointing toward the cell edge), indicating that, in many cases, the rate of retrograde flow exceeds the limit 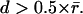 Fig. 2 b demonstrates that these limitations are largely overcome by GNN tracking.
Fig. 2 b demonstrates that these limitations are largely overcome by GNN tracking.
FIGURE 2.
Tracking of speckle trajectories probing the F-actin retrograde flow in a newt lung epithelial cell. (a) Speckle displacements at the leading edge tracked by global nearest-neighbor assignment. (b) Speckle displacements for the same region tracked by generalized nearest-neighbor (GNN) assignment. More links are extracted which reflect a directionally more coherent flow field. The asterisk in the inset highlights a pair of speckles, which is wrongly tracked in forward (toward the edge) direction in a, but is captured correctly in b. (c) Speckle trajectories initiated in the first 30 frames of the movie. Colors encode the frame number (dark blue, early time-point; light green, late time-points). The perimeters of the lamellipodium (Lp), lamella (La), and convergence zone (CZ), as defined by the ensemble kinematics and kinetics of the speckles in these regions in Ponti et al. (11), are overlaid.
We have to point out that our iterative scheme for building the motion model from single-speckle tracks converges only if the links of the first iteration are sufficiently accurate to reflect the main structure of the flow field. Although this was fulfilled for all data presented in this article, and generally holds for FSM data of F-actin, the solution does occasionally fail. In this case the initialization of the motion model has to be replaced by more sophisticated estimation of the large-scale motion. Algorithms to achieve this will be discussed in a separate, more technical article.
Fig. 2 c displays all speckle trajectories initiated in the first 30 frames of the movie. The proposed tracking methods allow us now to probe F-actin dynamics at submicron resolution (see also inset). The map is overlaid by the perimeters of the lamellipodium (Lp), lamella (La), and convergence zone (see Introduction). Differences in the kinetic and kinematic behavior of speckles in these cellular regions are documented in Table S1 in Supplementary Materials and Methods.
We validated the performances of ISE and GNN tracking on synthetic and experimental data, the results of which are summarized in the Supplementary Materials and Methods. For more details we refer to Ponti (21). Together, these analyses indicated that speckle trajectories can be tracked with high fidelity. The robustness of the method ensures that trajectory endpoints associate with true speckle birth and death events, which can be evaluated as events of local monomer exchange.
RESULTS AND DISCUSSION
Organization of turnover in lamellipodia and lamellae of migrating cells
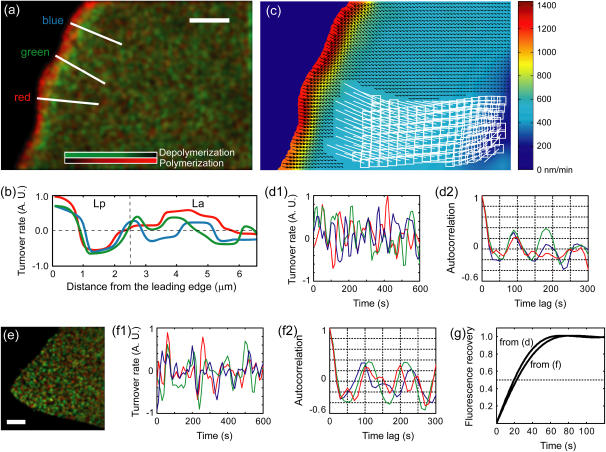
The goal of this study was to exploit single-speckle trajectories to analyze the F-actin dynamics in Lp and La of migrating cells. Fig. 3 a displays the map of F-actin turnover in the same cell for which speckle trajectories are shown in Fig. 2. Polymerization and depolymerization scores were obtained by analyzing ∼100,000 birth and death events collected over 60 frames (10 min).
FIGURE 3.
F-actin turnover in migrating and nonmigrating cells. (a) Steady-state map of assembly (red) and disassembly (green) in a migrating cell. Scale bar is 5 μm. Animated maps are available in Movie 3, Supplementary Materials and Methods. (b) Net assembly (= assembly – disassembly) along three profiles indicated in a. (c) Map of retrograde flow. Vectors show the flow velocity. Colors encode the vector magnitude, i.e., the flow speed. Animated speed/velocity maps are available in Movie 4, Supplementary Materials and Methods. White boxes display the end position of 1.2 μm × 1.2 μm probing windows, which move with the retrograde flow. For visual simplicity, their trajectories over 60 frames are approximated by the white line segments. (d1) Time evolution of turnover in three arbitrarily selected probing windows. (d2) Autocorrelation of the three curves, revealing a periodicity in turnover of 85–100 s. (e) Steady-state map of assembly (red) and disassembly (green) in a nonmigrating cell. Animated maps are available in Movie 5 in Supplementary Materials and Methods. Scale bar is 5 μm. (f1) Time evolution of turnover rate for three randomly selected probing windows for the cell in e. (f2) Autocorrelation, revealing a periodicity in turnover of 90–110 s. (g) Simulated FRAP curves using the primary periodicity extracted in d and f. They project a half-time of fluorescence recovery after complete photobleaching of 20–23 s.
At the cell edge, the map displays a narrow band of strong polymerization, juxtaposed to a similarly narrow band of depolymerization. Together, the two bands reflect the spatial organization of a fast treadmilling Lp. Profiles of Lp net turnover (Fig. 3 b) exhibited a surplus of assembly in the first 1 μm from the leading edge and a surplus of disassembly between 1 μm and 2–2.5 μm from the edge. Fast F-actin treadmilling in the Lp was accompanied by fast retrograde flow (Fig. 3 c). Beyond 2.5 μm, profiles of net assembly undergo random fluctuations between assembly and disassembly (Fig. 3 b). The transition between spatially organized bands and patterns of random net assembly and disassembly co-localizes with a steep gradient in retrograde flow (Fig. 3 c). In Ponti et al. (11) we combined these two criteria to automatically track the transition between the Lp and La.
Lamella networks of migrating cells assemble and disassemble in periodic patterns
The La is represented by a region with a punctate pattern of weaker polymerization/depolymerization contrast (Fig. 3 a). Animated turnover maps displayed cyclic La assembly, disassembly, and reassembly (compare to Movie 3 in Supplementary Materials and Methods). To quantify this behavior, we placed probing windows of 1.2 μm × 1.2 μm in the turnover map and measured the net rates of assembly and disassembly while the windows followed retrograde flow (Fig. 3 c). The size of the windows was determined by the average distance between assembly and disassembly peaks. As a consequence of the small window area only a few kinetic scores (1 to maximum 3) could be accumulated per window and time-point. To increase the robustness we integrated score values over time with a moving average over three frames. Fig. 3 d1 presents the net turnover rates of three sample windows. We manually counted the number of assembly and disassembly peaks per profile and found a mean period of 75 ± 8 s. This value was confirmed by autocorrelation analysis (Fig. 3 d2), which exhibited first-order side lobes at timelags of 85–100 s.
Average values of the turnover scores over 60 frames and the entire La region were negative, in agreement with our previous report of overall La disassembly (7). However, for the cell displayed in Fig. 3, the mean rate of disassembly was 1.7% of the mean amplitude of periodic turnover (difference between maximum assembly and maximum disassembly), indicating that oscillatory network assembly and disassembly dominate the kinetics of La turnover. We verified this behavior in three newt lung epithelial cells and three PtK1 cells and consistently found a mean rate of La disassembly 0.7–3.0% of the mean amplitude of assembly and disassembly cycles.
La turnover in random periodic patterns resembled the cortical F-actin turnover in contact-inhibited newt lung epithelial cells (14). To examine their similarity quantitatively, we repeated experiments in nonmigrating cells with marginal (threefold less) retrograde flow (Fig. 3 e; and see Movie 5 in Supplementary Materials and Methods). By losing polarity, nonmigrating cells also lose the band of polymerization and depolymerization associated with the leading edge Lp. The remaining pattern of random assembly and disassembly throughout the cortex exhibited the same local periodicity (83 ± 14 s; Fig. 3 f1) as La networks undergoing retrograde flow.
Periods of La turnover are consistent with fluorescence recovery after photobleaching (FRAP) and photoactivation of fluorophores (PAF) measurements of filament turnover
F-actin turnover in living cells had previously been analyzed by photoactivation of fluorophores (PAF) and fluorescence recovery after photobleaching (FRAP) (26–28). The numerical values reported in these studies varied from halftimes of ∼20–25 s in keratocytes and fibroblasts to rates that were one-order-of-magnitude slower in endothelial cells. To compare our measurements to FRAP and PAF data we converted the dynamics of assembly and disassembly cycles into simulated FRAP curves (Fig. 3 g). We obtained half-times of fluorescence recovery in the range 20–23 s.
In view of the apparently short lifetime of actin filaments, it was concluded in Theriot and Mitchison (26,27) that the network requires substantial repolymerization during the much longer period it travels from the leading edge to the cell center. Using single-fluorophore FSM, polymerization in more basal regions of the protruding network was indeed demonstrated (29). We confirmed this behavior based on multifluorophore FSM (7). Neither of the previous analyses, however, could capture the remarkable periodicity of turnover nor determine that hot spots of polymerization and depolymerization coexist at distances as short as 1.5–2 μm.
Separation of Lp network polymerization and depolymerization is unrelated to the periodicity of La turnover
We examined if the assembly and complete disassembly of the Lp network over 2.5 μm obeyed the same mechanisms as the turnover in the La, with the difference that the mediators of assembly and disassembly in the Lp were organized and synchronized in bands parallel to the cell edge while randomized in the La. If this were the case, the width of the Lp would be defined by the distance an Lp network patch could travel over the period of ∼75–85 s between its polymerization at the leading edge and its depolymerization at the Lp-La transition. Lp retrograde flow speeds reached maxima of 1.3 μm/min, which was insufficient to traverse a 2.5-μm-wide Lp in one turnover cycle (Fig. 3). We concluded that the stationary separation of juxtaposed bands of Lp assembly and disassembly and the periodicity of La F-actin turnover originate from different mechanisms.
Periodic patterns of La turnover are composed of a limited set of characteristic frequencies
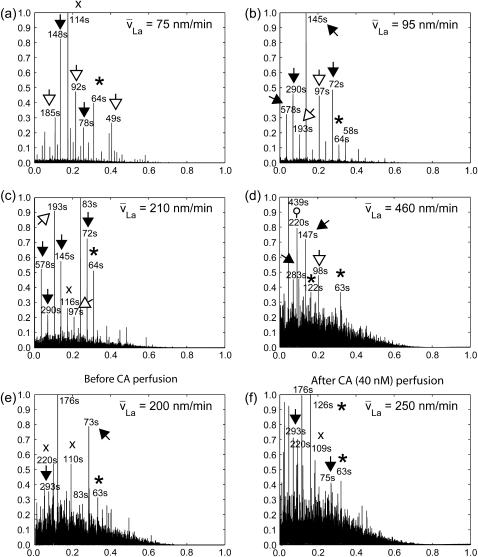
We refined the analysis of La turnover by computing power spectra. Because of the low number of scores per time-point and probing window, the turnover time series were very noisy. In addition, the duration of a movie (mostly 10 min) was short compared to the periodicity of turnover (1–2 min). Therefore, time series of at least 100 probing windows had to be integrated to obtain meaningful power spectra (see Supplementary Materials and Methods). Fig. 4, a–d, displays normalized power spectra of F-actin turnover in four cells with a sixfold variation in retrograde flow speed. All spectra display discrete spikes, indicating that the assembly and disassembly dynamics occurs with a finite number of frequencies. The dominant frequencies (or cycle periods) are similar in all cells, independent of the speed of retrograde flow. In agreement with the manual analysis of periodicity in Fig. 3, all power spectra contained a harmonic series ∼72 s, ∼144 s, and ∼288 s (solid arrow). A second series was found for ∼97 s and ∼184 s (open arrows), and a third peak at 63 s (asterisks), which rarely occurred with its lower harmonics. In longer movies, peaks were observable at ∼580 s, representing the third-order lower harmonics of 72 s (e.g., Fig. 4, b and c). Also, in some cells a fourth frequency emerged at 114 s. The fact that turnover could reproducibly be decomposed into three, and occasionally four, harmonic series with equal base frequencies causes us to speculate that F-actin assembly and disassembly follow an intrinsic clock associated with a kinetically very stable regulatory pathway. It will now take a large series of experiments and sophisticated spatiotemporal modeling to decipher the pathways that control this cyclic behavior. Also, it will eventually be desirable to examine the spatial and temporal heterogeneity of the spectra, both currently obscured by averaging. To achieve this goal, we require significantly more sensitive CCD cameras that allow us to reduce the sample exposure and to increase the frame rate and length of the movies.
FIGURE 4.
Power spectra of La turnover. (a–d) Comparison of spectra from four cells with different mean retrograde flow (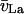 ). Arrows, open arrows, asterisks, and circle indicate frequencies above the noise floor and their harmonic relationships. (e and f) Spectra before and after perfusion of the cell with Calyculin A (CA).
). Arrows, open arrows, asterisks, and circle indicate frequencies above the noise floor and their harmonic relationships. (e and f) Spectra before and after perfusion of the cell with Calyculin A (CA).
Contraction augments the noise floor of power spectra but the characteristic frequencies of periodic turnover are preserved
The power spectra indicate a dependence of the noise floor on the speed of retrograde flow. Previously, we identified myosin contraction as the predominant driver of La retrograde flow (11). Therefore, we speculated that the increasing disorder of spectra at higher flow velocities was related to stronger network contractility. To test this hypothesis we examined the effect of Calyculin A (CA) (Fig. 4, e and f). CA is a type I phosphatase inhibitor known to inhibit myosin light-chain phosphatase and thus to promote contractility. As expected, power spectra after perfusion had a higher noise floor compared to spectra before perfusion. CA treatment induced a number of additional peaks in the low-frequency domain. Yet, none of the previously identified intrinsic frequencies of actin turnover was lost. We concluded that increased network contraction perturbs the dynamics of F-actin turnover but does not fundamentally affect the intrinsic periodicity.
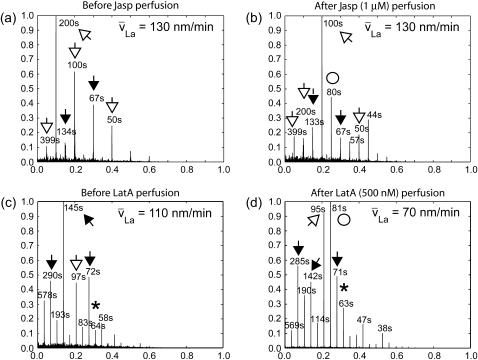
Perfusion of Jasplakinolide and Latrunculin A shift the dynamics to higher frequencies
Next, we tested the spectral responses to small molecule inhibitors of F-actin assembly and disassembly. We treated cells with 1-μM Jasplakinolide (Jasp, Fig. 5, a and b), which is known to stabilize actin filaments in vitro (30) and in vivo (31). In steady-state analysis, we observed the expected decrease of net disassembly by nearly two orders of magnitude. However, although average disassembly was inhibited by the drug, a new dynamic equilibrium of La assembly and disassembly with different frequencies was established (Fig. 5 b). Remarkably, when excluding the DC values the integrated power spectra of the two conditions were nearly identical (difference ∼10%). Hence, Jasp did, on average, inhibit La disassembly, but did not stall network turnover in periodic patterns. The spectra after drug application even contained peaks at frequencies higher than control cells (44 s accompanied by the lower harmonics at 80 s).
FIGURE 5.
Power spectra of La turnover in presence of known inhibitors of F-actin dynamics. (a and b) Spectra before and after perfusion of the cell with Jasplakinolide (Jasp), an inhibitor of actin depolymerization. (c and d) Spectra before and after perfusion of the cell with Latrunculin A (LatA).
In a second set of experiments, we treated cells with low concentrations (500 nM) of Latrunculin A (LatA, Fig. 5, c and d). LatA is known to sequester actin monomers and hence to deplete the pool of polymerization-competent subunits. At high concentrations LatA has been reported to depolymerize the network. In previous experiments with contact-inhibited cells, where the steady states of assembly and disassembly are equal, we found that LatA concentrations below 750 nM shifted the turnover to a new dynamic equilibrium, again with balanced rates of assembly and disassembly (14). We repeated these experiments in migrating cells where the La undergoes net disassembly. Mean disassembly rates before and after perfusion of LatA did not differ significantly (−2.7 × 10−3 A.U. before; −2.0 × 10−3 A.U. > 2 min after perfusion), indicating that the mild change in G-actin/F-actin ratio induced by low LatA concentrations do not profoundly shift the equilibrium toward actin disassembly. All characteristic frequencies of the periodic turnover pattern were preserved despite the drug treatment, but similar to the effects of Jasp, higher frequencies became more dominant (Fig. 6 d).
FIGURE 6.
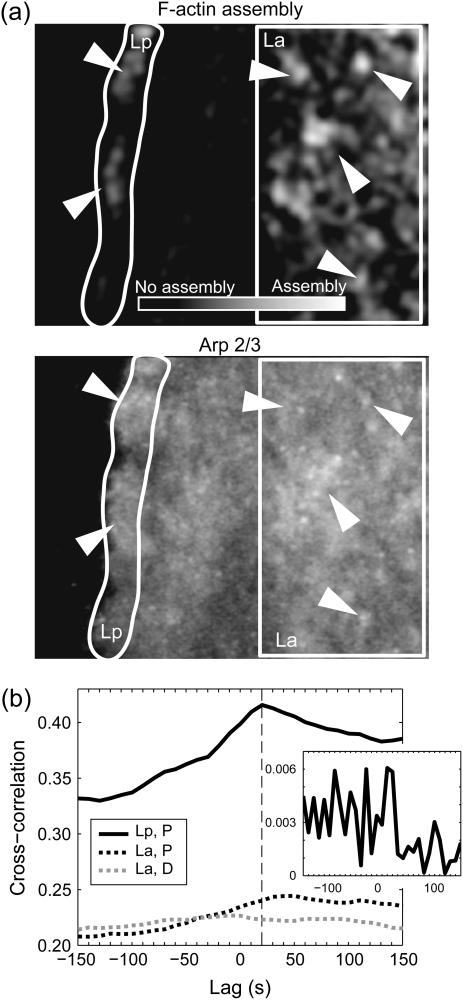
Correlation of F-actin assembly in Lp and La with dynamic clustering of Arp2/3 visualized by the GFP-p34 subcomplex. (a) Time-averaged map of F-actin assembly (top) and Arp2/3 accumulation (bottom). Arrowheads indicate positions where high assembly visually correlates with maxima in the Arp2/3 signal. (b) Cross-correlation analysis between Lp Arp2/3 signal and polymerization (solid line); La Arp2/3 signal and polymerization (dashed line); and La Arp2/3 signal and depolymerization (shaded dashed line). (Inset) Cross-correlation between a random field of polymerization scores and Arp2/3 signal.
Together, these experiments revealed a fundamental difference between the steady-state and dynamic responses of La networks to actin-affecting drugs. This find has major ramifications on the use of pharmacological agents for the perturbation of cytoskeleton function. First, as reported by other studies, many of the drugs induce dose-dependent responses. Second, this study suggests that drugs have to be characterized also in terms of the timescale of their effects. Although a specific drug, e.g., LatA, has very little effect on slow actin-dependent cell functions, faster processes may be critically accelerated with drug perfusion as the periodicity of turnover increases. Jasp, on the other hand, may stall slow processes that require actin disassembly, but—similar to LatA—boost faster processes. It may become a key application of qFSM to measure dose-response curves for the most popular perturbants of actin cytoskeleton functions and to complement them with a characterization of timescale-dependent responses.
La assembly and disassembly is spatially but not temporally correlated with Arp2/3
In search of a molecular mechanism of La assembly and disassembly in hot spots, we correlated time-resolved assembly maps with timelapse image sequences of the GFP-p34 component of the Arp2/3 complex (Fig. 6). Following Bretschneider et al. (32), we expected that Arp2/3 could be a direct promoter of La network assembly. By visual inspection bright signals of net assembly (Fig. 6 a, top panel) indeed appeared to co-localize with bright GFP-p34 signals (Fig. 6 a, bottom panel), especially in the La region (arrowheads). However, cross-correlation of the two two-dimensional signals over time revealed an average correlation of 0.22 with a weak maximum at ∼+50 s (Fig. 6 b, dashed line). To test the significance of the correlation value, we cross-correlated the GFP-p34 signal with a synthetic map of net assembly, where experimentally measured scores of network polymerization were randomly redistributed. The correlation dropped by three orders of magnitude (Fig. 6 b, inset), confirming that the correlation values in Fig. 6 b represent co-localization of Arp2/3 clusters with peaks of La network assembly. On the other hand, the lack of a strong correlation maximum at any particular timelag discounted the possibility of dynamically coupled Arp2/3 aggregation and assembly. To support this conclusion, we also cross-correlated the GFP-p34 signal and disassembly maps, leading to nearly the same correlation values (Fig. 6 b, shaded dashed line). Hence, our qFSM measurements of F-actin turnover agree with the data presented in Bretschneider et al. (32), in that hot spots of La network assembly and disassembly tend to co-localize with sites of Arp2/3 aggregation; but our dynamic analysis reveals that these events are independent in time. A possible explanation for the spatial, but not temporal relationships could rely on the assumption that Arp2/3-rich network region are more branched and thus are likely more dynamic. However, the initiation of polymerization involves a second mediator besides Arp2/3, for which the formin family members seem good candidates (33).
In the Lp, the level of cross-correlation between network assembly and GFP-p34 signal was clearly higher than in the La and we found a significant maximum, indicating a dynamic relationship between assembly and Arp2/3 aggregation (Fig. 6 b, solid line). The timelag of +20 s implies that the highest rates of assembly precede the maximum of the GFP-p34 signal. Although counterintuitive at the first sight, these data are compatible with a model of autocatalytic network assembly by dendritic nucleation: a localized burst of actin polymerization induces spatially confined Arp2/3 accumulation, as it increases the probability for Arp2/3-mediated filament side-binding. This will cause exponential network growth until the pool of polymerizable G-actin is locally depleted. At this point the assembly rate begins to taper off, while Arp2/3 continues to bind to preexisting filaments. The peak in Arp2/3 signal will be observed at the time-point the network turns from assembly into disassembly.
Lp assembles in cycles at the leading edge and disassembles in cycles at the base
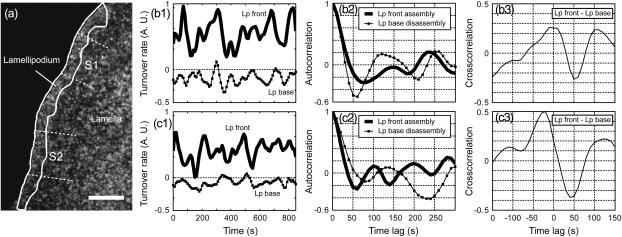
In view of the above observation that Lp assembly and disassembly over 2–3 μm could not be related to the periodicity of La turnover, we returned to the question whether the Lp network polymerizes and depolymerizes at space-dependent, yet temporally constant rates. The finding that assembly and Arp2/3 localization correlated spatially and temporally suggested that Lp polymerization is a nonstationary process. We investigated cells with monotonic forward protrusion (compare to Movie 6 in Supplementary Materials and Methods) and analyzed the qFSM scores located in the band 0–1 μm from the leading edge. Time-resolved analysis of the turnover in such a narrow region was challenging. On average, the number of scores per frame and μm2 was 1.3. To guarantee sufficient yield for data analysis we accumulated scores in sectors of 3-μm-length along the cell edge and filtered them in the time domain with a Gaussian, 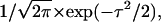 where τ denotes the difference between the time-point of the rate computation and the time of any contributing frame before and after. Fig. 7 a displays the position of two sectors S1 and S2, the results of which are shown in Fig. 7, b and c, respectively. Both sectors display periodic patterns of assembly. Very similarly to the assembly and disassembly cycles in the La, we assessed a dominant frequency of 88–100 s by manual counting of the number of extrema in the turnover curve (Fig. 7, b1 and c1). Yet, in stark contrast to La turnover curves, the network activity remained consistently positive, i.e., within the resolution limits of the spatial and temporal averaging, the Lp network oscillated only between faster and slower assembly. Autocorrelation analysis (Fig. 7, b2 and c2) of the assembly confirmed the periodicity. However, the maxima and minima of the side lobes were relatively weak due to fluctuations in the length of the period.
where τ denotes the difference between the time-point of the rate computation and the time of any contributing frame before and after. Fig. 7 a displays the position of two sectors S1 and S2, the results of which are shown in Fig. 7, b and c, respectively. Both sectors display periodic patterns of assembly. Very similarly to the assembly and disassembly cycles in the La, we assessed a dominant frequency of 88–100 s by manual counting of the number of extrema in the turnover curve (Fig. 7, b1 and c1). Yet, in stark contrast to La turnover curves, the network activity remained consistently positive, i.e., within the resolution limits of the spatial and temporal averaging, the Lp network oscillated only between faster and slower assembly. Autocorrelation analysis (Fig. 7, b2 and c2) of the assembly confirmed the periodicity. However, the maxima and minima of the side lobes were relatively weak due to fluctuations in the length of the period.
FIGURE 7.
Correlation of periodic F-actin assembly at the leading edge and disassembly at the Lp base. (a) Segmentation of the Lp region as described in Ponti et al. (11); compare to Movie 6 in Supplementary Materials and Methods. Scale bar is 5 μm. (b1) Turnover at the Lp front and at the Lp base for sector S1. Continuously positive values at the front indicate continuous net assembly. Continuously negative values at the base indicate continuous net disassembly. (b2) Autocorrelation of assembly and disassembly indicating a weak periodicity. (b3) Cross-correlation between assembly and disassembly, indicating that high rates of assembly at the Lp front is concurrent with a reduced disassembly at the Lp base. (c1–c3) The same analysis for sector S2.
Next, we speculated about the mechanism of oscillatory Lp network assembly. A direct and localized relationship between ADF/cofilin-mediated network disassembly and cell edge protrusion was proposed in Ghosh et al. (34). We thus hypothesized that peak disassembly at the Lp base would be in synchrony with peak assembly at the Lp front, implying that the recycling of ADP-actin into ATP-actin was sufficiently fast to explain the localized responses reported in Ghosh et al. (34). To measure turnover at the Lp base, we tracked the Lp-La transition using the method described in Ponti et al. (11), and integrated the scores falling into sectors of 1-μm depth and 3-μm width positioned in front of the transition line (compare to Supplementary Materials and Methods, Movie 6, constructed from 100 profiles of net assembly and flow speed perpendicular to the leading edge). The analysis required a minimum Lp width of 2 μm to avoid overlap between the analyses at Lp front and Lp base. The average width of the Lp in Fig. 7 was 2.4 μm. In the few occasions where the depth of an Lp sector fell temporarily below 2 μm we divided the scores equally between front and base half. Fig. 7, b1, b2, c1, and c2, display the disassembly at the Lp base for the two sectors S1 and S2. The curves evolve with a periodicity similar to the assembly at the Lp front, but are, in disagreement with our expectation, in phase with the leading-edge assembly, i.e., assembly peaks in the front coincided with low rates of disassembly in the back of the Lp. Cross-correlation analysis indicates a mutual relationship between assembly and disassembly with a maximum at timelags of 0 s (Fig. 7 b3) and −20 s (Fig. 7 c3; i.e., depolymerization at the Lp base lagged behind polymerization at the Lp front). The peak of negative correlation consistently occurred at +50 s, i.e., at a phase shift of ≈π relative to the dominant frequency of ≈100 s. In view of the rapid recycling and diffusion of G-actin (35), it seems unlikely that increases in polymerization of the Lp front are mediated by increases in network disassembly that take place 50 s earlier at the Lp base. Instead, we suspect that the coordination of F-actin turnover dynamics across the Lp results from signaling pathways that link Arp2/3 and cofilin activity. The investigation of these mechanisms is currently underway in our labs.
CONCLUSION
This article examines the dynamic assembly and disassembly of Lp and La networks at the front of migrating epithelial cells. We studied control cells with slow, continuous forward protrusion and perturbed F-actin dynamics by small molecule inhibitors of actin polymerization and depolymerization, and promoters of F-actin contraction. Similar to our previous study of cortical F-actin in nonmigrating cells, we discovered that turnover occurs in periodic patterns of a few and remarkably stable frequencies. Considering the kinematic, kinetic, and molecular differences between Lp and La (11), we performed separate analyses for both regions.
The La network assembles, disassembles, and reassembles in small, randomly distributed puncta. They move retrogradely at rates <500 nm/min, undergoing multiple cycles of polymerization and depolymerization with an average rate biased toward net disassembly, resulting in a F-actin concentration gradient from the leading edge toward the cell body. Perfusion of the network with the filament stabilizing drug Jasplakinolide inhibited the average disassembly, but did not cease periodic turnover. Similarly, treatment of cells with low doses of Latrunculin A had no effect on the net La disassembly and altered the periodicity of turnover only marginally. From these data we conclude that:
La undergoes continuous, periodic renewal similar to the actin cortex in stationary cells. The turnover is significantly more dynamic than is required for an overall disassembly of the network from the cell edge to the convergence zone. The high plasticity of the La has major implications on how the actin network integrates contraction-, adhesion-, and polymerization-induced forces as a mechanical entity to mediate cell protrusion.
Actin turnover occurs at different timescales, which are differentially affected by pharmacological manipulation. This finding shall caution our interpretation of small molecule inhibitor experiments. Obviously, actin-dependent cell functions are affected differently at different timescales.
The Lp network assembles and disassembles in a spatially organized fashion. Assembly is dominant in the first 1–1.5 μm from the leading edge, juxtaposed to a band of 1–1.5 μm of predominant disassembly. Despite the short Lp width and fast retrograde flow rate (>1 μm/min), it takes the network longer to flow from the leading edge to the Lp base than the time it takes the La to complete a full turnover cycle. We inferred that the spatially stationary distribution of Lp polymerization and depolymerization is unrelated to the mechanism causing periodic turnover of the La. However, also in the Lp, we measured cyclic modulation of assembly at the leading edge and of disassembly at the Lp-La transition. The phase shift of ≈π between peak assembly and peak disassembly suggested that the kinetic relationship between Lp front and base cannot be explained by simple concentration fluctuations in the G-actin pool but involves yet unknown regulation.
The two networks also differ in terms of their dependence on Arp2/3 function. Although Lp assembly correlates spatially and temporally with the aggregation of transient Arp2/3 clusters, La assembly appears to correlate only spatially with the distribution of Arp2/3 clusters. This suggests that the La network assembles preferentially in locations of elevated Arp2/3 concentration but that a secondary mechanism is required to mediate polymerization events.
To attain these results, we relied on maps of F-actin turnover with a spatial resolution <1 μm. This level of resolution represents a quantum step in the development of quantitative Fluorescent Speckle Microscopy, which builds on two algorithmic extensions: 1), a module for iterative speckle detection accounting for short-term interference between speckles; and 2), single-particle tracking combining local and global information of speckle flow to establish speckle correspondences between frames. Both modules critically enhance the performance in reconstructing complete speckle trajectories, and thus the determination of speckle appearance and disappearance events used to map the kinetics of F-actin turnover.
Our study leaves us with three remaining questions:
What are the molecular pathways that clock F-actin assembly and disassembly cycles so reproducibly?
What is the molecular pathway that couples periodic Lp assembly at the leading edge and periodic Lp disassembly at the Lp-La transition?
What is the molecular mechanism that mediates La assembly preferentially in regions with elevated Arp2/3 concentration?
Answers to these questions will greatly benefit from the here-described new possibilities of high-resolution qFSM, but require the development of additional molecular tools that reach beyond the scope of this article.
SUPPLEMENTARY MATERIALS AND METHODS
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org. SupplMaterial.pdf: This file contains additional information regarding the validation of the iterative speckle extraction and general nearest-neighbor tracking algorithms. Also, the document describes the approaches taken to analyze spectra of lamella F-actin turnover by averaging the time development of assembly and disassembly dynamics in hundreds of small sampling windows in combined Fourier analysis (compare with Fig. 3 c).
Supplementary Material
Acknowledgments
We are grateful to Dinah Loerke for the careful reading of the manuscript.
This work was funded by Human Frontiers Science Program Young Investigator award No. RGY5-2002 to C.M.W.-S. and G.D., and National Institutes of Health grant No. R01 GM67230 to C.M.W.-S and G.D.
References
- 1.Pollard, T. D., and G. B. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- 2.Wang, Y. 1985. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J. Cell Biol. 101:597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin, C. H., and P. Forscher. 1995. Growth cone advance is inversely proportional to retrograde F-actin flow. Neuron. 14:763–771. [DOI] [PubMed] [Google Scholar]
- 4.Danuser, G., and R. Oldenbourg. 2000. Probing F-actin flow by tracking shape fluctuations of radial bundles in lamellipodia of motile cells. Biophys. J. 79:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspi, A., O. Yeger, I. Grosheva, A. D. Bershadsky, and M. Elbaum. 2001. A new dimension in retrograde flow: centripetal movement of engulfed particles. Biophys. J. 81:1990–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramer, L. P. 1997. Molecular mechanism of actin-dependent retrograde flow in lamellipodia of motile cells. Front. Biosci. 2:260–270. [DOI] [PubMed] [Google Scholar]
- 7.Vallotton, P., S. L. Gupton, C. M. Waterman-Storer, and G. Danuser. 2004. Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc. Natl. Acad. Sci. USA. 101:9660–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin, C. H., E. M. Espreafico, M. S. Mooseker, and P. Forscher. 1996. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 16:769–782. [DOI] [PubMed] [Google Scholar]
- 9.Salmon, W. C., M. C. Adams, and C. M. Waterman-Storer. 2002. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 158:31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupton, S. L., W. C. Salmon, and C. M. Waterman-Storer. 2002. Converging populations of F-actin promote breakage of associated microtubules to spatially regulate microtubule turnover in migrating cells. Curr. Biol. 12:1891–1899. [DOI] [PubMed] [Google Scholar]
- 11.Ponti, A., M. Machacek, S. L. Gupton, C. M. Waterman-Storer, and G. Danuser. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786. [DOI] [PubMed] [Google Scholar]
- 12.Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- 13.Gupton, S. L., K. L. Anderson, T. P. Kole, R. S. Fischer, A. Ponti, S. E. Hitchcock-DeGregori, G. Danuser, V. M. Fowler, D. Wirtz, D. Hanein, and C. M. Waterman-Storer. 2005. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponti, A., P. Vallotton, W. C. Salmon, C. M. Waterman-Storer, and G. Danuser. 2003. Computational analysis of F-actin turnover in cortical actin meshworks using fluorescent speckle microscopy. Biophys. J. 84:3336–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danuser, G., and C. M. Waterman-Storer. 2003. Fluorescent speckle microscopy: where it came from and where it is going. J. Microsc. 211:191–207. [DOI] [PubMed] [Google Scholar]
- 16.Vallotton, P., A. Ponti, C. M. Waterman-Storer, E. D. Salmon, and G. Danuser. 2003. Recovery, visualization, and analysis of actin and tubulin polymer flow in live cells: a fluorescence speckle microscopy study. Biophys. J. 85:1289–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waterman-Storer, C. M. 2002. Fluorescent speckle microscopy (FSM) of microtubules and actin in living cells. In Current Protocols in Cell Biology. J. S. Bonifacino, M. Dasso, J. B. Harford, J. Lippincott-Schwartz, and K. M. Yamada, editors. Wiley, New York. [DOI] [PubMed]
- 18.Waterman-Storer, C. M., and E. D. Salmon. 1997. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J. Cell Biol. 139:417–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams, M. C., W. C. Salmon, S. L. Gupton, C. S. Cohan, T. Wittmann, N. Prigozhina, and C. M. Waterman-Storer. 2003. A high-speed multispectral spinning-disk confocal microscope system for fluorescent speckle microscopy of living cells. Methods. 29:29–41. [DOI] [PubMed] [Google Scholar]
- 20.Thomann, D., D. R. Rines, P. K. Sorger, and G. Danuser. 2002. Automatic fluorescent tag detection in 3D with super-resolution: application to the analysis of chromosome movement. J. Microsc. 208:49–64. [DOI] [PubMed] [Google Scholar]
- 21.Ponti, A. 2004. High-Resolution Analysis of F-Actin Meshwork Kinetics and Kinematics using Computational Fluorescent Speckle Microscopy. ETH Zurich, Zurich, Switzerland.
- 22.Blackman, S. S., and R. Popoli. 1999. Design and Analysis of Modern Tracking Systems. Artech House, Norwood, MA.
- 23.Burkard, K. E., and E. Cela. 1999. Linear assignment problems and extensions. In Handbook of Combinatorial Optimization. D.Z. Du and P.M. Pardalos, editors. Kluwer Academic Publishers, Dordrecht, The Netherlands. 75–149.
- 24.Labonté, G. 1999. A new neural network for particle-tracking velocimetry. Exp. Fluids. 26:340–346. [Google Scholar]
- 25.Kohonen, T. 1990. The self-organizing map. Proc. IEEE. 78:1464–1480. [Google Scholar]
- 26.Theriot, J. A., and T. J. Mitchison. 1991. Actin microfilament dynamics in locomoting cells. Nature. 352:126–131. [DOI] [PubMed] [Google Scholar]
- 27.Theriot, J., and T. Mitchison. 1992. Comparison of actin and cell surface dynamics in motile fibroblasts. J. Cell Biol. 119:367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGrath, J. L., J. H. Hartwig, Y. Tardy, and C. F. Dewey, Jr. 1998. Measuring actin dynamics in endothelial cells. Microsc. Res. Tech. 43:385–394. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe, Y., and T. J. Mitchison. 2002. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 295:1083–1086. [DOI] [PubMed] [Google Scholar]
- 30.Bubb, M. R., A. M. Senderowicz, E. A. Sausville, K. L. Duncan, and E. D. Korn. 1994. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 269:14869–14871. [PubMed] [Google Scholar]
- 31.Cramer, L. P. 1999. Role of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug Jasplakinolide. Curr. Biol. 9:1095–1105. [DOI] [PubMed] [Google Scholar]
- 32.Bretschneider, T., S. Diez, K. Anderson, J. A. Heuser, M. Clarke, A. Müller-Taubenberger, J. Köhler, and G. Gerisch. 2004. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr. Biol. 14:1–10. [DOI] [PubMed] [Google Scholar]
- 33.Higashida, C., T. Miyoshi, A. Fujita, F. Oceguera-Yanez, J. Monypenny, Y. Andou, S. Narumiya, and N. Watanabe. 2004. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 303:2007–2010. [DOI] [PubMed] [Google Scholar]
- 34.Ghosh, M., X. Song, G. Mouneimne, M. Sidani, D. S. Lawrence, and J. S. Condeelis. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746. [DOI] [PubMed] [Google Scholar]
- 35.Mogilner, A., and L. Edelstein-Keshet. 2002. Regulation of actin dynamics in rapidly moving cells: a quantitative analysis. Biophys. J. 83:1237–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.