Abstract
Alternative splicing of fibroblast growth factor receptor 2 (FGFR2) transcripts occurs in a cell-type-specific manner leading to the mutually exclusive use of exon IIIb in epithelia or exon IIIc in mesenchyme. Epithelial cell-specific exon choice is dependent on (U)GCAUG elements, which have been shown to bind Fox protein family members. In this paper we show that FGFR2 exon choice is regulated by (U)GCAUG elements and Fox protein family members. Fox-2 isoforms are differentially expressed in IIIb+ cells in comparison to IIIc+ cells, and expression of Fox-1 or Fox-2 in the latter led to a striking alteration in FGFR2 splice choice from IIIc to IIIb. This switch was absolutely dependent on the (U)GCAUG elements present in the FGFR2 pre-mRNA and required critical residues in the C-terminal region of Fox-2. Interestingly, Fox-2 expression led to skipping of exon 6 among endogenous Fox-2 transcripts and formation of an inactive Fox-2 isoform, which suggests that Fox-2 can regulate its own activity. Moreover, the repression of exon IIIc in IIIb+ cells was abrogated by interfering RNA-mediated knockdown of Fox-2. We also show that Fox-2 is critical for the FGFR2(IIIb)-to-FGFR2(IIIc) switch observed in T Rex-293 cells grown to overconfluency. Overconfluent T Rex-293 cells show molecular and morphological changes consistent with a mesenchymal-to-epithelial transition. If overconfluent cells are depleted of Fox-2, the switch from IIIc to IIIb is abrogated. The data in this paper place Fox-2 among critical regulators of gene expression during mesenchymal-epithelial transitions and demonstrate that this action of Fox-2 is mediated by mechanisms distinct from those described for other cases of Fox activity.
There are four well-characterized fibroblast growth factor receptors (FGFRs), which contain a single transmembrane domain, an intracellular tyrosine kinase domain, and an extracellular FGF binding domain composed of two or three immunoglobulin (Ig)-like domains. The transcripts encoding three FGFRs (FGFR1, -2, and -3) are alternatively spliced to produce isoforms that contain one of two different Ig-III domains. Alternative splicing of FGFR2 transcripts results in the production of two receptors that differ in the carboxy-terminal half of the Ig-III domain. This hemidomain is determined by the tissue-specific inclusion of either exon IIIb or exon IIIc, which ultimately controls ligand binding specificity (7, 14, 27, 52). FGFR2(IIIb) primarily binds FGF10 and FGF7 and is the isoform of choice in epithelial cells, whereas FGFR2(IIIc) binds FGF2 and is exclusively expressed in cells of mesenchymal origin (36, 49). FGF/FGFR2 signaling governs epithelial-mesenchymal interactions that are required for organogenesis in mouse embryos (3, 15, 16); therefore, it is critical for normal development to maintain the proper cell-type-specific expression of each receptor isoform. Mutations that alter the ligand binding specificity of FGFR2(IIIc) or those that lead to the inappropriate expression of exon IIIb in mesenchyme have been linked to developmental disorders in humans (3, 16, 35, 54). The importance of FGFR2 isoform choice is underscored by studies demonstrating a switch from FGFR2(IIIb) to FGFR2(IIIc) during the progression of prostate carcinomas (4, 49), where the loss of FGFR2(IIIb) appears to be required for this progression (51).
The regulation of FGFR2 alternative splicing depends on a complex interplay between cis-acting elements in the FGFR2 pre-mRNA and trans-acting factors, with some of the trans-acting factors appearing to be cell type specific. To study this regulation, we have employed two cell lines derived from Dunning rat prostate tumors. The DT3 cell line is a well-differentiated carcinoma that solely expresses endogenous FGFR2(IIIb), whereas the AT3 cell line is poorly differentiated and exclusively expresses FGFR2(IIIc) (49). We have also used human embryonic kidney 293T (HEK293T) cells, which although of uncertain cell type provenance include exon IIIc (46, 53). Repression of exon IIIb is mediated by the presence of weak splice sites (ss) flanking exon IIIb, an exonic silencing sequence in exon IIIb, and intronic silencing sequences upstream and downstream of exon IIIb (6, 9, 10, 43, 46). The exonic silencing sequence functions to recruit hnRNP A1 to exon IIIb, thereby repressing the inclusion of exon IIIb (13), while the upstream intronic silencing sequence and downstream intronic silencing sequence antagonize exon IIIb definition by binding the polypyrimidine tract binding protein (PTB) and other factors yet to be identified (6, 43, 45, 46). Silencing of exon IIIb is countered in epithelial cells by the action of several cis-acting elements. Intronic activating sequence 1 (IAS1), located downstream of exon IIIb, serves to recruit the splicing factor TIA-1. Binding of TIA-1 to IAS1 has been shown to activate the weak 5′ splice site of exon IIIb as well as weak splice sites of other exons (12). Two cell-type-specific cis-acting elements, intronic activating sequence 2 (IAS2) and intronic splicing activator and repressor (ISAR, also known as IAS3), serve to activate exon IIIb inclusion, while repressing inclusion of exon IIIc in an epithelial cell-specific manner (5, 11, 44). IAS2 and ISAR were postulated to function by forming a stem (11), and indeed, several lines of evidence support this proposal (1, 5, 11, 21, 29, 32). The major, if not sole, role of the stem in exon IIIb activation is to approximate sequences upstream of IAS2 with sequences downstream of ISAR (1). Recently, another sequence downstream of ISAR termed intronic splicing enhancer-intronic splicing silencer 3 (ISE/ISS-3) was shown to activate exon IIIb and repress exon IIIc, and the repression of exon IIIc required a noncanonical branch point sequence (18). Additionally, we have previously identified a GCAUG sequence element downstream of ISAR core and showed that it played a role in cell-type-specific exon IIIb activation and exon IIIc repression (1).
The GCAUG or UGCAUG sequence is a previously characterized splicing enhancer element that has been shown to be important for the proper splicing regulation of fibronectin, c-src, calcitonin/CGRP, nonmuscle myosin II heavy chain B (NMHC-B), and 4.1R transcripts (8, 17, 19, 23, 26, 30). In addition, a computational study demonstrated an overrepresentation of UGCAUG hexamers in the downstream intron of neural and muscle-specific alternatively spliced exons (2, 28). The factors responsible for recognizing the hexamers were first recognized by Jin et al. (20), who showed these proteins to be homologs of the Caenorhabditis elegans RNA binding protein feminizing on X (Fox-1). These authors demonstrated that overexpression of vertebrate homologs of Fox-1, called zebra fish Fox-1 (zFox-1) and mouse Fox-1 (mFox-1 or ataxin 2 binding protein 1 [A2BP1]), could regulate the alternative splicing of human mitochondrial ATP synthase γ subunit (F1γ), rat α-actinin, and rat fibronectin minigene constructs (20). Nakahata and Kawamoto identified brain- and muscle-specific isoforms of mouse Fox-1 and Fox-2 and demonstrated that expression of brain-specific isoforms of these proteins promoted the inclusion of the neuronal N30 cassette exon in NMHC-B transcripts (33). Additionally, Underwood et al. demonstrated that Fox-1 and Fox-2 are expressed in a number of mammalian cell lines to various degrees (41). They went on to show that Fox-1 and Fox-2 are specifically expressed in neurons and not glia in the brain and presented compelling evidence that these proteins are required for the neural cell-specific inclusion of the N1 exon in c-src transcripts (41).
In this study, we demonstrate that there are multiple (U)GCAUG elements in FGFR2 transcripts and these sites are essential for cell-type-specific regulation of exon choice. We investigated the role of vertebrate Fox proteins in this regulation and found that, while Fox-1 was not expressed in AT3 or DT3 cells, both of these expressed many Fox-2 transcripts. Additionally, we found that the expression levels of Fox-2 isoforms differed dramatically between DT3 cells, which express epithelial FGFR2(IIIb), and AT3 cells, which express mesenchyme FGFR2(IIIc). We demonstrated that overexpression of murine Fox-2 (mFox-2) in cells that normally include FGFR2 exon IIIc led to a dramatic switch from exon IIIc to exon IIIb in minigene reporters, and this change absolutely required the intact (U)GCAUG elements. We also determined that the RNA recognition motif (RRM) and the carboxy-terminal portion of the mFox-2 protein are vital for exon IIIb activation and exon IIIc repression. Most importantly, mFox-2 expression is capable of stimulating exon IIIb inclusion in endogenous FGFR2 transcripts in cells that normally include exon IIIc. Fox-2 expression also led to skipping of exon 6 among endogenous Fox-2 transcripts, which would lead to the synthesis of an inactive form of Fox-2 and can be used to autoregulate the levels of active Fox-2. Finally, a switch from exon IIIc to exon IIIb in endogenous FGFR2 transcripts was observed when T Rex-293 cells were induced to undergo what appears to be a mesenchymal-epithelial transition (MET) as the cells were grown to overconfluency. This switch from exon IIIc to exon IIIb inclusion was absolutely dependent on Fox-2 expression as interfering RNA (RNAi)-induced Fox-2 knockdown abrogated the switch. Complementation with an RNAi-resistant Fox-2 expression plasmid allowed for the IIIc-to-IIIb switch after knockdown of endogenous Fox-2. These data strongly suggest that Fox proteins, and most likely Fox-2, control the epithelial-mesenchymal choice between FGFR2 isoforms. Moreover, this epithelial-mesenchymal regulation is mediated by mechanisms distinct from those in previously described cases of Fox protein control, indicating that these proteins can modulate cell-type-specific splicing with remarkable versatility.
MATERIALS AND METHODS
Plasmid construction.
The plasmid DNA constructs used in this study were made by using standard cloning techniques described previously (5). The previously described minigene pI-11-FL (5) is hereafter referred to as pI12DE-WT FL (pI12 double exon wild-type [WT] FL). pI12DE-WT and pI12DE-C10-18 were previously described (1). pI12DE-WT FL and pI12DE-WT differ as follows: pI12DE-WT FL contains nearly all of intron 7 and intron 9, while pI12DE-WT contains only the portions of intron 7 and 9 that have greater than 90% intron homology between rat and human (5). pI12DE-IIIc Mut, made by chimeric PCR amplification, mutated the TGCATG sequence in exon IIIc to GTACGT, and pI12DE-C10-18 IIIc Mut, made in a similar manner, mutated the GCATGCATG sequence downstream of ISAR core to TACGTACGT as well as the TGCATG sequence in exon IIIc to GTACGT. The 5′ and 3′ ss of exon IIIb were mutated in pI12DE-IIIb ss Mut from AGGTAA to AGCCAA and from AGCACT to CCCACT, respectively. The 5′ and 3′ ss of exon IIIc were mutated in pI12DE-IIIc ss Mut from AGGTAC to AGCCAC and from AGGCCG to CCGCCG, respectively. pCS2+MT mFox-1 (NM_021477) and pCS2+MT zFox-1 (AB074763) were previously described (20). We used PCR amplification of pCS2+MT mFox-1 and zFox-1 to create pTracer EF mFox-1 and zFox-1, which contain the mFox-1 and zFox-1 cDNAs with a carboxy-terminal V5 epitope tag. pTracer EF mFxh (AF387322) was described previously (25). We used PCR amplification to create pTracer EF mFox-2. We used PCR amplification of pTracer EF mFox-2, followed by annealed oligonucleotide cloning of the nuclear localization signal (NLS), to create pTracer EF WT NLS mFox-2, Δ2-50 NLS, Δ2-102 NLS, Δ2-194 NLS, Δ2-281 NLS, Δ294-377 NLS, Δ206-377 NLS, Δ123-377 NLS, and Δ63-377 NLS expression constructs. To create pTracer EF rFox-2, we used PCR amplification of the rat gene from DT3 and AT3 cells. To create FX1 and FX3 RNAi-resistant pTracer EF rFox-2 plasmids, we used chimeric PCR amplification to introduce silent mutations at the third base of each codon targeted by the short interfering RNA (siRNA) sequence. pcDNA5/FRT/TO-Gint was described previously (42). To create pcDNA5/FRT/TO-mFox-2, pTracer EF mFxh was digested with BamHI and BclI and cloned into pcDNA5/FRT/TO digested with BamHI. We used PCR amplification of pTracer EF mFxh to create pRSET A mFox-2 and pGex2TK mFox-2, which expresses recombinant His-tagged mFox-2 protein and recombinant glutathione S-transferase (GST)-tagged mFox-2 protein, respectively. All constructs utilizing PCR amplification were sequenced for verification. Explanations on how plasmids were made and all oligonucleotide sequences will be provided upon request.
Cell culture and transfection.
AT3, DT3, and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (low glucose) supplemented with 10% fetal bovine serum (HyClone). Stable transfections were performed as previously described (1). Transient cotransfections were performed as previously described (6) with 50 ng of pI12DE-WT, pI12DE-WT FL, or pI12DE-C10-18 IIIc Mut reporter constructs and the indicated amounts of expression plasmids for mFox-2. The amount of plasmid DNA used in each transfection was kept constant by using the empty plasmid pTracer EF HisB (Invitrogen). Four wells of cells for each condition were plated at a density of 200,000 cells per well of a Falcon six-well plate (Becton Dickinson). At 48 h posttransfection, RNA was harvested for reverse transcription-PCR (RT-PCR) analysis from three separate wells and protein was harvested from the last well for Western blot analysis. Following the manufacturer's protocol, the T Rex-293 G-Int and mFox-2 cell lines were created by integrating the gene of interest (G-Int or mFox-2) into the FRT site in the T Rex-293 cells (Invitrogen). T Rex-293 cells were maintained in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% tetracycline minus fetal bovine serum (HyClone), 5 μg blasticidin/ml (Invitrogen), and 100 μg zeocin/ml (Invitrogen). T Rex-293 G-Int and mFox-2 cells were maintained in the same medium with 200 μg hygromycin/ml (Invitrogen) instead of 100 μg zeocin/ml (Invitrogen). The G-Int and mFox-2 cell lines were plated at a density of 200,000 cells per well in a Falcon six-well plate (Becton Dickinson) and when indicated were induced with tetracycline (Invitrogen) at a final concentration of 5 μg/ml. Five wells of each cell line were plated for each condition, and one well of cells was harvested and used for fluorescence-activated cell sorting (FACS) analysis by the Duke University Comprehensive Cancer Center fluorescence cell sorter shared resource facility. Protein was isolated for Western blot analysis from a second well, and RNA was isolated for RT-PCR analysis from three separate wells of cells.
siRNAs and RNA interference experiments.
The FX1 and FX3 siRNA duplexes were designed to target the coding region of Fox-2. The target sequences of these duplexes are GCAAATGGCTGGAAGTTAA and GAACATAACCTGACACTCT, respectively. The FX2 siRNA duplex was initially designed in the Black laboratory and was designed to target the 3′ untranslated region of Fox-2 (41). The target sequence of this oligonucleotide is CCTGGCTATTGCAATATTT. All siRNA duplexes were purchased from Dharmacon. RNA interference experiments in DT3 cells were performed as follows. DT3 cells were seeded at a density of 250,000 cells per well in a Falcon six-well plate (Becton Dickinson). The following day 6 μl of a 20 μM stock of siRNA was incubated in 250 μl of Opti-MEM medium (Invitrogen) for each well of cells to be treated. In a separate tube, 4 μl of Lipofectamine 2000 was incubated with 250 μl of Opti-MEM. After 7 min the two tubes were combined and allowed to incubate at room temperature. After 25 min the mixture was added directly to the cells and incubated with the cells for 30 h. At 30 h post-siRNA transfection, 50 ng of the minigene was transfected with 3 μl of the 20 μM siRNA stock using standard Lipofectamine (Invitrogen) protocols described previously (6). After 24 h, RNA was harvested from three separate wells for RT-PCR analysis and protein was pooled from each of the three separate wells for Western analysis. RNA interference experiments in T Rex-293 cells were performed as follows. T Rex-293 cells were seeded at a density of 500,000 cells per well in a Falcon six-well plate (Becton Dickinson). The following day each well was transfected with siRNAs in the same manner as described above for the DT3 cells with one exception: 500 ng of either pTracer EF B or pTracer EF mFox-2 was incubated with the siRNAs and transfected. After 48 h, RNA was harvested from three separate wells for RT-PCR analysis and protein was pooled from each of the three separate wells for Western analysis.
RNA isolation and RT-PCR assay of transfected minigenes.
Total cellular RNA, for RT-PCR and the Invader RNA assay, was isolated using Trizol reagent (Invitrogen). RT-PCRs using T7 and SP6 primers for minigene analysis, FGF-FB and FGF-RB for endogenous FGFR2 analysis, FibronF and FibronR for endogenous fibronectin analysis, and hFox-2 exon 4F and hFox-2 exon 7R for Fox-2 exon 6 analysis were performed as previously described (5). PCR products were digested with either AvaI or HincII (New England Biolabs), and analysis and quantification of PCR products from double-exon digests were performed as previously described (6). Analysis from double-exon splice site mutant constructs was performed exactly as previously described for single-exon constructs (1). Phosphoimager quantification of PCR bands was performed with ImageQuant (Molecular Dynamics). Each experiment is the result of an average of triplicate samples with error bars representing the standard deviations.
Invader RNA assay.
The Invader RNA assay (Third Wave Technologies, Madison, WI) was carried out as described previously (1). Briefly, to analyze double-exon minigenes, the Invader RNA assays were run in the biplex format using the probe set combinations IIIb-D/U-D and IIIb-IIIc/U-IIIc as previously described (44). Standard graphs comparing attomoles of RNA to fluorescence were created, and from the fluorescent readings, absolute levels of each splice variant were calculated.
Western blot analysis.
Transfected cells from overexpression studies were harvested, and cell lysates were prepared with a 3× freeze-thaw in 100 mM Tris, pH 7.5. Protein lysates were quantified using the Bradford assay, and 50 μg of each lysate was separated on a sodium dodecyl sulfate-polyacrylamide gel. The proteins were transferred overnight onto an Immuno-Blot polyvinylidene difluoride membrane (Bio-Rad). The membranes were blocked for 15 min at room temperature in blocking buffer (phosphate-buffered saline, 5% dry milk, 0.1% Tween 20), probed for 1 h at room temperature with the respective antibodies diluted in blocking buffer (1:5,000 for V5, 1:2,000 for CA150, and 1:100 for Fox-2), washed three times for 10 min each in blocking buffer, probed for 1 h at room temperature with the appropriate secondary antibody diluted in blocking buffer (1:5,000 for both ECL anti-mouse IgG and anti-rabbit IgG [Amersham Biosciences]), washed three times for 10 min each in blocking buffer, and washed one time for 10 min in phosphate-buffered saline, and signal was activated with ECL Western blotting detection reagents (Amersham Biosciences) and detected on Hyperfilm-MP (Amersham Biosciences). Whole-cell lysates for DT3, AT3, HEK293T, and T Rex-293 cells were made by lysing cells in 2× sodium dodecyl sulfate buffer.
Antibodies.
Antibodies to mFox-2 were made by Animal Farm Sciences, which immunized two rabbits with purified recombinant His-tagged mFox-2. Antigen-affinity-purified Fox-2 antibodies were made by coupling purified GST-Fox-2 to Actigel ALD Superflow resin and following the manufacturer's protocol (Sterogene Bioseparations, Inc.). GST-Fox-2 was purified as previously described for GST-human immunodeficiency virus type 1 Tat (39).
RESULTS
Intronic and exonic (U)GCAUG sequence elements are important for tissue-specific FGFR2 exon choice.
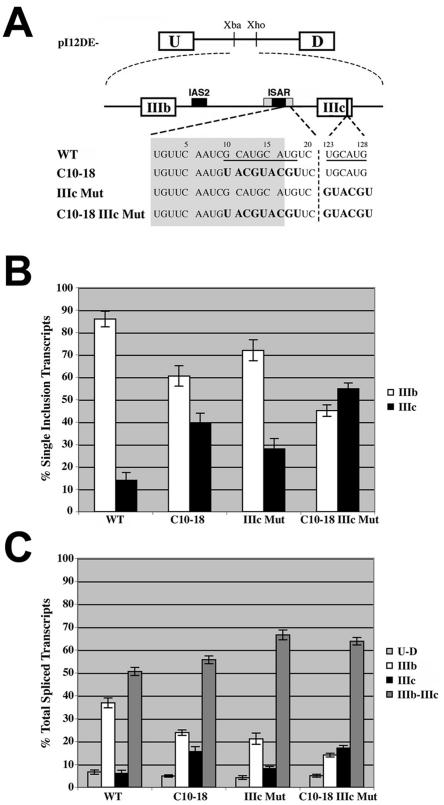
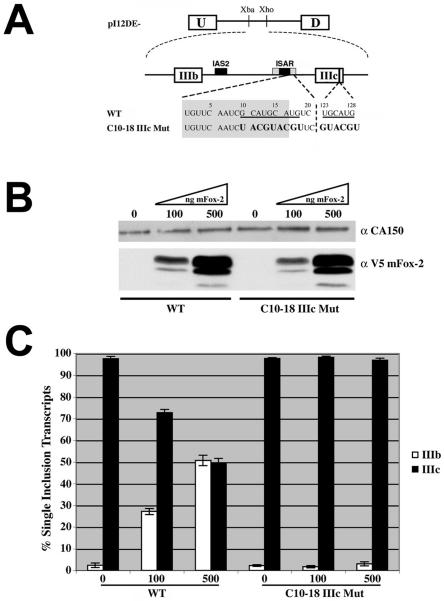
Previously we had identified two overlapping (U)GCAUG elements downstream of ISAR core that were important for both exon IIIb activation and exon IIIc repression (1). Understanding that (U)GCAUG elements generally are present in multiple copies in the same pre-mRNA (8, 17, 19, 20, 23, 26), we scanned the FGFR2 sequence for other (U)GCAUG elements and found a (U)GCAUG element present in exon IIIc. To determine if this sequence played a role in activating exon IIIb and repressing exon IIIc, we created minigenes where we mutated the overlapping GCAUG elements downstream of ISAR core (C10-18), the (U)GCAUG in exon IIIc (IIIc Mut), or both (C10-18 IIIc Mut) (Fig. 1A). We transfected these minigenes along with pI12DE-WT (Fig. 1A) into DT3 and AT3 cells, and RNA was isolated from stable cell populations and analyzed by RT-PCR and the Invader RNA assay. All of the minigenes transfected into AT3 cells resulted in nearly exclusive use of exon IIIc, suggesting that the (U)GCAUG elements do not play a role in these cells (data not shown). In DT3 cells, when analyzing single inclusion events, exon IIIb inclusion dropped from 85% for WT to 60% for C10-18 as was previously reported (Fig. 1B) (1). Interestingly, the IIIc Mut minigene showed 72% inclusion of exon IIIb (Fig. 1B), suggesting that this element is not as important for exon IIIb inclusion as is the C10-18 element. Nonetheless, when the IIIc Mut is combined with the C10-18 mutation in the C10-18 IIIc Mut minigene (Fig. 1A), exon IIIb levels drop to 45% (Fig. 1B), suggesting that these elements provide additive signals that promote cell-type-specific inclusion of exon IIIb. In contrast to the results with exon IIIb inclusion, exon IIIc repression was inhibited as well by each mutation. Analysis of all spliced products revealed that the double inclusion products increase from 50% for WT to 57% for C10-18, 68% for IIIc Mut, and 65% for C10-18 IIIc Mut (Fig. 1C), demonstrating that these elements repress exon IIIc in epithelial cells. These results indicate the following: (i) the (U)GCAUG elements mediate cell-type-specific activities, (ii) the elements downstream of ISAR core and in exon IIIc provide additive signals in promoting exon IIIb activation, and (iii) the (U)GCAUG elements both serve to repress exon IIIc.
FIG. 1.
Intronic and exonic (U)GCAUG elements are critical for exon IIIb activation and exon IIIc repression in DT3 cells. (A) Schematic of minigenes used to test the importance of (U)GCAUG elements located downstream of ISAR core and in exon IIIc. Wild-type (U)GCAUG elements are underlined. Mutated nucleotides are indicated in boldface. Numbers above nucleotides represent base pair numbers downstream of ISAR core and within exon IIIc, respectively. IAS2 and ISAR core are represented as black boxes; ISAR core resides within the full ISAR element (represented as a gray box). Nucleotides in the gray box are located within ISAR. (B) The percent inclusion among single inclusion transcripts (U-IIIb-D and U-IIIc-D) produced by minigenes in panel A, which were stably integrated in DT3 cells, was determined by Invader RNA assay [e.g., % IIIb = 100 × number of U-IIIb-D transcripts/(number of U-IIIb-D transcripts + number of U-IIIc-D transcripts)]. (C) Quantification of all spliced products for transcripts from minigenes in panel A, which were stably integrated into DT3 cells, was determined by Invader RNA assay [e.g., % U-IIIb-D = 100 × number of U-IIIb-D transcripts/(number of U-D transcripts + number of U-IIIb-D transcripts + number of U-IIIc-D transcripts + number of U-IIIb-IIIc-D transcripts)].
Western blot analysis reveals differences in expression levels of Fox-2 isoforms.
Having found that FGFR2 exon choice was dependent on (U)GCAUG elements, we sought to determine the identity of the trans factor(s) that recognizes these elements. The work of Jin et al. (20) and the recent work from the Kawamoto and Black laboratories (33, 41) made mammalian homologs of Fox-1 the most likely candidates. Examination of the literature and sequence comparisons revealed that humans and other mammals have three proteins that are highly homologous to the C. elegans Fox-1. We refer to these three proteins, which are encoded by three different genes, as Fox-1 (also known as A2BP1), Fox-2 (also known as RBM9, FXH, and RTA), and Fox-3 (see Table S1 in the supplemental material). We have concentrated our studies on Fox-1 and Fox-2. The RRMs of these two mammalian Fox proteins are identical; however, there is divergence in the amino- and carboxy-terminal portions of the proteins (see Fig. S1 in the supplemental material). Fox-1 is expressed predominantly in heart, brain, and skeletal muscle (20, 24, 38), whereas Fox-2 is expressed in a more ubiquitous manner (25, 34). Alternative splicing and alternative transcription start sites embellish the expression patterns for the two Fox proteins further by producing many different isoforms (reference 41 and data not shown). For instance the α and β isoforms include or skip exon 11, respectively, which results in two proteins with dramatically different carboxy-terminal sequences (see Table S1 in the supplemental material) (33, 34).
We wanted to determine if there were differences in Fox expression between DT3, AT3, and HEK293T cells. RT-PCR analysis revealed that these cells do not express detectable levels of Fox-1 mRNA; however, all three of these cell lines expressed Fox-2 transcripts. Nakahata and Kawamoto found that Fox-2 is alternatively spliced to include either a brain-specific exon, which they call B40, or a muscle-specific exon, which they call M43 (33). All of the clones that we obtained from these cell lines include exon B40, suggesting that the regulation of FGFR2 alternative splicing in DT3, AT3, and HEK293T cells was not due to a difference between expression of Fox-2 isoforms that include either exon B40 or exon M43 (data not shown). We obtained many different cDNAs from the AT3 and DT3 cells, including a DT3-specific isoform that skipped exon 6 (see below). Additionally, DT3 cells contain more transcripts that skip exon 11, which would give rise to the Fox-2β isoform (data not shown).
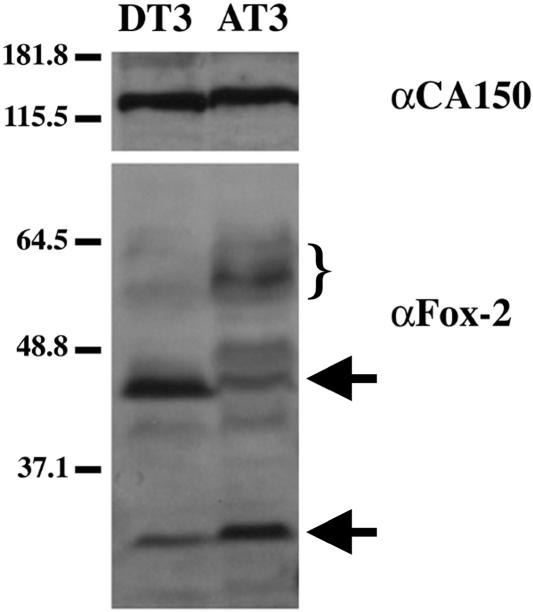
In order to characterize the expression and heterogeneity of Fox-2 proteins we obtained rabbit anti-murine Fox-2 antisera. After antigen affinity purification of the antisera, we probed total cellular extracts from DT3 and AT3 cells to determine if there were differences in the Fox-2 expression profile. Interestingly, the expression patterns between DT3 and AT3 cells were distinctly different (Fig. 2). Two major bands that have been confirmed to be Fox-2 by siRNA knockdown (see below and data not shown) differ in the level of expression between the two cell types (indicated by arrows in Fig. 2). Additionally there are higher-molecular-weight bands in AT3 cells that appear at lower levels or not at all in DT3 cells (bracketed in Fig. 2). These results suggest that the difference in expression levels of Fox-2 between the cell types could allow for the differences in regulating FGFR2 alternative splicing.
FIG. 2.
Expression levels of Fox-2 isoforms differ between DT3 and AT3 cells. Western blot analysis was performed on total cellular protein isolated from DT3 and AT3 cells. The blot was probed for CA150 as a loading control and Fox-2 to detect the endogenous expression of Fox-2 proteins in AT3 and DT3 cells. The arrows indicate the presence of confirmed Fox-2 isoforms. The bracket indicates the presence of presumed Fox-2 isoforms. Numbers at left are molecular masses in kilodaltons.
Overexpression of Fox proteins mediates exon IIIb inclusion in cells that normally include exon IIIc.
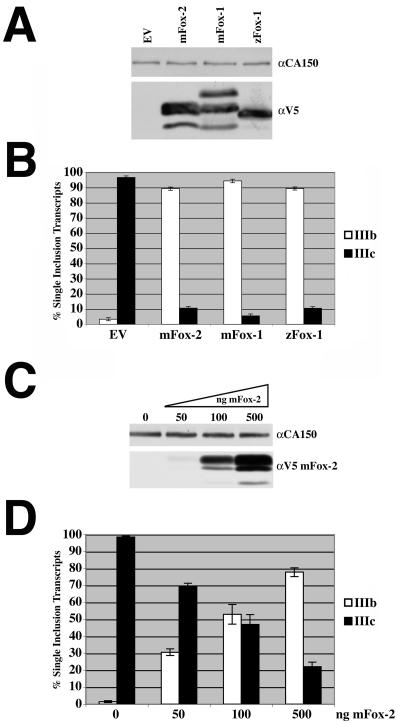
In order to evaluate the role of Fox proteins in FGFR2 alternative splicing we overexpressed murine and zebra fish Fox-1 (mFox-1 and zFox-1) expression constructs used in the work of Jin et al. (20), as well as a murine Fox-2 (mFox-2), which had previously been named mFXH (25), in cells that normally include exon IIIc. These three Fox cDNA plasmids express the α isoform of Fox. Because AT3 cells have low transfection efficiency, we decided to use HEK293T cells, which are readily transfectable. To determine the effect of Fox protein overexpression on FGFR2 alternative splicing, we cotransfected expression constructs encoding the Fox proteins with a constant amount of pI12DE-WT FL splicing reporter. pI12DE-WT FL was used to more fully resemble the endogenous gene as it contained almost the entirety of intron 7, upstream of exon IIIb, and intron 9, downstream of exon IIIc. RNA and protein were harvested from the cells to confirm zFox-1, mFox-1, or mFox-2 overexpression and the effect that the overexpression had on the WT minigene reporter. Western blot analysis revealed that the proteins were expressed at similar levels (Fig. 3A). RT-PCR analysis revealed that the empty-vector control lane resulted in no exon IIIb inclusion as expected, while zFox-1, mFox-1, and mFox-2 expression resulted in a dramatic, almost complete splicing switch from 95% exon IIIc inclusion to over 90% exon IIIb inclusion (Fig. 3B).
FIG. 3.
zFox-1, mFox-1, and mFox-2 overexpression switches splicing pattern of FGFR2 minigene construct. (A) Western blot analysis was performed following transfection of 1 μg of V5-tagged mFox-2, mFox-1, or zFox-1 expression plasmids in HEK293T cells. The blot was probed for CA150 as a loading control and V5 to detect expression of mFox-2, mFox-1, and zFox-1. (B) Quantification of single inclusion transcripts by RT-PCR analysis of transiently transfected minigene with empty vector (EV), mFox-2, mFox-1, or zFox-1 expression plasmids. (C) Western blot analysis was performed following transfection of 0, 50, 100, and 500 ng of mFox-2 expression plasmid in HEK293T cells. The blot was probed for CA150 as a loading control and V5 to detect increasing expression of mFox-2. (D) Quantification of single inclusion transcripts by RT-PCR analysis of transiently transfected minigene with increasing amounts of mFox-2 expression plasmid.
Having seen similar responses to zFox-1, mFox-1, and mFox-2 expression and noting the expression preference for these proteins, we chose to use mFox-2 to further investigate its role in FGFR2 alternative splicing regulation. We first wished to determine a range of protein expression in which to perform experiments. To do this, we cotransfected a constant amount of pI12DE-WT FL with increasing amounts of mFox-2. mFox-2 expression was demonstrated by Western blot analysis (Fig. 3C). When no mFox-2 cDNA is transfected, there is less than 1% exon IIIb inclusion; however, as the amount of mFox-2 expression was increased (Fig. 3C and D), the level of exon IIIb inclusion increased in a dose-dependent manner (Fig. 3D). With as little as 50 ng of transfected plasmid, we saw an increase to 30% exon IIIb inclusion, and when 500 ng of cDNA was transfected, we observed approximately 80% exon IIIb inclusion (Fig. 3D). In addition to increasing exon IIIb inclusion, overexpression of mFox-2 resulted in an increase in the skipped product, which does not include exon IIIb or exon IIIc. All of the results above demonstrated that zFox-1, mFox-1, and mFox-2 expression dramatically switched the FGFR2 alternative splicing pattern, resulting in near-complete exon IIIb inclusion.
Overexpression of mFox-2 coordinately activates exon IIIb inclusion and represses exon IIIc inclusion.
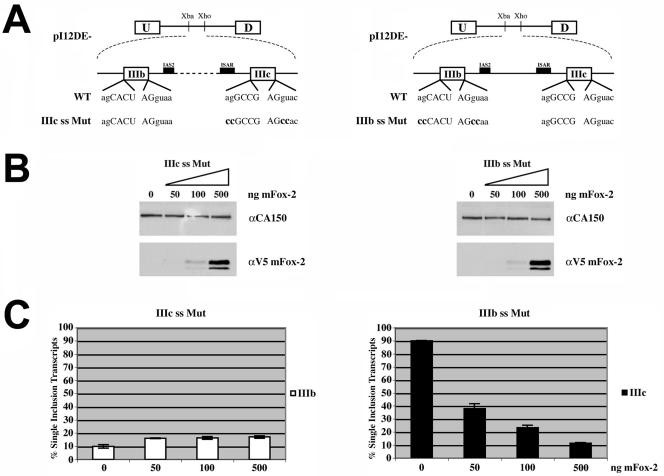
Having demonstrated that zFox-1, mFox-1, and mFox-2 overexpression switches FGFR2 exon choice, we wanted to determine the relative effects of mFox-2 on IIIb activation and IIIc repression. To do this, we used pI12DE-IIIc ss Mut, which has the 3′ and 5′ splice sites flanking exon IIIc mutated and allows the study of exon IIIb activation (Fig. 4A, left panel), and pI12DE-IIIb ss Mut, which has the 3′ and 5′ splice sites flanking exon IIIb mutated and allows the study of exon IIIc repression (Fig. 4A, right panel). We cotransfected a constant amount of each splicing reporter with increasing amounts of mFox-2. When increasing amounts of mFox-2 were cotransfected with pI12DE-IIIc ss Mut, we observed an increase in exon IIIb inclusion from 10% to 18% (Fig. 4B and C, left panels). When pI12DE-IIIb ss Mut was cotransfected with increasing amounts of mFox-2, we observed a dramatic dose-dependent decrease in exon IIIc inclusion from 90% to 10% (Fig. 4B and C, right panels). These results taken together demonstrate that mFox-2 can modestly activate exon IIIb and strongly repress exon IIIc. It is important to note that mFox-2 overexpression tends to favor the skipped splicing product, so the modest effect on IIIb activation may be an underrepresentation of the ability of mFox-2 to activate exon IIIb, and equally the strong effect on IIIc repression may be an overrepresentation of the ability of mFox-2 to repress exon IIIc inclusion.
FIG. 4.
mFox-2 overexpression activates exon IIIb and represses exon IIIc inclusion. (A) Schematic of minigenes used to test ability of mFox-2 expression to activate exon IIIb and repress exon IIIc independently of each other. (B) Western blot analysis was performed following transfection of increasing amounts of mFox-2 expression plasmid in HEK293T cells. The blot was probed for CA150 as a loading control and V5 to detect increasing amounts of mFox-2 expression. (C) Quantification of single inclusion transcripts by RT-PCR analysis of transiently transfected minigene reporters with increasing amounts of mFox-2 expression plasmid.
mFox-2 mediates exon switching through (U)GCAUG elements located downstream of ISAR core and within exon IIIc.
Given that mFox-1 and mFox-2 bind UGCAUG elements (20, 33), we wished to determine whether mFox-2 was mediating its effect through the FGFR2 (U)GCAUG elements that we identified above. To this end, we cotransfected increasing amounts of mFox-2 with constant amounts of pI12DE-WT, pI12DE-C10-18, pI12DE-IIIc Mut, and pI12DE-C10-18 IIIc Mut (Fig. 1A and 5A) in HEK293T cells. mFox-2 overexpression resulted in an increase in exon IIIb inclusion from 1% to 51% in a dose-dependent manner when cotransfected with pI12DE-WT (Fig. 5B and 5C). Overexpression of mFox-2 resulted in slightly diminished levels of exon IIIb when cotransfected with the C10-18 minigene in comparison to the WT minigene (data not shown). When pI12DE-IIIc Mut was cotransfected with increasing amounts of mFox-2, we observed much less exon IIIb inclusion in comparison to both the WT and C10-18 minigenes, suggesting that this element is more important for mFox-2-mediated exon IIIb inclusion and exon IIIc repression (data not shown). When both (U)GCAUG elements were mutated in pI12DE-C10-18 IIIc Mut (Fig. 5A), mFox-2 overexpression had no effect on FGFR2 alternative splicing as exon IIIb inclusion stayed at 2% with increased expression of mFox-2 (Fig. 5B and 5C). These results clearly demonstrate that mFox-2 absolutely requires both (U)GCAUG elements to activate exon IIIb and repress exon IIIc.
FIG. 5.
Intronic and exonic (U)GCAUG elements are critical for mFox-2-mediated FGFR2 exon IIIb activation and exon IIIc repression. (A) Schematic of minigenes used to test the role of (U)GCAUG elements in mFox-2-mediated exon IIIb activation and exon IIIc repression. Wild-type (U)GCAUG elements are underlined. Mutated nucleotides are indicated in boldface. Numbers above nucleotides represent base pair numbers downstream of ISAR core and within exon IIIc, respectively. IAS2 and ISAR core are represented as black boxes; ISAR core resides within the ISAR element (represented as a gray box). Nucleotides in the gray box reside within ISAR. (B) Western blot analysis was performed following transient transfection of increasing amounts of mFox-2 expression plasmid in HEK293T cells with WT and C10-18 IIIc Mut minigene reporters. The blot was probed for CA150 as a loading control and V5 to detect increasing expression of mFox-2. (C) Quantification of single inclusion transcripts by RT-PCR analysis of transiently transfected minigene reporters with increasing amounts of mFox-2 expression plasmid in HEK293T cells.
Interestingly, the product of skipping both exons IIIb and IIIc slightly increased when both (U)GCAUG elements were mutated (data not shown). It is possible that mFox-2 was capable of interacting with the transcripts produced from the mutant minigene construct, which still contains a UGCAUG and GCAUG element in the loop region between IAS2 and ISAR core. Even though the loop region had previously been determined to be dispensable for proper exon choice (1, 32), mFox-2 could bind the elements within the loop, resulting in an increase in exon skipping.
Nevertheless, the data above indicate that mFox-2 requires the (U)GCAUG elements downstream of ISAR core and in exon IIIc to mediate FGFR2 exon choice.
The C-terminal portion of mFox-2 is required for exon IIIb activation and exon IIIc repression.
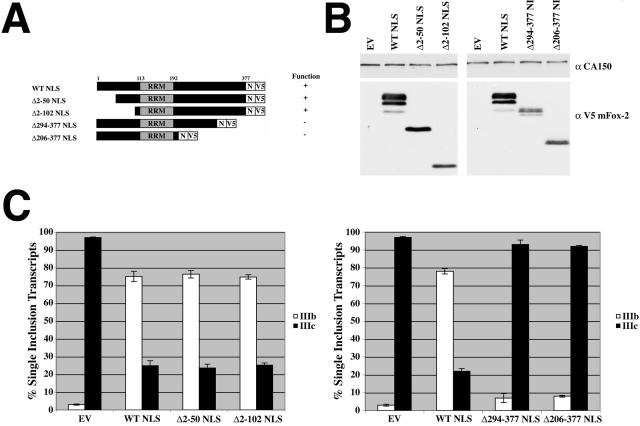
Having demonstrated the effect of mFox-2 expression in FGFR2 alternative splicing regulation, we wanted to determine which portions of the protein were required for regulating exon choice. Therefore, we created a series of deletion mutants of mFox-2, which included an NLS to ensure proper localization to the nucleus even in the absence of an endogenous mFox-2 NLS (Fig. 6A). The amino-terminally truncated and carboxy-terminally truncated mFox-2 expression constructs were cotransfected in separate experiments with the pI12DE-WT FL minigene reporter plasmid. The proteins were expressed to similar levels (Fig. 6B), and localization to the nucleus was determined by immunofluorescence (data not shown). Using RT-PCR analysis, we demonstrated that expression of Δ2-50 NLS and Δ2-102 NLS mFox-2 proteins resulted in the same amount (∼75%) of exon IIIb inclusion as that produced with expression of the WT NLS mFox-2 protein (Fig. 6C, left panel). The empty vector, used as a control, led to 1% exon IIIb inclusion (Fig. 6C, left panel). Two other amino-terminally truncated expression constructs were made deleting the entire RRM (Δ2-194 NLS and Δ2-281 NLS); however, expression from these constructs was not detected either by Western analysis or by immunofluorescence (data not shown). Expression of the carboxy-terminally truncated mFox-2 proteins (Δ294-377 NLS and Δ206-377 NLS), on the other hand, did not result in an increase in exon IIIb inclusion in comparison to expression of the WT NLS mFox-2 (Fig. 6C, right panel). When the empty vector was transfected, we observed 1% exon IIIb inclusion, and when WT NLS mFox-2 was expressed, we observed 78% exon IIIb inclusion; however, expression of both carboxy-terminally truncated mFox-2 proteins resulted in approximately 7% exon IIIb inclusion (Fig. 6C, right panel). Expression of the carboxy-terminal deletion constructs did result in an increase in the skipped splicing product, as did the WT NLS mFox-2, suggesting that these proteins were capable of interacting with the RNA. Two other carboxy-terminally truncated expression constructs were made deleting the entire RRM (Δ123-377 NLS and Δ63-377 NLS); however, they too were not detected either by Western analysis or by immunofluorescence, suggesting that deletions that destroy the RRM are not stable. The results taken together demonstrate that the amino-terminal 102 amino acids of the protein are dispensable for regulating FGFR2 exon choice, whereas the carboxy-terminal 84 amino acids of the protein are required for the proper regulation of FGFR2 exon choice. Assuming that the RRM solely mediates RNA binding, we propose that the carboxy-terminal region contains the elements critical for exon IIIb activation and IIIc repression.
FIG. 6.
The last 82 amino acids of mFox-2 are required for switching FGFR2 exon choice. (A) Schematic of WT, amino-terminal deletion, and carboxy-terminal deletion proteins. The RRM is shown in gray. The NLS (N) and V5 epitope tags are located at the carboxy-terminal portion of the protein. Functionality of the protein was determined by the ability of the expressed protein to change the FGFR2 splicing pattern. (B) Western blot analysis was performed following transient transfection of WT NLS and mutant NLS expression plasmids in HEK293T cells. The blot was probed for CA150 as a loading control and V5 to detect expression of WT NLS and mutant NLS proteins. (C) Quantification of single inclusion transcripts by RT-PCR analysis of transiently transfected minigene with empty vector (EV), WT mFox-2 NLS, and each mutant construct.
mFox-2 expression alters alternative splicing of endogenous FGFR2 transcripts.
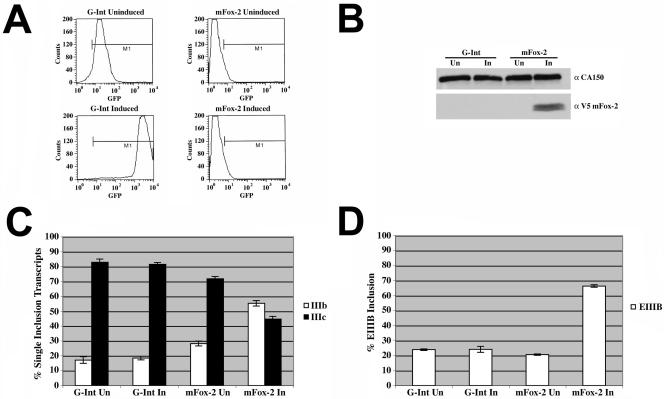
All of the data presented above relied on the use of minigene reporters to demonstrate the ability of mFox-2 to activate exon IIIb and repress exon IIIc. To further test a role of mFox-2 in FGFR2 alternative splicing, we sought to determine if expression of mFox-2 could switch the alternative splicing pattern of endogenous FGFR2 transcripts. To do this, we made the T Rex-293 mFox-2 cell line by incorporating the mFox-2 cDNA into the FRT site of the T Rex-293 cell line (Invitrogen). As a control, we used a previously described T Rex-293 G-Int cell line that expresses the green fluorescent protein (GFP) (42). After 3 days of 5-μg/ml tetracycline treatment, which is used to induce the expression of mFox-2 and GFP, the cells were harvested for FACS analysis and RNA and protein were isolated for RT-PCR and Western analysis. Using FACS analysis, we observed a small amount of GFP expression in cells that were uninduced; however, upon induction with tetracycline, we observed a significant increase in GFP expression (Fig. 7A). To show that mFox-2 was being expressed in the induced cells and not in uninduced cells or the control cell line, we detected its expression by Western analysis using the V5 antibody (Invitrogen) (Fig. 7B). Analysis of the alternative splicing of endogenous FGFR2 transcripts for the induced or uninduced T Rex-293 G-Int cell line revealed no change in the FGFR2 alternative splicing pattern (Fig. 7C). Induction of mFox-2 for 3 days, however, resulted in an increase of exon IIIb inclusion from 29% for the uninduced cells to 54% (Fig. 7C).
FIG. 7.
mFox-2 induction switches endogenous FGFR2 splicing pattern and promotes exon EIIIB inclusion in endogenous fibronectin transcripts. Expression of GFP and mFox-2 in T Rex-293 cells was induced with 5 μg/ml of tetracycline. (A) FACS analysis was used to demonstrate the tetracycline-dependent induction of GFP in the T Rex-293 G-Int cell line. (B) Western blot analysis was used to demonstrate induction of V5-tagged mFox-2. The blot was probed for CA150 as a loading control and V5 to detect mFox-2. (C) Quantification of RT-PCR analysis for endogenous FGFR2 exon IIIb or exon IIIc inclusion of uninduced (Un) or induced (In) T Rex-293 G-Int and mFox-2 cell lines. (D) Quantification of RT-PCR analysis for endogenous fibronectin exon EIIIB inclusion of uninduced (Un) or induced (In) T Rex-293 G-Int and mFox-2 cell lines.
To determine if mFox-2 expression could alter the alternative splicing of another endogenous gene, we examined the splicing pattern of endogenous fibronectin transcripts. Previously, it had been shown by Jin et al. that mFox-1 expression could weakly activate EIIIB inclusion in transcripts produced from a fibronectin minigene construct (20). We observed a dramatic increase to 67% exon EIIIB inclusion in response to mFox-2 induction in comparison to the 20 to 24% exon EIIIB inclusion in the GFP cell line and the uninduced mFox-2 cell line (Fig. 7D). These results clearly demonstrated that mFox-2 regulates the splicing of endogenous FGFR2 and fibronectin RNAs.
mFox-2 expression alters alternative splicing of endogenous hFox-2 transcripts: evidence for autoregulation.
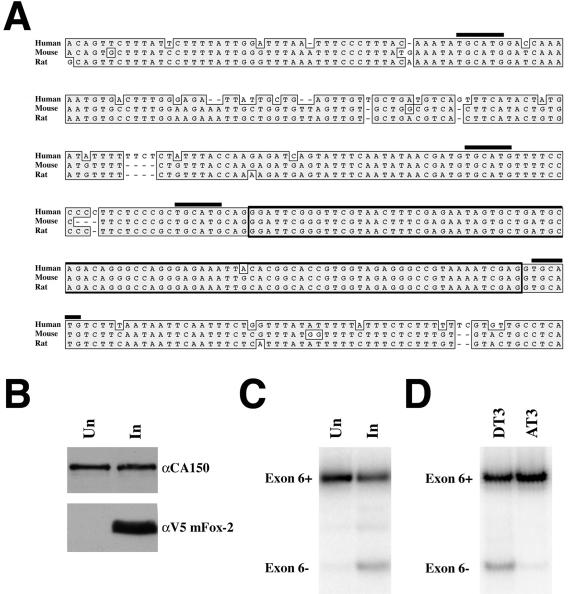
Upon scanning the genomic sequence of the human, mouse, and rat Fox-2 genes, we observed a preponderance of TGCATG and GCATG elements. The genomic region surrounding exon 6 of the transcript contained four UGCAUG elements that were highly conserved between the three species (Fig. 8A). Since exon 6 is surrounded by Fox binding sites, we believed that Fox-2 would be capable of regulating its own alternative splicing as has been shown previously for several alternative splicing factors (e.g., see reference 48 and references within). To determine if mFox-2 was capable of regulating inclusion of hFox-2 exon 6, we used the T Rex-293 mFox-2 cell line. After 3 days of 5-μg/ml tetracycline treatment the cells were harvested and RNA and protein were isolated for RT-PCR and Western analysis. Fox-2 induction was detected by Western analysis using the V5 antibody (Invitrogen) (Fig. 8B). Using primers specific for human Fox-2 exons 4 and 7, we were able to show that upon mFox-2 induction there was an increase in hFox-2 exon 6 skipping (designated exon 6− in Fig. 8C). Remarkably, this is what we observe in the DT3 cell line. The DT3 cell line contains a higher number of Fox-2 transcripts that are exon 6− than does the AT3 cell line (Fig. 8D). These results clearly demonstrate that Fox-2 is capable of autoregulating the inclusion of its own exon 6 and suggest that functional rFox-2 isoforms within the DT3 cell line regulate the alternative splicing of its own transcript, whereas rFox-2 isoforms in AT3 cells do not.
FIG. 8.
Induction of mFox-2 expression regulates alternative splicing of hFox-2 exon 6. Expression of mFox-2 in T Rex-293 cells was induced as described for Fig. 7. (A) ClustalW alignment of Fox-2 genomic sequence from human, mouse, and rat. TGCATG elements are indicated with black overlines, and exon 6 is enclosed with a box. (B) Western blot analysis was used to demonstrate induction of V5-tagged mFox-2. The blot was probed for CA150 as a loading control and V5 to detect mFox-2. (C) RT-PCR analysis of hFox-2 exon 6 inclusion. Induction of mFox-2 results in an increase in exon 6− transcripts. (D) RT-PCR analysis of rFox-2 exon 6 inclusion in DT3 and AT3 cells. DT3 cells contain more Fox-2 transcripts that are exon 6− compared to the AT3 cells.
Fox-2 is required for a IIIc-to-IIIb switch during a mesenchymal-to-epithelial transition.
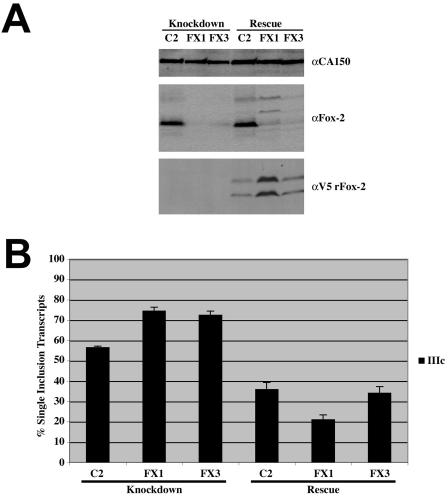
In order to establish that Fox-2 was involved in FGFR2 alternative splicing regulation, we sought to knock down Fox-2 in DT3 cells that include exon IIIb and repress exon IIIc. Cells were treated with a nonspecific siRNA and two separate siRNAs that target Fox-2. Western analysis demonstrates that the nonspecific C2 siRNA did not alter Fox-2 expression; however, the FX1 and FX3 siRNAs significantly reduced Fox-2 expression (Fig. 9A). RT-PCR analysis of the transfected minigene showed that, when the cells were treated with the C2 duplex, we saw 57% exon IIIc inclusion (Fig. 9B). When we knocked down expression of Fox-2 with either the FX1 or FX3 siRNA duplex, levels of exon IIIc increased to 75% and 73%, respectively (Fig. 9B). When we cotransfected RNAi-resistant Fox-2 expression plasmids, levels of exon IIIc inclusion dropped to 37% with the C2 siRNA, 20% with the FX1 siRNA, and 33% with the FX3 siRNA. Since Fox-2 overexpression has been shown to favor the skipped splicing product, it may explain the overall reduced levels of exon IIIc inclusion when cells were transfected with the RNAi-resistant expression plasmids. Furthermore, the expression level of the FX1-resistant Fox-2 construct may explain the even lower levels of exon IIIc inclusion (Fig. 9A).
FIG. 9.
RNAi-mediated Fox-2 knockdown allows for increased exon IIIc inclusion in DT3 cells. RNAi-resistant Fox-2 expression restores exon IIIc repression. (A) Western blot analysis was performed following transfection of siRNAs directed at no target (C2) or Fox-2 (FX1 and FX3). The blot was probed for CA150 as a loading control, Fox-2 to demonstrate knockdown and rescue of Fox-2 protein, and V5 to show expression of RNAi-resistant Fox-2 expression plasmid. (B) Quantification of single inclusion transcripts by RT-PCR analysis of transiently transfected pI12DE-IIIb ss Mut minigene reporter in cells treated with siRNAs and pTracer EF B (Knockdown) and cells treated with siRNAs and RNAi-resistant rFox-2 expression plasmids (Rescue).
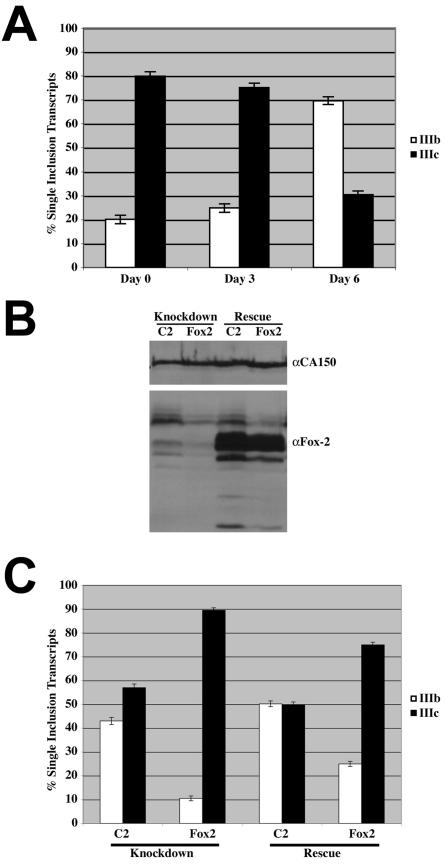
Having demonstrated that the Fox-2 knockdown affected the splicing of minigene reporter constructs, we next wanted to investigate whether Fox-2 knockdown would alter the alternative splicing of endogenous FGFR2 pre-mRNAs. To do this, we took advantage of a phenomenon that had been observed when the T Rex-293 cell line is grown to overconfluency. As these cells grow from a low confluence to a high confluence, the splicing pattern of the endogenous FGFR2 changes from primarily exon IIIc inclusion to exon IIIb inclusion. To demonstrate this phenomenon, we seeded 50,000 cells and harvested RNA the following day (day 0), at 3 days, and at 6 days. RT-PCR analysis demonstrates that exon IIIb inclusion increases from 20% at day 0 to nearly 70% at day 6 once the cells have reached maximal confluence (Fig. 10A). This change in FGFR2 splicing pattern is accompanied by other changes consistent with a mesenchyme-to-epithelium-like transition (A. P. Baraniak, S. Oltean, and M. A. Garcia-Blanco, unpublished results). Since siRNA transfections are transient in nature, we decided to increase the rate at which the cells grew to overconfluency by plating 500,000 cells per well and harvesting RNA and protein 48 h posttransfection. To investigate whether Fox-2 knockdown would alter the splicing pattern of endogenous FGFR2 pre-mRNAs, we treated these cells with the nonspecific C2 siRNA or FX2 siRNA, which targets the 3′ untranslated region of the human Fox-2 gene and was shown to knock down Fox-2 (41). Western analysis reveals that the FX2 siRNA knocks down expression levels in comparison to cells treated with the C2 siRNA (Fig. 10B). The knockdown in Fox-2 expression with the FX2 siRNA resulted in a significant decrease in the levels of exon IIIb inclusion as the cells reached overconfluency. Cells treated with the C2 duplex included exon IIIb in 42% of the transcripts, while cells that were treated with the FX2 siRNA included exon IIIb in only 10% of the transcripts. When the cells were transfected with siRNAs and also with an expression plasmid coding for a Fox-2 variant resistant to FX2 knockdown, levels of exon IIIb inclusion reached 50% with cells treated with the C2 siRNA and 25% with cells treated with the FX2 siRNA. An increase in endogenous exon IIIb inclusion from 10% in cells treated with the FX2 siRNA to 25% in cells treated with the FX2 siRNA and complemented with Fox-2 overexpression indicated that Fox-2 is involved in regulating the alternative splicing of endogenous FGFR2 transcripts.
FIG. 10.
Fox-2 is required for FGFR2 splicing switch during a mesenchymal-to-epithelial transition. (A) Quantification of RT-PCR analysis for endogenous FGFR2 exon IIIb or exon IIIc inclusion transcripts as T Rex-293 cells are grown to confluence. (B) Western blot analysis was performed following transfection of siRNAs directed at no target (C2) or Fox-2 (FX2). The blot was probed for CA150 as a loading control and Fox-2 to demonstrate knockdown of endogenous protein (Knockdown and Rescue) and expression of exogenous mFox-2 (Rescue). (C) Quantification of RT-PCR analysis for endogenous FGFR2 exon IIIb or exon IIIc inclusion transcripts in cells treated with siRNAs and pTracer EF B (Knockdown) and cells treated with siRNAs and the RNAi-resistant mFox-2 expression plasmid (Rescue).
DISCUSSION
(U)GCAUG elements abound in the proximity of regulated exons.
It is not surprising to find multiple and important GCAUG elements within alternatively spliced portions of FGFR2 transcripts. GCAUG or UGCAUG elements [hereafter (U)GCAUG] have been reported to play a role in the proper splicing regulation of fibronectin, c-src, NMHC-B, 4.1R, calcitonin/CGRP, the mitochondrial ATP synthase γ subunit (F1γ), and α-actinin RNAs (8, 17, 19, 20, 23, 26, 28, 30). Usually multiple copies of (U)GCAUG are found in the vicinity of regulated exons within these transcripts. Even in c-src, where only one (U)GCAUG element has been determined to be important for neural specific regulation of exon N1 (30), there are several (U)GCAUG elements that have been conserved from mouse to human (28) (data not shown). In a survey of 27 transcripts with alternative exons preferentially included in brain Minovitsky et al. showed that [UGCAUG] hexamers are overrepresented, particularly in the 400 nucleotides downstream of the exon (28). This overrepresentation was not observed when a similar analysis was carried out with 100 non-tissue-specific alternative exons (28). The intronic (U)GCAUG elements that we showed to be functionally significant in tissue-specific FGFR2 exon choice fit well with the bioinformatic data of Minovitsky et al. (28). The element downstream of ISAR core (CGCAUGCAUG) contains the preferred U at position 1, and even though it is found more than 900 nucleotides downstream of the regulated exon IIIb, the stem formed between the IAS2 and ISAR elements brings this (U)GCAUG element very close to the regulated exon (1). It is clear, however, that important (U)GCAUG elements may also be present outside intronic regions that immediately follow a regulated exon, and indeed we showed above that the UGCAUG sequence found within exon IIIc is critical for repression of this exon and for activation of the distant exon IIIb. Therefore, the location of these elements may be quite flexible.
Not all (U)GCAUG elements within a particular transcript have a demonstrable effect on splicing, and it is not immediately obvious what determines the difference. In the case of calcitonin/CGRP, it was shown that only two of the five (U)GCAUG elements, one intronic and one exonic, are needed for proper regulation (17). This mirrors the situation with FGFR2 splicing regulation where deletion of two (U)GCAUG elements within the loop region between IAS2 and ISAR core in FGFR2 transcripts has no effect on cell-type-specific exon inclusion (1, 32). It is possible that the elements within this large RNA loop are sequestered and thus cannot exert control over exon choice. Therefore, the number of (U)GCAUG repeats is not critical, but the position of these sequence elements within the pre-mRNA may be decisive.
The differential function of two (U)GCAUG elements in the same transcript neighborhood could be due to the influence of surrounding sequences; however, examination of the immediate localities in FGFR2 did not reveal discernible motifs that correlated with function. Moreover, bioinformatic analysis has shown that while (U)GCAUG elements occur within extended regions of conservation among homologous transcripts, these sequences are not shared between other (U)GCAUG-containing transcripts (28). Another possible explanation for the differential importance of (U)GCAUG elements is the existence of other bordering cis elements. In FGFR2 pre-mRNA, the functional (U)GCAUG element downstream of ISAR core is positioned next to multiple PTB binding sites upon formation of the IAS2-ISAR stem structure. This approximation could allow the Fox protein to counteract PTB-mediated repression of exon IIIb as proposed by Jin et al. for the α-actinin SM exon (20). In fact the functional (U)GCAUG elements in c-src, NMHC-B, 4.1R, calcitonin/CGRP, the mitochondrial ATP synthase γ subunit (F1γ), and fibronectin are also positioned near PTB binding sites (20) (data not shown). Since PTB has been reported to repress regulated exons in each of these transcripts, it is conceivable that proximity to PTB sites determines the function of (U)GCAUG elements. It is noteworthy that, in the case of Fox-2 transcripts, a (U)GCAUG element overlaps the 5′ splice site of exon 6. We do not favor the idea that the (U)GCAUG elements are part of a common larger cis element but rather that in each transcript the (U)GCAUG sequences neighbor unique subsets of other cis-acting elements. It must be noted that the rules that govern (U)GCAUG action will likely be complex given that different Fox proteins and isoforms recognize these sequences in different cell types (see above).
Fox proteins as regulators of cell-type-specific splicing.
We show here that overexpression of zFox-1, mFox-1, and mFox-2 in HEK293T cells, which normally include FGFR2 exon IIIc, resulted in a switch from exon IIIc to exon IIIb inclusion in FGFR2 minigene constructs. Our data suggest that this effect was due to a modest activation of exon IIIb and a strong repression of exon IIIc. These dual effects are similar to those observed for α-actinin upon overexpression of Fox-1 (20); however, the location of the (U)GCAUG elements relative to the regulated exons in these two transcripts does not lead to an obvious general model. In α-actinin the exon harboring a (U)GCAUG (SM) is activated, while in FGFR2 it (exon IIIc) is repressed. While both results from Jin et al. (20) and our own suggest a role for Fox proteins in alternative splicing of these transcripts, one has to cautiously interpret overexpression experiments that report effects on cotransfected minigene constructs. In order to assuage some of these concerns we used T Rex-293 mFox-2 cells where expression of mFox-2 at moderate levels switched the splicing pattern of endogenous FGFR2 transcripts from IIIc to IIIb. Additionally we observed a strong activation of exon EIIIB in fibronectin transcripts. These data indicate that Fox proteins can regulate exon choice and suggest that these proteins play such a role in determining tissue-specific alternative splicing decisions. Indeed such a function was confirmed by Fox-2 knockdown in this study and that of Underwood et al. (41).
How do Fox proteins control cell-type-specific FGFR2 exon choice? The simplest explanation would be that Fox proteins are expressed only, or are expressed at higher levels, in cells that include exon IIIb and repress exon IIIc, i.e., epithelial cells. Nonetheless, this simple model, which appears to explain neural specific effects of Fox proteins on the c-src N1 exon (41), cannot explain the differences between AT3 and DT3 cells. The data above demonstrate that Fox-2 isoforms are expressed in both the epithelial cell-like DT3 cells that include exon IIIb and repress exon IIIc and fibroblast-like AT3 cells that include exon IIIc. The Fox-2 isoforms expressed, however, are very different in these two cells, and the differences are not due to differential use of the B40 or M43 exon as in the system described by Nakahata and Kawamoto (33). Therefore, the regulation of FGFR2 by Fox-2 appears to be mediated by yet a different mechanism, and these observations suggest that the Fox proteins appear to be a remarkably diverse and versatile family of splicing regulators.
Fox-2 and EMT.
We show that Fox-2 is critical in mediating an alternative splicing switch of FGFR2 transcripts that accompanies an MET. We suggest that Fox-2 isoforms will play an important role in molding gene expression programs in transitions between epithelial and mesenchymal states. Epithelial-mesenchymal transitions (EMT) and the reverse MET are believed to play essential roles in embryonal morphogenesis and in the progression of cancers (22, 40). EMT are associated with dramatic changes in gene expression programs including the expected epithelial-mesenchymal expression of FGFR2(IIIb) and -(IIIc), respectively (37), and there are compelling data that suggest the importance of proper regulation of FGFR2 splicing in EMT. Indeed haploinsufficiency of the EMT regulator TWIST (50) leads to craniosynostosis syndromes very similar to those caused by defects in FGFR2 expression (31, 47). We postulate that Fox-2 will mediate a large number of alternative splicing changes during EMT and MET.
Supplementary Material
Acknowledgments
We thank Andrew Lieberman for providing the mFXH expression construct and Yui Jin for providing the mFox-1 and zFox-1 expression constructs. We thank Nicole Dixon for help in elucidating Fox-2 transcription start sites as well as critical reading of the manuscript. We thank Sebastian Oltean for help in purification of the Fox-2 antibodies and members of the Garcia-Blanco laboratory for many helpful discussions. We thank Doug Black for generously sharing data before publication and for providing us with a copy of his group's unpublished manuscript.
This research was supported by a PHS grant (RO1 GM063090) to M.A.G.-B.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baraniak, A. P., E. L. Lasda, E. J. Wagner, and M. A. Garcia-Blanco. 2003. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 23:9327-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brudno, M., M. S. Gelfand, S. Spengler, M. Zorn, I. Dubchak, and J. G. Conboy. 2001. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 29:2338-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns, R. C., T. J. Fairbanks, F. Sala, S. De Langhe, A. Mailleux, J. P. Thiery, C. Dickson, N. Itoh, D. Warburton, K. D. Anderson, and S. Bellusci. 2004. Requirement for fibroblast growth factor 10 or fibroblast growth factor receptor 2-IIIb signaling for cecal development in mouse. Dev. Biol. 265:61-74. [DOI] [PubMed] [Google Scholar]
- 4.Carstens, R. P., J. V. Eaton, H. R. Krigman, P. J. Walther, and M. A. Garcia-Blanco. 1997. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene 15:3059-3065. [DOI] [PubMed] [Google Scholar]
- 5.Carstens, R. P., W. L. McKeehan, and M. A. Garcia-Blanco. 1998. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol. Cell. Biol. 18:2205-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carstens, R. P., E. J. Wagner, and M. A. Garcia-Blanco. 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7388-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champion-Arnaud, P., C. Ronsin, E. Gilbert, M. C. Gesnel, E. Houssaint, and R. Breathnach. 1991. Multiple mRNAs code for proteins related to the BEK fibroblast growth factor receptor. Oncogene 6:979-987. [PubMed] [Google Scholar]
- 8.Deguillien, M., S. C. Huang, M. Moriniere, N. Dreumont, E. J. Benz, Jr., and F. Baklouti. 2001. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood 98:3809-3816. [DOI] [PubMed] [Google Scholar]
- 9.Del Gatto, F., and R. Breathnach. 1995. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 15:4825-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Gatto, F., M. C. Gesnel, and R. Breathnach. 1996. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 24:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Gatto, F., A. Plet, M. C. Gesnel, C. Fort, and R. Breathnach. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Gatto-Konczak, F., C. F. Bourgeois, C. Le Guiner, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Gatto-Konczak, F., M. Olive, M. C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dell, K. R., and L. T. Williams. 1992. A novel form of fibroblast growth factor receptor 2. Alternative splicing of the third immunoglobulin-like domain confers ligand binding specificity. J. Biol. Chem. 267:21225-21229. [PubMed] [Google Scholar]
- 15.De Moerlooze, L., B. Spencer-Dene, J. Revest, M. Hajihosseini, I. Rosewell, and C. Dickson. 2000. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127:483-492. [DOI] [PubMed] [Google Scholar]
- 16.Hajihosseini, M. K., S. Wilson, L. De Moerlooze, and C. Dickson. 2001. A splicing switch and gain-of-function mutation in FgfR2-IIIc hemizygotes causes Apert/Pfeiffer-syndrome-like phenotypes. Proc. Natl. Acad. Sci. USA 98:3855-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedjran, F., J. M. Yeakley, G. S. Huh, R. O. Hynes, and M. G. Rosenfeld. 1997. Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc. Natl. Acad. Sci. USA 94:12343-12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovhannisyan, R. H., and R. P. Carstens. 2005. A novel intronic cis element, ISE/ISS-3, regulates rat fibroblast growth factor receptor 2 splicing through activation of an upstream exon and repression of a downstream exon containing a noncanonical branch point sequence. Mol. Cell. Biol. 25:250-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh, G. S., and R. O. Hynes. 1994. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 8:1561-1574. [DOI] [PubMed] [Google Scholar]
- 20.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, R. B., R. P. Carstens, Y. Luo, and W. L. McKeehan. 2001. 5′- and 3′-terminal nucleotides in the FGFR2 ISAR splicing element core have overlapping roles in exon IIIb activation and exon IIIc repression. Nucleic Acids Res. 29:3557-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, Y., and J. Massague. 2004. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 118:277-279. [DOI] [PubMed] [Google Scholar]
- 23.Kawamoto, S. 1996. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J. Biol. Chem. 271:17613-17616. [PubMed] [Google Scholar]
- 24.Kiehl, T. R., H. Shibata, T. Vo, D. P. Huynh, and S. M. Pulst. 2001. Identification and expression of a mouse ortholog of A2BP1. Mamm. Genome 12:595-601. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman, A. P., D. L. Friedlich, G. Harmison, B. W. Howell, C. L. Jordan, S. M. Breedlove, and K. H. Fischbeck. 2001. Androgens regulate the mammalian homologues of invertebrate sex determination genes tra-2 and fox-1. Biochem. Biophys. Res. Commun. 282:499-506. [DOI] [PubMed] [Google Scholar]
- 26.Lim, L. P., and P. A. Sharp. 1998. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol. 18:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miki, T., D. P. Bottaro, T. P. Fleming, C. L. Smith, W. H. Burgess, A. M. Chan, and S. A. Aaronson. 1992. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc. Natl. Acad. Sci. USA 89:246-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minovitsky, S., S. L. Gee, S. Schokrpur, I. Dubchak, and J. G. Conboy. 2005. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 33:714-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mistry, N., W. Harrington, E. Lasda, E. J. Wagner, and M. A. Garcia-Blanco. 2003. Of urchins and men: evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA 9:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modafferi, E. F., and D. L. Black. 1997. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol. 17:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morriss-Kay, G. M., S. Iseki, and D. Johnson. 2001. Genetic control of the cell proliferation-differentiation balance in the developing skull vault: roles of fibroblast growth factor receptor signalling pathways. Novartis Found. Symp. 232:102-116. [DOI] [PubMed] [Google Scholar]
- 32.Muh, S. J., R. H. Hovhannisyan, and R. P. Carstens. 2002. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2). J. Biol. Chem. 277:50143-50154. [DOI] [PubMed] [Google Scholar]
- 33.Nakahata, S., and S. Kawamoto. 2005. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 33:2078-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norris, J. D., D. Fan, A. Sherk, and D. P. McDonnell. 2002. A negative coregulator for the human ER. Mol. Endocrinol. 16:459-468. [DOI] [PubMed] [Google Scholar]
- 35.Oldridge, M., E. H. Zackai, D. M. McDonald-McGinn, S. Iseki, G. M. Morriss-Kay, S. R. Twigg, D. Johnson, S. A. Wall, W. Jiang, C. Theda, E. W. Jabs, and A. O. Wilkie. 1999. De novo alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am. J. Hum. Genet. 64:446-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orr-Urtreger, A., M. T. Bedford, T. Burakova, E. Arman, Y. Zimmer, A. Yayon, D. Givol, and P. Lonai. 1993. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2). Dev. Biol. 158:475-486. [DOI] [PubMed] [Google Scholar]
- 37.Savagner, P., A. M. Valles, J. Jouanneau, K. M. Yamada, and J. P. Thiery. 1994. Alternative splicing in fibroblast growth factor receptor 2 is associated with induced epithelial-mesenchymal transition in rat bladder carcinoma cells. Mol. Biol. Cell 5:851-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata, H., D. P. Huynh, and S. M. Pulst. 2000. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 9:1303-1313. [DOI] [PubMed] [Google Scholar]
- 39.Sune, C., and M. A. Garcia-Blanco. 1995. Transcriptional trans activation by human immunodeficiency virus type 1 Tat requires specific coactivators that are not basal factors. J. Virol. 69:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiery, J. P. 2003. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15:740-746. [DOI] [PubMed] [Google Scholar]
- 41.Underwood, J. G., P. L. Boutz, J. D. Dougherty, P. Stoilov, and D. L. Black. 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 25:10005-10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner, E. J., A. Baines, T. Albrecht, R. M. Brazas, and M. A. Garcia-Blanco. 2004. Imaging alternative splicing in living cells. Methods Mol. Biol. 257:29-46. [DOI] [PubMed] [Google Scholar]
- 43.Wagner, E. J., A. P. Baraniak, O. M. Sessions, D. Mauger, E. Moskowitz, and M. A. Garcia-Blanco. 2005. Characterization of the intronic splicing silencers flanking FGFR2 exon IIIb. J. Biol. Chem. 280:14017-14027. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, E. J., M. L. Curtis, N. D. Robson, A. P. Baraniak, P. S. Eis, and M. A. Garcia-Blanco. 2003. Quantification of alternatively spliced FGFR2 RNAs using the RNA invasive cleavage assay. RNA 9:1552-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, E. J., and M. A. Garcia-Blanco. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10:943-949. [DOI] [PubMed] [Google Scholar]
- 47.Wilkie, A. O., M. Oldridge, Z. Tang, and R. E. Maxson, Jr. 2001. Craniosynostosis and related limb anomalies. Novartis Found. Symp. 232:122-133. [DOI] [PubMed] [Google Scholar]
- 48.Wollerton, M. C., C. Gooding, E. J. Wagner, M. A. Garcia-Blanco, and C. W. Smith. 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13:91-100. [DOI] [PubMed] [Google Scholar]
- 49.Yan, G., Y. Fukabori, G. McBride, S. Nikolaropolous, and W. L. McKeehan. 1993. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol. Cell. Biol. 13:4513-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, J., S. A. Mani, J. L. Donaher, S. Ramaswamy, R. A. Itzykson, C. Come, P. Savagner, I. Gitelman, A. Richardson, and R. A. Weinberg. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927-939. [DOI] [PubMed] [Google Scholar]
- 51.Yasumoto, H., A. Matsubara, K. Mutaguchi, T. Usui, and W. L. McKeehan. 2004. Restoration of fibroblast growth factor receptor 2 suppresses growth and tumorigenicity of malignant human prostate carcinoma PC-3 cells. Prostate 61:236-242. [DOI] [PubMed] [Google Scholar]
- 52.Yayon, A., Y. Zimmer, G. H. Shen, A. Avivi, Y. Yarden, and D. Givol. 1992. A confined variable region confers ligand specificity on fibroblast growth factor receptors: implications for the origin of the immunoglobulin fold. EMBO J. 11:1885-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeakley, J. M., J. B. Fan, D. Doucet, L. Luo, E. Wickham, Z. Ye, M. S. Chee, and X. D. Fu. 2002. Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol. 20:353-358. [DOI] [PubMed] [Google Scholar]
- 54.Yu, K., A. B. Herr, G. Waksman, and D. M. Ornitz. 2000. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc. Natl. Acad. Sci. USA 97:14536-14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.