Abstract
We have explored the electromechanical properties of DNA on a nanometer-length scale using an electric field to force single molecules through synthetic nanopores in ultrathin silicon nitride membranes. At low electric fields, E < 200 mV/10 nm, we observed that single-stranded DNA can permeate pores with a diameter ≥1.0 nm, whereas double-stranded DNA only permeates pores with a diameter ≥3 nm. For pores <3.0 nm diameter, we find a threshold for permeation of double-stranded DNA that depends on the electric field and pH. For a 2 nm diameter pore, the electric field threshold is ∼3.1 V/10 nm at pH = 8.5; the threshold decreases as pH becomes more acidic or the diameter increases. Molecular dynamics indicates that the field threshold originates from a stretching transition in DNA that occurs under the force gradient in a nanopore. Lowering pH destabilizes the double helix, facilitating DNA translocation at lower fields.
INTRODUCTION
DNA is an unusually stiff, highly charged polymer. The stiffness is due mainly to base stacking and the double helical structure, whereas most of the charge is accounted for by the phosphate backbone (1). These inimitable physical properties are vital to biology; for example, proteins exploit them to transcribe, package, regulate, and repair the information encoded in the sequence of nucleotides that comprises the DNA. The physical properties of DNA are exploited in a nascent strategy for rapidly sequencing DNA that would analyze the polymer as it translocates through a nanometer diameter pore in a membrane (2–7).
When an electric field is applied across a synthetic membrane with a nanometer diameter pore in it, polyanionic DNA immersed in electrolyte at the cathode migrates toward the anode and eventually permeates the membrane through the pore. The translocation of DNA through the pore induced by an electric field represents an especially stringent test of the electromechanical properties of the polymer: the force arising impels segments of the polymer to bend and stretch within the pore (8–10) on a length scale comparable to the size of a protein-binding site, which is ∼10–30 basepairs (bp) long (3–10 nm). Thus, a threshold field may be required to drive the DNA into the pore and keep it there despite repulsive forces and thermal agitation. For example, Henrickson et al. (8) discovered an 8 kT barrier to permeation of ssDNA through the proteinaceous pore formed by α-hemolysin in a lipid membrane, although the origin of the barrier was not determined unequivocally (11) essentially because the size of the pore and electrical charge on the amino acid side chains are not easily manipulated.
To explore the electromechanical properties of DNA on a length scale comparable to a protein-binding site, we have used an electric field to force single molecules through synthetic pores, ranging from 1.0 to 3.0 nm in diameter, in ultrathin (10–20 nm thick) Si3N4 membranes. The force due to the field acting on single nucleotides in a DNA strand during the translocation is approximately F ∼ q*ΔV/a, where q* is the effective charge per nucleotide, and ΔV is the electrostatic potential drop over the nucleotide separation a. At low fields, E < 200 mV/10 nm, we observed that single-stranded DNA (ssDNA) permeates pores with diameters as small as 1.0 nm, whereas double-stranded DNA (dsDNA) only permeates pores with a diameter ≥3.0 nm, presumably because the diameter of the double helix is ∼2 nm (1). On the other hand, for pores <3.0 nm in diameter we find a threshold for permeation of dsDNA that depends on the electric field and pH. For a 2 nm diameter pore, the threshold is E ≅ 3.1 V/10 nm at pH = 8.5 and it decreases as pH becomes more acidic. We used molecular dynamics (MD) to interpret the measurements with atomic detail. Matching the experimental setup, we built an all atom model of the nanopore, DNA, and electrolyte and simulated the field-induced permeation of ssDNA and dsDNA. We find that the electric field threshold for permeation is due to a stretching transition that occurs in the DNA double helix (12–14) and that acid pH mechanically destabilizes dsDNA, facilitating the translocation. This work extends a brief report in Heng et al. (15) providing a more detailed account of experimental and computational work as well as discussing the influence of the pH and the pore geometry on the electric field threshold for DNA translocation.
METHODS
Membrane fabrication
The fabrication of the nanopores in the Si3N4 membranes is described elsewhere (16). A single nanopore is created in a membrane by stimulated decomposition and sputtering using a tightly focused electron beam in a JEOL 2010F transmission electron microscope (TEM) operating at 200 keV. Fig. 1, a–c, shows TEM images taken at a tilt angle of 0° of circular pores with apparent diameter of d0 = 1.0 ± 0.2 nm, 1.9 ± 0.2 nm, and 3.0 ± 0.2 nm in a Si3N4 membrane nominally 10 nm thick. The thickness of similarly processed 10 nm membranes was found to be 10 ± 2 nm using scanning electron microscopy (SEM) and 11 ± 3 nm using electron energy loss spectroscopy. Fig. 1 d is a micrograph of a pore with an elliptical cross section 1.8 × 2.2 (±0.2) nm2 in a 20 nm thick Si3N4 membrane. The thickness of the 20 nm membrane is 20 ± 2 nm according to SEM. The TEM images shown in Fig. 1 represent a two-dimensional projection through the membrane; the shot noise observed in the central area identified with the pore indicates perfect transmission of electrons through the membrane. To investigate the three-dimensional structure, we tilted the membrane in the TEM about the pore axis and explored various defocus conditions. We have developed models for the pore geometry, one of which is represented schematically by two intersecting cones with a 10° cone angle in the inset to Fig. 1.
FIGURE 1.
(Top) TEM images of pores at 0° tilt with apparent diameter of (a) 1.0 ± 0.2, (b) 1.9 ± 0.2, (c) 3.0 ± 0.2 nm in a 10 nm thick Si3N4 membrane, and (d) 1.8 × 2.2 nm in Si3N4 20 nm thick. (Bottom) I/V characteristics in 1 M KCl solution of the same four pores. Line fits yield slopes of 0.63 ± 0.03 nS, 1.09 ± 0.03 nS, 1.24 ± 0.03 nS, and 0.68 ± 0.03 nS for the four pores, respectively.
Ionic current measurements
To characterize ion transport through the pore, we measured the electrolytic current as a function of the applied electrochemical potential at 23.5°C ± 1°C. The currents were measured using an Axopatch 200B amplifier (Axon, Union City, CA) in resistive feedback mode with a 10–100 kHz bandwidth. All experiments were carried out in a membrane transport bi-cell made of either acrylic or polydimethylsiloxane, where the membrane separates two identical compartments each containing ≈40 μL − 1 mL of 1 M KCl electrolyte and an Ag/AgCl electrode. Fig. 1 shows the current-voltage (I/V) characteristics of the pores shown in Fig. 1, a–d, measured over a range of ±1 V in 1 M KCl after >55 h of immersion in deionized water. In all cases the characteristics are approximately linear. A line fit to the data yields the conductance 0.63 ± 0.03 nS, 1.09 ± 0.03 nS, 1.24 ± 0.03 nS, and 0.68 ± 0.03 nS for the pores a–d, respectively. It is apparent that the conductance does not scale linearly with the diameter derived from TEM. We have analyzed these measurements using MD to estimate the ion mobility and then solve coupled Poisson-Nernst-Planck and the Stokes equations self-consistently for the ion concentration, velocity, and electrical potential. We find that the measurements are consistent with the presence of a small (relative to DNA) fixed negative charge in the pore and a reduction of the ion mobility due to the fixed charge and the ion proximity to the pore wall (16).
PCR/qPCR analysis
To establish unambiguously that the DNA injected at the cathode permeates a pore, the minute amount of the DNA near the anode was amplified using polymerase chain reaction (PCR) and analyzed by gel electrophoresis. The DNA was concentrated with a Microcone-10 centrifugal filter (Millipore, Bedford, MA), and the buffer was exchanged to water on the same filter. A total of 10 μL of the resulting solution was used for the reaction. The PCR reagent system was obtained from Invitrogen (Carlsbad, CA), the sequence specific primers were synthesized by Midland (Midland, TX). Amplified DNA was analyzed by agarose gel electrophoresis. A voltage applied across the gel provides a driving force separating molecules according to size and charge across a span of gel matrix. After staining with ethidium bromide, the DNA in each lane fluoresces under ultraviolet light and can be seen as bands spread according to the length of the strand or molecular weight from one end of the gel to the other.
The number of DNA copies that permeated through a pore was determined by quantitative real-time PCR (qPCR). Two PCR primers were designed to amplify a 72 bp region within a 622 base target sequence. A TaqMan probe was designed, mapping to this 72 bp target sequence and labeled with an fluorescin reporter dye at the 5′ end and with a tetramethylrhodamine quencher dye at the 3′ end. During amplification, the primers and the probe hybridize to the target sequence. Subsequently, during polymerization, the 5′ exonuclease activity of the DNA polymerase cleaves the probe, releasing the reporter dye and resulting in increased fluorescence, which is proportional to the amount of target.
To guarantee the validity of the results, concomitant with the gel arrays and qPCR results shown below, we also ran control experiments containing no DNA, an MW reference denoted as “100 bp ladder”, which contains a spread of DNA MW consisting of 15 blunt-ended fragments 100–1500 bp in multiples of 100 bp, and various dilutions of DNA ranging from 10 molecules to 100,000 molecules per batch to test the copy number.
MD modeling of Si3N4 pores
To simulate the translocation of DNA through the pore, a crystalline Si3N4 membrane was built by replicating a unit cell of β-Si3N4 crystal (17); the double-conical pore was then produced by removing silicon and nitrogen atoms. The narrowest part of each pore was located at the center of the Si3N4 membrane (z = 0); the diameter of the pore varied with z as d(z) = d0 + |z|· tan(γ), where d0 is the diameter of the pore at z = 0, and the conical angle γ is ∼10° (6,16). A 58 bp dsDNA helix was built from individual bp in the geometry suggested by Quanta (18); the sequence of DNA was identical to that used in the experiments. The helix was oriented normal to the membrane and placed 1 nm within the pore. ssDNA was obtained from that structure by removing one of the strands. The Si3N4/DNA complex was then solvated in a volume of preequilibrated TIP3P water molecules. K+ and Cl− ions were added in a desired proportion.
Phantom pores
To set up a phantom pore simulation, a 58-mer ssDNA strand was placed in a cylindrical volume of 1 M KCl. The strand was enveloped by a mathematical surface faithfully reproducing the shape of a double-conical (template) pore sculptured in a Si3N4 membrane, a so-called phantom pore. Initially, the phantom pore was made 2 nm wider in diameter than the Si3N4 pore such that the entire equilibrated 58-nucleotide single DNA strand could fit into it. The pore was then gradually shrunk in the simulation to its original shape (the shape of the Si3N4 pore). At the same time, 10 pN forces pushed the atoms lying outside of the shrinking surface toward the center of the pore. At the end of the simulation, the DNA strand adapted a straight conformation that conformed to the shape of the template Si3N4 pore. Two phantom pores were constructed: the first one imposed geometrical restraints on DNA atoms only, whereas in the second one, both DNA and electrolyte were subject to restraints.
MD methods
All MD simulations were carried out using the program NAMD2 (19), AMBER95 (20) force field describing nucleic acid, water, and ions, and a custom force-field describing the Si3N4 membrane. The van der Waals parameters and charges of Si3N4 atoms were taken from Wendel and Goddard (21); every Si3N4 atom was restrained by a harmonic force to its initial location in the crystalline membrane with the force constant of 0.01 or 0.1 kcal/(nm2 mol) for bulk and surface atoms, respectively. The spring constant of the harmonic bonds connecting silicon and nitrogen atoms in Si3N4 was adjusted to yield the relative permittivity of bulk Si3N4 of 7.5 (22). The distribution of partial charges on N1-protonated adenosine nucleoside was calculated using GAMESS (23) and refined with RESP (24). In all simulations, the temperature was kept at 295 K by applying Langevin forces; other simulation conditions and protocols are described in Aksimentiev et al. (17).
RESULTS AND DISCUSSION
Sorting DNA with a nanopore
After the conductance measurements, we tested the electric field driven permeability of ssDNA (58 mer) and dsDNA (58 bp) through the pores of Fig. 1, a–c. First, DNA in 1 M KCl electrolytic solution buffered at pH 8.5 with 10 mM Tris-Cl was injected at the compartment containing the (negative) Ag/AgCl cathode, then 200 mV was applied across the membrane, and the current through the pore was measured for 4 h to collect statistically significant data. Superimposed on the dc electrolytic current, we observed transients that we associated with the pore interacting with a single molecule. We have previously reported that field-driven translocations of the DNA cause a temporary blockade of the open current through the pore (4). However, the narrow bandwidth (100 kHz) of the amplifier precludes the observation of transients associated with interactions between the pore and the molecule that are shorter than 10–100 μs. Instead, we used PCR and gel electrophoresis to unequivocally measure permeability (2,25). Fig. 2, a–c, shows gel arrays illustrating the permeation of ssDNA (58 mer) and dsDNA (58 bp) through the pores shown in Fig. 1, a–c, respectively. ssDNA that is collected from the compartment housing the positive electrode is denoted by (+)58 mer. Notice that ssDNA is observed to permeate all three pores and gives the same amplified pattern as the ssDNA injected in the compartment housing the negative electrode, denoted by (−)58 mer. On the other hand, dsDNA only permeates the 3.0 nm ± 0.2 nm diameter pore at 200 mV.
FIGURE 2.
(a–c) Gel arrays showing five horizontal lanes with fluorescent bands that identify 58-mer ssDNA and 58 bp dsDNA in the compartments housing positive (+) and negative (−) electrodes testing the permeability of the 1 nm, 2 nm, and 3 nm pores shown in Fig. 1, a–c, respectively. The 100 bp ladder is included for reference. The applied voltage was 200 mV.
MD provides an atom level description of the translocation of DNA through these pores. A uniform electric field was applied across a 10 nm thick Si3N4 membrane producing a 1.3 V transmembrane bias driving DNA into 1.4, 2.0, and 3.0 nm diameter pores. Fig. 3 shows the position of the leading edge of the molecule inside the pore relative to the center of the membrane against the simulation time. We find that the 58 bp dsDNA fragment fails to permeate through 1.4 and 2.0 nm diameter pores; the translocation halts when the pore narrowed to a diameter of 2.5 ± 0.1 nm before the DNA arrived at the narrowest part of the pore. However, the same 58 bp fragment was observed to permeate a 3.0 nm diameter pore. Thus, the minimum diameter pore allowing dsDNA to permeate at ≤1.3 V is 2.5 nm.
FIGURE 3.
MD simulations of DNA translocation through pores in Si3N4 membranes. The pore profile is overlaid geometrically faithfully. The symbols indicate the location of the DNA leading edge as DNA permeates through the pore (from top to bottom); the transmembrane bias is 1.3 V. Animations illustrating these MD simulations are available at http://www.ks.uiuc.edu/Research/nanopore/stretching/.
Our MD simulations indicate that the translocation of dsDNA through a 3.0 nm diameter pore proceeds at the rate of ∼1 bp per nanosecond at a 1.3 V bias. This is in agreement with the recent experimental study (26) that reports the translocation velocity of a 6,600 bp DNA fragment at 1 bp per 20 ns at a 120 mV bias. In the case of DNA translocation through the transmembrane pore of α-hemolysin, the simulated velocity of DNA translocation is much smaller, <10 nucleotides/μs (27), which, again, is in agreement with experiment (28). We, therefore, conclude that the timescale covered by MD is adequate to describe translocation of dsDNA through synthetic nanopores. Thus, we believe that the “real world” timescale of DNA stretching and unzipping events described below is several tens of nanoseconds, i.e., within the reach of the MD method, when the electric field is high (1–6 V/10 nm).
As shown in Fig. 3, we observed only a partial permeation of a 58-nucleotide ssDNA through a 1.4 nm diameter pore within 20 ns. After 8 ns, the base of the leading nucleotide adheres to the surface of Si3N4, dramatically slowing the translocation. In addition to the driving electric force, three types of interactions affect the duration of the translocation: i), mechanical friction between DNA and the pore; ii), reduction of the driving force by the counterion condensation, decreasing the DNA effective charge; and iii), the hydrophobic adhesion of bases to the pore surface. On a timescale of 10 ns, hydrophobic adhesion is the predominate force affecting the duration of the translocation of ssDNA and RNA strands when the diameter of the pore is larger than the diameter of the strand (17,29).
Since we experimentally observe that ssDNA permeates a 1 nm diameter pore, we investigated the effect of these three interactions on the translocation kinetics. First, we evaluated the effect of steric friction between DNA and the pore when the surface adhesion forces are diminished. Our initial system was prepared by confining a 58-mer ssDNA in a 1.0 nm diameter phantom pore in which the steric forces representing the pore surface were applied to the DNA atoms only. In Fig. 4 (top), the center of mass (CoM) of the 10 DNA nucleotides, confined at the start of the simulation to the constriction (|z| < 2 nm) in the pore, is plotted against time. The DNA velocity is proportional to the total force driving the strand. When applying a force corresponding to a 0.43 V bias, the translocation velocity of ssDNA was 2 nm/ns.
FIGURE 4.
Translocation of ssDNA through a 1.0 nm diameter phantom pore. The DNA CoM is plotted against simulation time. (Top) Phantom pore restraints act on DNA only. (Bottom) Phantom pore restraints act on both DNA and the electrolyte. The simulated systems are shown as insets.
In the previous phantom pore simulations, water and ions were free to move around DNA, without being subject to the constraints imposed by the pore geometry. To investigate the effect of confining the electrolyte, we used a phantom pore to apply the steric constraints to both DNA and the KCl solution. When setting up the system by shrinking a 2.5 nm diameter pore to 1.0 nm, we observed condensation of K+ ions on the phosphate backbone of ssDNA. The presence of counterions dramatically reduced the total electrostatic force exerted on the DNA strand, drastically slowing the translocation. In Fig. 4 (bottom), we plot the CoM of the three nucleotides, confined at the beginning of the simulation to the center (|z| < 1 nm) of the pore. The three nucleotides were observed to move in the direction of the applied field, despite large stochastic displacements. From these simulations, we estimated that at a 1 V bias the translocation velocity should not exceed 1 nucleotide/10 ns (∼0.03 nm/ns), which is almost two orders of magnitude slower than the velocity observed when steric constraints are applied to DNA atoms only. Thus, the counterion condensation predominates over the steric friction in very narrow pores. Further improvements in the microscopic model of the Si3N4 surface have to be developed to quantitatively compare the effects of counterions condensation to the surface adhesion of DNA bases.
Stretching DNA with an electric field
To determine if a higher field could impel dsDNA through a pore with a diameter <3.0 nm, we tested permeability as a function of voltage using the 2.0 nm diameter pore in a 10 nm membrane shown in Fig. 1 a, and the 1.8 × 2.2 nm2 cross-section pore in the 20 nm thick membrane shown in Fig. 1 d. The gel array in Fig. 5 a, illustrating the permeability of the 2 nm diameter pore, shows a fluorescent band in the lane corresponding to dsDNA at the compartment housing the positive electrode, (+)58 bp, only for an applied bias of 3 V, indicating that 58 bp dsDNA can be forced through a pore only if the applied voltage is V > 2.75 V. The gel array of Fig. 5 b indicates that the threshold depends on the electric field. A fluorescent band is observed in the lane corresponding to dsDNA collected at the compartment housing the positive electrode, (+)58 bp, only for an applied bias of 6 V, indicating that 58 bp dsDNA can be forced through a 1.8 × 2.2 nm2 pore in a 20 nm membrane if the applied voltage is V > 5.75 V. Fig. 5 c represents the results of two qPCR analyses showing the number of copies permeating as a function of the applied potential through the same two pores used in Fig. 5, a and b. In correspondence with the >2.75 V threshold obtained from the gel array for 58 bp, the 2.0 nm diameter pore exhibits a threshold voltage of >2.5 V for 622 bp dsDNA. Similarly, the 1.8 × 2.2 nm2 pore shows a threshold voltage >5 V. Taken all together these data indicate that the threshold for permeation occurs at nearly the same electric field, E = 3.1 ± 0.7 MV/cm, which is estimated assuming a uniform voltage drop across the membrane. Thus, we attribute the ×2 shift in threshold voltage to the difference in the thickness of the membranes.
FIGURE 5.
(a) Gel arrays containing three horizontal lanes with fluorescent bands indicating 58 bp dsDNA found in the compartments housing the negative (−) and positive (+) electrodes in a bi-cell with a 2 nm diameter pore in a 10 nm thick membrane as shown in Fig. 1 b (along with a 100 bp ladder.) The 58 bp permeates the pore only for V > 2.75 V. (b) Similar to a but instead using a 1.8 × 2.2 nm2 cross-section pore in a 20 nm thick membrane shown in Fig. 1 d. The 58 bp permeates this pore only for V > 5.75V. (c) qPCR results obtained for the same pores showing the copy number versus voltage. A total of 622 bp dsDNA permeates the 1 nm pore for V > 2.5 V, and the 1.8 × 2.2 nm2 pore for V > 5 V.
To discover the microscopic origin of the field threshold for permeation, we carried out MD simulations of dsDNA translocations through a 2.0 nm diameter pore in both 10 and 20 nm thick membranes; the latter pore has an elliptic 1.8 × 2.2 nm2 cross section in correspondence with the experiment. Fig. 6 shows the position of the DNA leading edge relative to the center of the membrane against the simulation time; the symbols identify simulations carried out at different biases. For both pores, we observed a voltage threshold for permeation of dsDNA that agrees quantitatively with experiment. Although the average diameter of both pores is 2.0 nm, the cross section of the pore in the 20 nm thick membrane is an ellipse. The major axis of the ellipse determines the effective diameter of the pore for permeation of dsDNA. Therefore, the scaling of the threshold voltages with the membrane thickness is slightly sublinear.
FIGURE 6.
Simulated permeation of dsDNA through 2.0 nm diameter pores in 10- (blue) and 20- (yellow) nm thick Si3N4 membranes. The latter pore has an elliptic 1.8 × 2.2 nm2 cross section. The pore profiles are overlaid geometrically faithfully. The symbols indicate the location of the DNA leading edge as DNA permeates through the pore (from top to bottom).The voltage threshold for dsDNA permeation is between 2.6 and 3.2 V for the 10 nm thick membrane (blue) and between 4 and 5 V for the 20 nm thick one (yellow). Animations illustrating these MD simulations are available at http://www.ks.uiuc.edu/Research/nanopore/stretching/.
Fig. 7 illustrates a typical translocation observed at a voltage greater than threshold. Initially, dsDNA was driven into a pore by applying a 1.3 V bias, but the molecule halted when the pore narrowed to 2.5 nm in diameter; this conformation is shown in Fig. 7 a. Subsequently a 3.2 V bias was applied, which generated a force sufficient to stretch the double helix, enabling it to reach the center of the membrane as illustrated in Fig. 7 b. When passing by the narrowest part of the pore, the DNA ends unzip as shown in Fig. 7 c. The length of the unzipped fragment was found to vary from one simulation to the other, but typically was from 2 to 12 nucleotides. After both strands had passed through the pore constriction as shown in Fig. 7 d, the translocation reached a steady-state regime in which the DNA strand is stretched in the middle of the pore but relaxes outside of the membrane. Geometric restraints of the pore greatly reduce structural fluctuations destabilizing stretched DNA conformation (30), and so the pattern of hydrogen bonds is preserved when dsDNA permeates a 2.0 nm diameter pore.
FIGURE 7.
Stretching of a DNA helix by an electric field in a nanopore. The snapshots illustrate an MD simulation of a translocation through a 2.0 nm diameter pore in a 10 nm thick membrane driven by 3.2 V bias. (a) The starting conformation originated from a 6.4 ns simulation at 1.3 V bias; the DNA helix is only slightly stretched. A 3.2 V bias is applied. (b) After 1.6 ns, DNA stretches, reaching the center of the membrane. (c) DNA ends fray after ∼2.1 ns. (d) Both DNA ends pass through the pore constriction; the translocation is in a steady-state regime.
As each MD simulation is, in essence, a single molecule in silico experiment, large variations of the outcome due to changing starting conditions can be expected. To verify the reproducibility of our quantitative prediction about the voltage threshold for DNA translocation, we carried out three additional simulations with the 2.0 nm diameter pore in the 10 nm thick membrane at 2.6, 3.2, and 3.9 V using starting conformations that is different from the previous choices. The results of these test simulations are in agreement with our previous estimate of the translocation threshold, although the translocation trajectories differ significantly.
To further investigate the origin of this sharp voltage threshold, we plotted the electrostatic potential along the pore axis V(z) against the distance from the center of the membrane (z-z0) in a pore containing only electrolyte and no DNA for all simulation conditions studied (we have previously determined that taking DNA into account does not alter significantly the distribution of the electrostatic potential (15)). Scaling V(z) with the applied bias Vbias and (z-z0) with the membrane width Lmem converged all data to one universal curve, Fig. 8 a. We find that the expression V(z)/Vbias = 1/π × arctan(−b(z-z0)/Lmem) approximates the variation of the electrostatic potential across the membrane, where the geometrical factor b was found to vary from 8 to 12 at 1 M KCl. Fitting this function to all data points yielded b = 9.4. The scaled force acting on an elementary charge (q = 1.6 × 10−19) is plotted in Fig. 8 b. The force drops off very abruptly away from the membrane center, which explains why the threshold for dsDNA permeation is so sharp. Thus, at a 1.3 V bias, permeation of dsDNA halts when the pore narrows to 2.5 nm in diameter (Fig. 6), the force acting on the leading edge of DNA at that conformation is <20 pN. At 2.6 V, the location of the DNA leading edge is closer to the membrane center than at 1.3 V (Fig. 6); the force acting on the leading edge of DNA is ∼45 pN, still insufficient to stretch the helix. As the bias increases, the force eventually exceeds 60 pN, stretching DNA in the pore toward the membrane center. As the stretching proceeds, the force on the leading edge increases as abruptly so that once the DNA passes the threshold determined by the force acting on a leading edge of the molecule, it is unlikely to pull back from the pore.
FIGURE 8.
The transmembrane potential V(z) (a) and the force F(z) acting on a probe charge (b) against the distance from the membrane center. Both plots are in dimensionless units. The colors indicate different simulation conditions; solid lines represent the best fit of the analytical expressions (see text) to all data points. The inset in a illustrates the distribution of the electrostatic potential in a 1.4 nm diameter pore at a 1.3 V bias.
Influence of the pH on permeation threshold
Finally, to test the hypothesis that the origin of the sharp field threshold for permeation is due to the stretching transition, we destabilized the double helix by varying the pH. The stability of the double-helical structure of DNA is derived from hydrogen-bonding, base-stacking interactions and the shell of polarized water molecules that envelops it. To denature or melt the DNA, these forces have to be nullified. It has already been shown (1) that dsDNA denatures for pH > 12 or pH < 2 due to ionization of the bases that disrupts the normal Watson-Crick hydrogen bonding, and so we explored a range of pH from 3.0 to 8.5. Following Rouzina and Bloomfield (31,32), if dsDNA melts during the stretching transition, the force required should decrease as the helix is destabilized. And so, we tested the permeability as a function of both pH and voltage using the 1.8 × 2.2 nm2 in the 20 nm thick membrane shown in Fig. 1 d.
We varied the pH by adding 10 mM of potassium citrate (K3C6H5O7·H2O) and 10 mM of potassium phosphate (K2HPO4) in 1 M KCl to form a buffer of pH 4 and pH 3, respectively. After injecting dsDNA in the compartment housing the cathode, a voltage ranging 3–6 V was applied across the membrane for 4 h while the current through the pore was measured. Subsequently, the minute amount of the DNA near the anode that permeated the membrane was analyzed. The results of the test are summarized in Fig. 9. Notice that in the gel array shown in Fig. 9 a, which was obtained for dsDNA immersed in a pH 3 buffer, a fluorescent band is observed in the lane corresponding to dsDNA collected in the compartment housing the positive electrode, (+)58 bp, only for an applied bias of V > 4.0 V, indicating that 58 bp dsDNA can be forced through a 1.8 × 2.2 ± 0.2 nm pore in a 20 nm membrane if the applied voltage is V > 4.0 V. Similarly, Fig. 9 b shows results obtained in a pH 4 buffer, indicating that dsDNA can be forced through the same pore if the applied voltage is V > 5.0 V. And finally, to establish that the pore was not affected by the pH 3 or pH 4 buffers, we repeated the experiment at pH 8.5 to reproduce the results shown in Fig. 5 b. After the experiments at pH 3 and pH 4, the membrane with the same pore was immersed in a pH 8.5 buffer, and once again, we injected dsDNA into the compartment housing the cathode, applied a voltage ranging 3–6 V for 4 h, and measured (the same) electrolytic current through the pore. Subsequently, as shown in Fig. 9 c, we detected DNA near the anode only for V > 5.75 V, just as we did for the pristine pore represented by Fig. 5 b.
FIGURE 9.
(a) Gel array containing three horizontal lanes with fluorescent bands indicating 58 bp dsDNA found at pH 3 in the compartments housing the negative (−) and positive (+) electrodes in a bi-cell with a 1.8 × 2.2 nm2 pore in a 20 nm thick membrane as shown in Fig. 1 d (along with a 100 bp ladder.) The 58 bp permeates the pore only for V > 4.00 V for pH 3. (b) Similar to a but instead using pH 4. The 58 bp permeates this pore only for V > 5.0 V for pH 4. (c) Similar to a but instead using pH 8.5. The 58 bp permeates this pore only for V > 5.75 V for pH 8.5. (d) qPCR results obtained for the same pore showing the copy number versus voltage. A total of 622 bp dsDNA permeates the pore for V > 3.75 V for pH 3, V > 4.75 V for pH 4, and V > 5.0 V for pH 8.5.
We also measured the number of DNA copies translocating near threshold by qPCR. Fig. 9 d is a plot of the results of two qPCR analyses showing the number of copies permeating the same pore as a function of the applied potential associated with the pH 3 and pH 4 buffers. In the same figure, we have superimposed the data of Fig. 5 c, taken on the same pore in a pH 8.5 buffer for reference. In correspondence with the V = 4.0 V and 5.0 V thresholds measured for 58 bp at pH 3 and pH 4, respectively, using a gel array, qPCR indicates a threshold voltage of 3.75 V and 4.75 V for 622 bp dsDNA. Both of these values are shifted to a lower field relative to the data obtained at pH 8.5, which shows a threshold voltage of V = 5.0 V. The systematic shift of the critical field from a value of E = 3.1 (±0.7) MV/cm at pH 8.5 to E = 2.3 (±0.6) MV/cm at pH 3, corresponds to a decrease in the tensile force on an elementary charge from F = qE ∼ 50 pN to ∼37 ± 9 pN as the pH becomes more acidic.
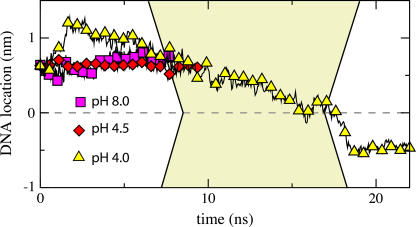
At low pH, atom N1 of the A nucleoside and atom N3 of C nucleoside are likely to accept a proton from the solution, neutralizing the charge of the entire nucleotide (1); the pKa values of these protonation sites are ∼3.8 and 4.5, respectively (1). The protonation of the nucleosides disrupts a delicate balance of the forces holding together the double helix; at very low pH, the helix denatures. At moderate pH (3–5), one can expect that only a fraction of A and C nucleosides is protonated, favoring the double helical organization of DNA but altering physical properties of the helix if compared to the pH 8.5 conditions. To investigate how these affect the electromechanics of DNA in a nanopore, we set up two additional MD simulations in which all or a half of all A nucleotides in our 58 bp helix were protonated. This effectively corresponds to a simulation carried out at pH 4 (20% of all nucleosides deprotonated) and pH 4.5 (10%), respectively. For the initial conformation of DNA we chose the final state from the 6.4 ns simulation in which a 1.3 V bias drove dsDNA into a 2.0 nm diameter pore (Fig. 3). We have previously established that, starting from this conformation, a 2.6 V bias was not sufficient to drive dsDNA through the pore at the pH 8 conditions (Fig. 6). After adding protons to the selected nucleosides, we resumed simulations at a 2.6 V bias. Initially, the helix adjusted its conformation in reaction to the change of the nucleoside protonation and partially retreated from the pore; only one hydrogen bond was observed between nucleotide T and the protonated nucleotide A. However, as indicated in Fig. 10, we observed that the leading edge of the DNA passes through the narrowest part of the pore at the pH 4 conditions, whereas at the pH 4.5 and pH 8 conditions, the translocation halted before reaching the pore constriction. Lowering the pH, and so lowering the overall charge of the helix, was observed to decrease the translocation velocity. At pH 4, the weakening of the hydrogen bond network due to the protonation of the A nucleotides was observed to induce unzipping of the DNA ends in the nanopore before reaching the center of the membrane. The unzipping, resulting from the destabilization of the DNA structure, is apparently the microscopic origin of the experimentally observed voltage threshold shift upon lowering the pH.
FIGURE 10.
MD simulation of DNA translocation through a 2.0 nm diameter pore at a 2.6 V bias. The pH values correspond to the percentage of protonated nucleotides of type A (see text). The DNA strand was found to permeate the pore only at pH 4.0. Animations illustrating these MD simulations are available at http://www.ks.uiuc.edu/Research/nanopore/stretching/.
CONCLUSIONS
In summary, we have explored the electromechanical properties of DNA, using an electric field to force dsDNA and ssDNA through synthetic pores, ranging from 1 to 3 nm in diameter, in ultrathin (10–20 nm thick) Si3N4 membranes. Although we have observed that ssDNA can permeate a 1 nm diameter pore even at low fields, E < 200 mV/10 nm, we find a threshold for permeation of dsDNA through pores with a diameter <3 nm that depends on the electric field and pH. For a 2 nm diameter pore, the threshold is ∼3.1 MV/cm at pH 8.5, corresponding to a force per nucleotide of ∼50 ± 11 pN. We attribute threshold field to the stretching transition observed in measurements of long, single molecules of dsDNA near 60 pN tension at pH = 8. The threshold field decreases as the pH becomes more acidic, consistent with the destabilization of the double helix and suggesting that dsDNA melts as it translocates through the pore.
Acknowledgments
We thank Dr. Susan Power of Cellular Molecular Technologies for assistance in conducting qPCR and Marcus Dittrich for carrying out GAMESS calculations.
We gratefully acknowledge the use of the Center for Microanalysis of Materials supported by a Department of Energy grant (DEFG02-91-ER45439), and the supercomputer time at the National Center for Supercomputer Applications provided through Large Resource Allocation Committee grants MCA93S028 and MCA05S028. This work was funded by grants from the National Science Foundation (0210843), the Air Force (FA9550-04-1-0214), the National Institutes of Health (R01-HG003713-01 and P41-PR05969).
References
- 1.Saenger, W. 1984. Principles of Nucleic Acid Structure. Springer-Verlag, New York.
- 2.Kasianowicz, J. J., E. Brandin, D. Branton, and D. W. Deamer. 1996. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA. 93:13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayley, H., and P. S. Cremer. 2001. Stochastic sensors inspired by biology. Nature. 413:226–230. [DOI] [PubMed] [Google Scholar]
- 4.Heng, J. B., C. Ho, T. Kim, R. Timp, A. Aksimentiev, Y. V. Grinkova, S. Sligar, K. Schulten, and G. Timp. 2004. Sizing DNA using a nanometer-diameter pore. Biophys. J. 87:2905–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akeson, M., D. Branton, J. J. Kasianowicz, E. Brandin, and D. W. Deamer. 1999. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 77:3227–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heng, J. B., A. Aksimentiev, C. Ho, V. Dimitrov, T. Sorsch, J. Miner, W. Mansfield, K. Schulten, and G. Timp. 2005. Beyond the gene chip. Bell Labs Technical Journal. 10:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, H., F. Kosari, G. Andreadakis, M. A. Alam, G. Vasmatzis, and R. Bashir. 2004. DNA-mediated fluctuations in ionic current through silicon oxide nanopore channels. Nano Lett. 4:1551–1556. [Google Scholar]
- 8.Henrickson, S. E., M. Misakian, B. Robertson, and J. J. Kasianowicz. 2000. Driven DNA transport into an asymmetric nanometer-scale pore. Phys. Rev. Lett. 85:3057–3060. [DOI] [PubMed] [Google Scholar]
- 9.Li, J., M. Gershow, D. Stein, E. Brandin, and J. A. Golovchenko. 2003. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2:611–615. [DOI] [PubMed] [Google Scholar]
- 10.Storm, A. J., J. H. Chen, H. W. Zandbergen, and C. Dekker. 2005. Translocation of double-strand DNA through a silicon oxide nanopore. Phys. Rev. E. 71:051903–051912. [DOI] [PubMed] [Google Scholar]
- 11.Ambjörnsson, T., S. P. Apell, Z. Konkoli, E. A. DiMarzio, and J. J. Kasianowicz. 2002. Charged polymer membrane translocation. J. Chem. Phys. 117:4063–4073. [Google Scholar]
- 12.Bustamante, C., S. B. Smith, J. Liphardt, and D. Smith. 2000. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 10:279–285. [DOI] [PubMed] [Google Scholar]
- 13.Cluzel-Schaumann, H., M. Rief, C. Tolksdorf, and H. E. Gaub. 2000. Mechanical stability of single DNA molecules. Biophys. J. 78:1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith, S. B., Y. Cui, and C. Bustamante. 1996. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 271:795–799. [DOI] [PubMed] [Google Scholar]
- 15.Heng, J. B., A. Aksimentiev, C. Ho, P. Marks, Y. V. Grinkova, S. Sligar, K. Schulten, and G. Timp. 2005. Stretching DNA using the electric field in a synthetic nanopore. Nano Lett. 5:1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho, C., R. Qiao, J. B. Heng, A. Chatterjee, R. Timp, N. R. Aluru, and G. Timp. 2005. Electrolytic transport through a synthetic nanometer-diameter pore. Proc. Natl. Acad. Sci. USA. 102:10445–10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aksimentiev, A., J. B. Heng, G. Timp, and K. Schulten. 2004. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys. J. 87:2086–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polygen. 1988. Quanta Polygen Corporation, Waltham, MA.
- 19.Kalé, L., R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan, and K. Schulten. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151:283–312. [Google Scholar]
- 20.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- 21.Wendel, J. A., and W. A. Goddard. 1992. The Hessian biased force-field for silicon nitride ceramics: predictions of the thermodynamic and mechanical properties for a- and b-Si3N4. J. Chem. Phys. 97:5048–5062. [Google Scholar]
- 22.Xu, D., J. C. Phillips, and K. Schulten. 1996. Protein response to external electric fields: relaxation, hysteresis and echo. J. Phys. Chem. 100:12108–12121. [Google Scholar]
- 23.Schmidt, M. W., K. K. Baldridge, J. A. Boatz, S. Elbert, M. Gordon, J. H. Jensen, S. Koseki, N. Matsunaga, K. A. Nguyen, S. Shujun, T. Windus, M. Dupuis, and J. A. Montgomery. 1993. General atomic and molecular electronic structure system. J. Comput. Chem. 14:1347–1363. [Google Scholar]
- 24.Bayly, C. I., P. Cieplak, W. D. Cornell, and P. A. Kollman. 1993. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97:10269–10280. [Google Scholar]
- 25.Szabò, I., G. Bàthori, F. Tombola, A. Coppola, I. Schmehl, M. Brini, A. Ghazi, V. De Pinto, and M. Zoratti. 1998. Double-stranded DNA can be translocated across a planar bilayer containing purified mitochondrial porin. FASEB J. 12:495–502. [DOI] [PubMed] [Google Scholar]
- 26.Storm, A. J., C. Storm, J. Chen, H. Zandbergen, J.-F. Joanny, and C. Dekker. 2005. Fast DNA translocation through a solid-state nanopore. Nano Lett. 5:1193–1197. [DOI] [PubMed] [Google Scholar]
- 27.Mathe, J., A. Aksimentiev, D. R. Nelson, K. Schulten, and A. Meller. 2005. Orientation discrimination of single-stranded DNA inside the α-hemolysin membrane channel. Proc. Natl. Acad. Sci. USA. 102:12377–12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meller, A., L. Nivon, and D. Branton. 2001. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 86:3435–3438. [DOI] [PubMed] [Google Scholar]
- 29.Yeh, I. C., and G. Hummer. 2004. Diffusion and electrophoretic mobility of single-stranded RNA from molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 101:12177–12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris, S. A., Z. A. Sands, and C. A. Laughton. 2005. Molecular dynamics simulations of duplex stretching reveal the importance of entropy in determining the biomechanical properties of DNA. Biophys. J. 88:1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, M. C., J. R. Wenner, I. Rouzina, and V. A. Bloomfield. 2001. The effect of pH on the overstretching transition of dsDNA: evidence of force-induced DNA melting. Biophys. J. 80:874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouzina, I., and V. A. Bloomfield. 2001. Force-induced melting of the DNA double helix. II. Effect of solution conditions. Biophys. J. 80:894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]