Abstract
The 17 putative RNA helicases required for pre-rRNA processing are predicted to play a crucial role in ribosome biogenesis by driving structural rearrangements within preribosomes. To better understand the function of these proteins, we have generated a battery of mutations in five putative RNA helicases involved in 18S rRNA synthesis and analyzed their effects on cell growth and pre-rRNA processing. Our results define functionally important residues within conserved motifs and demonstrate that lethal mutations in predicted ATP binding-hydrolysis motifs often confer a dominant negative phenotype in vivo when overexpressed in a wild-type background. We show that dominant negative mutants delay processing of the 35S pre-rRNA and cause accumulation of pre-rRNA species that normally have low steady-state levels. Our combined results establish that not all conserved domains function identically in each protein, suggesting that the RNA helicases may have distinct biochemical properties and diverse roles in ribosome biogenesis.
In the nucleolus, RNA polymerase I transcribes a single polycistronic pre-rRNA that is processed to obtain the mature 18S, 5.8S, and 25S-28S rRNAs. In the yeast Saccharomyces cerevisiae, the primary 35S pre-rRNA is packaged into a large RNA-protein complex (RNP) termed the small ribosomal subunit (SSU) processome-90S preribosome (8, 15, 16). The SSU processome is essential for processing at sites A0, A1, and A2, thereby releasing the 20S and 27SA2 pre-rRNAs from the primary transcript (Fig. 1) (8). The U3 small nucleolar RNA (U3 snoRNA) is an integral component of the SSU processome and base pairs with the pre-rRNA for assembly and function of the SSU processome (8). Processing at sites A0, A1, and A2 initiates the formation of 43S and 66S preribosomal particles (12, 32, 46). Alternatively, cleavage at site A3 occurs first, leading to the formation of the 23S and 27SA3 pre-rRNAs. Subsequent processing at sites A0 (22S), A1 (21S), and A2 yields the 20S pre-rRNA (43S preribosome) (Fig. 1).
FIG. 1.
(A) Pre-rRNA processing in Saccharomyces cerevisiae. RNA polymerase I transcribes a 35S pre-rRNA that contains external (ETS) and internal (ITS) transcribed spacers. The pre-rRNA is subjected to chemical modifications and cleaved at several sites to produce the mature 18S rRNA, 5.8S rRNA, and 25S rRNA. Pre-rRNA processing can start with cleavage at site A0, yielding the 33S pre-rRNA, followed by cleavages at sites A1 and A2 or A3. Alternatively, cleavage at site A3 occurs first, leading to the formation of the 23S and 27SA3 pre-rRNAs, which are subsequently processed at sites A0 (22S), A1 (21S), and A2 (20S), yielding the 20S pre-rRNA (43S preribosome). The 43S preribosome is exported to the cytoplasm, where it is cleaved at site D to generate the mature 18S rRNA and the SSU. Within the 66S preribosomes, the 27SA2 is subjected to processing steps that lead to the production of 5.8S and 25S rRNAs. The 27SA2 can be cleaved at site A3, leading to the production of the 25S rRNA and the short form of 5.8S [5.8S(S)]. Alternatively, the 27SA2 pre-rRNA is processed at another site, yielding the long form of 5.8S [5.8S(L)] and the 25S rRNA. These rRNAs join with 5S rRNA to form the mature 60S LSU. The oligonucleotides used in this study are indicated. (B) Schematic representation of conserved motifs in DEXD/H box helicases and explanation of their proposed function. The mutations that were introduced are indicated by arrows.
The 27SA2 and 27SA3 precursors, already packaged in 66S large ribosomal subunit (LSU) preribosomes, are then processed to the mature 5.8S and 25S rRNAs. The 20S precursor, part of the 43S preribosomes, is transported to the cytoplasm where it is cleaved at site D, generating the mature 18S rRNA and 40S small ribosomal subunits.
Ribosome biogenesis in yeast requires over 150 trans-acting protein factors, not including the small nucleolar RNAs that are also involved (12, 32, 46). As might be expected, many of the nonribosomal proteins harbor motifs that are characteristic for enzymes. These include numerous P-loop-type NTPases, protein kinases, and putative remodeling enzymes, like DEXD/H box proteins and AAA-ATPases, underscoring the complexity of ribosome biogenesis (12, 13, 17, 46). The DEXD/H box proteins function in a variety of aspects of RNA metabolism, including pre-mRNA splicing, nuclear transcription, translation, and pre-rRNA processing (45). These proteins are often referred to as RNA helicases or unwindases, since they can unwind small duplexes of RNA in an NTP-dependent, generally ATP-dependent, manner in vitro. However, many DEXD/H box proteins appear to have more diverse or distinct functions than just unwinding RNA (7, 45) and are therefore generally referred to as DEXD/H box NTPases or DEXD/H box proteins. For simplicity, we will refer to these proteins as RNA helicases.
In pre-mRNA splicing, RNA helicases play a pivotal role either by unwinding or weakening RNA duplexes or by altering RNA-protein interactions (41, 45). In addition, they may also function as “RNPases.” Active and processive unwinding of RNA duplexes can result in dissociation of RNA-bound proteins, as has been demonstrated in vitro (20). Like pre-mRNA splicing, ribosome biogenesis is a highly dynamic process that involves numerous RNA-RNA and RNA-protein interactions and structural rearrangements. The RNA helicases may mediate or disrupt base-pairing interactions between the large number of snoRNAs and the pre-rRNA or initiate structural rearrangements within the pre-rRNP to facilitate protein binding or dissociation. Thus, it is very likely that they play a central regulatory role in ribosome biogenesis.
A total of 17 RNA helicases have been implicated in ribosome biogenesis, each with a unique function; many of these are conserved in humans (45). Seven of these proteins (Dhr1, Dhr2, Dbp8, Rok1, Fal1, Rrp3, and Dbp4) (4, 6, 21, 24, 27, 47) have been shown to be specifically involved in synthesis of the 18S rRNA and thus the assembly of the SSU. Has1 is the only putative RNA helicase that is required for both SSU and LSU synthesis (11). These proteins belong to the class of DEAD (Dbp8, Rok1, Fal1, Rrp3, and Dbp4) and DEAH (Dhr1 and Dhr2) box RNA helicases and are essential for cell growth. A characteristic feature of these proteins is the presence of a conserved core within the polypeptides, which generally consists of seven to eight motifs (Fig. 1B). Motif I (Walker A), motif II (Walker B; DEXD or DEXH), and motif III (SAT) have been implicated in binding of ATP and the cation cofactor (motifs I and II) and in coupling ATP hydrolysis with the mechanical function (3, 45). The recently discovered Q motif, which appears to be specific for DEAD box-type RNA helicases, has been proposed to be required for ATP binding, ATP hydrolysis, and binding of the RNA substrate (5, 44).
To gain more insight into the role of the conserved motifs, we have performed a comprehensive characterization of almost all the DEXD/H box RNA helicases involved in ribosome biogenesis. Based on previous in vivo and in vitro studies with RNA helicases involved in pre-mRNA splicing, our study focused on generating dominant negative mutations to freeze the biochemical reactions in which these proteins are involved, thereby leading to accumulation of preribosomal complexes. The function of the endogenous helicase is thus inhibited by overexpression of an inhibitory variant of the same protein (18). For example, a mutant enzyme with an inactivated catalytic site retains its ability to bind substrate but can no longer release it, thereby sequestering the limited substrate. Such an approach has been successfully applied to precisely determine the function of many RNA helicases in energy-dependent steps of spliceosome assembly-disassembly. In almost all cases, these proteins were trapped in inactive spliceosomes (2, 10, 19, 26, 31, 39, 40). This strategy has also been adapted to characterize the recently discovered Q motif (44).
Here, we present our results on the RNA helicases involved in SSU biogenesis. We have found that mutations in domains required for ATP binding (motifs I and II) generally caused a dominant negative growth defect, while mutations in the Q motif and motif III did not. For each dominant negative protein, we have determined the effects of the mutations on pre-rRNA processing and found that yeast strains expressing these mutants accumulate pre-rRNA species that under normal conditions are barely detectable. Our results suggest that overexpression of dominant negative helicases stalls processing of pre-rRNA, leading to defects in processing at sites A0, A1, and A2. Since all the tested RNA helicases are essential for growth, we also addressed whether the mutants can support growth. Collectively, our results show that conserved motifs do not function identically in each protein and imply that these RNA helicases may have distinct or diverse biochemical properties. Our results also define functionally important residues in the SSU RNA helicases.
MATERIALS AND METHODS
Yeast strains and media.
YPH499 (mata ura3-52 lys2-80 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) was used as the parental strain to construct the strains described here. Galactose-inducible, triple hemagglutinin (3HA)-tagged genes and HA- and tandem affinity purification (TAP) carboxyl-tagged genes (Kanr marker and Kluyveromyces lactis TRP marker, respectively) were generated as previously described (25, 34). Strains with doxycycline (DOX)-repressible DEXD/H box RNA helicases alleles (tetO7; Kanr marker) were generated as described previously (14). Strains expressing 3HA carboxyl-tagged proteins were grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C. Strains expressing 3HA amino-terminal-tagged proteins (GAL::3HA-DHR1; GAL::3HA-FAL1) were grown in YPG/R (1% yeast extract, 2% peptone, 2% galactose, and 2% raffinose). For the serial dilution experiment (see Fig. 5), 2-μg/ml doxycycline (Sigma) was used. Strains carrying pYES2 plasmids (pYES2 or pYES2.1, 2μm URA; Invitrogen) were grown in synthetic complete medium lacking uracil (-URA; Clontech), supplemented with 2% dextrose (SD) or with 2% galactose and 2% raffinose (SG/R).
FIG. 5.
Motif I and II SSU RNA helicase mutations are generally lethal, whereas the Q motif and motif III mutations are frequently tolerated. SSU RNA helicases controlled by the tetO7 promoter are efficiently depleted when doxycycline is added to medium. Cells were grown in dextrose-containing media (YPD) to exponential phase, and serial dilutions (10-fold) of tetO7 strains were spotted on either glucose-containing plates (−DOX) (left panels) or glucose containing plates with doxycycline (+DOX) (right panels) and incubated at 30°C. Serial dilutions (10-fold) of SSU RNA helicase tetO7 conditional strains harboring pYES2 plasmids encoding TAP-tagged wild-type and mutant RNA helicases (indicated on the left of each panel) were grown in synthetic dextrose medium (SD-URA) and spotted on glucose-containing plates (SD-URA; DEX) and galactose-containing plates with doxycycline (SG/R-URA plus DOX; GAL + DOX). Growth was monitored at 17°C, 23°C, and 30°C.
DNA manipulations.
The TAP-tagged alleles were amplified using PCR from yeast genomic DNA prepared from a strain expressing the TAP carboxyl-tagged DEXD/H proteins. The pGAL constructs were generated by cloning the PCR products into the pYES2 vector (2μm URA; Invitrogen) using the BamHI and NotI restriction sites (DBP8-TAP and FAL1-TAP) or into the pYES2.1 TOPO vector (DHR1-TAP, DHR2-TAP, and RRP3-TAP). All mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's procedures. The sequence of all mutants was confirmed by automated DNA sequencing (W. M. Keck DNA sequencing facility at Yale University).
RNA manipulations and immunoprecipitations.
For the Northern blot analyses, cells transformed with pYES2 RNA helicase-DNA constructs were grown in SD-URA at 23°C to exponential phase and shifted to SG/R-URA, and growth was monitored for 24 or 48 h. Depending on when the rate of growth changed, RNA was extracted from cells harvested at either 0, 10, or 24 h (Dbp8, Dhr1, and Dhr2) after the shift or 0, 24, or 48 h after the shift (Fal1 and Rrp3). Northern blot analysis of high-molecular-weight RNA species on formaldehyde-1.25% agarose gels was performed as described previously (9). Immunoprecipitations and analysis of coimmunoprecipitated RNAs were performed as described previously (23).
Serial dilutions.
To determine whether proteins were dominant negative, YPH499 strains carrying pYES2 RNA helicase plasmids were grown in SD-URA to exponential phase and subsequently shifted to SG/R-URA for 6 h to induce protein expression. Ten-fold dilutions were made and spotted on SD-URA and SG/R-URA solid media. For the complementation experiments, tetO7 strains carrying pYES2 plasmids were grown in SD-URA. Ten-fold dilutions were made and spotted on SD-URA and doxycycline-supplemented SG/R-URA. Plates were incubated at 17°C, 23°C, and 30°C.
Western blot analysis.
Mpp10 was detected using a rabbit polyclonal antibody (9). For the Western blot analysis (see Fig. 3B and C), strains harboring pYES2 RNA helicase plasmids were grown in SD-URA to exponential phase. Protein expression was induced for 6 h in SG/R-URA. TAP-tagged proteins were detected with the peroxidase antiperoxidase antibody (PAP; Sigma) using procedures described previously (23).
FIG. 3.
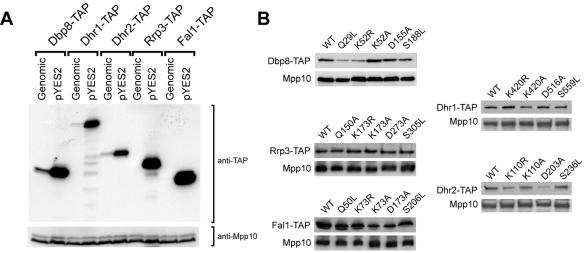
Overexpression of SSU RNA helicases from pYES2 plasmids. (A) SSU RNA helicases expressed from pYES2 plasmids accumulate at levels 5- to 50-fold higher than those of genomically encoded SSU RNA helicases. Yeast expressing genomically encoded TAP-tagged SSU RNA helicases under the control of the endogenous promoter (Genomic) or from pYES2 plasmids (pYES2) were grown in synthetic medium containing galactose and raffinose (SG/R-URA) to exponential phase. Extracts prepared from an equal amount of cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and TAP-tagged SSU RNA helicases (indicated on top of each lane) were detected by Western blot analysis with an antibody (PAP) that recognizes the protein A portion of the TAP tag. Mpp10 was used as a control for loading of equal amounts of protein. (B) Mutations in conserved motifs in SSU RNA helicases do not dramatically affect protein expression and stability. Strains carrying SSU RNA helicase pYES2 plasmids were grown in synthetic medium containing dextrose to exponential phase and subsequently grown in synthetic medium containing galactose (SG/R-URA) for 6 h to induce protein expression. TAP-tagged wild-type or mutant RNA helicases (as indicated on top of each panel) were detected by Western blot analysis using an antibody (PAP) that recognizes the protein A domain in the TAP tag. In each lane, extracts prepared from an equal number of cells were loaded, and Mpp10 was used as a control for loading equal amounts of protein.
RESULTS
Rrp3, Rok1, Dhr1, Dhr2, and Dbp8 interact with the SSU processome.
Many of the RNA helicases involved in SSU biogenesis (SSU RNA helicases) have been frequently detected in tandem-affinity-purified SSU processomes-90S preribosomes (8, 15, 16, 22). To verify their association with the SSU processome-90S preribosome, we assessed whether tagged proteins coimmunoprecipitated two known components of the SSU processome, the U3 snoRNA and the Mpp10 protein (Fig. 2). For this purpose, we constructed strains in which 3HA tags were fused to the amino- or carboxy-terminal end of chromosomally encoded genes. Immunoprecipitation experiments were performed using anti-HA antibodies. To assess the efficiency of coimmunoprecipitation and to make sure that a lack of coprecipitation was not due to RNA degradation, 10% of the input material and 10% of the supernatant were also analyzed (Fig. 2, lanes 1, 3, 4, and 6). Dbp8, Rrp3, Dhr2, and Rok1 coimmunoprecipitated both U3 snoRNA and Mpp10, albeit to various degrees, while Fal1 did not coprecipitate significant amounts of U3 and Mpp10 (Fig. 2, lanes 2 and 5). The DEAH box- and SSU processome-associated protein Dhr1 was used as a positive control (4, 8), whereas the parental strain (YPH499) and a factor involved in LSU biogenesis (Rpf2) were used as negative controls (49). Probing of the Western blots with anti-HA antibodies revealed that all 3HA-tagged SSU RNA helicases were enriched in the immunoprecipitates (data not shown). Taken together, our results establish that almost all tested SSU DEXD/H box proteins associate with the SSU processome-90S preribosome.
FIG. 2.
SSU RNA helicases associate with the SSU processome. The parental strain (YPH499) and strains expressing 3HA-tagged SSU RNA helicase proteins (Dhr1, Dhr2, Dbp8, Rrp3, Fal1, and Rok1) or 3HA-tagged LSU protein Rpf2 were grown in YP medium to exponential phase. Tagged proteins were immunoprecipitated from extracts using mouse anti-HA monoclonal antibodies (12CA5). Coimmunoprecipitated proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and coimmunoprecipitated RNA was resolved on 6% polyacrylamide-8 M urea gels. Mpp10 was detected with a rabbit anti-Mpp10 polyclonal antibody (9), whereas the U3 snoRNA was detected by Northern blotting with a radiolabeled antisense U3 oligonucleotide (9).
Construction and expression of mutant RNA helicase alleles.
To better understand the function of the SSU RNA helicases in ribosome biogenesis, we performed a large-scale mutational analysis to assess the importance of predicted motifs for protein function. We focused on generating dominant negative mutations, because if a protein is dominant negative it efficiently competes with the endogenous protein for RNA or RNP substrate binding. Also, it provides a strong indication that the protein is assembled into or interacts with preribosomes. As templates for the mutagenesis experiments, high-copy-number (2μm plasmid) constructs harboring TAP epitope-tagged alleles were generated, which were placed under the conditional GAL1 promoter (pYES2). This plasmid and promoter was used to ensure high levels of expression and therefore to increase the probability that the mutant proteins would outcompete the wild-type proteins. For our analysis, we specifically targeted amino acids that are predicted to be involved in ATP binding and/or ATP hydrolysis and that were most likely to cause a dominant negative phenotype, based on others' previous studies (1a, 2, 6, 10, 19, 26, 28, 29, 31, 35-37, 39, 40, 44, 50). We focused on the Q motif, motif I (Walker A), motif II (Walker B; DEXD/H), and motif III (SAT). A schematic overview of the motifs and the mutations that were introduced in each SSU RNA helicase is shown in Fig. 1B. The highly conserved glutamine in the Q motif, common only to DEXD box proteins (44), was mutated to either an alanine (Rrp3) or a leucine (Dbp8; Fal1) (44). The mutation in the invariant lysine in the predicted motif I (Walker A) was mutated to either an alanine or an arginine. Based on studies performed with DNA helicases-recombinases (33, 42, 43), the alanine substitution should abrogate ATP binding and hydrolysis, while the more conservative arginine mutation generally blocks only ATP hydrolysis. The first aspartic acid in motif II (DEXD/H box), which may coordinate binding of the Mg2+ cofactor (3), was replaced by alanine. The first amino acid in motif III was mutated to a leucine, which, based on in vivo and in vitro studies, may uncouple ATP hydrolysis and RNA helicase activity (30, 31, 40). Overall, we predicted that these mutations would specifically affect ATP binding and hydrolysis and would have the lowest probability of affecting RNA or RNP substrate binding.
To determine the degree of overexpression of the mutated helicases, we compared the helicase protein levels in strains expressing the wild-type TAP-tagged RNA helicases from the pYES2 plasmid to strains expressing the genomically encoded TAP-tagged RNA helicases. Strikingly, expression from the pYES2 plasmid yielded approximately 5- to 50-fold-higher expression levels than expression from the endogenous promoter (Fig. 3A; Genomic versus pYES2). In this exposure, the genomically expressed Rrp3-TAP and Fal1-TAP were not detectable and only became visible in longer exposures (data not shown). We next analyzed the expression levels of the mutant RNA helicases and compared them to the wild-type proteins expressed from the pYES2 plasmids (Fig. 3B). Protein expression was induced for 6 h by growing the cells in galactose-containing medium (SG/R). The results show that, except for Dbp8 Q29L, Dbp8 K52R, and Dhr2 D203A, the expression levels of the mutant RNA helicases were comparable to those of the wild-type proteins, demonstrating that the mutations did not dramatically affect their accumulation in the cell.
While we had wanted to include the SSU helicases Dbp4 and Rok1 in our analyses, we were unable to do so for the following reasons. In contrast to the other helicases, TAP-tagged Dbp4 was expressed only at low levels from the pYES2 plasmid (data not shown) and so was not a good candidate for the generation of dominant negative mutants. Strains expressing a genomically encoded Rok1-TAP protein grew much slower than the parental strain (approximately twofold; data not shown), suggesting that the tag alone impaired Rok1 function. We therefore focused on the other putative RNA helicases required for SSU biogenesis.
RNA helicases carrying mutations in motifs I and II are generally dominant negative.
To determine whether the mutations conferred a dominant negative growth defect when overexpressed in the presence of the endogenous protein, we made 10-fold serial dilutions of strains carrying pYES2 plasmids harboring the wild-type or mutant RNA helicases. Yeasts were spotted on galactose-containing medium to express the helicases and on dextrose-containing medium to ascertain equal spotting of cells. Growth was monitored at 17°C, 23°C, and 30°C (Fig. 4 and Table 1). For most RNA helicases, overexpression of motif I and motif II mutants, which we predicted would block ATP binding and/or hydrolysis, caused reduced growth rates, although to varying degrees. This was frequently most prominent at 17°C and indicates that the RNA helicases harboring mutations in ATP binding-hydrolysis motifs are dominant negative in vivo. There were, however, some exceptions including Dhr1 K420R, Dbp8 K52R (at 17°C and 23°C), and Dhr2 D203A. Furthermore, motif I and II mutants in Rrp3 and Fal1 caused only very moderate dominant negative growth defects. However, in general (also based on results discussed below) (Table 1), we conclude that mutations in motifs I and II are dominant negative.
FIG. 4.
Motifs I and II of SSU RNA helicase mutants are frequently dominant negative. YPH499 strains carrying pYES2 plasmids encoding TAP-tagged wild-type or mutant SSU RNA helicases (indicated on the left of each panel) were grown in synthetic medium containing dextrose (SD-URA) to exponential phase and subsequently grown in synthetic medium containing galactose (SG/R-URA) for 6 h to induce protein expression. Serial dilutions (10-fold) were spotted on either dextrose-containing plates (SD-URA; DEX) to verify even spotting of the culture and on galactose-containing plates (SG/R-URA; GAL) to induce expression from the pYES2 plasmids. Growth was monitored at 17°C, 23°C, and 30°C.
TABLE 1.
Summary of resultsa
The column “Dominant negative?” summarizes the data shown in Fig. 4 and 6; +++, ++, +, and +/− signs indicate the degree of dominant negative growth observed on plates or in culture. The column “Supports growth?” summarizes the data shown in Fig. 5; +++, ++, and + signs indicate the degree of complementation from the mutated RNA helicases. The column “Processing defects caused by dominant negative mutants (23°C)” summarizes data shown in Fig. 7.
In contrast, mutations in both the Q and motif III (SAT) of the SSU helicases did not cause dominant negative growth defects. It has been proposed that the Q motif is required for ATP binding, ATP hydrolysis, and RNA-RNP substrate binding, whereas motif III (SAT) may couple ATP hydrolysis and mechanical activity (5, 44, 45). Our results show that none of the Q motif mutations caused a dominant negative effect on growth. Furthermore, except for the Dhr2 S236L mutant, overexpression of motif III mutants also did not dramatically affect the growth rate at all the temperatures tested. Thus, SSU helicases with mutations that are predicted to affect RNA binding or helicase activity generally were not dominant negative when overexpressed.
In summary, our results show that for most of the RNA helicases tested, mutations in motif I and motif II caused dominant negative growth defects, whereas mutations in motif III and the Q motif did not.
Highly conserved residues in motif I and motif II are essential for function; however, substitutions in the Q motif and motif III are not always deleterious.
All of the RNA helicases required for SSU biogenesis are essential for viability. To assess the importance of the predicted motifs for function in vivo, we tested whether the mutated proteins could support growth in strains where the endogenous proteins were depleted. For this purpose, the chromosomally encoded genes were placed under the control of the tetracycline-doxycycline-regulatable promoter (tetO7). When these conditional strains transformed with the pYES2 RNA helicase constructs are grown on DOX-supplemented galactose-containing plates, they were dependent on the RNA helicase protein expressed from the pYES2 plasmid. In this way, we could test whether the RNA helicase mutants were required for cell viability and thus whether they were functional in vivo. The results of the complementation assays are shown in Fig. 5 and summarized in Table 1. The first panel on the left in Fig. 5 shows that the addition of DOX to solid medium caused a significantly reduced growth rate in tetO7 conditional strains, although with some variability, demonstrating that the antibiotic causes efficient RNA helicase depletion. As expected, we observed that mutations in RNA helicases that conferred a dominant negative growth defect were generally not functional in vivo. Conversely, a large number of the RNA helicase mutants that were not dominant negative sustained growth as well as their wild-type counterpart and were therefore functional. Exceptions were the Dhr2 D203A and Dhr1 K420R mutants, which either partially (Dhr2 D203A) or completely (Dhr1 K420R) (Table 1) failed to support growth but were not dominant negative.
In contrast, mutations in the Q motif had little effect on growth. Both the glutamine-to-alanine or glutamine-to-leucine mutations in the Q motif supported growth, indicating that an intact Q motif is not essential for function in SSU RNA helicases. However, mutation of the Q in Dbp8 and Rrp3 did cause slight temperature sensitivity (Fig. 5; Table 1), suggesting that it is important to a limited extent. In contrast to previously published work (44), the glutamine-to-leucine mutation in the Fal1 Q motif did not appear dramatically impair its function (Fal1 Q50L) (Fig. 5).
Mutations in motif III had diverse effects on RNA helicase function. Substitutions in motif III (SAT-LAT) affected the function of the DEAH box RNA helicases Dhr1 and Dhr2, while the DEAD box RNA helicase motif III mutants (Dbp8, Rrp3, and Fal1) were capable of supporting growth in depleted cells (Fig. 5; Table 1).
Although we have not tested other substitutions, these results suggest that mutations in highly conserved regions are very often tolerated and that many of these conserved residues are not absolutely essential for function. Furthermore, our results demonstrate that substitutions in motifs I and II generally disrupted SSU RNA helicase function, whereas mutations in the Q motif and motif III were not always deleterious. However, because the mutant proteins were overexpressed in our assay, it is possible that our assay did not allow us to distinguish between fully functional and partially defective enzymes. Nevertheless, considering that the same mutations in different RNA helicases gave different results, we can conclude that not all of the conserved residues are equally important for SSU RNA helicase function in vivo.
Overexpression of dominant negative SSU RNA helicases delays pre-rRNA processing of the 35S pre-rRNA and the synthesis of 18S rRNA.
We examined the effect of the dominant negative SSU helicases on pre-rRNA processing. To obtain RNA for analysis, yeast was grown in liquid culture at 23°C, since the growth defect was usually most prominent at low temperatures. At mid-log phase, the cells were shifted to galactose-containing medium to induce expression of the mutant RNA helicases. Growth was monitored for 48 h. In most cases, mutations in helicases that were dominant negative on plates also conferred growth defects in culture, although the effect was sometimes not dramatic (Fig. 6). Yeasts were harvested at several time points after the shift to galactose, depending on when the growth rate changed, and RNA was extracted for Northern blot analysis. For the strains expressing dominant negative mutations that caused a strong growth defect on solid medium (mutations in Dbp8, Dhr1, and Dhr2), we analyzed RNA from cells harvested at 0, 10, and 24 h following the shift to galactose (Fig. 7 and Table 1). For the analysis of strains expressing dominant negative mutants that caused a moderate or weak growth defects on solid medium and in culture (Rrp3 and Fal1), we isolated RNA from cells harvested at 0, 24, and 48 h after the shift (Fig. 7 and Table 1).
FIG. 6.
Growth curves of strains expressing SSU RNA helicases from pYES2 plasmids. Growth rates of the wild type (diamonds), motif I K-A mutants (squares), motif I K-R mutants (triangles), and motif II D-A mutants (stars) in SG/R-URA at 23°C are shown. Strains carrying pYES2 RNA helicase plasmids (indicated in the legends on the figure) were grown in synthetic dextrose-containing medium (SD-URA) to exponential phase and subsequently shifted to galactose-based medium (SG/R-URA); growth was monitored for 24 to 48 h at 23°C. The absorbance at an optical density of 600 nm is plotted on the y axis, whereas the time in galactose-based medium (in hours) is plotted on the x axis.
FIG. 7.
Overexpression of dominant negative SSU RNA helicases causes aberrant pre-rRNA processing. Strains carrying pYES2 RNA helicase plasmids (indicated on the top of each panel) were grown in synthetic dextrose-containing medium (SD-URA) to exponential phase and subsequently shifted to galactose-based medium (SG/R-URA). Yeasts were grown for 24 to 48 h at 23°C. RNA was extracted from cells harvested at the indicated time points (in hours), and equal amounts of RNA was resolved by 1.25% denaturing agarose gels. Northern blots were hybridized with various radiolabeled oligonucleotides (indicated on the sides of the panels). The positions of the various pre-rRNAs on the membrane are depicted on the left of each panel.
Our Northern analysis results suggest that overexpression of the Dbp8, Fal1, and Dhr2 dominant negative mutants affected processing at sites A0, A1, and A2 and therefore the production of 18S rRNA. The defects in processing at A0, A1, and A2 are illustrated by the accumulation of 23S pre-rRNAs (Fig. 1 and Fig. 7, lanes 1 to 18 and 31 to 51). The 23S pre-rRNA is an 18S rRNA precursor that contains the 5′ external and internal transcribed spacers up to the A3 cleavage site. Therefore, its accumulation points to defects in processing at A0, A1, and A2. In some cases, we also observed concomitant reduction in levels of the 32S and 20S pre-rRNAs in cells expressing the dominant negative proteins. Notably, overexpression of the Dbp8 dominant negative mutants also caused a modest accumulation of 22S pre-rRNAs, indicating delays in A1 and A2 processing (Fig. 1A and Fig. 7, lanes 4 to 12). Similarly, the dominant negative Rrp3 mutants delayed processing at sites A1 and A2, as indicated by the accumulation of the 22S pre-rRNA species (Fig. 1A and Fig. 7, lanes 22 to 30). Indeed, this is consistent with the pre-rRNA processing defects observed upon genetic depletion of Rrp3 (27). In most cases, the dominant negative mutants did not completely block but seemed to kinetically delay pre-rRNA processing, leading to accumulation of intermediates (in these experiments, this was consistently the 23S pre-rRNA) that were usually barely detectable. Also, except for the Dbp8 and Dhr2 mutants, we observed little effect on the accumulation of the mature 18S rRNA, also indicating a slowing in pre-rRNA processing. These results are consistent with previously published genetic depletion results (4, 6, 21), except that genetic depletion of Dhr1 has been shown to primarily affect cleavages at sites A1 and A2 (4).
DISCUSSION
Here, we present a detailed and comprehensive mutational analysis of DEXD/H box RNA helicases required for 18S rRNA synthesis, which we call the SSU RNA helicases. We have verified that many of the SSU RNA helicases interact with the SSU processome-90S preribosomes and have defined functionally important amino acids within these proteins. Interestingly, many of the lethal mutations in motifs predicted to be involved in ATP binding conferred a dominant negative growth defect when overexpressed in a wild-type strain, suggesting that ATP binding is essential for in vivo function but likely not required for binding of RNA or RNP substrate. Analysis of pre-rRNA processing in strains expressing dominant negative mutants revealed kinetic delays in 35S pre-rRNA processing and in some cases reduced accumulation of the 18S rRNA, defects that in most cases were similar to what was observed upon genetic depletion of the RNA helicases.
Most of the SSU RNA helicases associate with the SSU processome.
Our immunoprecipitation experiments confirmed that almost all of the tested SSU RNA helicases interact with the SSU processome-90S preribosome, as judged by their association with the SSU processome components Mpp10 and U3 snoRNA. Prior to our work, the SSU helicases could be divided into two groups: those that had been frequently copurified with preribosomes in tandem affinity purifications (Dbp8, Dhr1, Dbp4, and Rok1) and those that had been rarely or never detected in preribosomes (Rrp3, Dhr2, and Fal1) (15, 22, 48). Consistent with this, we found that the SSU helicases in the first group all coimmunoprecipitated SSU processome components. Also consistent with this, we found that Fal1 (in the second group) did not coimmunoprecipitate SSU processome components. However, both Rrp3 and Dhr2, which havenever been found in tandem affinity purifications, did coimmunoprecipitate small amounts of SSU processome components. It may be that enzymes such as the SSU RNA helicases transiently interact with preribosomes and thus are not always detectable in purified particles or yield small amounts of coimmunoprecipitating SSU processome components. In addition, while we have found that Dhr2 coimmunoprecipitates SSU processome components Mpp10 and U3 snoRNA, another group found that it did not (4). Consistent with our results, the pattern of pre-rRNAs obtained upon Dhr2 depletion implies a function in the SSU processome-90S preribosome (4).
The DEAD box protein-specific Q motif.
It has been proposed that the highly conserved glutamine in the Q motif is required for ATP binding, ATP hydrolysis, and binding of RNA substrate (5, 44). Glutamine-to-alanine or -leucine mutations in the DEAD box proteins eIF4-A, Ded1, Prp5, and Fal1 were previously shown to be lethal, suggesting that this conserved amino acid has an important function in these proteins (44). Therefore, we were surprised that analogous mutations in the Q motifs of the DEAD box SSU RNA helicases Dbp8, Rrp3, and Fal1 did not appear to markedly affect function in vivo. We do not understand why we have obtained different results upon mutation of the Q motif of Fal1 than others have found previously (44), but it is consistent with our overall results. Of the 13 RNA helicases tested in our combined analyses, only the Dbp10, Drs1, and Dbp6 Q motif mutations were lethal. The Dbp6 and Drs1 Q motif mutants also conferred a dominant negative growth defect (1). Although our analysis of the Q motif was not exhaustive because we did not test other substitutions, our results suggest that an intact glutamine in the Q motif in most cases is not essential for function of the DEAD box RNA helicases involved in ribosome biogenesis.
Elements implicated in ATP binding: motifs I and II.
Our results show that the mutations in motifs I and II, which are predicted to affect ATP binding and hydrolysis, were generally dominant negative when expressed in yeast. Moreover, of these, none of the motif I lysine mutants were able to support growth. Likewise, for many RNA helicases required for splicing, analogous mutations in splicing RNA helicases in motif I (Prp16 and Prp22) and motif II (Prp16, Prp22, and Prp43) are lethal and also cause dominant negative growth defects (19, 26, 38). Since a dominant negative effect is a very strong indication that these mutants have retained their ability to bind their RNA and/or RNP substrates, it is tempting to speculate that for most RNA helicases required for ribosome biogenesis, ATP binding is not essential for RNA-RNP substrate binding. Indeed, almost all of the dominant negative splicing factor mutants that have been characterized interact with the spliceosome and are often more stably bound than their wild-type counterparts (10, 31, 36, 39).
Substitutions in highly conserved residues in motif III are frequently tolerated.
Previous results suggest that motif III has a role in coupling ATP hydrolysis with helicase activity but is not required for binding ATP. In the prototypical DEAD box protein eIF4-A and the DEAH box protein Prp22, substitution of the serine (SAT) in motif III abrogated RNA helicase activity but did not dramatically affect ATP hydrolysis (30, 40); in the case of Prp2, it also did not interfere with ATP hydrolysis (31). We showed that the serine-to-leucine substitutions in motif III only affected the DEAH box RNA helicases Dhr1 and Dhr2; the latter mutant also conferred a strong dominant negative growth defect when overexpressed (Table 1). Introduction of this mutation in the motif III of DEAH box LSU RNA helicases showed similar results, but the Mtr4 motif III mutant was the only one that conferred a strong dominant negative effect in vivo (1). Thus, it appears that an intact first amino acid in motif III (SAT or TAT for Dbp8) is not essential for function for many of the RNA helicases required for ribosome biogenesis.
The use of dominant negative RNA helicases as tools to study RNA helicase-RNP complex interactions in vivo.
One of the goals of this project was to use the dominant negative mutant proteins to purify stalled or intermediate preribosomal complexes because dominant negative mutants are predicted to stably bind their RNA or RNP substrates. However, when we analyzed the association of TAP-tagged wild-type and dominant negative proteins with pre-rRNA precursors, both the wild-type and mutant proteins coimmunoprecipitated many diverse pre-rRNAs (data not shown). After testing different negative controls in the immunoprecipitations, we concluded that the observed interactions were probably related to overexpression of the helicases yielding nonspecific interactions. Expression of SSU RNA helicase mutants from a CEN-based low-copy-number plasmid (pGAD3) did not cause dominant negative growth defects (data not shown), indicating that very high levels of expression are absolutely essential to outcompete the wild-type proteins.
Collectively, our results with 23 mutations in the SSU RNA helicases, with 4 to 5 mutations per helicase, show that lethal mutations in RNA helicases involved in ribosome biogenesis predicted to block ATP binding and/or ATP hydrolysis are very often also dominant negative when overexpressed in yeast. This indicates that ATP binding and hydrolysis are essential for function but not always a prerequisite for RNA or RNP substrate binding. Moreover, mutation of residues that are conserved among all the tested RNA helicases did not have the same effect on every protein, strongly indicating that they do not all share the same biochemical properties. Thus, it is very likely that these RNA helicases have distinct functions in ribosome biogenesis.
Acknowledgments
We are grateful to Alicia Lee, Paul Stone, and Neal Janson for expert technical assistance. We thank the members of our laboratory for critically reading the manuscript.
This work was supported by Leslie H. Warner and Anna Fuller Postdoctoral Cancer Research fellowships (S.G.) and the National Institutes of Health (S.J.B.; GM52581).
REFERENCES
- 1.Bernstein, K. A., S. Granneman, A. V. Lee, S. Manickam, and S. J. Baserga. 2006. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis. Mol. Cell. Biol. 26:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Blum, S., S. R. Schmid, A. Pause, P. Buser, P. Linder, N. Sonenberg, and H. Trachsel. 1992. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89:7664-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campodonico, E., and B. Schwer. 2002. ATP-dependent remodeling of the spliceosome: intragenic suppressors of release-defective mutants of Saccharomyces cerevisiae Prp22. Genetics 160:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 4.Colley, A., J. D. Beggs, D. Tollervey, and D. L. Lafontaine. 2000. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol. Cell. Biol. 20:7238-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordin, O., N. K. Tanner, M. Doere, P. Linder, and J. Banroques. 2004. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 23:2478-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daugeron, M. C., and P. Linder. 2001. Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis. Nucleic Acids Res. 29:1144-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 8.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, D. A., S. Wormsley, T. M. Agentis, and S. J. Baserga. 1997. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 17:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwalds-Gilbert, G., D. H. Kim, S. H. Kim, Y. H. Tseng, Y. Yu, and R. J. Lin. 2000. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. RNA 6:1106-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emery, B., J. de la Cruz, S. Rocak, O. Deloche, and P. Linder. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 52:141-158. [DOI] [PubMed] [Google Scholar]
- 12.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 13.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 14.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulating gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 15.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 16.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 17.Granneman, S., and S. J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43-50. [DOI] [PubMed] [Google Scholar]
- 18.Herskowitz, I. 1987. Functional inactivation of genes by dominant negative mutations. Nature 329:219-222. [DOI] [PubMed] [Google Scholar]
- 19.Hotz, H. R., and B. Schwer. 1998. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics 149:807-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291:121-125. [DOI] [PubMed] [Google Scholar]
- 21.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:7283-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S. J., and S. J. Baserga. 1999. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19:5441-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, W. Q., J. A. Clark, and M. J. Fournier. 1997. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol. Cell. Biol. 17:4124-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 26.Martin, A., S. Schneider, and B. Schwer. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277:17743-17750. [DOI] [PubMed] [Google Scholar]
- 27.O'Day, C. L., F. Chavanikamannil, and J. Abelson. 1996. 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res. 24:3201-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh, J. Y., and J. Kim. 1999. ATP hydrolysis activity of the DEAD box protein Rok1p is required for in vivo ROK1 function. Nucleic Acids Res. 27:2753-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pause, A., N. Methot, and N. Sonenberg. 1993. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 13:6789-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pause, A., and N. Sonenberg. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11:2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plumpton, M., M. McGarvey, and J. D. Beggs. 1994. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 13:879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raue, H. A. 2004. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae, p. 199-222. In M. O. Olson (ed.), The nucleolus. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 33.Rehrauer, W. M., and S. C. Kowalczykowski. 1993. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 268:1292-1297. [PubMed] [Google Scholar]
- 34.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 35.Schmid, S. R., and P. Linder. 1991. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol. 11:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider, S., H. R. Hotz, and B. Schwer. 2002. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 277:15452-15458. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, S., and B. Schwer. 2001. Functional domains of the yeast splicing factor Prp22p. J. Biol. Chem. 276:21184-21191. [DOI] [PubMed] [Google Scholar]
- 38.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwer, B., and C. Guthrie. 1992. A dominant negative mutation in a spliceosomal ATPase affects ATP hydrolysis but not binding to the spliceosome. Mol. Cell. Biol. 12:3540-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwer, B., and T. Meszaros. 2000. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 19:6582-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwer, B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8:113-116. [DOI] [PubMed] [Google Scholar]
- 42.Sung, P., D. Higgins, L. Prakash, and S. Prakash. 1988. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 7:3263-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sung, P., and S. A. Stratton. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983-27986. [DOI] [PubMed] [Google Scholar]
- 44.Tanner, K. N., O. Cordin, J. Banroques, M. Doere, and P. Linder. 2003. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11:127-138. [DOI] [PubMed] [Google Scholar]
- 45.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251-262. [DOI] [PubMed] [Google Scholar]
- 46.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 47.Venema, J., C. Bousquet-Antonelli, J. P. Gelugne, M. Caizergues-Ferrer, and D. Tollervey. 1997. Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol. 17:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vos, H. R., R. Bax, A. W. Faber, J. C. Vos, and H. A. Raue. 2004. U3 snoRNP and Rrp5p associate independently with Saccharomyces cerevisiae 35S pre-rRNA, but Rrp5p is essential for association of Rok1p. Nucleic Acids Res. 32:5827-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wehner, K. A., and S. J. Baserga. 2002. The sigma 70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9:329-339. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, M., and M. R. Green. 2001. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev. 15:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]