Abstract
The cytoplasmic trafficking of the prototype strain of minute virus of mice (MVMp) was investigated by analyzing and quantifying the effect of drugs that reduce or abolish specific cellular functions on the accumulation of viral macromolecules. With this strategy, it was found that a low endosomal pH is required for the infection, since bafilomycin A1 and chloroquine, two pH-interfering drugs, were similarly active against MVMp. Disruption of the endosomal network by brefeldin A interfered with MVMp infection, indicating that viral particles are routed farther than the early endocytic compartment. Pulse experiments with endosome-interfering drugs showed that the bulk of MVMp particles remained in the endosomal compartment for several hours before its release to the cytosol. Drugs that block the activity of the proteasome by different mechanisms, such as MG132, lactacystin, and epoxomicin, all strongly blocked MVMp infection. Pulse experiments with the proteasome inhibitor MG132 indicated that MVMp interacts with cellular proteasomes after endosomal escape. The chymotrypsin-like but not the trypsin-like activity of the proteasome is required for the infection, since the chymotrypsin inhibitors N-tosyl-l-phenylalanine chloromethyl ketone and aclarubicin were both effective in blocking MVMp infection. However, the trypsin inhibitor Nα-p-tosyl-l-lysine chloromethyl ketone had no effect. These results suggest that the ubiquitin-proteasome pathway plays an essential role in the MVMp life cycle, probably assisting at the stages of capsid disassembly and/or nuclear translocation.
Minute virus of mice (MVM) is a member of the genus Parvovirus of the family Parvoviridae. MVM is a small, nonenveloped, icosahedral virus that replicates in the nucleus of actively dividing cells (10). The genome is linear, single stranded, and approximately 5 kb long and is organized into two overlapping transcription units. The left gene, driven by the P4 promoter, encodes the nonstructural proteins NS1 and NS2, and the right gene, driven by the P38 promoter, encodes the structural proteins VP1 and VP2 (8). During the infection process and in DNA-containing particles, most VP2 molecules are cleaved to VP3 by the removal of some amino acids from the N terminus (9, 41, 58). Although this cleavage can be mimicked in vitro with trypsin, it seems that the in vitro tryptic cleavage site does not correspond to the natural cleavage site in MVM (58), and it is not essential for the proteolytic processing of VP2 into VP3 (61).
The process of penetration for many enveloped viruses has been quite well characterized (27, 29, 70). In contrast, the mechanisms of viral entry of nonenveloped viruses into the cytoplasm and nuclear targeting of the virus or its genome prior to replication are still poorly understood. However, the use of certain members of the parvovirus family as vectors for gene therapy has prompted research in this field. A common uptake principle of these viruses involves receptor-mediated endocytosis; however, cell surface attachment differs among parvoviruses. MVM has been reported to use sialic acid moieties of cell surface glycoproteins as the receptor (10). Canine parvoviruses (CPV) and feline parvoviruses use the transferrin receptor (43). For B19, one glycolipid, globotetraosylceramide, or globocide is the receptor (6, 7). Bovine parvovirus binds to sialated erythrocyte membrane glycoproteins and attaches to the major membrane glyprotein, glycophorin A (60). Adeno-associated viruses (AAVs), of the Dependovirus genus of parvoviruses, were reported to employ membrane-associated heparan sulfate proteoglycans as cellular receptors (AAV-2) (56) or sialic acid (AAV-4 and -5) (25, 68). Acidification is known to be essential for the entry of AAV and CPV, since drugs that interfere with the endosomal pH are able to block the infection (1, 2, 15, 42, 65). The infectious entry of CPV could also be blocked by the disruption of microtubules and by low temperatures, suggesting the involvement of microtubule-dependent transport (65). Functional microtubules and microfilaments are also needed for AAV translocation to the nucleus (47).
The release of these viruses may be directly linked to the acidification of the vesicle. However, the exact mechanism and time course of this release from the endosomal compartments remain unclear. Sequence analysis of the VP1 revealed phospholipase A2 motifs in the capsid proteins of parvoviruses, an activity that was not known to exist in viruses and that might be responsible for parvovirus entry (14, 32, 72). The mechanism and time course by which the viral particles, once released into the cytoplasm, translocate to the nucleus is not known. Nuclear localization signals present in VP1 and VP2 sequences (62) are thought to be essential for the transport of the intact or partially uncoated capsids to the nucleus for viral replication (64, 67). Interaction between proteasomes and AAV has been previously reported and suggested to occur after endosomal escape (15, 16, 71). In these reports, a significant enhancement of recombinant AAV transduction was observed when the proteasome activity was reduced or abolished by a proteasome inhibitor coadministered with the virus. In addition, it was found that AAV-2 and -5 capsids are ubiquitinated during the infection, a process that was suggested to be enhanced by the endosomal maturation of the viral capsids. These observations gave rise to the hypothesis that the cellular proteasomes would represent an obstacle for AAV infections.
In the present studies we have investigated different aspects of the cytoplasmic trafficking of particles of the prototype strain of MVM (MVMp) with special regard to endosomal trafficking and proteasome interaction. The results indicate that MVMp uptake involves the endocytic pathway in which acidification and several endosomal vesicles are required. MVMp capsids interact with cellular proteasomes after endosomal escape. In this interaction, the chymotrypsin-like but not the trypsin-like activity of the proteasome plays a crucial role. We hypothesize that the interaction of incoming MVMp particles with the cellular proteasomes represents a natural feature of the life cycle of the virus, most probably linked to capsid disassembly and/or nuclear translocation.
MATERIALS AND METHODS
Cells and viruses.
Mouse A9 fibroblast cells (34), previously described as a host of MVMp (59), were propagated in Dulbecco's modified Eagle's medium supplemented with 10% of heat-inactivated fetal bovine serum. Stocks of MVMp were propagated on A9 cell monolayers, titrated by using a standard 50% tissue culture infective dose technique, and stored at −70°C.
Chemicals.
Brefeldin A (BFA), bafilomycin A1 (BA), chloroquine, nocodazole (ND), cytochalasin D (CD), N-tosyl-l-phenylalanine chloromethyl ketone (TPCK), Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK), aclarubicin, and lactacystin were purchased from Sigma (St. Louis, Mo.), while MG132, epoxomicin, and PR39 were obtained from Calbiochem-Novabiochem (La Jolla, Calif.). Dimethyl sulfoxide was used to dissolve lactacystin, MG132, epoxomicin, CD, ND, and TPCK. TLCK, PR39, and chloroquine were dissolved in water and aclarubicin; BFA and BA were dissolved in ethanol. All drugs were stored at −20°C.
Viral DNA replication kinetics.
Quantification of viral DNA through 24 h postinfection was performed in an attempt to determine the time point at which there is a maximal viral DNA accumulation in a minimal period of time and that would ultimately be used as the end point for quantification with the drug assays. A9 cells were seeded in 12-well plates at 105 cells per well in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and were incubated at 37°C in a 10% CO2 atmosphere. Twenty-four hours later, cells were infected at a multiplicity of infection (MOI) of 10 infectious particles/cell for 1 h at 4°C. The cells were subsequently washed to remove unbound virus and were further incubated until 24 h. At 2-h intervals, cells were trypsinized and collected by low-speed centrifugation. Total DNA was extracted from the cell pellet by using the DNAeasy tissue kit (Qiagen, Valencia, Calif.) and was quantified by real-time PCR.
Drug treatments.
A9 cells were propagated and infected as specified above. The infection was performed in the presence of the drugs followed by washing to remove unbound virus and was further incubated for 3 h with the reversible drugs BFA, BA, chloroquine, and MG132; for 1 h with the irreversible drugs lactacystin, epoxomicin, TPCK, and TLCK; and constantly with the reversible drugs aclarubicin, PR39, ND, and CD. When the incubation with the drugs was finished, the cells were washed with Dulbecco's modified Eagle's medium and were further incubated at 37°C until 18 h. In the case of the cytoskeleton-disrupting agents ND and CD, cells were pretreated with the drugs 1 h before the infection. At 18 h postinfection, cells were trypsinized and collected by low-speed centrifugation. Total DNA was extracted as indicated above. Control cells were similarly infected with MVMp but did not receive any inhibitor; instead, they were treated with dimethyl sulfoxide, ethanol, or water, depending on the dissolvent used. Negative controls were included that were not infected or treated with any compound.
Pulse treatments.
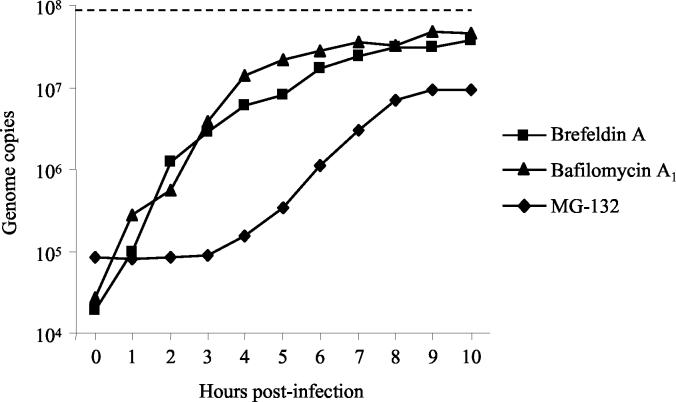
The time course of MVMp endosomal trafficking was examined for the bulk of internalized viral particles by performance of a postinfection pulse treatment with the endosome-interfering drugs BFA and BA. In addition, MG132 was used to determine the period during which MVMp interacts with cellular proteasomes. A9 cells were infected with MVMp at an MOI of 10 for 1 h at 4°C to allow viral attachment but not internalization, followed by washing to remove unbound virus, and were subsequently incubated at 37°C. At progressive postinfection times, as described in the Fig. 3 legend, BFA (5 μg/ml), BA (150 nM), or MG132 (25 μM) was added to the medium. The cells were incubated until 18 h postinfection and collected, and their total DNA was extracted as specified above.
FIG. 3.
Pulse experiments with endosome- and proteasome-interfering drugs. A9 cells were infected with MVMp at an MOI of 10 for 1 h at 4°C to allow viral attachment but not internalization, followed by washing to remove unbound virus, and were further incubated at 37°C. At 1-h intervals from 0 to 10 h postinfection, BFA (5 μg/ml), BA (150 nM), or MG132 (25 μM) was added to the medium. Control cells were not treated with any drug. The cells were incubated until 18 h postinfection and trypsinized, and total DNA was extracted and quantified as described in Materials and Methods. The MVM DNA content in nontreated cells is represented by a dotted line. Values represent the mean of three independent experiments.
Viral binding and internalization assay.
A possible effect of the drugs on virus internalization was assessed. A9 cells were infected at an MOI of 10 for 1 h at 4°C in the presence of the highest doses of the different drugs. Cells were then washed two times to remove unbound virus and were then incubated in the presence of the drugs at 37°C for 2 h to allow internalization. Subsequently, cells were trypsinized and collected by low-speed centrifugation. The cell pellet from each well was resuspended in 500 μl of trypsin-EDTA solution and was further incubated at 37°C for 10 min to remove viral particles that had not internalized. The cells were pelleted and washed three times with Dulbecco's modified Eagle's medium. Total DNA was extracted from the cell pellet as previously described.
Cell DNA synthesis assay.
A potential effect of the drugs on cell DNA synthesis was investigated. A9 cells were seeded in 12-well plates at 105 cells per well in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and were incubated at 37°C in a 10% CO2 atmosphere. Twenty-four hours later, the cells were treated with the highest doses of the different drugs as specified above and were incubated at 37°C. At 0 and 18 h of incubation, cellular DNA was measured by quantifying the copy numbers of the mouse β-actin gene (GenBank accession no. M12481) by real-time PCR. Primers used were as follows: forward primer, 5′-TGCTGTC CCTGTATGCCTCTG-3′, and reverse primer, 5′-AATGCCTGGGTACATGGTGGT-3′.
Real-time PCR.
Quantitative PCR was used to detect and quantify viral DNA and NS1 transcript (cDNA) sequences as well as the cellular DNA content. Primers for MVM DNA amplification were as follows: forward, 5′-GACGCACAGAAAGAGAGTAACCAA-3′ (nucleotide positions, 231 to 254); and reverse, 5′-CCAACCATCTGCTCCAGTAAACAT-3′ (nucleotide positions, 709 to 732). Primers for cDNA amplification were as follows: forward, 5′-AAATGGGGCAAAGTTCCTGAT-3′ (nucleotide positions, 1917 to 1937); and reverse, 5′-CCGATGCAAGTGGAGTTAGTG-3′ (nucleotide positions, 2067 to 2047). cDNA synthesis was performed with the reverse primer.
Amplification and real-time detection of PCR products were performed on the DNA and cDNA samples using the Lightcycler system (Roche Diagnostics, Rotkreuz, Switzerland) with SYBR Green (Roche). The fluorescent dye SYBR Green I binds to double-stranded DNA. At the end of the extension step of every cycle, the fluorescence was measured. The cycle number at which the fluorescence starts to increase is related to the initial number of target copies. A melting-curve analysis was performed for specific product identification. PCR was performed using the FastStart DNA SYBR Green kit (Roche) following the manufacturer's instructions. Cycling conditions consisted of a step at 95°C for 10 min to activate the polymerase enzyme followed by 35 cycles with the following thermal profile: 94°C and 15 s, 66°C and 5 s, and 72°C and 30 s. As external standards, the PCR products generated by the above primers were cloned into the pGEM-T vector (Promega, Madison, Wis.) and were used in 10-fold dilutions. The amount of input cells used for every sample was standardized by determining the exact cellular DNA content. This was achieved by quantifying the copy numbers of the mouse β-actin gene as previously described.
Detection of viral structural proteins.
A9 cells (n = 105) were infected and treated with drugs as previously described. At 18 h postinfection, cells were collected and were subsequently lysed in protein-loading buffer. Proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. After transfer to a polyvinylidene difluoride membrane, the blot was probed with a mouse anti-VP2 antibody (1:2,000 dilution; kindly provided by J. Almendral), followed by a horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; Dako Diagnostics, Zug, Switzerland). The viral structural proteins were visualized with a chemiluminescence system (Pierce, Rockford, Ill.).
RESULTS
Viral DNA replication kinetics.
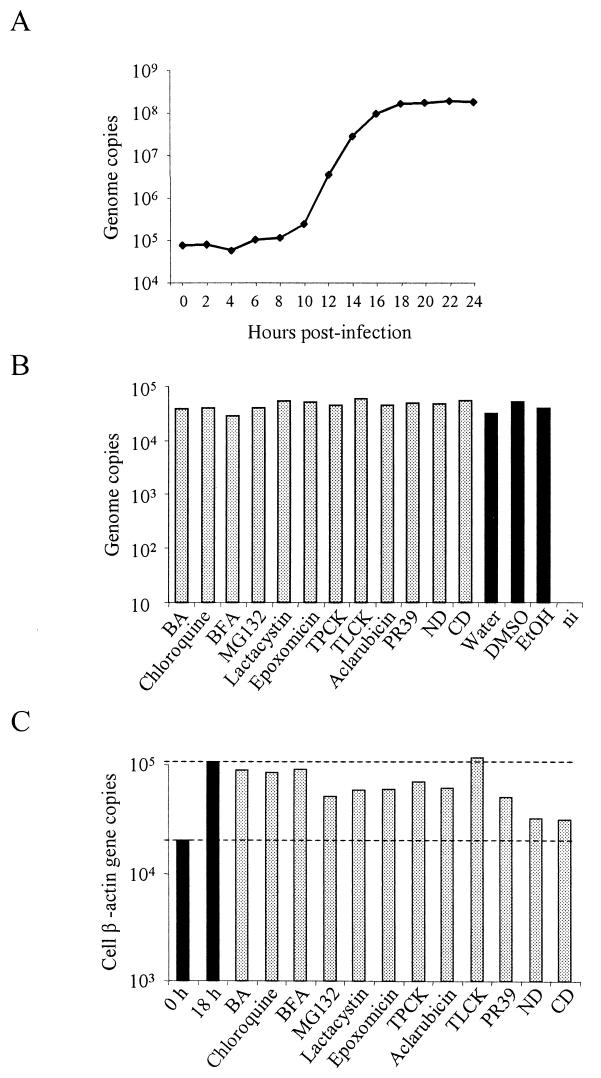
As shown in Fig. 1A, viral DNA started to accumulate in the nucleus by 8 to 10 h. Between 10 and 14 h the accumulation was exponential and reached a plateau by 18 h postinfection.
FIG.1.
(A) MVM DNA replication kinetics. A9 cells were infected with MVMp at an MOI of 10 for 1 h at 4°C, followed by washing to remove unbound virus, and were further incubated at 37°C. At 2-h intervals, total DNA was extracted and quantified as described in Materials and Methods. (B) Viral binding and internalization assay. A9 cells were infected in the presence of the highest doses of the different drugs. Cells were then washed and incubated in the presence of the drugs at 37°C for 2 h to allow internalization. Subsequently, cells were collected and were further incubated in a trypsin-EDTA solution to remove viral particles that have not internalized. The cells were pelleted, and after final washings, total DNA was extracted and MVM DNA was quantified by real-time PCR. DMSO, dimethyl sulfoxide. (C) Cell DNA synthesis assay. A9 cells were treated with the highest doses of the different drugs and were incubated at 37°C. At 0 and 18 h of incubation, cellular DNA was measured by quantifying the copy numbers of the mouse β-actin gene (GenBank accession no. M12481) by real-time PCR. Values represent the mean of three (two for panels B and C) independent experiments. ni, noninfected.
MVMp requires low endosomal pH.
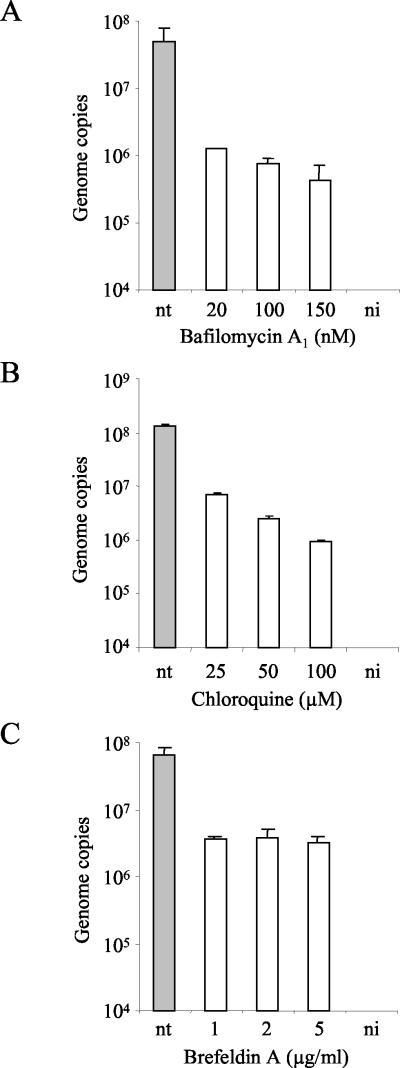
Acidification inside the endosomes is required by certain viruses to escape from this compartment and enter the cytosol. To investigate a requirement of MVMp for low endosomal pH, cells were treated with BA, a potent and selective inhibitor of vacuolar ATPases (3, 5, 20), or chloroquine, a lysosomotropic weak base, which raises the pH of intracellular compartments (52). A9 cells were incubated with increasing doses of BA or chloroquine in the medium for 4 h as described above. Treatment with BA caused a dose-dependent reduction in the accumulation of viral DNA (Fig. 2A). At 20, 100, and 150 nM concentrations, the amounts of viral DNA were 39-, 64-, and 114-fold smaller than in untreated cells, respectively. When applied at the time of the infection and maintained in the medium, the effect of BA was much more evident (Fig. 3). Viral transcription (NS1 transcripts) and expression of structural proteins were also affected by treatments with BA (see Fig. 7A and B). Treatment with chloroquine similarly reduced viral DNA accumulation in a dose-dependent manner (Fig. 2B). At 25, 50, and 100 μM concentrations the amounts of accumulated viral DNA were 19-, 54-, and 140-fold smaller than in untreated cells, respectively. Therefore, the low pH that MVMp encounters inside the endosomes plays an important role in the course of the infection.
FIG. 2.
Sensitivity of MVMp to treatment of cells with BA, chloroquine, or BFA. A9 cells were infected for 1 h at 37°C with MVMp at an MOI of 10 in the presence of increasing doses of BA, chloroquine, or BFA. Subsequently, cells were washed to remove an excess of virus and were incubated at 37°C for an additional 3 h in the presence of the inhibitors. Cells were washed to remove the drug and were further incubated. The amount of viral DNA was quantified 18 h postinfection. Values represent the mean of three independent experiments. nt, nontreated; ni, noninfected.
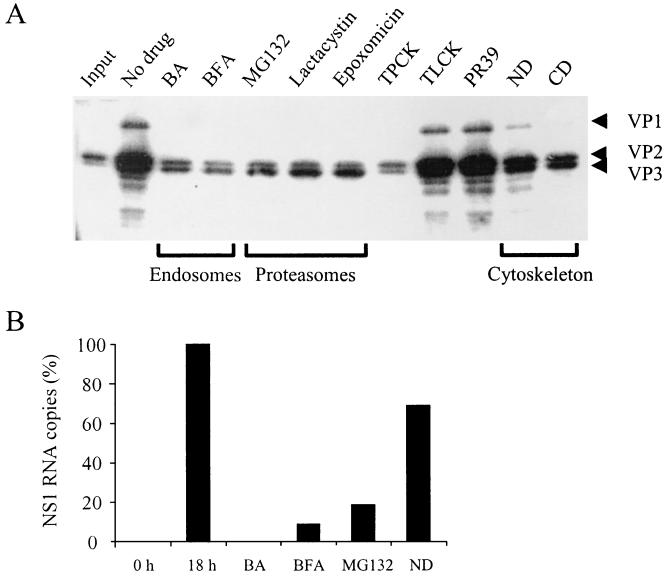
FIG. 7.
(A) Effect of drugs on viral structural protein synthesis. A9 cells were infected and treated with drugs as described in Materials and Methods. At 18 h postinfection, cells were collected and lysed in protein-loading buffer. Proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. After transfer to a polyvinylidene difluoride membrane, the blot was probed with a mouse anti-VP2 antibody followed by a horseradish peroxidase-conjugated secondary antibody. (B) Effect of some representative drugs on NS1 transcription. Values represent the mean of two independent experiments.
MVMp particles are routed toward late elements of the endosomal pathway.
Viruses that enter by endocytosis can be directly released to the cytosol or routed to other endosomal vesicles (26). To examine whether MVMp particles require further vesicles within the endosomal pathway, A9 cells were treated with BFA, a fungal antibiotic that causes the tubulation of the endosomal system blocking the transition between early and late endosomes (33). A9 cells were incubated with increasing doses of BFA in the medium for 4 h as described above. Treatment with BFA led to a decrease in the accumulation of viral DNA up to 21-fold at 5 μg/ml (Fig. 2C). As in BA-treated cells, when BFA was applied at the time of the infection and maintained in the medium, the effect on viral DNA accumulation was much more intense (Fig. 3). Treatments with BFA were also effective in blocking NS1 transcription and expression of MVM structural proteins (see Fig. 7A and B). These results indicated that MVMp particles are routed toward the late elements of the endosomal pathway.
Slow endosomal trafficking of the bulk of MVMp particles.
In order to study the time course of MVMp endosomal trafficking for the bulk of internalized viral particles, cells were infected and at different times postinfection the endosome-interfering drugs BFA and BA were added as previously described. The activity of BFA and BA against MVMp followed a similar pattern (Fig. 3). Already 1 h after internalization, the activity of the two endosome-blocking drugs decreased, indicating that a small proportion of MVMp particles had already been released to the cytosol. However, it was only after 7 to 8 h postinfection that the endosome-blocking drugs no longer had a significant effect on viral DNA accumulation, indicating that endosomal trafficking for the bulk of viral particles was concluded by that time.
Cytoskeleton-dependent transport of MVMp.
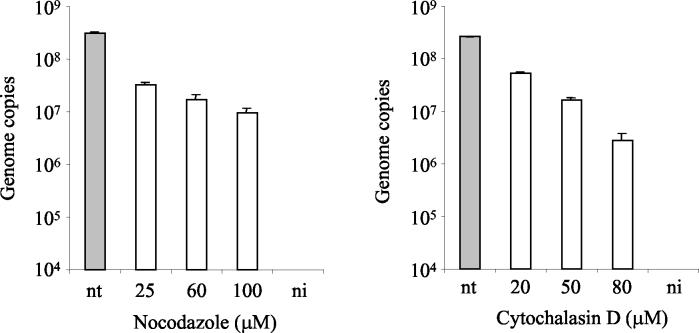
To determine whether transport of MVMp through the cytoplasm towards the nucleus requires an intact cytoskeleton network, A9 cells were treated with increasing amounts of ND and CD, as described above. ND has highly specific antimicrotubular activity for mammalian cells in culture, promoting tubulin depolymerization, and CD inhibits actin filament function. Both compounds have been widely used to study the cytoskeleton-virus interaction (4, 24, 46, 53, 65). When added together with MVMp, ND and CD had a moderate decreasing effect on viral DNA accumulation (Fig. 4). This observation implies that MVMp intracellular movement depends on a functional cytoskeleton and that both, microtubules and microfilaments, are required. Interestingly, the blocking effect of the cytoskeleton-disrupting drugs, kept in the medium during the infection, was clearly inferior to that obtained with BFA and BA when these drugs were also maintained in the medium during the infection (Fig. 3). Similarly, synthesis of viral structural proteins was moderately affected by treatments with ND and CD (see Fig. 7A). For ND-treated cells, transcription was also investigated. The result showed likewise a moderate decrease in the level of NS1 transcripts (see Fig. 7B).
FIG. 4.
Effect of cytoskeleton-disrupting agents on MVMp infection. A9 cells were pretreated with ND or CD for 1 h at 37°C. Subsequently, the cells were infected with MVMp for 1 h at 37°C at an MOI of 10 in the presence of ND or CD. Cells were washed to remove unbound virus and were further incubated at 37°C in the presence of the inhibitors. The amount of viral DNA was quantified 18 h postinfection. Values represent the mean of three independent experiments. nt, nontreated; ni, noninfected.
MVMp infection requires functional proteasomes.
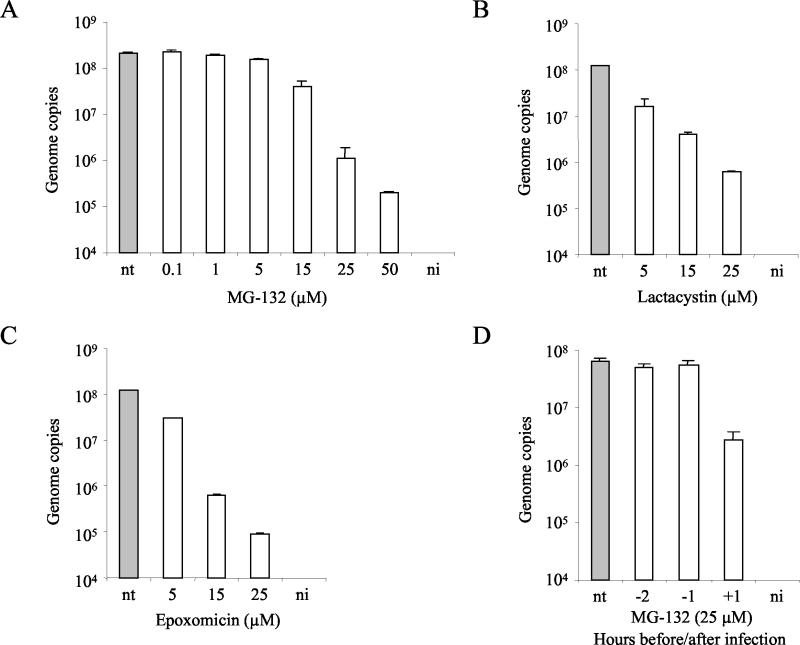
Proteasomes are large multicatalytic proteinase complexes implicated in the degradation of most cellular proteins (39). Proteasomes are involved in major histocompatibility complex class I antigen presentation (11, 30), and degradation of incoming virus by the ubiquitin-proteasome machinery has been previously described for AAV (15, 16, 50, 71). Hence, the possibility that cellular proteasomes could also be a degradative factor for MVMp virions was investigated. Cells were treated with increasing doses of the peptide aldehyde MG132, a potent and reversible inhibitor of the chymotrypsin-like activity of the proteasome (31) as indicated above. Interestingly, the blockage of cellular proteasome activity caused a remarkable dose-dependent reduction of viral DNA accumulation that reached 1,056-fold at 50 μM (Fig. 5A). Furthermore, viral protein synthesis and NS1 transcription were also affected by treatments with MG132 (see Fig. 7A and B).
FIG. 5.
Sensitivity of MVMp to proteasome inhibitors. (A to C) Cells were infected with MVMp for 1 h at 37°C at an MOI of 10 in the presence of increasing doses of MG132, lactacystin, or epoxomicin. Subsequently, cells were washed to remove unbound virus particles and were incubated at 37°C for an additional 1 h (3 h for MG132) in the presence of the inhibitors. Cells were washed and were further incubated at 37°C. (D) A9 cells were treated for 2 h with MG132 (25 μM), 2 and 1 h before and 1 h after virus exposure. Cells were washed to remove the drug and were further incubated at 37°C. The amount of viral DNA was quantified 18 h postinfection. Values represent the mean of three independent experiments. nt, nontreated; ni, noninfected.
With the aim of ruling out a major alteration on A9 cells caused by the proteasome inhibitor and because the action of MG132 on the proteasome can be fully reversed within 1 h after the withdrawal of the compound (11, 40), a 2-h incubation with MG132 at 2 and 1 h before infection and 1 h after virus exposure was performed. MVMp was only sensitive if the drug was applied after the infection and not before (Fig. 5D).
To further confirm these results, other proteasome inhibitors with different mechanisms of action were used as well. Lactacystin, which inhibits the three best-characterized peptidase activities of the proteasome, i.e., trypsin-like, chymotrypsin-like, and peptidyl glutamyl-peptide-hydrolyzing activities (17), and epoxomicin, which is a potent specific inhibitor of the chymotrypsin-like activity of the proteasome (37), were added at the time of infection and were kept in the medium for 2 h, as described in the Fig. 5 legend. The inhibition of the proteasome activity by lactacystin or epoxomicin caused a large dose-dependent reduction in viral DNA accumulation that at 25 μM reached 198- and 1,302-fold, respectively (Fig. 5B and C). In addition, MVM structural protein synthesis was also disturbed by treatments with lactacystin or epoxomicin (see Fig. 7A).
MVMp interacts with proteasomes after endosomal escape.
With the aim of defining the period at which MVMp particles interact with the cellular proteasomes, A9 cells were infected and, at different times postinfection, MG132 was added as specified in the Fig. 3 legend. The results showed that, while the endosome-interfering drugs BFA and BA already lost activity during the first hours postinfection, the proteasome inhibitor had full interfering activity during the first 3 h postinfection (Fig. 3), indicating that, during this period in which endosomal trafficking is in progress, there was no significant interaction of MVMp with the proteasomes. From 3 to 9 h postinfection the inhibitory activity of MG132 declined from 1,120- to 10-fold and reached a plateau, suggesting that by 9 h postinfection most of the viral particles had already interacted with the cellular proteasomes.
The chymotrypsin-like but not the trypsin-like activity of the proteasome is necessary for MVMp infection.
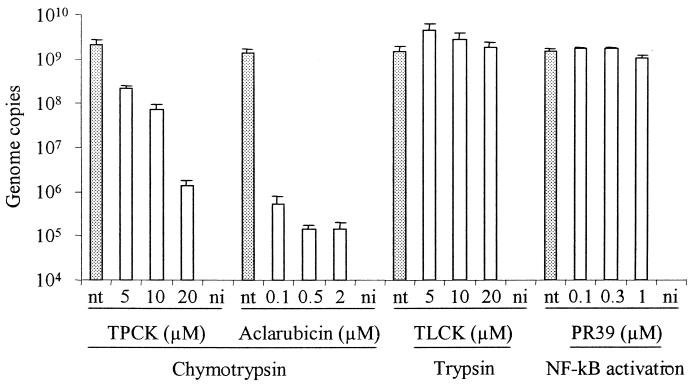
The three peptidase activities of the proteasome are differentially blocked by the proteasome inhibitors used in this study. Lactacystin inhibits the three proteolytic activities (17), and MG132 mainly inhibits chymotrypsin-like activity (31), while epoxomicin almost exclusively inhibits chymotrypsin-like activity (37). With the purpose to identify which activity or activities of the proteasome are required for MVMp infection, the chymotrypsin-like and the trypsin-like activities, proteasomal and nonproteasomal, were specifically inhibited with TPCK and TLCK, respectively, as specified above. The results showed that, while the trypsin inhibitor TLCK had no effect on viral DNA accumulation, a large dose-dependent decrease was observed with the chymotrypsin inhibitor TPCK, reaching 1,352-fold at 20 μM (Fig. 6). Because the inhibitory action of TPCK is not limited to the proteasome, aclarubicin (also known as aclacinomycin), a nonpeptidic compound isolated from Streptomyces that specifically and reversibly inhibits the chymotrypsin activity of the 20S proteasome (18) was used. Aclarubicin turned out to be the most active compound against MVMp, reaching at 2 μM a 9,915-fold reduction of viral DNA accumulation (Fig. 6). The inhibitory effect of aclarubicin was also evident when it was present at 0.5 μM for only the first 4 h of the infection. In this case MVM DNA nuclear accumulation was 1,846-fold lower than in untreated cells (data not shown).
FIG. 6.
Effect of TPCK, TLCK, aclarubicin, and PR39 on MVMp infection. A9 cells were infected for 1 h at 37°C with MVMp at an MOI of 10 in the presence of increasing doses of TPCK, TLCK, aclarubicin, or PR39. Subsequently, cells were washed and incubated at 37°C for an additional 1 h in the presence of the irreversible inhibitors TPCK and TLCK or constantly in the case of the reversible drugs aclarubicin and PR39. Cells were then washed, and the amount of viral DNA was quantified 18 h postinfection. Values represent the mean of three independent experiments. nt, nontreated; ni, noninfected.
It is known that proteasome inhibitors block activation of NF-κB, a transcription factor required for the expression of many genes (40). Expression of the viral gene encoding NS1 protein is required for MVM DNA replication. Hence, a possible effect of NF-κB inhibition on viral DNA replication was explored. NF-κB activation was specifically inhibited with PR39, which does not affect the overall proteasome activity (21). MVMp was not affected by treatment of cells with PR39 at any dose (Fig. 6).
Accordingly with these results, viral protein synthesis was also blocked by treatments with TPCK, while TLCK and PR39 had no effect (Fig. 7A).
Binding and internalization assay.
The virus binding and internalization process was investigated in the presence or absence of drugs. A9 cells were infected in the presence of the highest doses of the different drugs as previously described. The results showed that the process of binding and internalization of the virus was not disturbed in the presence of the drugs (Fig. 1B).
Cell DNA synthesis assay.
The effects of drugs on cell DNA proliferation were assessed. As shown in Fig. 1C, the drugs that were more active against cell DNA synthesis were ND and CD. Interestingly, these drugs had only a limited effect on MVM infection, as judged by viral DNA, RNA, and protein synthesis.
DISCUSSION
Drugs that block specific cellular functions and their effect on virus infection have been extensively studied to elucidate different aspects of the infection mechanisms of viruses, including members of the Parvoviridae family (1, 2, 15, 42, 47, 65). Most of these approaches use viral expression of reporter enzymes, viral infectivity, and/or microscopy as the end point. Care has to be taken to avoid major cell damage or interaction of the drug with the reporter molecule itself. Recently, it has been observed that treatment of cells with proteasome inhibitors interferes with luciferase and beta-galactosidase reporter assays (13). Ideally, the method used should be highly sensitive and performed early after the infection. This would allow the use of low doses of the drug and during short periods of time, thus minimizing cell damage due to the inhibition of essential cellular functions. In our studies, the nuclear accumulation of the viral DNA was quantified 18 h postinfection, at a time point when the viral DNA accumulation reached a plateau (Fig. 1A). If a given compound disturbs or totally blocks the viral nuclear translocation, the replication and accumulation of the viral DNA in the nucleus will be delayed or abolished. Given that the reporter molecule is the viral DNA itself, it has the possibility to be accurately quantified by real-time PCR. Since MVM DNA replication is only possible during the S phase of the cell cycle, cell DNA synthesis was assessed by the exact quantification of β-actin gene copies in the presence of maximum doses of the different drugs. The different compounds did not significantly inhibit cellular DNA synthesis, the only exceptions being ND and CD (Fig. 1C). Interestingly, the blocking effect of the cytoskeleton-disrupting agents on MVM DNA replication was only limited.
One of the questions addressed was whether MVMp requires low endosomal pH, as has been previously shown for other parvoviruses (1, 2, 15, 42, 65). To answer this question, the effect of BA, a specific inhibitor of the vacuolar H+-ATPase and therefore of endosomal-lysosomal acidification, was analyzed. The results showed that MVMp is affected by BA in a dose-dependent manner (Fig. 2A and 7A and B). This observation suggests that MVMp entry requires acidification. However, BA could also cause secondary effects depending on the cell type, such as inhibition of receptor-ligand dissociation (23), altered trafficking of transmembrane proteins (44), inhibition of late endosome-lysosome fusion (63), and fragmentation of early endosomes (12). Considering this limitation, we examined the effects of the weak base chloroquine, which interferes with the pH of intracellular vesicles through a different mechanism (36, 52). The effect obtained with chloroquine was similar to that obtained with BA (Fig. 2B), confirming that MVMp enters via an endocytic pathway and requires low pH.
Regarding endosomal transport, another question addressed was whether MVMp particles are routed farther than the early endosomal compartment, as has been suggested for CPV, or are directly released to the cytosol (42, 55, 65, 69). To address this question, the sensitivity of MVMp to BFA, a drug that disrupts the endosomal network (33), was examined. In addition, a pulse experiment with the two endosome-interfering drugs BFA and BA was performed in an attempt to elucidate the time that the bulk of MVMp particles are confined in the endosomal compartment. The results showed that BFA is active against MVMp (Fig. 2C and 7A and B), suggesting that MVMp is routed farther than the early endosomal compartment toward late endosomes. MVMp endosomal transport is slow and takes up to 7 to 8 h to be completed for the bulk of the internalized viral particles (Fig. 3). These observations strongly suggest that MVMp passes through several endosomal compartments before its release to the cytosol. Previsani et al. (45) reported the presence of viral DNA in the nuclear fraction of infected cells already 1 h after the infection; however, it was stated that the approach employed did not make a distinction between viral DNA inside the purified nucleus and that which could be attached to the outside. In our studies, viral DNA replication was not detected until 8 to 10 h postinternalization (Fig. 1A). In contrast to the slow penetration of MVMp, a very rapid endosomal trafficking has been suggested for AAV (1, 53). However, it should be emphasized that the slow endosomal trafficking reported here refers to the bulk of MVMp particles. For instance, already 1 h after internalization, some MVMp particles have already escaped from endosomes (Fig. 3).
It has been shown that transport between early and late endosomes requires the cytoskeleton (22). In accordance with the results obtained with endosome- interfering drugs, MVMp was also sensitive to treatments of cells with ND or CD (Fig. 4 and 7A and B). Interestingly, the inhibitory effect of either of the cytoskeleton-disrupting drugs was clearly inferior to that obtained with BFA or BA when these two drugs were permanently kept in the media (Fig. 3). A similar result was obtained when viral protein synthesis or viral transcription was analyzed (Fig. 7A and B). Since it is highly improbable that movement of virions, subviral particles, or viral genome-protein complexes relies on passive diffusion (54), this result could most probably be due to a limited effect on the cytoskeleton of A9 cells exerted by these drugs.
Once the virus is released to the cytosol from the endosomal compartment, the virus targets the nucleus by a process that is poorly understood. Somehow, the viral particle has to disassemble and expose the DNA. It has been suggested that CPV is still intact after it is released from endosomes, since antibodies that recognize intact capsids microinjected in the cytosol were able to block the infection (66). Therefore, if uncoating has not been accomplished after endosomal trafficking, it should take place during the free cytosolic transport of the virus to the nucleus and/or inside the nucleus. In an attempt to identify cellular factors that interact with viral particles immediately after endosomal release and that could potentially modify the capsid structure, the cellular proteasomes were investigated. The proteasome is a multicatalytic proteinase complex implicated in the degradation of most cellular proteins (39). In mammalian cells, proteasomes are localized in both the nucleus and the cytoplasm (57). Proteasome-virus interaction has been previously reported. Proteasome inhibitors blocked interference with human immunodeficiency virus gag polypeptide processing and decreased the infectivity and release of secreted virions (49). Human immunodeficiency virus type 1-encoded Env and Vpu protein-mediated degradation of the CD4 receptor is also dependent on proteasomes (19, 48). Miller and Pintel (38) demonstrated that the NS2 protein of MVM is degraded by the proteasome. Interaction between proteasomes and AAV has been previously reported (15, 16, 71). In these reports, a considerable enhancement of recombinant AAV transduction was demonstrated when proteasome inhibitors were coadministered with the virus. In addition, it was found that AAV-2 and -5 capsids are actually ubiquitinated during the infection process. These results led to the hypothesis that the ubiquitin-proteasome pathway would actually represent a barrier for AAV to complete its latent life cycle. In accordance with these observations, we have also observed that MVMp interacts with proteasomes. However, this interaction does not represent a barrier. On the contrary, interaction of MVMp with cellular proteasomes was required for the infection process, as shown by the sensitivity of MVMp to different proteasome inhibitors with specific and distinct mechanisms of action (Fig. 5 and 7A and B). Moreover, the action of the proteasome inhibitor MG132 was found to be time dependent. Pulse experiments with MG132 demonstrated that MVMp interacts with cellular proteasomes during a well-defined period of time after internalization, notably between 3 and 9 h. No significant proteasome interaction was observed during the first 3 h after internalization (Fig. 3). The reason for this observation would be that, as is shown in Fig. 3, during the first 3 h from the time of internalization, a large proportion of MVMp particles was still confined to the endosomal compartment. Therefore, the proteasomes are required only after the endosomal escape of MVMp.
Considering that the proteasome exhibit multiple proteolytic functions, the specific activity necessary for MVMp was further examined. A “bite-chew” model for the proteasome action has been proposed in which a protein that interacts with the proteasome first encounters the chymotryptic site, which takes a “bite” out of the protein. This causes the proteasome to change the structure in such a way as to activate other protease sites that would “chew” on the protein (28). In view of this model, it would be possible that the proteasome and, more particularly, chymotrypsin activity are required for a proteolytic processing of MVM capsids, similar to that of VP2 cleavage to VP3. Interestingly, the N terminus of VP2 contains chymotryptic cleavage sites (35) and is externalized in DNA-containing particles. The inhibition of the chymotryptic activity of the proteasome with epoxomicin, which does not inhibit nonproteasomal proteases, such as trypsin, chymotrypsin, papain, calpain, and cathepsin B (37, 51), was in fact a very potent inhibitor of MVMp (Fig. 5C and 7A). These observations were further confirmed because the protease inhibitor TPCK, an irreversible inhibitor of chymotrypsin-like serine proteases, including that of the 20S proteasome, and aclarubicin, a reversible inhibitor of chymotrypsin activity exclusive to the 20S proteasome (18), were both very active against MVMp at low doses. In contrast, TLCK, which inhibits trypsin-like serine proteases, without affecting those of chymotrypsin-like nature, had no effect on MVMp infection (Fig. 6 and 7A).
In summary, different aspects of the cytoplasmic trafficking of MVMp were elucidated in the present studies. It was observed that infection of A9 cells by MVMp can be disturbed by cell treatments that raise the endosomal pH or block endosomal trafficking or disrupt the cytoskeleton, indicating the involvement of an endocytic pathway in which acidification is required. The endosomal trafficking of the bulk of MVMp viral particles is slow and is followed by the interaction with cellular proteasomes. The chymotrypsin-like but not the trypsin-like activity of the proteasome is critical for the infection to progress. These data strongly suggest that MVMp makes use of the cellular proteasome machinery as part of its life cycle, a new pathway that has hitherto not been described.
REFERENCES
- 1.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74:2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basak, S., and H. Turner. 1992. Infectious entry pathway for canine parvovirus. Virology 186:368-376. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, N., D. Schober, E. Prchla, R. F. Murphy, D. Blaas, and R. Fuchs. 1998. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J. Virol. 72:9645-9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose, S., A. Malur, and A. K. Banerjee. 2001. Polarity of human parainfluenza virus type 3 infection in polarized human lung epithelial A549 cells: role of microfilament and microtubule. J. Virol. 75:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, E. J., A. Siebers, and K. Altendorf. 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA 85:7972-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, K. E., S. M. Anderson, and N. S. Young. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262:114-117. [DOI] [PubMed] [Google Scholar]
- 7.Chipman, P. R., M. Agbandje-McKenna, S. Kajigaya, K. E. Brown, N. S. Young, T. S. Baker, and M. G. Rossmann. 1996. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc. Natl. Acad. Sci. USA 93:7502-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens, K. E., and D. J. Pintel. 1988. The two transcription units of the autonomous parvovirus minute virus of mice are transcribed in a temporal order. J. Virol. 62:1448-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinton, G. M., and M. Hayashi. 1976. The parvovirus MVM: a comparison of heavy and light particle infectivity and their density conversion in vitro. Virology 74:57-63. [DOI] [PubMed] [Google Scholar]
- 10.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91-174. [DOI] [PubMed] [Google Scholar]
- 11.Coux, O., K. Tanaka, and A. L. Goldberg. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801-847. [DOI] [PubMed] [Google Scholar]
- 12.D'Arrigo, A., C. Bucci, B. H. Toh, and H. Stenmark. 1997. Microtubules are involved in bafilomycin A1-induced tubulation and Rab5-dependent vacuolation of early endosomes. Eur. J. Cell Biol. 72:95-103. [PubMed] [Google Scholar]
- 13.Deroo, B. J., and T. K. Archer. 2002. Proteasome inhibitors reduce luciferase and beta-galactosidase activity in tissue culture cells. J. Biol. Chem. 277:20120-20123. [DOI] [PubMed] [Google Scholar]
- 14.Dorsch, S., G. Liebisch, B. Kaufmann, P. von Landenberg, J. H. Hoffmann, W. Drobnik, and S. Modrow. 2002. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 76:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douar, A. M., K. Poulard, D. Stockholm, and O. Danos. 2001. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 75:1824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan, D., Y. Yue, Z. Yan, J. Yang, and J. F. Engelhardt. 2000. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Investig. 105:1573-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 18.Figueiredo-Pereira, M. E., W. E. Chen, J. Li, and O. Johdo. 1996. The antitumor drug aclacinomycin A, which inhibits the degradation of ubiquitinated proteins, shows selectivity for the chymotrypsin-like activity of the bovine pituitary 20 S proteasome. J. Biol. Chem. 271:16455-16459. [DOI] [PubMed] [Google Scholar]
- 19.Fujita, K., S. Omura, and J. Silver. 1997. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J. Gen. Virol. 78:619-625. [DOI] [PubMed] [Google Scholar]
- 20.Furuchi, T., K. Aikawa, H. Arai, and K. Inoue. 1993. Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, blocks lysosomal cholesterol trafficking in macrophages. J. Biol. Chem. 268:27345-27348. [PubMed] [Google Scholar]
- 21.Gao, Y., S. Lecker, M. J. Post, A. J. Hietaranta, J. Li, R. Volk, M. Li, K. Sato, A. K. Saluja, M. L. Steer, A. L. Goldberg, and M. Simons. 2000. Inhibition of ubiquitin-proteasome pathway-mediated I kappa B alpha degradation by a naturally occurring antibacterial peptide. J. Clin. Investig. 106:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruenberg, J., G. Griffiths, and K. E. Howell. 1989. Characterization of the early endosome and putative endocytic carrier vesicles in vivo and with an assay of vesicle fusion in vitro. J. Cell Biol. 108:1301-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada, M., S. Shakado, S. Sakisaka, S. Tamaki, M. Ohishi, K. Sasatomi, H. Koga, M. Sata, and K. Tanikawa. 1997. Bafilomycin A1, a specific inhibitor of V-type H+-ATPases, inhibits the acidification of endocytic structures and inhibits horseradish peroxidase uptake in isolated rat sinusoidal endothelial cells. Liver 17:244-250. [DOI] [PubMed] [Google Scholar]
- 24.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaludov, N., K. E. Brown, R. W. Walters, J. Zabner, and J. A. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasamatsu, H., and A. Nakanishi. 1998. How do animal DNA viruses get to the nucleus? Annu. Rev. Microbiol. 52:627-686. [DOI] [PubMed] [Google Scholar]
- 27.Kielian, M., and S. Jungerwirth. 1990. Mechanisms of enveloped virus entry into cells. Mol. Biol. Med. 7:17-31. [PubMed] [Google Scholar]
- 28.Kisselev, A. F., T. N. Akopian, V. Castillo, and A. L. Goldberg. 1999. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol. Cell 4:395-402. [DOI] [PubMed] [Google Scholar]
- 29.Klasse, P. J., R. Bron, and M. Marsh. 1998. Mechanisms of enveloped virus entry into animal cells. Adv. Drug Delivery Rev. 34:65-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kloetzel, P. M. 2001. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2:179-187. [DOI] [PubMed] [Google Scholar]
- 31.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 32.Li, Y., Z. Zadori, H. Bando, R. Dubuc, G. Fediere, J. Szelei, and P. Tijssen. 2001. Genome organization of the densovirus from Bombyx mori (BmDNV-1) and enzyme activity of its capsid. J. Gen. Virol. 82:2821-2825. [DOI] [PubMed] [Google Scholar]
- 33.Lippincott-Schwartz, J., L. Yuan, C. Tipper, M. Amherdt, L. Orci, and R. Klausner. 1991. Brefeldin A's effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 67:601-616. [DOI] [PubMed] [Google Scholar]
- 34.Littlefield, J. W. 1964. Three degrees of guanylic acid-inosinic acid pyrophosphorylase deficiency in mouse fibroblasts. Nature 203:1142-1144. [DOI] [PubMed] [Google Scholar]
- 35.Maroto, B., J. C. Ramírez, and J. M. Almendral. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J. Virol. 74:10892-10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marsh, M., and A. Helenius. 1989. Virus entry into animal cells. Adv. Virus Res. 36:107-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, C. L., and D. J. Pintel. 2001. The NS2 protein generated by the parvovirus minute virus of mice is degraded by the proteasome in a manner independent of ubiquitin chain elongation or activation. Virology 285:346-355. [DOI] [PubMed] [Google Scholar]
- 39.Myung, J., K. B. Kim, and C. M. Crews. 2001. The ubiquitin-proteasome pathway and proteasome inhibitors. Med. Res. Rev. 21:245-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palombella, V. J., O. J. Rando, A. L. Goldberg, and T. Maniatis. 1994. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 78:773-785. [DOI] [PubMed] [Google Scholar]
- 41.Paradiso, P. R. 1981. Infectious process of the parvovirus H-1: correlation of protein content, particle density and viral infectivity. Virology 39:800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker, J. S., and C. R. Parrish. 2000. Cellular uptake and infection by canine parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J. Virol. 74:1919-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker, J. S., W. J. Murphy, D. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Presley, J. F., S. Mayor, T. E. McGraw, K. W. Dunn, and F. R. Maxfield. 1997. Bafilomycin A1 treatment retards transferrin receptor recycling more than bulk membrane recycling. J. Biol. Chem. 272:13929-13936. [DOI] [PubMed] [Google Scholar]
- 45.Previsani, N., S. Fontana, B. Hirt, and P. Beard. 1997. Growth of the parvovirus minute virus of mice MVMp3 in EL4 lymphocytes is restricted after cell entry and before viral DNA amplification: cell-specific differences in virus uncoating in vitro. J. Virol. 71:7769-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, P. C., and R. W. Compans. 1998. Host cell dependence of viral morphology. Proc. Natl. Acad. Sci. USA 95:5746-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanlioglu, S., P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds, and J. F. Engelhardt. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by Rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74:9184-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubert, U., L. C. Anton, I. Bacik, J. H. Cox, S. Bour, J. R. Bennink, M. Orlowski, K. Strebel, and J. W. Yewdell. 1998. CD4 glyprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 72:2280-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz, O., V. Marechal, B. Friguet, F. Arenzana-Seisdedos, and J. M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwarz, K., R. de Giuli, G. Schmidtke, S. Kostka, M. van den Broek, K. B. Kim, C. M. Crews, R. Kraft, and M. Groettrup. 2000. The selective proteasome inhibitors lactacystin and epoxomicin can be used to either up- or down-regulate antigen presentation at nontoxic doses. J. Immunol. 164:6147-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seglen, P. O., B. Grinde, and A. E. Solheim. 1979. Inhibition of the lysosomal pathway of protein degradation in isolated rat hepatocytes by ammonia, methylamine, chloroquine and leupeptin. Eur. J. Biochem. 95:215-225. [DOI] [PubMed] [Google Scholar]
- 53.Seisenberger, G., M. U. Ried, T. Endress, H. Buning, M. Hallek, and C. Brauchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science 294:1929-1932. [DOI] [PubMed] [Google Scholar]
- 54.Sodeik, B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8:465-472. [DOI] [PubMed] [Google Scholar]
- 55.Suikkanen, S., K. Sääjärvi, J. Hirsimäki, O. Välilehto, H. Reunanen, M. Vihinen-Ranta, and M. Vuento. 2002. Role of recycling endosomes and lysosomes in dynein-dependent entry of canine parvovirus. J. Virol. 76:4401-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka, K., A. Kumatori, K. Ii, and A. Ichihara. 1989. Direct evidence for nuclear and cytoplasmic colocalization of proteasomes (multiprotease complexes) in liver. J. Cell. Physiol. 139:34-41. [DOI] [PubMed] [Google Scholar]
- 58.Tattersall, P., A. J. Shatkin, and D. C. Ward. 1977. Sequence homology between the structural polypeptides of minute virus of mice. J. Mol. Biol. 111:375-394. [DOI] [PubMed] [Google Scholar]
- 59.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thacker, T. C., and F. B. Johnson. 1998. Binding of bovine parvovirus to erythrocyte membrane sialylglyproteins. J. Gen. Virol. 79:2163-2169. [DOI] [PubMed] [Google Scholar]
- 61.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1992. The trypsin-sensitive RVER domain in the capsid proteins of minute virus of mice is required for efficient cell binding and viral infection but not for proteolytic processing in vivo. Virology 191:846-857. [DOI] [PubMed] [Google Scholar]
- 62.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J. Virol. 67:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Weert, A. W., K. W. Dunn, H. J. Gueze, F. R. Maxfield, and W. Stoorvogel. 1995. Transport from late endosomes to lysosomes, but not sorting of integral membrane proteins in endosomes, depends on the vacuolar proton pump. J. Cell Biol. 130:821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vihinen-Ranta, M., L. Kakkola, A. Kalela, P. Vilja, and M. Vuento. 1997. Characterization of a nuclear localization signal of canine parvovirus capsid proteins. Eur. J. Biochem. 250:389-394. [DOI] [PubMed] [Google Scholar]
- 65.Vihinen-Ranta, M., A. Kalela, P. Mäkinen, L. Kakkola, V. Marjomäki, and M. Vuento. 1998. Intracellular route of canine parvovirus entry. J. Virol. 72:802-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vihinen-Ranta, M., W. Yuan, and C. R. Parrish. 2000. Cytoplasmic trafficking of the canine parvovirus capsid and its role in infection and nuclear transport. J. Virol. 74:4853-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 76:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walters, R. W., S. M. Yi, S. Keshavjee, K. E. Brown, M. J. Welsh, J. A. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610-20616. [DOI] [PubMed] [Google Scholar]
- 69.Weichert, W. S., J. S. Parker, A. T. Wahid, S. F. Chang, E. Meier, and C. R. Parrish. 1998. Assaying for structural variation in the parvovirus capsid and its role in infection. Virology 250:106-117. [DOI] [PubMed] [Google Scholar]
- 70.White, J., M. Kielian, and A. Helenius. 1983. Membrane fusion proteins of enveloped viruses. Q. Rev. Biophys. 16:151-195. [DOI] [PubMed] [Google Scholar]
- 71.Yan, Z., R. Zak, G. W. Luxton, T. C. Ritchie, U. Bantel-Schaal, and J. F. Engelhardt. 2002. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 76:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]