Abstract
F1-ATPase, a water-soluble portion of FoF1-ATP synthase, is a rotary motor driven by ATP hydrolysis. The central γ-subunit rotates in the α3β3 cylinder by repeating four stages of rotation: ATP-binding dwell, rapid 80° substep rotation, catalytic dwell, and rapid 40° substep rotation. In the catalytic dwell, at least two catalytic reactions occur—cleavage of the enzyme-bound ATP and presumably release of the hydrolyzed product(s) from the enzyme—but we found that a slow ATP cleavage mutant of F1-ATPase from thermophilic Bacillus PS3 rotates at low ATP concentration without substeps and the catalytic dwell. Analysis indicates that in this alternative reaction pathway the two catalytic reactions occur during the preceding long ATP-binding dwell. Thus, F1-ATPase can operate through (at least) two competing reaction pathways, not necessarily through a simple consecutive reaction.
INTRODUCTION
F1 is a molecular rotary motor driven by ATP hydrolysis (1). In vivo, F1 is connected with its partner Fo, a membrane-embedded portion, and constitutes a holoenzyme FoF1-ATP synthase. FoF1-ATP synthase synthesizes ATP from ADP and inorganic phosphate (Pi) using proton-motive force across membranes (2,3). The F1 portion (α3β3γδɛ) has catalytic sites responsible for ATP synthesis reaction, and the Fo portion (ab2c10) has a path for proton flow. A downhill proton flow through Fo drives the rotation of F1 in a direction reverse to hydrolysis, resulting in production of ATP. When isolated, F1 only catalyzes the ATP hydrolysis reaction and is often called F1-ATPase. The crystal structure of F1 shows that the rod-shaped γ-subunit is surrounded by a cylinder made of three α- and three β-subunits arranged alternately (4). The catalytic sites are located mainly on β-subunits, and some residues from adjacent α-subunits also contribute.
Rotation of the γ-subunit against the α3β3 cylinder by ATP hydrolysis was first proposed by Boyer (5), was supported by results of biochemical (6) and spectroscopic experiments (7), and was finally visualized by single-molecule observation under an optical microscope (8). The mechanism of rotary catalysis by F1 is known to have four principal features: First, the γ-subunit makes a 120° step when F1 hydrolyzes one ATP molecule (9). Second, this 120° step is further divided into four stages: ATP-binding dwell, 80° substep rotation, catalytic dwell, and 40° substep rotation (10). F1 rotates by repeating these four stages. Third, binding of ATP occurs during the ATP-binding dwell, and at least two catalytic events occur during the catalytic dwell (10). One of these has been shown to be cleavage of ATP at a catalytic site (11), and the other is currently assumed to be release of hydrolyzed product(s) from F1 (10). Fourth, in a single 120° rotation, ATP binding and ATP cleavage occur in two different β-subunits (12). Three β-subunits probably cooperate with each other to complete one 120° rotation; each of three β-subunits carries out one of the elementary reactions in ATP hydrolysis including ATP binding, ATP cleavage, and release of the hydrolyzed product(s).
In the above scenario, three β-subunits carry out their individual roles, faithfully obeying the order of sequential catalytic events in a single reaction pathway. Indeed, a single mechanism seems able to account for rotation of F1 at any [ATP], because time-averaged rotation rates of F1 show simple Michaelis-Menten dependence on [ATP] from the nanomolar to the millimolar range (10,13) and the duration of the catalytic dwell is constant (∼2 ms) at [ATP] from the micromolar to the millimolar range (10). However, in the course of studying the mutant F1 from thermophilic Bacillus PS3 that can hydrolyze ATP only at a very low rate, we noticed that this was not always the case. This mutant F1 has a very long catalytic dwell during which ATP cleavage (time constant (τ0) = 320 ms) and another event (τ0 = 20 ms), presumably the product release, occur (11). When we observed rotations of this mutant at low [ATP] where the ATP-binding dwell was more than several seconds, substeps and catalytic dwell disappeared, and the mutant F1 rotation included a repetition of the ATP-binding dwell and a rapid 120° rotation. ATP cleavage and product release can probably occur in the period when F1 is still in the ATP-binding dwell. Therefore, against the previous assumption that the order of sequential catalytic events of F1 is always maintained in a single reaction pathway, F1 can take at least two catalytic pathways in ATP hydrolysis.
MATERIALS AND METHODS
Materials
ATP, phosphoenolpyruvate, and bovine serum albumin (BSA) were purchased from Sigma (St. Louis, MO). Biotin-PEAC5-maleimide and Maleimide-C3-NTA were from Dojindo (Kumamoto, Japan). Pyruvate kinase, streptavidin, and gold colloid (diameter = 80 nm) were from Roche Diagnostics, Wako (Osaka, Japan) and British Biocell International (Cardif, UK), respectively. We used α3β3γ subcomplex derived from thermophilic Bacillus strain PS3. Here, we call this subcomplex F1. The mutant F1 (F1(β-E190D)) containing a replacement of Glu-190 with Asp in the β-subunit was purified as reported previously (11). Streptavidin-coated gold colloid was prepared as previously described (14).
Rotation assay
For the rotation assay, two cysteines of the γ-subunit were biotinylated (11). A flow chamber was made of a Ni2+-NTA-coated coverglass (16 × 16 mm2; Matsunami Glass, Osaka, Japan) and a slide glass (76 × 26 mm2) with a spacer of 50-μm thickness. Biotinylated F1(β-E190D) (0.5–2 pM) in solution A (50 mM MOPS-KOH, 50 mM KCl) containing 10 mg/ml BSA was infused into the flow chamber, and after 10 min, F1(β-E190D) unbound to the glass surface was washed away by 100 μl of solution A. Then 2 pM streptavidin-conjugated gold colloid in solution A containing 10 mg/ml BSA was infused. After a 10-min incubation, unbound gold colloids were washed away by 100 μl of solution A containing 2 mM MgCl2, ATP-regenerating system (50 μg/ml pyruvate kinase and 2.5 mM phosphoenolpyruvate), and the indicated amount of ATP. The rotating gold colloid was observed by dark-field microscopy (IX-70; Olympus, Tokyo, Japan) with a condenser (numerical aperture, 1.2–1.4) and ×100 objective lens (numerical aperture, 0.5–1.35). The images of the rotating gold colloid were recorded directly to the hard disc of a computer as an eight-bit Audio Video Interleave file with a fast-framing camera (Hi-Dcam II; NAC Image Technology, Tokyo, Japan) at 500–1000 frames per second. Custom software (created by R. Yasuda, Duke University) was used for analyses of the bead movements and dwells.
RESULTS
Rotation at saturating [ATP]
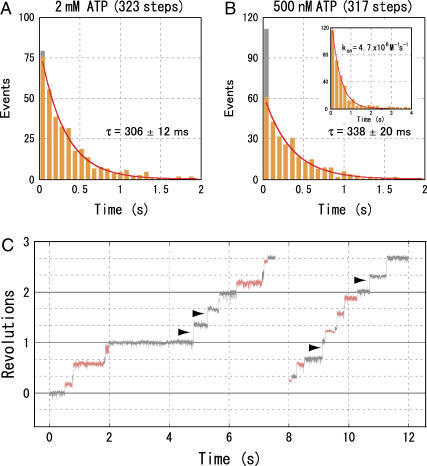
F1(β-E190D) hydrolyzes ATP slowly because of its sluggish ATP cleavage. It exhibits a characteristic rotation where the duration of the catalytic dwell is 140 times longer than that of the wild-type (11). Using this mutant, we can therefore observe the catalytic dwell more easily at the recording rate of 500–1000 frames per second. At 2 mM ATP, a Vmax condition where ATP binding should take place in 150 μs (= (kon × 2 mM)−1), we observed the rotation with discrete 120° steps pausing at the position of the catalytic dwell. (Some mutant F1 molecules showed the unusually long pauses (>100 ms) at the angle of the ATP-binding dwell. Since the average duration of these long pauses varied from molecule to molecule, we considered this pause not to be an intrinsic property of F1 and did not use these molecules for analysis.) The histogram of the dwelling time corresponded to a single exponential curve (Fig. 1 A). We attribute the time constant obtained, 305 ± 11 ms (mean ± SE), mainly to the slow cleavage of the bound ATP in F1(β-E190D) (11). To simplify, we set the bin width of the histogram to 80 ms in this case, and the other time constant of 20 ms, which we have presumed to represent the product release, was not resolved because of the 80-ms bin size (11). At [ATP]s above 20 μM, the histograms showed a similar single exponential decay (data not shown).
FIGURE 1.
Catalytic dwell of an F1(β-E190D) molecule at 2 mM (A) and 500 nM (B) ATP. In the first bin, the number of catalytic dwells <3 ms is indicated by gray. Inset in B shows the ATP-binding dwell of the same molecule as in B. Red lines show a fit with y = C × exp(−t/τ0). C and τ0 are a proportional constant and a time constant, respectively. (C) Typical time courses of the stepwise rotation of an F1(β-E190D) molecule at 500 nM ATP at 1000 frames per second. Horizontal lines are 120° apart. The ATP-binding dwells are shown in gray and the catalytic dwells in orange. The 120° rotations without catalytic dwells are indicated by arrowheads.
Rotation at [ATP] below Km
As [ATP]s decreased under Km (1.4 μM), however, the histograms started to deviate from a single exponential decay. At 500 nM ATP, for example, the first bin (<80 ms) contained many more events than expected from a single exponential decay that were fitted well with the data after the second bin (Fig. 1 B). A closer inspection of the rotation trajectory revealed that 120° step rotations without catalytic dwell occurred at significant frequency (Fig. 1 C, indicated by arrowheads). Out of 317 120° rotations, 51 showed no obvious (<3 ms) catalytic dwells (gray portion of the bar in Fig. 1 B). This result is in sharp contrast with the rotation at 2 mM ATP, where only 4 out of 324 catalytic dwells were <3 ms. Therefore, rotation of F1(β-E190D) appears to proceed through two parallel reaction pathways, one with and one without catalytic dwell. We named the former the conventional pathway and the latter an alternative pathway. The time constant of the dwell-time histogram in Fig. 2 B is 338 ± 20 ms, a value close to that of rotation at 2 mM ATP, and corresponds to the time constant for cleavage of ATP of the conventional pathway.
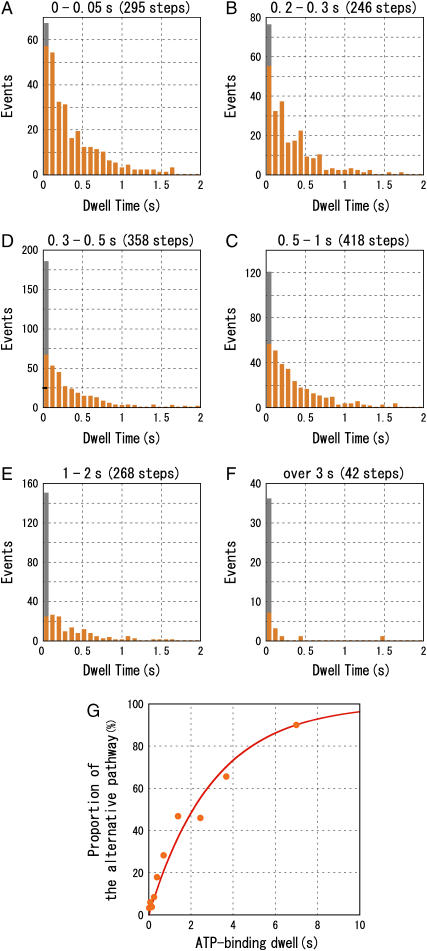
FIGURE 2.
Correlation between the catalytic dwell and the ATP-binding dwell. Durations of the ATP-binding dwells are (A), 0–0.05 s; (B), 0.2–0.3 s; (C), 0.3–0.5 s; (D), 0.5–1 s; (E), 1–2 s; and (F), >3 s. In the first bin, the number of catalytic dwells <3 ms is indicated in gray. (G). The proportion of the alternative pathway as a function of the duration of the ATP-binding dwell. Sample numbers for the points are, left to right, 295, 235, 368, 246, 358, 418, 268, 63, 32, and 10. The red line shows a fit with a single exponential, y = 100 × (1 − exp(−t/τ0)). We assumed that occurrence of the conventional pathway is 100% at ATP-binding dwell = 0 s.
Unlike those of the catalytic dwell, histograms of the ATP-binding dwell at low [ATP]s were fitted well by a single exponential curve (Fig. 1 B, inset) and were not affected by the duration of the preceding catalytic dwell (see below). The rate constant kon for ATP binding was determined to be (3.3 ± 0.3) × 106 M−1 s−1 (data for seven molecules). This value is one order of magnitude smaller than that of the wild-type and is consistent with the previous estimates (11). This result suggests that the ATP-binding dwell is terminated by the binding of ATP as reported previously in both of the two pathways (10).
Dependence on the ATP-binding dwell
The alternative pathway was more frequently observed as [ATP] decreased. As [ATP] decreased, ATP came to the catalytic site less frequently, and the ATP-binding dwell became longer. We therefore assume that duration of the ATP-binding dwells determines the population of F1(β-E190D) that takes the alternative pathway in the following 120° rotation. To determine whether this was in fact the case, we analyzed the correlation between the duration of the ATP-binding dwell and occurrence frequency of the alternative pathway (Fig. 2). We classified the ATP-binding dwells (2311 dwells, 7 molecules, 500 nM–2 μM ATP) into 10 groups according to their durations and made histograms of the catalytic dwells that followed the individual ATP-binding dwells. The catalytic dwells longer than 5 s (6 out of 2311 dwells) were neglected because they were probably attributable to the ADP-inhibited state, in which F1 fails to release ADP, lapsing into pauses of rotation over several tens of seconds (15). When the ATP-binding dwell was short, <50 ms (Fig. 2 A), the histogram showed a single exponential decay (τ = 337 ± 19 ms) as observed at 2 mM ATP. As the ATP-binding dwell became longer, the first bin of the histogram became larger than the values extrapolated from a single exponential (Fig. 2, A–F). When the ATP-binding dwell was over 3 s, most catalytic dwells (36 out of 42) were included in the first bin (Fig. 2 F). We defined catalytic dwells <3 ms (gray portion of the first bin) as belonging to the alternative pathway and plotted their proportion relative to the total counts of catalytic dwells as a function of the ATP-binding dwell duration (Fig. 2 G). As shown, the proportion of the alternative pathway increases as the duration of the preceding ATP-binding dwell becomes longer. This increase can be simulated with a single exponential curve with a time constant of 3.0 ± 0.3 s. The enzyme therefore tends to choose the alternative pathway rather than the conventional pathway when the enzyme must wait for ATP for longer than 3 s.
DISCUSSION
Can wild-type F1 also take the alternative pathway?
Our results clearly show that the mutant F1(β-E190D) can rotate according to either of two reaction pathways. In the newly discovered alternative pathway of the mutant F1(β-E190D), rotation does not pause at the 80° position but proceeds with a single 120° step rotation. The rotation with the alternative pathway is currently observed only for the sluggish F1(β-E190D) mutant at low [ATP]. The immediate question is therefore whether this alternative pathway is allowed only for the sluggish mutant or is a general feature for the wild-type F1. In addition, the apparent contradiction between our results and those of two previous reports on rotation of the wild-type F1 should be explained (10,13). Sakaki et al. recently reported that time-averaged rotation rates of F1 show simple Michaelis-Menten dependence on [ATP] from the nanomolar to the millimolar range, suggesting that a single mechanism governs rotation over this [ATP] range (13). However, at low [ATP], the binding of ATP is rate-limiting in turnover of catalysis both for conventional and alternative pathways, and the interim between 120° rotations is governed by the ATP-binding dwell with little difference between the two pathways. Therefore apparent simple Michaelis-Menten kinetics does not exclude the possibility of the alternative pathway at low [ATP]. Yasuda et al. showed that the duration of the catalytic dwell is not affected by the duration of the ATP-binding dwell and is constant at [ATP] from 2 μM to 6 mM (10). In our case, [ATP] was below μM, and kon for ATP of the mutant was∼1 order of magnitude smaller than that of the wild-type. As a consequence, the ATP-binding dwell we observed, for example, 6.1 × 102 ms at 500 nM ATP, was longer by 40-fold than that of the wild-type at 2 μM ATP (17 ms). Therefore, almost all rotations of the wild-type should proceed through the conventional pathway in the previous experiments at 2 μM ATP. Detection of the alternative pathway for the wild-type F1 at lower [ATP], for example 20 nM, might be practically difficult because the ATP-binding dwell would be 1000-fold longer than the catalytic dwell and the two dwells would be difficult to distinguish by ordinary measurement methods. Altogether, we think it likely that the alternative pathway exists in the wild-type.
What is the alternative pathway?
The alternative pathway is characterized by the rotation without the catalytic dwell at the 80° position (Fig. 1). It is unlikely that the two events, ATP cleavage and product release, occur at the 80° position too rapidly for the time resolution of our measurement because this behavior would not explain why the alternative pathway is observed only when the preceding ATP-binding dwell is long. More likely is that ATP hydrolysis can take place during the ATP-binding dwell even though very rarely. The time constant for the transition from the conventional pathway to the alternative pathway is 3.0 s (Fig. 2 G). This value corresponds to the time constant for the cleavage of ATP when the mutant is at the ATP-binding dwell. We assume that product release also occurs during the ATP-binding dwell because the 20-ms event, which is presumably the product release, also disappeared from the catalytic dwell in the alternative pathway.
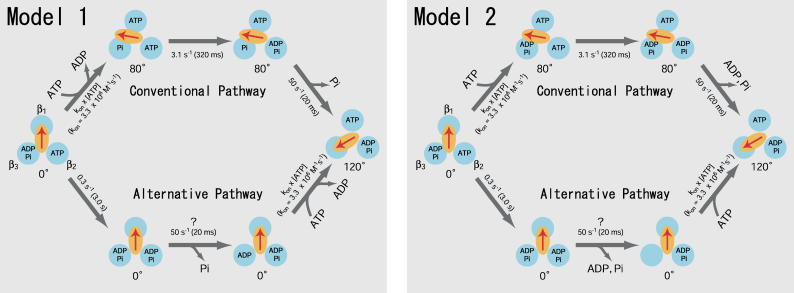
As discussed in Sakaki et al. (13), we currently assume that the rotation in the conventional pathway proceeds as follows (Fig. 3, model 1, upper path). ATP binds to the β1 subunit of F1 in the ATP-binding dwell and drives the 80° substep rotation of the γ-subunit, which accompanies ADP-release from the β3 subunit. During the catalytic dwell, cleavage of the previously bound ATP to ADP and Pi occurs at the β2 subunit. After the cleavage, the β3 subunit releases Pi, an action coupled with the 40° substep rotation. In the alternative pathway (Fig. 3, model 1, lower path), hydrolysis of ATP at the β2 subunit and release of Pi from the β3 subunit occur during the ATP-binding dwell, and ADP is released from the β3 subunit during the 120° rotation that is triggered by the ATP-binding to the β1 subunit. Another conventional pathway in which release of ADP is coupled with the 40° substep rotation is also possible (Fig. 3, model 2, upper path). In this pathway, three catalytic sites are occupied by nucleotides during the catalytic dwell (trisite catalysis). The sequence of events in the alternative pathway corresponding to this conventional pathway is as follows (Fig. 3, model 2, lower path): ATP cleavage at the β2 subunit, product release from the β3 subunit, ATP-binding to the β1 subunit, and 120° rotation. In this pathway, one or two catalytic sites are occupied by ATP or ADP during catalysis (bisite catalysis). If model 2 is really functional, F1 works with the trisite mode under usual conditions but does so with the bisite mode when [ATP] is extremely low.
FIGURE 3.
Two models of the reaction pathways of the ATP-hydrolysis reaction in F1(β-E190D). F1(β-E190D) are represented as three β-subunits (blue) and a γ-subunit (orange oval with an arrow). The rate constants obtained from this work and the previous works (11) are shown. The rate constant 50 s−1 is assumed to be unchanged between the conventional and alternative pathway. The models represent the pathways of F1(β-E190D) but the wild-type F1 probably has similar alternative pathways as well (see the text). (Model 1) ADP is released during the rotary motion upon ATP-binding. (Upper path) The conventional pathway. (Lower path) The alternative pathway. (Model 2) ADP release and ATP binding occur at a different stage in the reaction pathway. (Upper path) The conventional pathway. The 40° substep rotation is coupled with release of the products. (Lower path) The alternative pathway. The products are released without accompanying rotation. Note that occupancy of the catalytic sites by ATP (or ADP) during catalysis in this alternative pathway is one or two (bisite catalysis).
F1 reaction is not a simple consecutive reaction
This work revealed that F1 can operate through (at least) two pathways. We have long assumed the faithfulness of F1 in keeping the order of sequential catalytic events tracking a single reaction pathway. Actually, we stated in the previous work (16) “During a long period in which one slow β-subunit waits for ATP, other β-subunits are also just waiting without letting the next catalytic event start.” But the 80° dwell with short lifetime (∼2 ms) was not detectable with the experimental setup in Ariga et al. (16) and this statement should be corrected. Our finding implies that catalytic events of all three catalytic sites are coordinated not as a single, linear consecutive reaction but as diverging-converging reaction pathways. Recently, two pathways were revealed in myosin V as well (17). Two heads of myosin V work cooperatively, and existence of two pathways may not be rare among motor proteins in which the catalytic subunits work cooperatively. Two reaction pathways are competing with each other and slowing down one of them increases the weight of other pathways, which may originally make small contributions to the overall flux. The weight of each reaction pathway may also change with substrate concentrations. Characterization of multiple reaction pathways and their physiological relevance need to be further clarified.
Acknowledgments
We thank our colleagues, especially Drs. H. Noji, T. Masaike, R. Iino, and R. Watanabe, for critical discussions, and Dr. H. Ueno and M. Takeda for technical advice. We are grateful to Drs. K. Kinosita Jr. and R. Yasuda for development of methods and custom software.
Katsuya Shimabukuro's present address is Dept. of Biological Science, Florida State University, Tallahassee, FL 32306.
References
- 1.Yoshida, M., E. Muneyuki, and T. Hisabori. 2001. ATP synthase—a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2:669–677. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell, P. 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 191:144–148. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, P. D. 2000. Catalytic site forms and controls in ATP synthase catalysis. Biochim. Biophys. Acta. 1458:252–262. [DOI] [PubMed] [Google Scholar]
- 4.Abrahams, J. P., A. G. Leslie, R. Lutter, and J. E. Walker. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 370:621–628. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, P. D. 1993. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta. 1140:215–250. [DOI] [PubMed] [Google Scholar]
- 6.Duncan, T. M., V. V. Bulygin, Y. Zhou, M. L. Hutcheon, and R. L. Cross. 1995. Rotation of subunits during catalysis by Escherichia coli F1-ATPase. Proc. Natl. Acad. Sci. USA. 92:10964–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabbert, D., S. Engelbrecht, and W. Junge. 1996. Intersubunit rotation in active F-ATPase. Nature. 381:623–625. [DOI] [PubMed] [Google Scholar]
- 8.Noji, H., R. Yasuda, M. Yoshida, and K. Kinosita Jr. 1997. Direct observation of the rotation of F1-ATPase. Nature. 386:299–302. [DOI] [PubMed] [Google Scholar]
- 9.Yasuda, R., H. Noji, K. Kinosita Jr., and M. Yoshida. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell. 93:1117–1124. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda, R., H. Noji, M. Yoshida, K. Kinosita Jr., and H. Itoh. 2001. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 410:898–904. [DOI] [PubMed] [Google Scholar]
- 11.Shimabukuro, K., R. Yasuda, K. Y. Hara, E. Muneyuki, K. Kinosita Jr., and M. Yoshida. 2003. Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. USA. 100:14731–14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishizaka, T., K. Oiwa, H. Noji, S. Kimura, E. Muneyuki, M. Yoshida, and K. Kinosita Jr. 2004. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 11:142–148. [DOI] [PubMed] [Google Scholar]
- 13.Sakaki, N., R. Shimo-Kon, K. Adachi, H. Itoh, S. Furuike, E. Muneyuki, M. Yoshida, and K. Kinosita Jr. 2005. One rotary mechanism for F1-ATPase over ATP concentrations from millimolar down to nanomolar. Biophys. J. 88:2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueno, H., T. Suzuki, K. Kinosita Jr., and M. Yoshida. 2005. ATP-driven stepwise rotation of FoF1-ATP synthase. Proc. Natl. Acad. Sci. USA. 102:1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirono-Hara, Y., H. Noji, M. Nishiura, E. Muneyuki, Y. K. Hara, Y. Yasuda, K. Kinosita Jr., and M. Yoshida. 2001. Pause and rotation of F1-ATPase during catalysis. Proc. Natl. Acad. Sci. USA. 98:13649–13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ariga, T., T. Masaike, H. Noji, and M. Yoshida. 2002. Stepping rotation of F1-ATPase with one, two, or three altered catalytic sites that bind ATP only slowly. J. Biol. Chem. 277:24870–24874. [DOI] [PubMed] [Google Scholar]
- 17.Uemura, S., H. Higuchi, A. O. Olivares, E. M. De La Cruz, and S. Ishiwata. 2004. Mechanochemical coupling of two substeps in a single myosin V motor. Nat. Struct. Mol. Biol. 11:877–883. [DOI] [PubMed] [Google Scholar]