Abstract
The N gene conditions for resistance to Tobacco mosaic virus (TMV) but only below 28°C. However, a TMV-based vector expressing green fluorescent protein (TMV-GFP) showed only limited movement at 33°C in tobacco plants harboring the N gene and other genes cointrogressed from Nicotiana glutinosa. TMV-GFP moved efficiently in tobacco plants that either lacked these genes or that contained the N gene but were transgenic for RNA1 of Cucumber mosaic virus. These findings identified novel temperature-independent resistance to the movement of TMV-GFP which could be neutralized by a different viral transgene. Using the N gene and nahG gene-transgenic tobacco, we show that this novel resistance is manifested specifically by the N gene itself and operates via a pathway independent of salicylic acid.
The N gene is the best-characterized gene for resistance to a plant virus, namely, Tobacco mosaic virus (TMV) (1, 2, 19, 20, 22, 40, 41, 51). It constitutes a classical example of a gene-for-gene interaction between a resistance gene and a pathogen avirulence gene (23). The locus containing this resistance gene as well as other linked sequences was introgressed in tobacco from Nicotiana glutinosa in the 1930s (32). This resistance is still in widespread use in field tobacco cultivars. In the early 1990s, the N gene was the first plant virus resistance gene to be cloned (51), and the nature of the encoded gene products and their pattern of expression upon infection with TMV have been analyzed extensively (19, 20). The TMV avirulence factor also has been mapped to the helicase domain of the viral RNA-dependent RNA polymerase (RdRp) (1, 2, 22, 40, 41). The genome of TMV, a plus-sense single-stranded RNA, acts as an mRNA for the synthesis of two RdRp-associated proteins (a 126-kDa protein and a 183-kDa readthrough protein of an amber termination codon), both containing this helicase domain. The virus encodes another three polypeptides expressed via subgenomic RNAs (42).
Infection of tobacco containing the N gene by TMV results in a hypersensitive response (HR) accompanied by generic systemic acquired resistance (SAR) that confines TMV to cells surrounding the initial site of infection (reviewed in reference 46). SAR is induced by salicylic acid (SA), which also inhibits TMV replication (13). At temperatures above 28°C, the HR and the restriction response associated with the N gene are inactive, and so TMV spreads throughout the plant. Reducing the temperature of incubation to below 28°C allows activation of the N gene, resulting in necrosis in all tissues containing TMV. The local physiological and cellular events that take place during the induction of the HR in the TMV-N gene system have been described in detail (35, 52).
The use of plant viruses as vectors for the transient expression of foreign genes in plants has attracted considerable interest (33, 48). In some instances, the reduced ability of the modified virus to spread successfully throughout the plant constitutes a limitation to this technology (16, 21, 25, 28). Considerable effort has been devoted to understanding and eliminating this constraint, often by using green fluorescent protein (GFP) as a reporter (44, 49). TMV-based vectors expressing GFP (TMV-GFP) also have been used as tools in studies of virus movement (7, 8, 9, 30, 31) as well as N gene-mediated resistance to TMV (52).
Some plant viruses can interact with each other synergistically (37). In some situations, such an interaction can lead to breakage of resistance against one of the viruses in virus pairs (reviewed in reference 4). Synergy and resistance breakage also have been observed in some instances in transgenic plants expressing genes from one of the virus pairs. In some situations, the interaction is a complementation of function associated with virus movement (14, 15, 26). In other situations, the viral transgene has been shown to encode proteins involved in the suppression of gene silencing responses and probably other plant resistance responses (3, 34, 50). In the course of studies on viral synergy, in which the effects of viral transgenes on infection by other, unrelated viruses were assessed, TMV-GFP was used to delineate the effects of RNA1 transgenes derived from Cucumber mosaic virus (CMV) on virus movement in tobacco. These studies led to the discovery of a novel component of N gene-mediated resistance manifested against TMV-based vectors. Moreover, the effect of this resistance response on the movement of TMV-GFP was overcome by several CMV RNA1 transgenes.
MATERIALS AND METHODS
Plants.
Tobacco plants (Nicotiana tabacum) of cultivars Samsun NN and Samsun nn were used. The generation of full-length CMV RNA1-transgenic tobacco cultivar Samsun NN plants was described previously (11). Tobacco cultivar Samsun nn plants were transformed with the same CMV RNA1 transgene construct as that used for cultivar Samsun NN plants by following previously described procedures (11). The generation and characterization of transgenic tobacco cultivar Samsun NN plants expressing either an intact CMV strain LS RNA1 construct or a chimeric CMV strain LS-CMV strain Fny RNA1 (LF1) construct will be described elsewhere (L. Zhang, W. J. Chang, and P. Palukaitis, unpublished data). Constructs containing a deletion of helicase motif III or VI in LF1, specifically, an in-frame deletion of nucleotides 2508 to 2546 or nucleotides 2975 to 3022, respectively, were obtained from intact, chimeric LF1 cDNA by using PCR-based standard mutagenesis techniques (47).
Other plants used here were tobacco cultivar Petite Havana SR1 nn transformed with the N gene (SR1::NN plants) (51), tobacco cultivar Samsun NN plants transformed with the nahG gene of Pseudomonas putida (17, 39), tobacco resulting from the fertilization of CMV RNA1-transgenic tobacco cultivar Samsun nn plants with pollen from SR1::NN plants, tobacco resulting from the fertilization of CMV RNA1-transgenic tobacco cultivar Samsun NN plants with pollen from nahG-transgenic tobacco cultivar Samsun NN plants, and nontransgenic Nicotiana benthamiana plants. All plants were homozygous for the above genes before crosses were made. The progeny of the NN cross between SR1::NN tobacco and CMV RNA1-transgenic tobacco cultivar Samsun nn were first assessed biologically to verify the presence of a functional N gene and a CMV RNA1 transgene (11). Plants were grown in a greenhouse or in a growth chamber at either 25 or 33°C, respectively, depending on the experiment.
Viruses.
Wild-type (WT) TMV infectious transcript RNA was generated from full-length cDNA in clone pTMV004 (36). All the TMV-based expression vectors are derivatives of p30B.GFP, which expresses a chimeric virus containing a subgenomic RNA promoter for the capsid protein (CP) gene derived from strain U5 of the tobamovirus Tobacco mild green mosaic virus. The use of a duplicated subgenomic RNA promoter from a heterologous tobamovirus prevents homologous recombination between duplicated TMV CP subgenomic RNA promoters (one of them directing the expression of the inserted gene) in the expression vector (44, 49, 52). Infectious RNA transcripts from a TMV-based vector expressing free GFP (designated here TMV-GFP) were obtained from TMV-GFP 1056, a proprietary construct obtained from Large Scale Biology Corporation (Vacaville, Calif.) and derived from TMV p30B.GFP with modifications in the virus movement protein gene. Plasmid TMV.DsRed was derived from TMV-GFP 1056 after replacement of the GFP gene with the red fluorescent protein (DsRed; ClonTech) gene (obtained from S. Chapman, Scottish Crop Research Institute [SCRI]). Plasmid TMV-GFP 5725 is similar to clone p30B.GFP, except that the GFP gene was inserted into the XhoI site of the vector polylinker (obtained from S. Santa Cruz, Horticultural Research Institute, East Malling, United Kingdom). The GFP gene was deleted from plasmid TMV-GFP 1056 after linearization with XhoI and PacI and filling in with a linker fragment obtained from plasmid TMV-GFP 5725 digested with the same enzymes. This procedure gave rise to plasmid TMV 1056, harboring no additional gene but still containing two CP subgenomic RNA promoters. All plasmids were linearized with KpnI, and RNA transcripts were made by using T7 RNA polymerase.
Transcript RNAs were inoculated onto leaves of N. benthamiana. Four days after inoculation, the inoculated leaves were collected and ground in 50 mM sodium phosphate (pH 7; 1/10 [wt/vol]). The extracts were clarified by centrifugation for 5 min at 5,000 × g, and the resulting supernatants were divided into aliquots and kept at −20°C. These crude virus preparations were used for the inoculation of tobacco plants after dilution in 50 mM sodium phosphate (pH 7; 1/100 for WT TMV and 1/10 for TMV-GFP). Plants were inoculated mechanically by gently rubbing the inocula (crude virus preparations or transcript RNA) on aluminium oxide-dusted leaves. Infectious transcript RNAs of potato virus X (PVX) and PVX expressing GFP (PVX-GFP) were obtained from full-length cDNA clones pTX.P3C2 and pTXS.GFP, respectively (5, 6), by using T7 RNA polymerase after linearization with SpeI.
Detection of GFP and DsRed fluorescence.
GFP-derived fluorescence images were obtained and processed as described previously (10). Whole leaves were viewed by using a Black Ray long-wave UV lamp (model B; 100 A; UV Products, Upland, Calif.). Experiments in which virus movement in the whole plant was assessed were performed at least twice, and at least five plants per treatment were used in each experiment. Higher-magnification fluorescence was detected with an epifluorescence microscope (Nikon) equipped with a fluorescein isothiocyanate-tetramethyl rhodamine isothiocyanate multiband filter (Chroma; catalog no. 51004). DsRed fluorescence was monitored with the aid of a Leica MZ FLIII fluorescence stereomicroscope equipped with a fluorescence G filter (excitation filter, 456/10 nm; barrier filter, 590 nm) (Leica, Heerbrugg, Switzerland) and coupled to a digital video camera (model KY-F55B; Photonic Science, Milham, United Kingdom).
Protoplast electroporation and nucleic acid analysis.
Tobacco plants were kept for 5 days in a growth chamber at a constant temperature of 25°C and with 16 h of daylight prior to the isolation and electroporation of mesophyll protoplasts as described previously (24). Approximately 5 μg of transcript RNA was electroporated with 106 protoplasts. Protoplast cultures were kept undisturbed in a growth chamber for 24 h prior to their harvest by centrifugation at low speed. Protoplast pellets were resuspended in 300 μl of 50 mM Tris-HCl (pH 8.0)-10 mM EDTA- 2% sodium dodecyl sulfate- 0.5% 2-mercaptoethanol. The samples were then extracted with phenol, and the RNA was precipitated with ethanol.
For Northern blot hybridization analysis, total nucleic acid samples were fractionated by electrophoresis in 1% agarose gels under denaturing conditions (6% formaldehyde), blotted onto nitrocellulose membranes, and hybridized to digoxigenin-labeled RNA probes against TMV or against PVX. A probe complementary to nucleotides 999 to 2144 of the TMV genome was obtained from a subclone encompassing nucleotides 256 to 2144 (provided by P. Boevink, SCRI) by using T7 RNA polymerase after linearization with XbaI. A probe complementary to the last 157 nucleotides of the PVX genome was obtained by using T7 RNA polymerase after linearization of full-length clone pTX.P3C2 with XhoI. Blots were incubated with digoxigenin-labeled probes by following the manufacturer's instructions (Roche Diagnostics, Lewes, United Kingdom).
RESULTS
A CMV RNA1 transgene enhances the local movement of TMV-GFP in NN tobacco.
In the course of synergy studies, the effects of viral transgenes on infection by unrelated viruses were assessed. To analyze the effect of a CMV RNA1 transgene on virus movement in tobacco, a TMV-based vector expressing GFP (derived from plasmid TMV-GFP 1056 and referred to here as TMV-GFP) was used to monitor in vivo the effect of the transgene on virus movement.
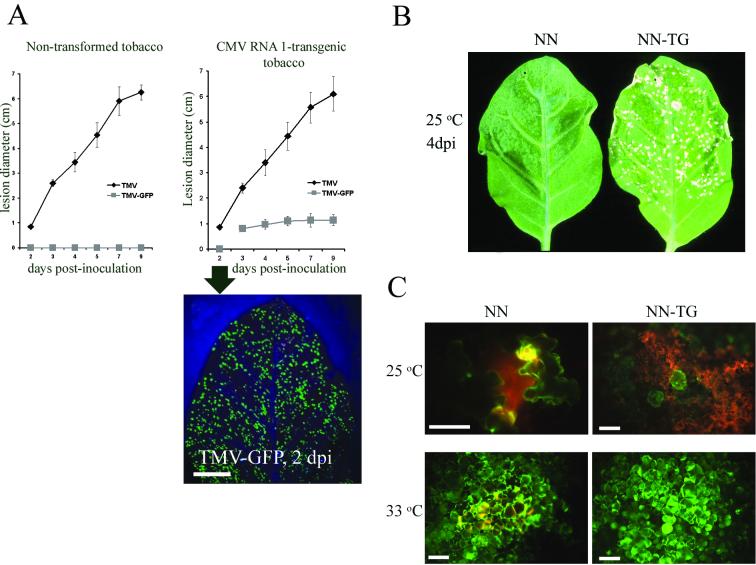
Infection by TMV-GFP failed to induce visible necrotic local lesions (NLL) in nontransformed tobacco cultivar Samsun NN at 25°C (Fig. 1A, left graph, and 1B, left leaf). In contrast, in tobacco cultivar Samsun NN transgenic for CMV RNA1, TMV-GFP induced visible NLL (Fig. 1A, right graph, and 1B, right leaf). The latter lesions were, however, smaller than those induced by WT TMV (Fig. 1A, right graph). On the other hand, the NLL induced by WT TMV were of comparable sizes in both types of plants (Fig. 1A, left graph versus right graph; samples labeled TMV). Furthermore, visible NLL were apparent in tobacco cultivar Samsun NN inoculated with WT TMV as early as 2 days postinoculation (dpi) and increased in diameter over the next week (Fig. 1A, both graphs, curves labeled TMV). In contrast, the NLL induced by TMV-GFP in CMV RNA1-transgenic tobacco cultivar Samsun NN were visible only at 3 dpi and did not increase substantially in diameter on subsequent days (Fig. 1A, right graph). However, analysis at 2 dpi of GFP fluorescence with a UV lamp showed that TMV-GFP had already spread from the initially infected cells (Fig. 1A, image below right graph). Therefore, the visible NLL induced by TMV-GFP in CMV RNA1-transgenic tobacco cultivar Samsun NN not only were smaller than those induced by WT TMV but also appeared later (delayed by ca. 24 h) (Fig. 1A, right graph).
FIG. 1.
A CMV RNA1 transgene enhances the local movement of TMV-GFP in NN tobacco. (A) Graphic plot of the rate of growth (mean and standard deviation) at 25°C of NLL in leaves of tobacco cultivar Samsun NN not transformed (left graph) or transformed with RNA1 of CMV (right graph) and inoculated with either WT TMV or TMV-GFP. The image below the right graph shows the detection under a UV lamp of GFP-fluorescent foci in tobacco cultivar Samsun NN transformed with CMV RNA1 at 2 dpi with TMV-GFP, despite the absence of visible necrotic lesions (right graph). (B) Infection by TMV-GFP at 25°C induced visible NLL in inoculated leaves of CMV RNA1-transgenic tobacco cultivar Samsun NN at 4 dpi (right leaf, labeled NN-TG), whereas inoculated leaves of nontransformed tobacco cultivar Samsun NN remained asymptomatic (left leaf, labeled NN). (C) Microscopic detection of GFP fluorescence under UV light in tobacco leaves 4 dpi with TMV-GFP and kept at 25°C (upper row) or at 33°C (lower row). Left and right panels show foci of infection in tobacco cultivar Samsun NN either not transformed or transgenic for CMV RNA1 (labeled NN and NN-TG, respectively). A fluorescein isothiocyanate-rhodamine filter was used to show green fluorescence from GFP, while autofluorescence from necrotized tissue appears orange. Healthy tissue appears as a dark background. Bars, 100 μm.
It is known that in the TMV-tobacco cultivar Samsun NN system as well as in other virus-host systems, the HR fails to occur in the total absence of virus movement (12, 52). Therefore, to determine whether the absence of visible lesions was due to a complete lack of movement of the virus, the cell-to-cell movement of TMV-GFP in inoculated leaves was examined by using fluorescence microscopy. In tobacco cultivar Samsun NN, at 25°C and at 4 dpi, the movement of TMV-GFP was limited to a few cells, resulting in microscopic lesions not detectable by the naked eye (Fig. 1C, upper left panel). In contrast, in tobacco cultivar Samsun NN expressing RNA1 of CMV, TMV-GFP spread to a larger number of cells, but further movement was restricted by activation of the N gene-mediated resistance response. This response led to the necrosis of most of the infected cells by 4 dpi and resulted in visible NLL (Fig. 1C, upper right panel, and 1B, right leaf). At 33°C, N gene-mediated resistance to TMV was inactive, and TMV-GFP showed movement to numerous cells in both types of plants (Fig. 1C, lower panels). These data indicated that (i) the absence of visible NLL in nontransformed tobacco cultivar Samsun NN (Fig. 1B, left leaf) was not due to the absence of an HR but rather was due to the lesions being microscopic (Fig. 1C, upper left panel) and (ii) the CMV RNA1 transgene greatly enhanced the movement of TMV-GFP that was highly restricted by the N gene at 25°C (Fig. 1).
Novel temperature-independent resistance to the movement of TMV-GFP specific to the NN genotype.
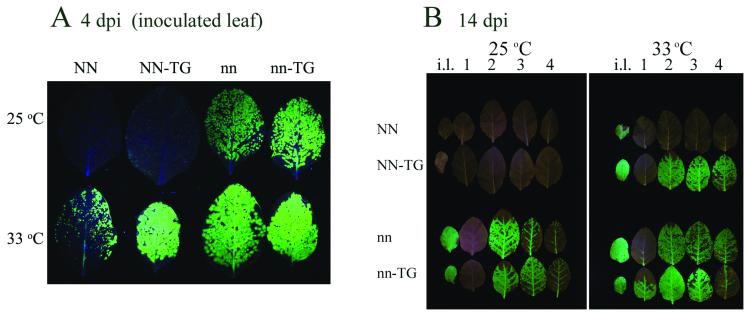
The enhancement by the CMV RNA1 transgene of the cell-to-cell movement of TMV-GFP in NN tobacco also was observed at 33°C, at which the N gene-mediated induction of HR and SAR, which restricts the cell-to-cell movement of WT TMV, is inactive. The effect of the CMV RNA1 transgene on the movement of TMV-GFP was demonstrated by using a UV lamp to detect the presence of TMV-GFP (Fig. 2A, compare two leaves at lower left). To ascertain whether the N allele was associated with the restriction of TMV-GFP in nontransgenic tobacco, plants of tobacco cultivar Samsun nn, lacking the N allele, were tested for their ability to restrict the cell-to-cell movement of TMV-GFP. Such plants also were made transgenic for CMV RNA1 to compare the effects of the CMV RNA1 transgene on TMV-GFP movement in both cultivars (Samsun NN and Samsun nn). In fact, in the absence of the N alelle, TMV-GFP moved from cell to cell as efficiently in the presence as in the absence of the CMV RNA1 transgene (Fig. 2A, compare two leaves at upper right). This lack of a difference in movement by TMV-GFP was observed at both 25 and 33°C (Fig. 2A, compare two leaves at upper right with two leaves at lower right). Thus, the restriction of cell-to-cell movement in tobacco cultivar Samsun NN plants at 25 and 33°C (Fig. 1 and 2A) was associated in both cases with the presence of either the N gene or other genes tightly linked to the N gene.
FIG. 2.
A CMV RNA1 transgene neutralizes novel resistance to TMV-GFP specific for the NN genotype. The patterns of GFP fluorescence were observed under a UV lamp in tobacco cultivar Samsun NN, either not transformed or transgenic for CMV RNA1 (labeled NN and NN-TG, respectively), and tobacco cultivar Samsun nn, either not transformed or transgenic for CMV RNA1 (labeled nn and nn-TG, respectively). (A) Leaf at 4 dpi with TMV-GFP and kept at either 25°C (upper row) or 33°C (lower row). (B) Whole plant at 14 dpi with TMV-GFP and kept at either 25°C (left panel) or 33°C (right panel). Each row of leaves shows the detached inoculated leaf (i.l.) followed by the detached four consecutive leaves (numbered 1 to 4 in ascending order).
A CMV RNA1 transgene enhances both local movement and systemic movement of TMV-GFP by neutralizing the novel resistance associated with the NN genotype.
The tobacco cultivar Samsun nn and Samsun NN plants used above were also assessed at both 25 and 33°C to determine the effects of the N allele and of the CMV RNA1 transgene on the long-distance movement of TMV-GFP. TMV-GFP was visualized by using a UV lamp (Fig. 2B). At 25°C, there was no systemic movement of TMV-GFP in tobacco cultivar Samsun NN,either not transformed or transgenic for CMV RNA1, due to the activation of N gene-mediated HR and SAR (Fig. 2B, left panel, upper two rows). At 33°C, TMV-GFP movement in nontransformed tobacco cultivar Samsun NN plants was restricted mostly to within the inoculated leaf and sometimes to isolated areas on the main veins of one or two leaves above the inoculated leaf (Fig. 2B, right panel, first row). However, in tobacco cultivar Samsun NN plants transgenic for CMV RNA1, infection of leaves above the inoculated leaf occurred with TMV-GFP (Fig. 2B, right panel, second row). Interestingly, both nontransgenic and CMV RNA1-transgenic tobacco cultivar Samsun nn plants supported the long-distance movement of TMV-GFP at 25°C as well as at 33°C (Fig. 2B, both panels, lower two rows). Thus, the restriction of both cell-to-cell movement and long-distance movement of TMV-GFP is linked to the presence of the N allele and is independent of the temperature. The presence of the CMV RNA1 transgene in tobacco cultivar Samsun NN plants had the effect of altering the phenotype of TMV-GFP movement (at 33°C, when N gene-mediated HR and SAR are inactive) to the phenotype shown by TMV-GFP in tobacco cultivar Samsun nn plants, i.e., promoting both cell-to-cell movement (Fig. 2A, lower row; compare the second leaf from the left with the third and fourth leaves), and long-distance movement (Fig. 2B, right panel; compare the second row from the top with the third and fourth rows).
The demonstration that the CMV RNA1 transgene does not enhance the movement of TMV-GFP in an n allele context indicates that the CMV RNA1 transgene operates by neutralizing the novel temperature-independent resistance to the movement of TMV-GFP conditioned by the NN genotype rather than by generically improving virus movement. This specificity also was demonstrated in experiments showing that neither PVX nor tobacco rattle virus vectors expressing GFP were retarded in their movement in tobacco cultivar Samsun NN versus tobacco cultivar Samsun nn and that no enhancement of the movement of either virus was observed in tobacco transgenic for CMV RNA1 (data not shown).
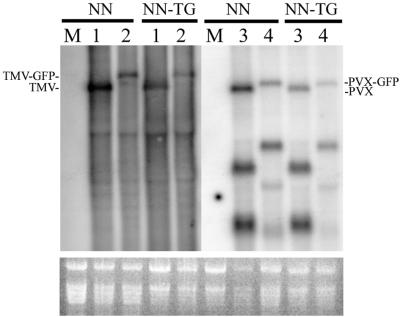
To confirm that the above observations were due to effects on virus movement rather than virus replication, the accumulation of TMV and the accumulation of TMV-GFP in tobacco protoplasts were compared. The accumulation of WT TMV or TMV-GFP was similar in isolated tobacco mesophyll cells from both nontransgenic and CMV RNA1-transgenic tobacco cultivar Samsun NN (Fig. 3, lanes 1 and 2), as was the accumulation of PVX or PVX-GFP (Fig. 3, lanes 3 and 4). The latter viruses were used as an internal control, since no synergy for the accumulation of PVX has been observed in CMV RNA1-transgenic plants (reference 43 and data not shown).
FIG. 3.
A CMV RNA1 transgene does not enhance the replication of TMV-GFP in NN tobacco. The upper panel shows Northern blot hybridization analysis of the accumulation of viral RNA in protoplasts isolated from tobacco cultivar Samsum NN, either not transformed or transgenic for CMV RNA1 (labeled NN and NN-TG, respectively), 24 h after electroporation with water (lane M), WT TMV transcript RNA (lane 1), TMV-GFP transcript RNA (lane 2), PVX transcript RNA (lane 3), or PVX-GFP transcript RNA (lane 4). The lower panel shows rRNA accumulation.
To assess whether the novel resistance and the neutralizing effect of the CMV RNA1 transgene affected other TMV-based constructs, we used a vector expressing red fluorescent protein (TMV.DsRed). The gene encoding red fluorescent protein shares no sequence homology with the GFP gene (38). The results obtained with TMV.DsRed were similar to those obtained with TMV-GFP (data not shown). As a further control, the GFP gene was deleted from TMV-GFP 1056. The resulting vector, which still contained the two CP subgenomic RNA promoters, induced small NLL in nontransformed tobacco cultivar Samsun NN plants at 25°C but also induced larger NLL in CMV RNA1-transgenic Samsun NN plants at 25°C than in nontransformed tobacco plants (data not shown). The movement of a different TMV-based vector expressing GFP (TMV-GFP 5725) also was enhanced by the CMV RNA1 transgene in an NN context at temperatures above 33°C (data not shown). Therefore, the results observed with TMV-GFP can be extended to other TMV-based vectors and are not related to the nature of the insert or the particular vector construct.
It is highly unlikely that our results are due to somaclonal variations in the CMV RNA1-transgenic plants, since the effect attributed here to the CMV RNA1 transgene was observed in independently transformed lines (Table 1), irrespective of whether the lines exhibited susceptibility or systemic resistance to infection by CMV (11). All of these lines supported the replication of CMV RNA2 and RNA3. In addition, other transgenic lines generated independently to express RNA1 of the LS strain of CMV or chimeric RNA1 derived from the 5′ half of RNA1 of the LS strain and the 3′ half of the Fny strain of CMV (45, 53) also showed the replication of CMV strain Fny RNA2 and RNA3 as well as enhancement of the local movement of TMV-GFP (Table 1). These data also show that the effects observed are not specific to only CMV strain Fny RNA sequences.
TABLE 1.
Analysis of the abilities of different CMV RNA1 transgenes to enhance the local movement of TMV-GFP in tobacco cultivar Samsun NN at 25°C
| RNA1 transgene used to transform tobacco cultivar Samsun NN | Transgenic line tested | Induction of visible necrotic local lesionsa | Complementation of replication of CMV RNA2 and RNA3b |
|---|---|---|---|
| CMV strain Fnyc | A1 | + | + |
| B1C | + | + | |
| CMV strain LS | A16 | + | Not tested |
| A17 | + | Not tested | |
| A19.1 | + | + | |
| A19.4 | + | Not tested | |
| CMV strain LS-CMV strain Fny chimera | B21 | + | + |
| CMV strain LS-CMV strain Fny chimera with deletion of helicase motif III | C1 C2 C4 C5 C24 | Not tested + + + + | − Not tested Not tested Not tested Not tested |
| CMV strain LS-CMV strain Fny chimera with deletion of helicase motif VI | D1.1 D5.6 D8.2 D14.7 D36 | Not tested + + + + | − Not tested Not tested Not tested Not tested |
| None | − | − |
+ and −, presence and absence, respectively, of visible necrotic local lesions 5 days after inoculation of the leaf with TMV-GFP.
+ and −, presence and absence, respectively, of GFP-derived fluorescence in single epidermal cells of leaves inoculated with CMV transcript RNA2 and a derivative of CMV RNA3 in which the GFP gene substituted for the viral coat protein gene (10). Inculated leaves were scanned with a fluorescences microscope.
Line A1 is susceptible to systemic infection by CNMV strain Fny whereas line B1C ia resistant to systemic infection by CMV strain Fny (11).
The novel resistance is specifically associated with the N gene.
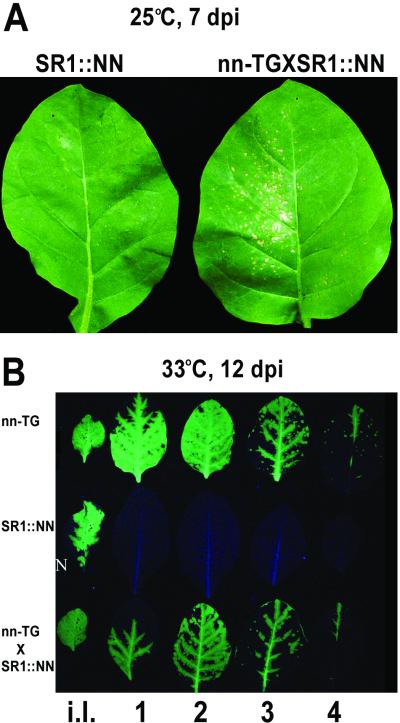
It is possible that the observed temperature-independent resistance against the movement of TMV-GFP is not a function of the N gene itself but rather is a function of one of the other genes closely linked to the N gene cointrogressed from N. glutinosa. To discriminate between these alternatives, nn tobacco plants transformed with the N gene (line SR1::NN) (51) were inoculated with TMV-GFP. At 25°C, these plants restricted the cell-to-cell movement of TMV-GFP (Fig. 4A, left leaf), as did tobacco cultivar Samsun NN plants (Fig. 1B, left leaf). Very small lesions were formed, although they were larger than the microscopic lesions observed with tobacco cultivar Samsun NN (compare Fig. 4A, left leaf, with Fig. 1B, left leaf). On the other hand, in the progeny from the cross between CMV RNA1-transgenic tobacco cultivar Samsun nn plants and SR1::NN plants, the cell-to-cell movement of TMV-GFP was facilitated considerably, resulting in larger, clearly visible NLL (Fig. 4A, right leaf). These plants were tested to verify the presence of both the N gene and the CMV RNA1 transgene (data not shown).
FIG. 4.
The novel resistance is specifically associated with the N gene. (A) At 25°C and at 7 dpi with TMV-GFP, NLL were observed in inoculated leaves of the progeny from the cross between tobacco cultivar Samsun nn plants transgenic for CMV RNA1 (nn-TG) and plants transgenic for the N gene (SR1::NN), yielding nn-TG × SR1::NN (right leaf). In the inoculated leaf of parental CMV RNA1-transgenic tobacco cultivar Samsun nn, no visible NLL were produced (not shown), whereas in the inoculated leaf of parental SR1:: NN tobacco, very small NLL were visible (SR1:: NN) (left leaf). (B) At 33°C and at 12 dpi with TMV-GFP, the spread of virus in the whole plant was assessed with a UV lamp. Each row of leaves shows the detached inoculated leaf (i.l.) followed by the detached four consecutive leaves (numbered 1 to 4 in ascending order).
At 33°C, the systemic movement of TMV-GFP in the progeny from the genetically crossed plants was similar to that observed in the parental CMV RNA1-transgenic tobacco cultivar Samsun nn plants (Fig. 4B, bottom and top rows, respectively), whereas the systemic movement of TMV-GFP in the parental SR1::NN tobacco plants was very poor or not detectable (Fig. 4B, middle row). These results are similar to those shown for tobacco cultivar Samsun NN and cultivar Samsun nn plants in Fig. 1B and 2B. Thus, the resistance phenotype is specifically associated with the N gene and not with genes linked to the N gene cointrogressed from N. glutinosa.
The novel resistance mechanism operates via a pathway independent of SA.
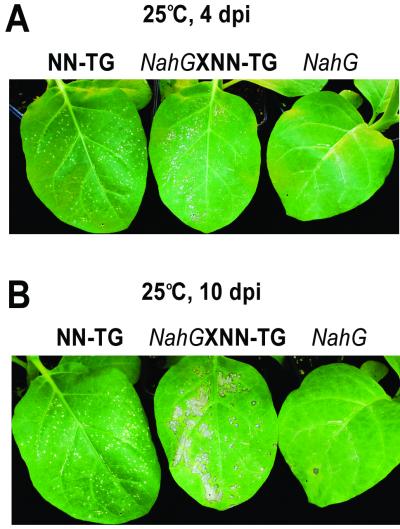
The effect of the CMV RNA1 transgene on the movement of TMV-GFP could be due to an interaction with either the N gene itself or some factor downstream of the known pathways activated by the N gene. N gene-activated SAR is mediated by SA. To determine whether the novel resistance to the movement of TMV-GFP is associated with the SA-mediated pathway, transgenic tobacco cultivar Samsun NN plants expressing the bacterial nahG gene (39), which causes hydrolysis of SA, were infected with WT TMV and TMV-GFP. When such plants were infected with WT TMV at 25°C, they developed necrotic lesions. However, as the SA-mediated resistance response was not activated, the virus continued to move slowly throughout the plant, inducing a phenotype of spreading necrosis, due to the activated HR (17, 39). However, infection with TMV-GFP at 25°C was confined to subliminal, microscopic lesions in nahG-transgenic tobacco cultivar Samsun NN, which did not grow further (Fig. 5, right leaves, and data not shown). In contrast, in the progeny resulting from the cross between nahG-transgenic tobacco cultivar Samsun NN and CMV RNA1-transgenic tobacco cultivar Samsun NN, TMV-GFP induced visible NLL (Fig. 5A, middle leaf) and a phenotype of spreading necrosis (Fig. 5, middle leaves). In parental tobacco cultivar Samsun NN transgenic for CMV RNA1, visible NLL were also induced (Fig. 5A, left leaf), but there was no phenotype of spreading necrosis (Fig. 5, left leaves). Thus, it appears that the novel resistance associated with the N gene follows a pathway that is not dependent on SA for its induction.
FIG. 5.
The novel resistance operates independently of SA. (A) At 25°C and 4 dpi with TMV-GFP, NLL of comparable sizes were induced in inoculated leaves of the progeny from the cross between tobacco cultivar Samsun NN transformed with the nahG gene (NahG) and tobacco cultivar Samsun NN transgenic for CMV RNA1 (NN-TG), yielding nahG-NN × NN-TG (central leaf), and in inoculated leaves of parental tobacco cultivar Samsun NN transgenic for CMV RNA1 (NN-TG) (left leaf). In the inoculated leaves of parental tobacco cultivar Samsun NN transformed with the nahG gene, infection remained subliminal and no visible NLL were induced (NahG) (right leaf). (B) At 10 dpi, NLL in inoculated leaves of nahG-NN × NN-TG plants (central leaf) displayed a phenotype of spreading necrosis, whereas NLL in inoculated leaves of tobacco cultivar Samsun NN transgenic for CMV RNA1 (left leaf) had barely increased in diameter from 4 dpi and no macroscopic NLL were visible in inoculated leaves of nahG-NN plants (right leaf).
The enhancement of TMV-GFP local movement by a CMV RNA1 transgene does not depend on the putative helicase activity of the encoded 1a protein.
The effects observed above are probably mediated by the CMV 1a protein, encoded by the CMV RNA1 transgene. However, this notion cannot be unequivocally shown, since alteration of the initiator methionine could lead to translation initiation from another of the more than 30 methionine residues present in the 111-kDa 1a protein. Nevertheless, to attempt to delimit CMV RNA1 or CMV 1a sequences involved in the ability to neutralize the effect of the N gene on the movement of TMV-GFP, we analyzed the movement of TMV-GFP in transgenic lines containing deletions in the putative helicase domain of the CMV 1a protein. The putative helicase activity, thought to be required for viral replication (29), is localized in the C-terminal half of the 1a protein of CMV. The helicase domain consists of six motifs (27). We had available tobacco cultivar Samsun NN lines transformed with chimeric CMV RNA1 from which motif III or VI of the helicase domain had been deleted. In these lines, the transgene could not sustain the replication of inoculated CMV RNA2 and RNA3 (Table 1). However, these transgenic lines were still able to enhance the local movement of TMV-GFP (Table 1). That is, visible NLL were induced at 25°C in the inoculated leaves but were absent from nontransformed plants (Table 1).
DISCUSSION
We have shown that the movement of TMV-GFP is restricted in tobacco cultivar Samsun NN compared to the movement of this virus vector in tobacco cultivar Samsun nn. This NN context-associated resistance affects both cell-to-cell movement (Fig. 1 and 2A) and long-distance movement (Fig. 2B). Furthermore, it remains operative at 33°C, a temperature nonpermissive for the inhibition of TMV movement (Fig. 2B). In contrast, in tobacco cultivar Samsun NN plants transgenically expressing RNA1 of CMV (11), both the cell-to-cell and the long-distance movement phenotypes of TMV-GFP were restored at 33°C to that shown by TMV-GFP in tobacco cultivar Samsun nn plants (Fig. 2). At 25°C, the presence of the CMV RNA1 transgene induced a phenotype of visible NLL in inoculated leaves; in comparison, microscopic, nonvisible NLL were induced in nontransformed tobacco cultivar Samsun NN plants (Fig. 1). The latter observation indicates that the local movement of TMV-GFP was also enhanced by the transgene, even in the presence of N gene-activated HR and SAR. Therefore, the CMV RNA1 transgene neutralized the effect of novel N gene-mediated, temperature-independent resistance on the movement of TMV-GFP without preventing the onset of NLL at temperatures below 28°C. Moreover, the movement of TMV-GFP in nn tobacco plants transformed with the N gene (line SR1::NN) (51) was also inhibited relative to that in nontransformed nn tobacco plants (Fig. 4). This result indicates that the novel resistance to TMV-GFP is caused by the N gene itself and not by other genes that were introgressed into tobacco from N. glutinosa together with the N gene locus (32). Small differences were observed in the phenotypes of local movement of TMV-GFP in tobacco cultivar Samsum NN and SR1::NN at 25°C. That is, the NLL were macroscopic in the latter plants (compare left leaves in Fig. 4A and 1B). This result may be related to the absence in the latter plants of a family of genes containing sequence similarities to the N gene (51). In any event, the presence of a CMV RNA1 transgene in these SR1::NN plants enhanced the systemic movement of TMV-GFP to the same extent as that seen in tobacco cultivar Samsun nn plants (Fig. 4B).
The enhancing effect of the CMV RNA1 transgene on the movement of TMV-GFP was not observed for WT TMV (Fig. 1A and data not shown). This result suggests that the novel N gene-mediated, temperature-independent resistance described in this report did not affect detectably the ability of the WT virus to move and accumulate. WT TMV either may have moved on before the resistance could be expressed or may not have activated the resistance.
How does this novel resistance to TMV-based vectors operate? It is clear that the N gene mediates the resistance, which is temperature independent. The resistance operates by inhibiting both cell-to-cell movement and systemic movement without causing reduced viral accumulation in the infected cell (Fig. 3). Could the resistance affect the plasmodesmatal gating function of the TMV movement protein? It is known that the N gene affects the ability of the TMV movement protein to alter the gating capacity of plasmodesmata and therefore the efficiency of virus movement, but this effect occurs in a temperature-dependent manner (18). The demonstration that the enhancing effect of the CMV RNA1 transgene on the movement of TMV-GFP was temperature independent makes it unlikely that CMV RNA1 or the CMV 1a protein acts by suppressing the temperature-dependent effect of the N gene on the gating ability of the TMV movement protein (18).
How does the CMV RNA1 transgene neutralize this novel resistance? We suggest that the CMV 1a protein may interfere with a hypothetical gene-for-gene interaction between the viral elicitor and the receptor that activates this novel aspect of the N gene response described in this report. The nature of the viral elicitor is not known. Direct interference between the CMV RNA1 transgene product and the receptors that trigger N gene-mediated HR and SAR is unlikely, since the CMV transgene does not prevent the activation of HR and SAR at 25°C. However, it is possible that the CMV RNA1 transgene somehow alters the specific, time-dependent pattern of differential expression of the two N gene products, resulting in the activation of HR and SAR. Alterations in the pattern of expression of these products have been shown to compromise the size and timing of the appearance of NLL and the eventual establishment of SAR (19, 20). In this regard, we observed a 24-h delay in the appearance of HR, despite the occurrence of some cell-to-cell movement (Fig. 1A, right graph and the image below it). On the other hand, a reduction in the ability of a mutant virus to move from cell to cell (as in the case of TMV-GFP versus WT TMV in nn tobacco plants) also resulted in a reduction in the size of the NLL as well as a delay in the appearance of the NLL and the establishment of SAR in NN tobacco plants (18, 40).
With regard to the possible relationship between this novel resistance and the SAR pathway (reviewed in reference 44), we show that in NN tobacco plants transgenic for the bacterial nahG gene, which causes hydrolysis of SA, infection by TMV-GFP remains confined to very few cells, with no induction of visible NLL (Fig. 5, right leaves). Therefore, the novel resistance to TMV-GFP movement is not dependent on SA. While it seems unlikely that the temperature-independent N gene product could interact with other components of the pathway leading to SAR beyond the stimulation of SA, this possibility cannot be excluded. Such activation may manifest itself more slowly than HR and may not be able to restrict the movement of WT TMV.
It would be interesting to determine whether the same domains of the N gene are associated with the SA-mediated resistance response and the novel temperature-independent, CMV RNA1-suppressible resistance response. In this regard, the viral elicitor of the N gene response to TMV was mapped to the helicase domain (22, 40, 41), and CMV RNA1 encodes the 1a protein, a component of the viral RdRp that also contains a putative helicase domain (29). We show that the effect of the CMV RNA1 transgene on the movement of TMV-GFP is maintained even when the encoded 1a protein contains deletions of motifs that would abolish its putative helicase activity. However, the enzymatic activity of TMV helicase is not a feature required for the activation of N gene-dependent HR by the latter (22). Thus, the nature of the interaction remains unknown.
Testing of transgenic plants expressing various N gene mutants as well as alternative splicing of N gene transcripts could lead to a clearer understanding of the nature of the elicitors of the two distinct resistance responses. Such studies may also provide more information on how CMV RNA1 or the encoded 1a protein is able to suppress a resistance mechanism that has an inhibitory effect on the movement of a TMV-based vector.
Acknowledgments
We thank L. A. J. Mur for providing the seeds of the nahG gene-transgenic NN tobacco plants; Barbara Baker, Department of Plant Pathology, University of California, for providing seeds of the SR1::NN plants; and Large Scale Biology Corporation for providing the clone expressing TMV-GFP 1056. We also thank Sean Chapman, SCRI, for providing us with vector TMV.DsRed and S. Santa Cruz, Horticultural Research Institute, for providing us with vector TMV-GFP 5725.
This work was supported (in part) by contract QLK3-CT-2000-00361 from the European Commission and by a grant-in-aid from the Scottish Executive Environment and Rural Affairs Department.
REFERENCES
- 1.Abbink, T. E. M., J. de Vogel, J. F. Bol, and H. J. M. Linthorst. 1998. Tobacco mosaic virus helicase domain induces necrosis in N gene-carrying tobacco in the absence of virus replication. Mol. Plant-Microbe Interact. 11:1242-1246. [Google Scholar]
- 2.Abbink, T. E. M., J. de Vogel, J. F. Bol, and H. J. M. Linthorst. 2001. Induction of a hypersensitive response by chimeric helicase sequences of tobamoviruses U1 and Ob in N-carrying tobacco. Mol. Plant-Microbe Interact. 14:1086-1095. [DOI] [PubMed] [Google Scholar]
- 3.Anandalakshmi, R., G. J. Pruss, R. Marathe, A. C. Mallory, T. H. Smith, and B. V. Vance. 1998. A viral suppressr of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atabekov, J. G., and M. F. Taliansky. 1990. Expression of a plant virus-coded transport function by different viral genomes. Adv. Virus Res. 38:201-248. [DOI] [PubMed] [Google Scholar]
- 5.Baulcombe, D. C., S. Chapman, and S. Santa Cruz. 1995. Jellyfish green fluorescent protein as a reporter for virus infection. Plant J. 7:1045-1053. [DOI] [PubMed] [Google Scholar]
- 6.Boevink, P., S. Santa Cruz, C. Hawes, N. Harris, and K. J. Oparka. 1996. Virus-mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. Plant J. 10:935-941. [Google Scholar]
- 7.Boyko, V., J. Ferralli, and M. Heinlein. 2000. Cell-to-cell movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubules. Plant J. 22:315-325. [DOI] [PubMed] [Google Scholar]
- 8.Boyko, V., J. Laak, J. Ferralli, E. Suslova, M.-O. Kwon, and M. Heinlein. 2000. Cellular targets of functional and dysfunctional mutants of tobacco mosaic virus movement protein fused to green fluorescent protein. J. Virol. 74:11339-11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyko, V., J. Ferralli, J. Ashby, P. Schellenbaum, and M. Heinlein. 2000. Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 11:826-832. [DOI] [PubMed] [Google Scholar]
- 10.Canto, T., D. A. M. Prior, D. K.-H. Hellwald, K. J. Oparka, and P. Palukaitis. 1997. Characterization of cucumber mosaic virus. IV. Movement protein and coat protein are both essential for cell-to-cell movement of cucumber mosaic virus. Virology 237:237-248. [DOI] [PubMed] [Google Scholar]
- 11.Canto, T., and P. Palukaitis. 1998. Transgenically expressed cucumber mosaic virus RNA 1 simultaneously complements replication of cucumber mosaic virus RNAs 2 and 3 and confers resistance to systemic infection. Virology 250:325-336. [DOI] [PubMed] [Google Scholar]
- 12.Canto, T., and P. Palukaitis. 1999. The hypersensitive response to cucumber mosaic virus in Chenopodium amaranticolor requires virus movement outside the initially infected cell. Virology 265:74-82. [DOI] [PubMed] [Google Scholar]
- 13.Chivasa, S., A. M. Murphy, M. Naylor, and J. P. Carr. 1997. Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 9:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper, B., M. Lapidot, J. A. Heick, A. Dodds, and R. Beachy. 1995. A defective movement protein of TMV in transgenic plants confers resistance to multiple viruses whereas the functional analog increases susceptibility. Virology 206:307-313. [DOI] [PubMed] [Google Scholar]
- 15.Dasgupta, R., B. H. Garcia, and R. M. Goodman. 2001. Systemic spread of an RNA insect virus in plants expressing plant viral movement protein genes. Proc. Natl. Acad. Sci. USA 98:4910-4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson, W. O., D. J. Lewandowski, M. E. Hilf, P. Bubrick, A. J. Raffo, J. J. Shaw, G. L. Grantham, and P. R. Desjardins. 1989. A tobacco mosaic virus-hybrid expresses and loses an added gene. Virology 172:285-292. [DOI] [PubMed] [Google Scholar]
- 17.Delaney, T. P., S. Uknes, B. Vernooij, L. Friedrich, K. Weyman, D. Negrotto, T. Gaffney, M. Gut-Rella, H. Kessmann, E. Ward, and J. Ryals. 1994. A central role of salicylic acid in plant disease resistance. Science 266:1247-1250. [DOI] [PubMed] [Google Scholar]
- 18.Deom, C. M., S. Wolf, C. A. Holt, W. J. Lucas, and R. N. Beachy. 1991. Altered function of the tobacco mosaic virus movement protein in a hypersensitive host. Virology 180:251-256. [DOI] [PubMed] [Google Scholar]
- 19.Dinesh-Kumar, S. P., W. H. Tham, and B. J. Baker. 2000. Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc. Natl. Acad. Sci. USA 97:14789-14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinesh-Kumar, S. P., and B. J. Baker. 2000. Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97:1908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolja, V. V., K. L. Herndon, T. P. Pirone, and J. C. Carrington. 1993. Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J. Virol. 67:5968-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson, F. L., S. Holzberg, A. Calderon-Urrea, V. Handley, M. Axtell, C. Corr, and B. Baker. 1999. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18:67-75. [DOI] [PubMed] [Google Scholar]
- 23.Flor, H. 1971. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9:275-296. [Google Scholar]
- 24.Gal-On, A., I. B. Kaplan, and P. Palukaitis. 1995. Differential effects of satellite RNA on the accumulation of cucumber mosaic virus RNAs and their encoded proteins in tobacco vs zucchini squash with two strains of CMV helper virus. Virology 208:58-66. [DOI] [PubMed] [Google Scholar]
- 25.German-Retana, S., T. Candresse, E. Alias, R.-P. Delbos, and O. LeGall. 2000. Effects of green fluorescent protein of β-glucuronidase tagging on the accumulation and pathogenicity of a resistance-breaking Lettuce mosaic virus isolate in susceptible and resistant lettuce cultivars. Mol. Plant-Microbe. Interact. 13:316-324. [DOI] [PubMed] [Google Scholar]
- 26.Giesmann-Cookemeyer, D., S. Silver, A. A. Vaewhongs, S. A. Lommel, and C. M. Deom. 1995. Tobamovirus and dianthovirus movement proteins are functionally homologous. Virology 213:38-45. [DOI] [PubMed] [Google Scholar]
- 27.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in relication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo, H. S., J. J. López-Moya, and J. A. García. 1998. Susceptibility to recombination rearrangements of a chimeric plum pox potyvirus genome after insertion of a foreign gene. Virus Res. 57:183-195. [DOI] [PubMed] [Google Scholar]
- 29.Hayes, R. J., and K. W. Buck. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63:363-368. [DOI] [PubMed] [Google Scholar]
- 30.Heinlein, M., B. L. Epel, H. S. Padgett, and R. N. Beachy. 1995. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270:1983-1985. [DOI] [PubMed] [Google Scholar]
- 31.Heinlein, M., H. S. Padgett, J. S. Gens, B. G. Pickard, and S. J. Casper. 1998. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10:1107-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes, F. O. 1938. Inheritance of resistance to tobacco mosaic virus in tobacco. Phytopathology 28:553-561. [Google Scholar]
- 33.Johnson, J., T. Lin, and G. Lomonossoff. 1997. Presentation of heterologous peptides on plant viruses: genetics, structure, and function. Annu. Rev. Phytopathol. 35:67-86. [DOI] [PubMed] [Google Scholar]
- 34.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 35.Lacomme, C., and S. Santa Cruz. 1999. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA 96:7956-7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewandowski, D. J., and W. O. Dawson. 1998. Deletion of internal sequences results in tobacco mosaic virus defective RNAs that accumulate to high levels without interfering with replication of the helper virus. Virology 251:427-437. [DOI] [PubMed] [Google Scholar]
- 37.Matthews, R. E. F. 1991. Plant virology, 3rd ed. Academic Press, Inc., San Diego, Calif.
- 38.Matz, M. V., A. F. Fradkov, Y. A. Labas, A. P. Savitsky, A. G. Zaraisky, M. L. Markelov, and S. A. Lukyanov. 1999. Fluorescent proteins from nonbioluminiscent Anthozoa species. Nat. Biotechnol. 17:969-973. [DOI] [PubMed] [Google Scholar]
- 39.Mur, L. A. J., Y.-M. Bi, R. M. Darby, S. Firek, and J. Draper. 1997. Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. Plant J. 12:1113-1126. [DOI] [PubMed] [Google Scholar]
- 40.Padgett, H. S., and R. N. Beachy. 1993. Analysis of a tobacco mosaic virus strain capable of overcoming N gene-mediated resistance. Plant Cell 5:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padgett, H. S., Y. Watanabe, and R. N. Beachy. 1997. Identification of the TMV replicase sequence that activates the N gene-mediated hypersensitive response. Mol. Plant-Microbe Interact. 10:709-715. [Google Scholar]
- 42.Palukaitis, P., and M. Zaitlin. 1986. Infectivity and replication, p. 105-131. In M. H. V. Van Regenmorten and H. Fraenkel-Conrat (ed.), The plant viruses: tobacco mosaic virus. Plenum Press, New York, N.Y.
- 43.Palukaitis, P., and I. B. Kaplan. 1997. Potential ecological impact, p. 77-94. In M. Tepfer and E. Balazs (ed.), Virus resistant transgenic plants. Springer-Verlag KG, Berlin, Germany.
- 44.Rabindran, S., and W. O. Dawson. 2001. Assessment of recombinants that arise from the use of a TMV-based transient expression vector. Virology 284:182-189. [DOI] [PubMed] [Google Scholar]
- 45.Rizzo, T. M., and P. Palukaitis. 1990. Construction of full-length cDNA clones of cucumber mosaic virus RNAs 1, 2 and 3: generation of infectious RNA transcripts. Mol. Gen. Genet. 222:249-256. [DOI] [PubMed] [Google Scholar]
- 46.Ryals, J. A., U. H. Neuenschwander, M. G. Willits, A. Molina, H.-Y. Steiner, and M. Hunt. 1996. Systemic acquired resistance. Plant Cell 8:1809-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Scholthof, H. B., K.-B. G. Scholthof, and A. O. Jackson. 1996. Plant virus gene expression vectors for transient expression of foreign proteins in plants. Annu. Rev. Phytopathol. 34:299-323. [DOI] [PubMed] [Google Scholar]
- 49.Shivprasad, S., G. P. Pogue, D. J. Lewandowski, J. Hidalgo, J. Donson, L. K. Grill, and W. O. Dawson. 1999. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255:312-323. [DOI] [PubMed] [Google Scholar]
- 50.Vance, V. B., P. H. Berger, J. C. Carrington, A. H. Hunt, and X. M. Shi. 1995. 5′ Proximal potyviral sequences mediate potato virus X/potyviral synergistic disease in transgenic tobacco. Virology 206:583-590. [DOI] [PubMed] [Google Scholar]
- 51.Whitham, S., S. P. Dinesh-Kumar, D. Choi, R. Hehl, C. Corr, and B. Baker. 1994. The product of the tobacco mosaic virus resistance gene N: similarity to Toll and interleukin-1 receptor. Cell 78:1101-1115. [DOI] [PubMed] [Google Scholar]
- 52.Wright, K. M., G. H. Duncan, K. S. Pradel, F. Carr, S. Wood, K. J. Oparka, and S. Santa Cruz. 2000. Analysis of the N gene hypersensitive response induced by a fluorescently tagged tobacco mosaic virus. Plant Physiol. 123:1375-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, L., K. Hanada, and P. Palukaitis. 1994. Mapping local and systemic symptom determinants of cucumber mosaic cucumovirus in tobacco. J. Virol. 75:3185-3191. [DOI] [PubMed] [Google Scholar]