Abstract
DEXD/H box putative RNA helicases are required for pre-rRNA processing in Saccharomyces cerevisiae, although their exact roles and substrates are unknown. To characterize the significance of the conserved motifs for helicase function, a series of five mutations were created in each of the eight essential RNA helicases (Has1, Dbp6, Dbp10, Mak5, Mtr4, Drs1, Spb4, and Dbp9) involved in 60S ribosomal subunit biogenesis. Each mutant helicase was screened for the ability to confer dominant negative growth defects and for functional complementation. Different mutations showed different degrees of growth inhibition among the helicases, suggesting that the conserved regions do not function identically in vivo. Mutations in motif I and motif II (the DEXD/H box) often conferred dominant negative growth defects, indicating that these mutations do not interfere with substrate binding. In addition, mutations in the putative unwinding domains (motif III) demonstrated that conserved amino acids are often not essential for function. Northern analysis of steady-state RNA from strains expressing mutant helicases showed that the dominant negative mutations also altered pre-rRNA processing. Coimmunoprecipitation experiments indicated that some RNA helicases associated with each other. In addition, we found that yeasts disrupted in expression of the two nonessential RNA helicases, Dbp3 and Dbp7, grew worse than when either one alone was disrupted.
Virtually every cellular process involving RNA requires the function of an ATP-dependent DEXD/H box RNA helicase. These enzymes are known to be involved in mRNA splicing, transcription, RNA editing, and ribosome biogenesis. RNA helicases are conserved from bacteria to humans and require eight conserved motifs for ATP binding and hydrolysis, substrate binding, and conformational changes related to their functions (Fig. 1A) (11, 15, 16, 42, 45, 48). In particular, motif I, motif II (DEXD/H box), and motif VI are required for ATPase activity; motif III (SAT) couples ATP hydrolysis with helicase function and motif VI is also required for RNA interactions. Recently, it has been shown that RNA helicases, in addition to their ability to bind and unwind RNA, can also modify ribonucleoprotein complexes (RNPs) (19, 25).
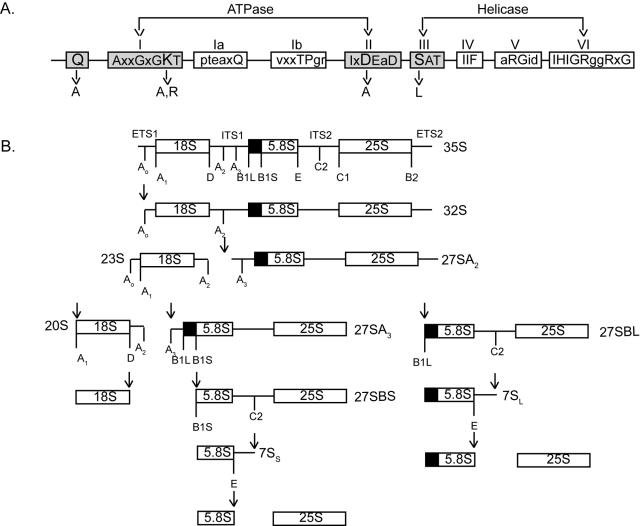
FIG. 1.
(A) Conserved motifs in DEXH/D box RNA helicases. Motifs I and II (DEXD/H box) are required for ATP binding and hydrolysis, whereas motifs III (SAT) and VI are required for RNA unwinding and RNP remodeling functions. The mutations that we made are shown. (B) Schematic of pre-rRNA processing in yeast. Pre-rRNA processing of the 18S, 5.8S, and 25S rRNAs begins on the 35S pre-rRNA primary transcript. Cleavage at A2 in internal transcribed spacer 1 (ITS1) in the 35S pre-rRNA separates the 18S rRNA (which is incorporated into the SSU) from the 5.8S and 25S rRNAs (which are incorporated into the LSU). Processing of the 5.8S and 25S rRNAs occur through two distinct pathways. The main difference is that in the major pathway, the 5′ end of 5.8S is cleaved at B1S, whereas in the minor pathway the 5′ end of 5.8S is extended to B1L (the long form). ETS, external transcribed spacer.
Pre-rRNA processing begins with transcription of the 35S pre-rRNA in the nucleolus. Through a stepwise series of cleavage events, this transcript is processed into the mature 18S, 5.8S, and 25S rRNAs (Fig. 1B). The 5S rRNA is transcribed and processed separately. The 5S, 5.8S, and 25S rRNAs become incorporated into the large ribosomal subunit (LSU), while the 18S becomes incorporated into the small ribosomal subunit (SSU). Processing in internal transcribed spacer 1 separates the small and large ribosomal precursors and forms the 20S and 27SA2 pre-rRNAs (Fig. 1B). The 20S pre-rRNA is subsequently processed at site D to form the mature 18S rRNA. Processing of the 5.8S and 25S rRNAs occurs through two distinct pathways. In the major pathway, the 27SA2 pre-rRNA is subsequently cleaved into the 27SA3 pre-rRNA at site A3. The 27SA3 pre-rRNA undergoes an additional cleavage step at B1S to mature the 5′ end of the 5.8S rRNA into its shorter form. Subsequently, the 27SBS pre-rRNAs undergo cleavage at C2, where the 5.8S and 25S rRNA are separated. The 3′ end of the 7S pre-rRNA is then processed at site E to give rise to the mature 5.8S, whereas cleavage at C1 matures the 5′ end of the 25S rRNA. The main difference between the major and minor processing pathways is that in the major pathway, which occurs 70% of the time, the 5′ end of the 5.8S rRNA is cleaved into a shorter form.
Recent genomic and proteomic data have suggested that there are >200 nonribosomal proteins that are required for pre-rRNA processing, including putative RNA helicases (21, 35, 51). RNA helicases are thought to be important for mediating RNA-RNA and RNA-protein interactions required for both pre-rRNA processing and ribosome assembly. In the yeast Saccharomyces cerevisiae, 17 of them are involved in pre-rRNA processing, 7 for SSU biogenesis, and 9 for LSU biogenesis. One RNA helicase is required for both LSU and SSU biogenesis. Of the 10 helicases involved in LSU biogenesis, Dbp6, Dbp7, Dbp9, Mak5, and Drs1 have previously been described as involved in both 25S and 5.8S rRNA maturation, whereas Dbp3, Dbp10, Spb4, and Mtr4 are necessary for processing of the 5.8S rRNA alone (5, 8, 10, 12, 13, 28, 40, 54, 56). Has1 has previously been shown to be required for SSU biogenesis; here, we confirm that it has an additional role in LSU biogenesis (18). Of the 10 LSU putative RNA helicases, only 2 have been previously shown to have ATPase and helicase activities in vitro (27, 41). Although previous studies have shown that these helicases are required for pre-rRNA processing at different sites, the substrates of these helicases and their precise roles in pre-rRNA processing are currently unknown.
To better understand the cellular roles of RNA helicases and their motifs in each step of pre-rRNA processing, we created a series of mutations in the conserved regions of eight essential RNA helicases involved in LSU biogenesis. Five individual mutations were made in motifs predicted to be required for putative ATPase (Q motif, motif I, and motif II) and unwinding (motif III) functions (Fig. 1A). We subsequently assayed yeasts carrying these mutated helicases for dominant negative growth defects and for complementation. Unlike other types of mutations, dominant negative mutants likely retain their protein structure. Therefore, helicases with dominant negative mutations might be used to specifically inhibit helicase enzymatic functions, while maintaining the ability to bind (but not release) RNA or RNP substrate.
By this approach, we determined that mutation of specific residues in motifs required for ATPase activity frequently caused dominant negative growth defects, suggesting that these regions are essential for function but not needed for RNA or RNP substrate binding. In addition, helicases expressing mutations in motifs required for substrate binding infrequently caused dominant negative growth. Pre-rRNA processing defects accompanied the dominant negative growth defects. Coimmunoprecipitation experiments showed that mutated RNA helicases with similar effects on pre-rRNA processing associated together. Our results suggest that RNA helicases that function in early or later steps of pre-rRNA processing do indeed associate together. Finally, we have evidence that is consistent with the hypothesis that yeasts with disruptions in the two nonessential RNA helicases, Dbp3 and Dbp7, grow worse than when either one is disrupted.
MATERIALS AND METHODS
Yeast strains and media.
All yeast strains were derived from YPH499 (mata ura3-52 lys2-80 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1). Yeast was grown in rich medium YPD (1% yeast extract, 2% peptone, and 2% glucose), YPG/R (1% yeast extract, 2% peptone, 2% galactose [GAL], and 2% raffinose), or synthetic medium lacking uracil or lacking tryptophan (-URA or -TRP) and supplemented with dextrose (SD-URA and SD-TRP) or galactose (SG-URA and SG-TRP), where specified. Yeast strains were created with RNA helicase genes under a tetracycline (TET)-repressible promoter (tetO7) as described previously (2). Repression of the TET promoter was achieved with the addition of 2 μg of doxycycline (DOX)/ml to SG-URA or SG-TRP medium (SG-URA plus DOX; SG-TRP plus DOX). Yeasts with C-terminal triple hemagglutinin (3HA)-tagged proteins or N-terminal GAL-3HA-tagged proteins were created as described previously (33). The tandem affinity purification (TAP) tag was genomically integrated at the C terminus of each RNA helicase gene (39). The pGM435L plasmid containing the MTR4 gene sequence flanked by 5′ and 3′ untranscribed regions under a GAL10 promoter with a TRP marker was a gift from Alan Tartakoff (32).
Cloning of DEXD/H box helicase genes into yeast expression plasmids and mutagenesis.
TAP-tagged RNA helicase alleles were PCR amplified from yeast genomic DNA and cloned into the pYES2 2μm plasmid (Invitrogen; DBP6, DBP9, DRS1, HAS1, MAK5, and SPB4) or the pGAD3 centromere-based plasmid (DBP10) using BamHI and NotI restriction sites. Both plasmids have a galactose-inducible and dextrose-repressible promoter and a uracil-selectable marker. Site-directed mutagenesis of cloned RNA helicases in plasmids and the pGM435L MTR4 plasmid was conducted with the QuikChange kit (Stratagene). Sequencing of plasmids was carried out by the Yale University Keck Sequencing Facility to confirm mutagenesis.
Assay for dominant negative growth defects on plates.
Growth was assayed by plating serial dilutions of yeast with and without mutated RNA helicases. Yeasts transformed with plasmids containing wild-type or mutated RNA helicases were grown in 10 ml of SD-URA (SD-TRP for pGM435L plasmids) medium to early log phase. Yeasts were pelleted and resuspended in SG-URA (SG-TRP for pGM435L plasmids) medium for 3 h. Two hundred microliters of yeast grown to an optical density at 600 nm (OD600) of 0.4 was pelleted and resuspended in 1 ml of water. Yeasts were then diluted 1-, 10-, 100-, and 1,000-fold; 7 μl was spotted onto SD-URA, SG-URA, or SG-URA plus doxycycline (DOX, 2 μg/ml) plates where specified (pGM435L plasmids were plated on TRP plates).
Western blots.
Yeasts harboring genomically integrated C-terminally TAP-tagged RNA helicases or plasmids with wild-type or mutated RNA helicases were grown to early log phase in 10 ml of YPD or SD-URA medium, respectively. Yeasts harboring plasmids with wild-type or mutated RNA helicases were shifted into SG-URA medium for 6 h. Two milliliters of culture at an OD600 value of 0.4 was pelleted, and protein was extracted as previously described (29). Protein was analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotted with peroxidase-antiperoxidase antibodies (PAP; Sigma) to detect the protein A portion of the TAP tag and anti-Mpp10 antibodies as previously described (3, 4).
Assay for dominant negative growth defects in liquid medium.
Yeasts transformed with plasmids containing wild-type or mutated RNA helicases were grown to early log phase in 10 ml of SD-URA medium (SD-TRP for pGM435L plasmids). Yeasts were diluted to an OD600 of 0.2 and resuspended in SG-URA medium (SG-TRP for pGM435L plasmids), and growth was recorded at intervals of 6 h for 30 h total. Yeasts were diluted as necessary to maintain them at an OD600 of 0.05 to 0.6.
Northern blots.
RNA was extracted from yeast containing plasmids with clones of wild-type or mutated RNA helicases. RNA was extracted from yeast containing but not expressing the mutated RNA helicases from 25 ml of culture grown to an OD600 of 0.4 in SD-URA medium. Yeasts expressing plasmids containing wild-type or mutated RNA helicases were grown to early log phase in SD-URA medium (not expressed), pelleted, washed with water, and resuspended in SG-URA medium (expressed) for 12 or 24 h. pGM435L plasmids were grown in SD-TRP medium. RNA was extracted after 24 h of expression from 25 ml of yeast grown to an OD600 of 0.4. Twenty micrograms of RNA was run on 1.25% agarose-formaldehyde gels as previously described (31).
Coimmunoprecipitation.
Coimmunoprecipitation experiments were conducted on yeast strains with double-tagged proteins as indicated in the figures. Coimmunoprecipitation experiments were carried out with 200 μl (each) of anti-HA (12CA5 culture supernatant) with glass bead extracts (31) and blotted with PAP antibodies (1:6,666) as previously described (39). The blots were stripped and reprobed with anti-HA antibodies (12CA5 culture supernatant, 1:100).
RESULTS
Strategy for creation of dominant negative mutant proteins.
To further define where RNA helicases required for large-subunit biogenesis function in pre-LSU rRNA processing, we sought to create dominant negative mutations that would function by sequestering the endogenous substrate from the wild-type protein. The mutant helicase would therefore be predicted to seize its substrate and “freeze” reaction intermediates. Dominant negative mutations are different from other types of mutations, since the protein maintains the ability to bind its substrate but is unable to release it, thereby sequestering the substrate and hindering the function of the concurrently expressed wild-type helicase. Since the substrates for these RNA helicases are currently unknown, we hypothesized that these mutations could be used to further pinpoint where they function in pre-rRNA processing and what their substrates may be. The helicases required for pre-LSU processing tested here are all essential for cell growth. The types of mutations we introduced in the LSU RNA helicases were based upon many previous studies performed on RNA helicases required for other cellular processes (http://www.medecine.unige.ch/∼linder/mutant_rna_helicases.html). Based upon these results, individual mutations were made in the putative Q motif (Q to A in DEAD box but not DEXH box RNA helicases) (50), motif I (K to A or R) (9, 18, 24, 34, 36, 43, 46, 49), motif II (D to A) (9, 24, 34, 36, 44, 47), and motif III (S to L) (34, 37, 38, 57) of each essential RNA helicase (Fig. 1A). Using this strategy, we would be able to infer which motifs are required for ATPase and unwinding activity and which may inhibit substrate binding.
Creation and protein expression levels of mutant RNA helicases.
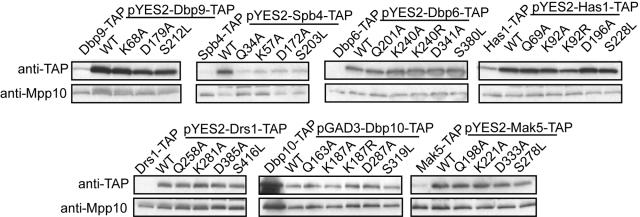
The mutations outlined above were made in RNA helicase genes cloned into either the pGAD3 (DBP10) or pYES2 (DBP6, DBP9, DRS1, HAS1, MAK5, and SPB4) plasmids, which contain a C-terminal TAP tag and a galactose-inducible and dextrose-repressible promoter. In the case of MTR4, we used the previously published and extensively studied pGM435L plasmid containing the MTR4 gene sequence under a GAL10 promoter (32). We first investigated whether the plasmid-borne wild-type or mutated RNA helicases were equally expressed or overexpressed relative to the endogenously TAP-tagged helicases, because overexpression is important for obtaining a dominant negative growth defect (23). Yeasts containing the RNA helicase plasmids were grown to early log phase in DEX medium and then shifted into GAL medium for 6 h. Protein expression was determined by Western blotting with anti-TAP (PAP) antibodies. In most cases, the wild-type or mutated helicases expressed from plasmids were greatly overexpressed compared to the endogenously TAP-tagged RNA helicases. The results show that, except for the Spb4 and Dbp10 mutants, all plasmid-encoded RNA helicases were expressed at comparable levels (Fig. 2).
FIG. 2.
Comparison of the protein levels of genomically tagged RNA helicases to the levels of wild-type and mutated RNA helicases expressed from plasmids. TAP-tagged RNA helicases created by genomic integration and yeast transformed with wild-type or mutated TAP-tagged RNA helicase plasmids (pGAD3 or pYES2) were grown to early log phase. Yeasts transformed with the helicase plasmids were shifted into SG-URA medium for 5 h. Protein was extracted, and equal amounts were run on a 10% SDS-PAGE gel, transferred to a nitrocellulose membrane, and blotted with PAP to detect the TAP tag. The blot was reprobed with anti-Mpp10 rabbit antibodies.
Mutations in motifs I and II are frequently dominant negative.
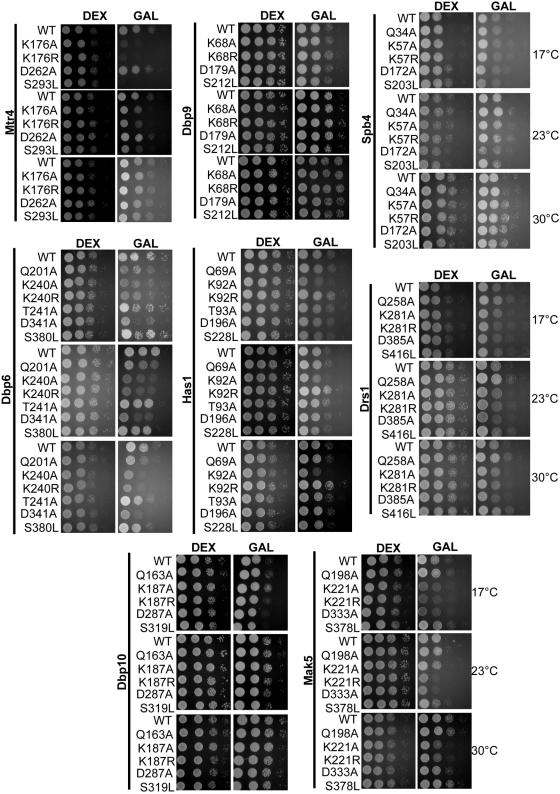
Yeasts expressing wild-type or mutated RNA helicase plasmids were screened for defects in growth. If expression of the mutated plasmid confers reduced growth in comparison to yeast expressing only the wild-type protein, then the mutated RNA helicase conferred a dominant negative growth defect. Yeasts were grown in dextrose medium to early log phase, serially diluted onto GAL or DEX plates, and incubated at 17°C, 23°C, and 30°C. Growth on dextrose plates, when the plasmid-borne helicases are not expressed, was used to ascertain that equal amounts of yeasts were plated. The results show that identical mutations made in the same conserved motifs yielded different results among the various RNA helicases tested (Fig. 3; Table 1). Overall, RNA helicases with mutations in the motif I and motif II box most frequently caused dominant negative growth defects (Fig. 1A and 3; Table 1). In contrast, yeasts expressing mutations in the Q motif or motif III occasionally caused dominant negative growth defects (Fig. 1A and 3; Table 1). For example, only Dbp6 Q201A had dominant negative growth defects; similarly, only Mtr4 S293L displayed dominant negative growth at lower temperatures (Fig. 3; Table 1). Dominant negative growth defects were observed in all RNA helicases tested except Dbp9 and Dbp10 (Fig. 3). The low protein expression levels of Spb4 and Dbp10 helicases could explain why few dominant negative growth defects were seen with Spb4 (only D172A in liquid culture) (Fig. 4) and none were observed with Dbp10.
FIG. 3.
Screen by serial dilution on plates for dominant negative growth defects in yeasts overexpressing mutant RNA helicases. The yeast parent strain, YPH499, was transformed with plasmids containing wild-type or mutated RNA helicases, as indicated. Yeasts were grown to early log phase, serial diluted (1, 10, 100, and 1,000 times) onto GAL or DEX plates, and incubated at 17°C, 23°C, or 30°C. Gene expression from the plasmid bearing the helicase was observed when plating on GAL was carried out.
TABLE 1.
Summary of resultsa
| Protein | Processing defects caused by dominant negative mutants at 30°C? | Processing defects upon genetic depletion? (reference) | Motif | Mutation | Dominant negative on plates or in liquid culture?
|
Supports growth?
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 17°C | 23°C | 30°C | 17°C | 23°C | 30°C | |||||
| Mtr4 | Cleavage at A0, A1, A2, and C2 | 7S accumulation (13) | I | K176A | +++ | ++ | + | − | − | − |
| K176R | +++ | ++ | + | − | − | − | ||||
| II | D262A | − | − | − | +++ | +++ | +++ | |||
| III | S293L | +++ | ++ | + | − | − | − | |||
| Dbp9 | Not determined | Cleavage at A0, A1, and A2 (8) | I | K68A | − | − | − | +++ | +++ | +++ |
| K68R | − | − | − | + | +++ | +++ | ||||
| II | D179A | − | − | − | + | +++ | +++ | |||
| III | S212L | − | − | − | +++ | +++ | +++ | |||
| Spb4 | Cleavage at A0, A1, A2, and C2 | Cleavage at A0, A1, A2, C1, and C2 (12) | Q | Q34A | − | − | − | +++ | +++ | +++ |
| I | K57A | − | − | − | + | ++ | +++ | |||
| K57R | − | − | − | + | ++ | +++ | ||||
| II | D172A | − | + | + | + | ++ | +++ | |||
| III | S203L | − | − | − | + | ++ | +++ | |||
| Dbp6 | None observed | Cleavage at A0, A1, and A2 (28) | Q | Q201A | +++ | + | + | − | − | − |
| I | K240A | +++ | +++ | +++ | − | − | − | |||
| K240R | +++ | +++ | +++ | − | − | − | ||||
| T241A | − | − | − | +++ | +++ | +++ | ||||
| II | D341A | +++ | +++ | +++ | − | − | − | |||
| III | S380L | − | − | − | +++ | +++ | +++ | |||
| Has1 | Cleavage at A0, A1, and A2 | Cleavage at A0, A1, and A2 (18) | Q | Q69A | − | − | − | − | − | − |
| I | K92A | − | − | + | − | − | − | |||
| K92R | − | − | + | − | − | − | ||||
| T93A | − | − | − | + | + | + | ||||
| II | D196A | − | − | + | − | − | − | |||
| III | S228L | − | − | − | ++ | ++ | ++ | |||
| Drs1 | Cleavage at A0, A1, and A2 | Not determined | Q | Q258A | − | − | + | − | + | + |
| I | K281A | + | + | + | − | − | − | |||
| K281R | + | + | + | − | − | − | ||||
| II | D385A | + | + | + | − | − | − | |||
| III | S416L | − | − | + | − | − | − | |||
| Dbp10 | Not determined | Cleavage at A0, A1, and A2 (5) | Q | Q163A | − | − | − | − | − | − |
| I | K187A | − | − | − | − | − | − | |||
| K187R | − | − | − | − | − | − | ||||
| II | D287A | − | − | − | − | − | − | |||
| III | S319L | − | − | − | +++ | +++ | +++ | |||
| Mak5 | Cleavage at A0, A1, and A2 | Not determined | Q | Q198A | − | − | − | +++ | +++ | +++ |
| I | K221A | +++ | ++ | ++ | − | − | − | |||
| K221R | +++ | +++ | ++ | − | − | − | ||||
| II | D333A | +++ | +++ | ++ | − | − | − | |||
| III | S378L | − | − | − | +++ | +++ | +++ | |||
The column “Dominant negative on plates or in liquid culture?” summarizes data shown in Fig. 3 and 4; +++, ++, and + indicate the degrees of dominant negative growth. The column “Supports growth?” summarizes data shown in Fig. 6; +++, ++, and + indicate the degrees of complementation from the mutated RNA helicases. The column “Processing defects caused by dominant negative mutants at 30°C?” summarizes data shown in Fig. 5.
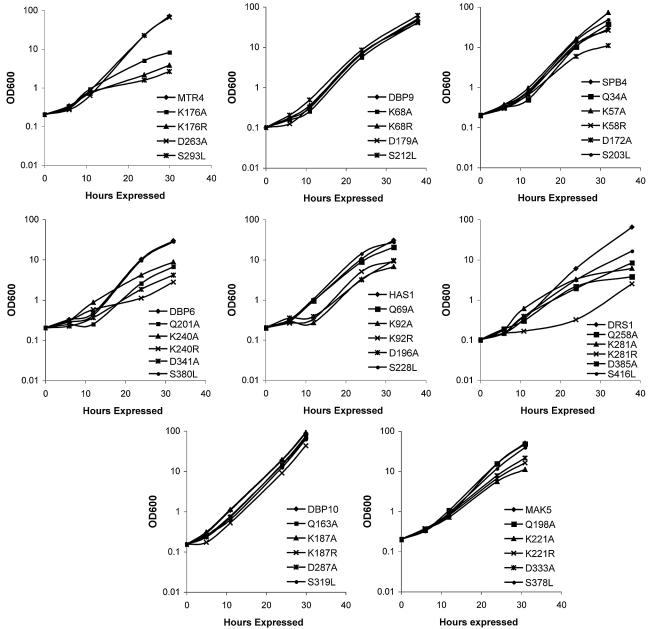
FIG. 4.
Growth curves of yeast overexpressing mutant RNA helicases. The yeast parent strain, YPH499, was transformed with plasmids containing wild-type or mutated RNA helicases. Yeasts were grown to early log phase at 30°C in DEX medium and then shifted into GAL medium for 30 h. Growth was recorded with a spectrophotometer at 0, 6, 12, 24, and 30 h of expression in GAL medium.
To verify the dominant negative growth defects observed and to determine if the mutations made in the RNA helicases have similar growth defects in liquid medium, growth curves were determined (Fig. 4; Table 1). In all cases, the dominant negative growth defects observed on plates were also present in yeasts grown in liquid medium. Interestingly, some mutations that did not cause dominant negative growth defects on agarose plates did cause dominant negative growth defects in liquid medium. For example, mutations in Has1 in motif I (K92A and K92R) and in motif II (D196A) caused a stronger dominant negative growth defect in culture than on plates (Fig. 4; Table 1). Similarly, mutations in Drs1 in the Q motif (Q258A) and motif III (S416L) caused dominant negative growth defects in culture, while we did not observe them on plates. Growth in liquid medium was therefore more sensitive in detecting growth defects than was monitoring growth on plates by serial dilution.
Overexpression of dominant negative LSU RNA helicases leads to pre-rRNA processing defects.
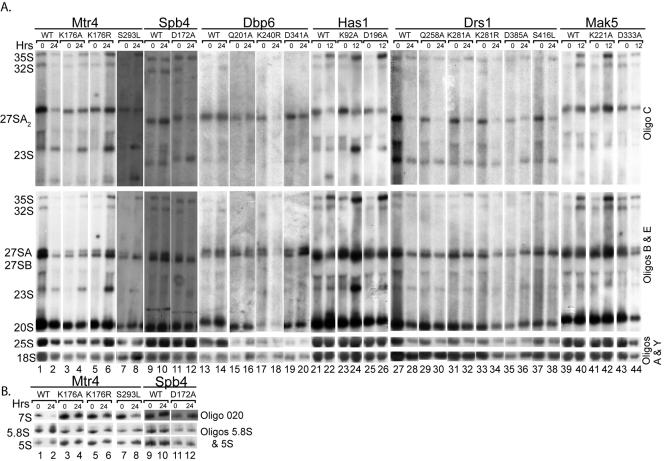
Northern blot analysis of steady-state RNA harvested for strains expressing the dominant negative mutants was used to further pinpoint pre-rRNA processing steps where the helicases are required. Yeasts containing wild-type or mutant RNA helicase plasmids were grown in DEX medium until early log phase and then shifted into GAL medium to express the plasmids for 12 or 24 h. RNA was extracted from yeast grown in DEX (0 h of expression) or GAL (24 h of expression) medium. Upon Northern blot analysis, different pre-rRNA processing defects were observed for each wild-type RNA helicase and its dominant negative mutants. In all cases, overall levels of the mature 25S, 18S, 5.8S, and 5S rRNAs were relatively unaffected by overexpression of the dominant negative helicases (Fig. 5 and data not shown), likely due to the mild nature of the growth defects. However, precursors to the mature rRNAs did accumulate. For Has1 and Spb4, the pre-rRNA processing defects observed by expression of the dominant negative mutations were similar to those observed by steady-state analysis from protein depletion (Fig. 5). Expression of the dominant negative mutants in Has1 caused processing defects at A0, A1, and A2 cleavage sites, since the 35S and 23S pre-rRNAs accumulated. Similarly, the dominant negative D172A Spb4 mutant caused very weak pre-rRNA processing defects, as seen by slight accumulation of the 23S pre-rRNA and modest reduction of the 27SA2 pre-rRNA, indicating defects at A0, A1, and A2. In addition, a very modest 7S pre-rRNA accumulation was evident, indicating a defect in cleavage at site C2 (Fig. 5, lanes 9 to 12; Table 1) (12, 18). For Drs1, an RNA helicase where steady-state pre-rRNA levels from depletion was not previously determined, defects at A0, A1, and A2 cleavage sites as determined by accumulation of the 35S and 23S pre-rRNAs and reduction of the 27SA2 and 20S pre-rRNAs were observed (Fig. 5A, lanes 27 to 38; Table 1). Since defects in these processing sites were generally observed when SSU processome proteins are depleted, we suggest that these results may be due to indirect effects on pre-rRNA processing. In some cases, no obvious pre-rRNA processing defects were observed from expression of the dominant negative mutations (i.e., Dbp6 and Mak5), suggesting that the effects from overexpression of these plasmids result in only a minor delay in pre-rRNA processing that would not be detectable in this RNA steady-state analysis but that would cause growth defects (Fig. 5A, lanes 13 to 20 and 39 to 44).
FIG. 5.
Analysis of pre-rRNA processing in yeast expressing wild-type or mutant RNA helicase plasmids. Yeasts were grown in DEX medium until early log phase (0 h of expression) and then shifted into GAL medium for 12 or 24 h (12 or 24 h of expression). RNA was extracted, run on a 12.5% agarose-formaldehyde gel, and transferred to a Hybond N+ membrane. The membrane was then probed with oligonucleotide C (which detects 35S, 32S, 27SA, and 23S pre-rRNAs), oligonucleotides B and E (which detect 35S, 27SA2, 27SB, 23S, and 20S pre-rRNAs), and oligonucleotides A and Y (which detect 18S and 25S rRNAs). RNAs extracted from strains expressing Mtr4 and Spb4 plasmids were also run on 10% polyacrylamide gels and transferred to a Hybond N+ membrane. These membranes were probed with oligonucleotides to detect 7S pre-rRNA, 5.8S rRNA, and 5S rRNA.
The pre-rRNA processing defects observed with expression of dominant negative mutations in Mtr4 also mirrored those previously observed upon protein depletion. Mtr4 was shown to be required for 3′ end processing of the 7S to the 5.8S rRNA, since yeasts depleted of Mtr4 accumulate the 7S pre-rRNA (1). Indeed, overexpression of the dominant negative mutations K176A and K176R in Mtr4 led to a modest increase of the 7S pre-rRNA compared to overexpression of the wild-type protein (Fig. 5B, compare lanes 2 with 4 and 6; Table 1). Furthermore, in all the dominant negative Mtr4 mutants, processing delays at A0, A1, and A2 were observed as determined by accumulation of the 35S and 23S pre-rRNAs. Since Mtr4 has previously been shown to be required for 3′ end formation of snoRNAs and/or snRNAs (1, 53) and as a component of an exosome quality control polyadenylation complex (30, 52), it is possible that the dominant negative mutants may inhibit essential functions of Mtr4 in these other processes as well.
Mutations in motifs I and II cause growth defects.
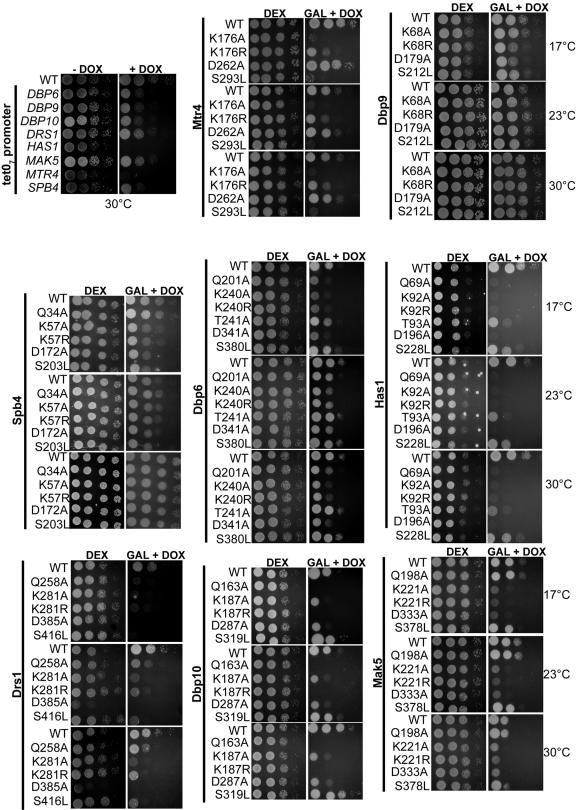
To determine whether the mutated RNA helicases could support growth when the endogenous RNA helicase was depleted, we created conditional strains in which the endogenous RNA helicase gene was under the control of a tetracycline-regulatable (tetO7) promoter. In the presence of the drug doxycycline, expression of the endogenous protein was repressed (Fig. 6). Since all the RNA helicases tested are essential, we can assess whether the mutant proteins can support growth in the presence of DOX (Fig. 6). Plasmids containing wild-type or mutant RNA helicases were transformed into tetO7-helicase strains. Yeasts were grown to early log phase inDEX, serially diluted onto plates containing DEX or GAL plus DOX, and incubated at 17°C, 23°C, and 30°C. Growth on DEX, when the plasmids were not expressed, was used to ascertain equal plating of the yeast. Lack of growth on GAL plus DOX, when the plasmids were expressed and the endogenous helicases were repressed, indicates that the mutated residue in the RNA helicase was essential for function.
FIG. 6.
The function of the mutant RNA helicases in vivo. The ability of the wild-type or mutant RNA helicase expressed from plasmids to restore growth when the endogenous helicase is depleted was assayed by serial dilution. Wild-type or mutant RNA helicase plasmids were transformed into yeast strains containing each respective helicase under a TET-repressible promoter. Yeasts were grown to early log phase in DEX medium, serially diluted (1, 10, 100, and 1,000 times) onto GAL plates with doxycycline (GAL + DOX), and incubated at 17°C, 23°C, or 30°C.
In all cases, proteins containing mutations that caused a strong dominant negative growth defect were unable to support growth in the absence of the wild-type protein. Vice versa (in most cases), proteins containing mutations that did not cause a strong dominant negative growth defect were able to support growth in the absence of wild-type protein. Dbp10 is an exception in that mutations in the Q motif, motif I, and motif II did not confer dominant negative growth defects but were unable to support growth in the absence of the wild-type protein. These results can be explained by insufficient overexpression of the Dbp10 mutant plasmids (Fig. 2). Similarly, for Has1, mutations in the Q motif and in motif I (T93A) did not cause dominant negative growth defects but were unable to fully complement depletion of the endogenous HAS1 gene. Collectively, mutations in the Q motif, motif I, and motif II most frequently were unable to complement depletion of the wild-type protein. In contrast, most RNA helicases with mutations in motif III were able to support growth when the endogenous RNA helicase was depleted, indicating that mutation of this residue to alanine does not interfere with function. Our results suggest that the residues required for ATP hydrolysis are most often required for function, whereas mutations in the first residue of motif III, which is thought to be required for RNA unwinding or RNA remodeling, are frequently tolerated.
Some RNA helicases coimmunoprecipitate nonribosomal proteins and each other.
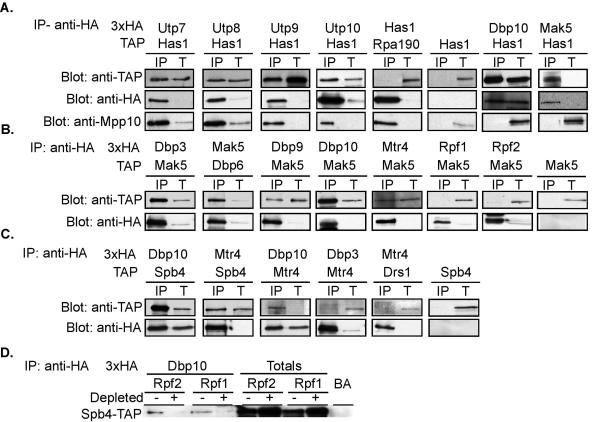
Considering the results of the mutational analysis shown and previously published genetic depletion experiments, we found that a subset of RNA helicases function at the same pre-rRNA processing steps; therefore, we hypothesized that these RNA helicases may be stably associated in vivo (Table 1). To assess whether different RNA helicases are in complexes together, we carried out coimmunoprecipitation experiments with strains in which two helicases were differentially tagged. Has1 has been previously shown to be also required for small ribosomal subunit biogenesis (18). Since depletion of Has1 resulted in pre-rRNA processing defects similar to defects observed during depletion of components of the SSU processome (4, 17), we determined whether it associates with components of the SSU processome. Yeasts with TAP-tagged Has1 and 3xHA-tagged SSU processome components (Utp7, Utp8, Utp9, or Utp10) were immunoprecipitated with beads conjugated to HA antibodies. The immunoprecipitates were run on a 10% SDS-PAGE gel, transferred to a nitrocellulose membrane, and blotted with anti-TAP and anti-HA antibodies. The results indicated that Utp7, Utp8, Utp9, and Utp10 were able to coimmunoprecipitate Has1-TAP (Fig. 7A, lanes anti-TAP and IP). As expected, Utp7, Utp8, Utp9, and Utp10 also immunoprecipitated another SSU processome component, Mpp10 (Fig. 7A, lanes anti-Mpp10 and IP). In yeast with double-tagged Has1-3xHA and Rpa190-TAP (a subunit of RNA polymerase I, used here as an unrelated protein), Has1 did not immunoprecipitate Rpa190 but Has1-3xHA was enriched and did immunoprecipitate Mpp10 (Fig. 7A, lanes anti-HA and anti-Mpp10 IP). Similarly, single-tagged Has1-TAP was negative for signals in the anti-TAP and anti-Mpp10 IP lanes (Fig. 7A). Enrichment of the HA-tagged SSU processome protein in the immunoprecipitation was verified by blotting with anti-HA antibodies (Fig. 7A, lanes anti-HA and IP). These results suggest that Has1 may be a component of the SSU processome.
FIG. 7.
Some RNA helicases coimmunoprecipitate other proteins required for ribosome biogenesis or other helicases. Protein extracts were made from the indicated yeast strains and immunoprecipitated using anti-HA antibodies conjugated to beads. Immunoprecipitated protein (IP) and total protein extracted (T; 5% of total extract) were run on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were Western blotted with PAP antibodies which detect the protein A part of the TAP tag (anti-TAP), anti-Mpp10 antibodies (anti-Mpp10) which detects a protein component of the SSU processome, and anti-HA antibodies (anti-HA) which detect the immunoprecipitated protein. BA, beads alone. (A) SSU processome proteins (Utp7, Utp8, Utp9, and Utp10) immunoprecipitate Has1; proteins required for LSU biogenesis (Dbp10 and Mak5) immunoprecipitate Has1. (B) Mak5 is immunoprecipitated by other RNA helicases required for LSU biogenesis (Dpb3, Dpb6, Dbp9, Dbp10, and Mtr4) but not other nonribosomal proteins required for large subunit biogenesis (Rpf1 and Rpf2). (C) Dbp10, Spb4, and Mtr4, three RNA helicases required for synthesis of the 5.8S rRNA, coimmunoprecipitate each other but not RNA helicases required for early steps in pre-LSU-rRNA processing (Dbp3 and Drs1). (D) When nonribosomal proteins required for LSU biogenesis (Rpf2 and Rpf1) are depleted, Dbp10 no longer immunoprecipitates Spb4.
Similarly, Has1 has also been implicated in LSU biogenesis (18, 22). We therefore determined if HA-tagged RNA helicases required for LSU biogenesis were able to coimmunoprecipitate Has1. Dbp10 and Mak5 were able to immunoprecipitate Has1 (Fig. 7A, lanes IP). As expected, Dbp10 and Mak5 did not immunoprecipitate Mpp10 (Fig. 7A, lanes anti-Mpp10 and IP). The stable association of Has1 with proteins required for both LSU and SSU biogenesis suggests that Has1 may indeed have a dual role in LSU and SSU pre-rRNA processing.
We also tested if RNA helicases required for both early and later steps of processing of the LSU are associated by coimmunoprecipitation. Two RNA helicases, Mak5 and Mtr4, were chosen for in-depth analysis as representatives of proteins required for early and later steps of pre-LSU rRNA processing, respectively (Fig. 7B) (1, 56). First, we determined if RNA helicases required for LSU biogenesis (Dbp3, Dbp9, Dbp10, and Mtr4) could coimmunoprecipitate Mak5. Protein extracts were made from yeast with double-tagged RNA helicases (either Mak5-TAP with 3HA-tagged Dbp3, Dbp9, Dbp10, or Mtr4 or Mak5-3xHA with Dbp6-TAP). The HA-tagged proteins were immunoprecipitated with beads conjugated to HA antibodies, run on an SDS-PAGE gel, and transferred to a nitrocellulose membrane. The membrane was Western blotted with anti-TAP and anti-HA antibodies. Dbp3, Dbp9, Dbp10, and Mtr4 were able to coimmunoprecipitate Mak5 (Fig. 7B, lanes anti-TAP and IP). Mak5 was able to coimmunoprecipitate Dbp6 (Fig. 7B, lanes anti-TAP and IP). As expected, HA-tagged protein was enriched in comparison to the total (Fig. 7B, lanes anti-HA and IP). Similarly, yeasts expressing only Mak5-TAP were negative for signals in the anti-TAP IP lanes (Fig. 7B). However, Rpf1 and Rpf2, two nonribosomal proteins required for early and later steps of large-subunit pre-rRNA processing, were unable to coimmunoprecipitate Mak5 (Fig. 7B, lanes anti-TAP and IP). These results suggest that Mak5 is able to stably associate with other RNA helicases required for both early (Dbp6, Dbp9, and Dbp3) and later (Mtr4 and Dbp10) steps of pre-rRNA processing. In contrast, Mak5 was unable to stably associate with Rpf1 and Rpf2, two nonribosomal proteins that function at the same pre-rRNA processing steps. To expand upon these results, we determined if other RNA helicases could coimmunoprecipitate Mtr4 in the same manner as did Mak5. Dbp10, a protein required for processing of 5.8S rRNA, was able to immunoprecipitate Mtr4; Mtr4 was able to immunoprecipitate Spb4, another protein required for processing of the 5.8S rRNA (Fig. 7C, lanes anti-TAP and IP). In contrast, Dbp3, a RNA helicase required for an earlier pre-rRNA processing step, did not immunoprecipitate Mtr4, while Mtr4 did not immunoprecipitate Drs1, another RNA helicase required for earlier pre-rRNA processing steps (Fig. 7C, lanes anti-TAP and IP). These results suggest that Mtr4 specifically interacts with a subset of RNA helicases involved in later steps of LSU biogenesis, as might be expected from its processing defects upon depletion. As expected, yeast expressing only Spb4-TAP was negative for signal in the anti-TAP IP lanes (Fig. 7C, lanes anti-HA and IP). Although a subset of RNA helicases were coimmunoprecipitable, we were not able to detect the association of the Mak5 RNA helicase with nonribosomal proteins Rpf1 and Rpf2, proteins also required for large-subunit biogenesis (Fig. 7B and data not shown). This suggests that in some instances, some helicases may not be stable components of the pre-LSU processing machinery, which is not unexpected since they are enzymes.
Conversely, we asked whether depletion of nonribosomal proteins required for LSU biogenesis would alter the ability of RNA helicases to coimmunoprecipitate each other. We therefore created strains in which two RNA helicases were differentially tagged, Dbp10-3xHA and Spb4-TAP, where nonribosomal protein Rpf2 or Rpf1 was under a galactose-inducible and dextrose-repressible promoter. Rpf2 and Rpf1 are nonribosomal proteins that have been previously shown to function in early and later steps of pre-LSU processing (20, 55). Yeasts were grown to early log phase in YPG/R medium (undepleted) and shifted into YPD for 24 h (depleted). Protein extracts were made from yeast grown in YPG/R and YPD medium; protein was immunoprecipitated with beads conjugated to HA antibodies, run on 10% SDS-PAGE gels, transferred to a nitrocellulose membrane, and blotted with anti-TAP antibodies. When yeasts were grown in YPD (undepleted), Dbp10 immunoprecipitated Spb4 (Fig. 7D, − lanes). However, when either Rpf1 or Rpf2 were depleted (Fig. 7D, + lanes), Dbp10 could no longer immunoprecipitate Spb4. These results suggest that the presence of Rpf1 and Rpf2 was required for the association of RNA helicases Dbp10 and Spb4 with each other. These results imply that RNA helicase interactions may be dependent on formation of large pre-LSU complexes.
Disruption of DBP3 and DBP7 is synthetically sick.
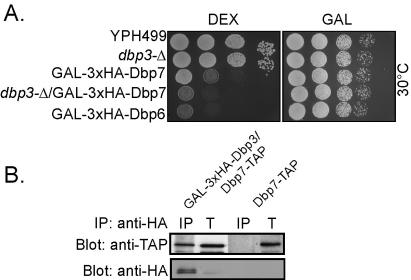
Because Dbp3 and Dbp7 are nonessential helicases involved in LSU biogenesis, we asked whether they were functionally redundant. We created yeast strains in which expression of both helicases could be interrupted and tested whether their growth was affected. Several haploid yeast strains were created to test this: one where the DBP3 gene is disrupted (dbp3-Δ), one where DBP7 gene is under the control of a galactose promoter (GAL-3xHA-DBP7), one that combined these two manipulations (dbp3-Δ/GAL-3xHA-DBP7), and one where DBP6 gene is under the control of a galactose-inducible promoter (GAL-3xHA-DBP6). The last one served as a control for the effect on growth of depletion of an essential RNA helicase. These strains were serially diluted onto YPD or YPG plates and incubated at 30°C. As expected, the parent strain, YPH499, and yeast disrupted in DBP3 grew on YPD plates to roughly the same extent (Fig. 8A). In contrast, yeasts depleted of Dbp7 (GAL-3xHA-DBP7) grew much more slowly than these two strains (Fig. 8A). However, when both DBP3 and DBP7 expression was interrupted, yeasts grew worse than when only one was disrupted. Indeed, growth was equivalent to yeast depleted of the essential RNA helicase Dbp6 (Fig. 8A). Yeasts serially diluted onto YPG, where all proteins are expressed, were used to demonstrate equal distribution of yeast onto plates (Fig. 8A). Therefore, disruption of expression of the Dbp3 and Dbp7 proteins results in a synthetically sick phenotype.
FIG. 8.
Disruption of DBP3 and DBP7, encoding two nonessential RNA helicases, is synthetically sick. (A) Yeast strains YPH499, Δdbp3, GAL-3xHA-Dbp7, Δdbp3/GAL-3xHA Dbp7, and GAL-3xHA Dbp6 were grown to early log phase, serially diluted (1, 10, 100, and 1,000 times) onto DEX (YPD) and GAL (YPG) plates, and incubated at 30°C. (B) Protein extracts made from yeast strains GAL-3xHA-Dbp3/Dbp7-TAP and Dbp7-TAP were immunoprecipitated (IP) using anti-HA antibodies conjugated to beads. Immunoprecipitated protein and total yeast extract (T; 5% of total protein extracted) were run on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were Western blotted with PAP antibodies (anti-TAP) and anti-HA antibodies (anti-HA).
We asked whether Dbp3 and Dbp7 were in a complex together by coimmunoprecipitation. Protein extracts were made from yeast with double-tagged Dbp7-TAP/GAL-3xHA-Dbp3 or Dbp7-TAP. The HA-tagged protein was immunoprecipitated by beads conjugated with HA antibodies; the immunoprecipitate was run on a 10% SDS-PAGE gel, transferred to a membrane, and Western blotted with anti-TAP and anti-HA antibodies. The results indicate that Dbp3 coimmunoprecipitates Dbp7 (Fig. 8B, lane IP) and that Dbp3 was enriched during the immunoprecipitation (Fig. 8B, lane anti-HA). Our results were consistent with those of Jansen et al., who conducted a genome-wide approach to predict protein-protein interactions and predicted that Dbp3 and Dbp7 can interact (26).
DISCUSSION
Here, we have created a comprehensive series of mutations in conserved residues in eight putative RNA helicases required for LSU biogenesis to further define motifs that are required for their function. In general, we show that the conserved ATPase motifs in the LSU RNA helicases but not in the helicase motifs are essential for function and that when these proteins are overexpressed they confer a dominant negative phenotype. In addition, we determined that some helicases coimmunoprecipitate and are therefore complexed together and that coassociation is required for assembly of the pre-LSU RNP. Finally, we determined that yeasts genetically depleted of the two nonessential RNA helicases, Dbp3 and Dbp7, grow worse than when either is individually disrupted.
In analyzing all of the LSU RNA helicases, we were able to compare the effects of mutating the same conserved residues. Surprisingly, mutations made in the conserved motifs did not consistently cause dominant negative phenotypes among all the helicases tested. These results suggest that although these residues are invariant, these motifs may not be functioning identically in vivo. Consistent with our results, recent structural analyses of related helicases suggest that although these proteins share a conserved structure, they do not necessarily have identical biochemical properties (6).
Although we were successful in creating many dominant negative mutations in most of the RNA helicases tested, we only tested a subset of residues for each motif in each helicase. Therefore, it is possible that in cases where we did not observe dominant negative growth defects, other mutations could be made that would result in reduced growth. In addition, due to the massive protein overexpression in the mutant RNA helicases, we may not have been able to detect weak growth defects for mutant protein complementation. The analysis presented here is therefore comprehensive but not exhaustive. We initially sought to determine the associated RNA substrates of the helicases by coimmunoprecipitation with the dominant negative helicases; however, attempts with the endogenously tagged RNA helicases did not reveal any associating pre-rRNAs (data not shown), which may be due to the transient association of these enzymes with their substrates. In addition, immunoprecipitation of the dominant negative helicases did not reveal specific association of pre-rRNAs either (data not shown). These results were also true of SSU RNA helicases (22a).
Q Motif.
Recent work by Tanner et al. identified a 17-amino-acid motif with an invariant glutamine located upstream of motif I, called the Q motif (50). In the case of Ded1, a translation-initiation factor, this motif was shown to be critical for ATP binding, substrate affinity, and helicase activity (7). Furthermore, mutations in the Q motif made in RNA helicases required for mRNA splicing and translation have previously been shown to have growth defects (eILFA, Ded1, and Prp5) (51). Here, we have mutated the invariant glutamine to an alanine in the six DEAD box LSU RNA helicases (Dbp6, Dbp10, Drs1, Has1, Mak5, and Spb4). In only two cases, Dbp6 and Drs1, did mutations in the Q motif confer dominant negative growth (Fig. 3 and 4; Table 1). However, in four of six RNA helicases (Dbp6, Dbp10, Drs1, and Has1), mutations in the Q motif could not support growth in the absence of the wild-type protein (Fig. 6 and Table 1). These results are not surprising, since recent work by Cordin et al. has shown that this motif is essential for RNA substrate binding in the Ded1 helicase (7). Our results suggest that the conserved glutamine in the Q motif is often essential for protein function in LSU helicases. In contrast, this is not the case for SSU helicases, where mutations in this motif did not cause dominant negative growth defects and the Q motif mutants were still functional in the absence of the wild-type protein (22a), suggesting that the function of the Q motif in SSU and LSU helicases may be different.
Motifs I and II.
Motif I (Walker A) is present in a large family of proteins, including RNA helicases; it binds and hydrolyzes ATP (6). Motif II, the DEXD/H box (Walker B) from which this class of RNA helicases derives its name, is specific to helicases and is also thought to be important for ATP binding and hydrolysis (6). LSU helicases containing mutations made in these two motifs most frequently caused dominant negative growth defects and were unable to complement depletion of the wild-type protein. Since dominant negative mutations were frequently identified, it suggests that these motifs are not required for binding to the RNA-RNP substrate. However, the ability to bind and hydrolyze ATP is critical for their function in pre-rRNA processing, since alterations in these motifs frequently caused pre-rRNA processing defects. Similar results were observed with proteins required for SSU biogenesis, suggesting that these motifs in most of the RNA helicases required for pre-rRNA processing function in a similar manner (22a). These results are consistent with those of Rocak et al., who found that mutations in motif I (K92A) of the Has1 protein inhibited ATP hydrolysis and helicase functions in vitro and were unable to complement depletion of the wild-type protein (41). Consistent with their results, we have shown here that the K92A mutation in Has1 is dominant negative. In addition, mutations made in motifs I and II in the SSU helicase Rok1 similarly were unable to complement depletion of the endogenous protein (36). Likewise, mutations made in RNA helicases required for pre-rRNA splicing and translation (Prp16, Prp22, Prp43, and eIF4A) also caused dominant negative phenotypes (24, 34, 43, 44, 46, 47). Collectively, these results indicate that the motifs predicted to be required for ATP hydrolysis are required for helicase function.
Motif III.
Motif III (SAT) is thought to be located at a hinge region that connects the ATPase and RNA recognition domains and is therefore essential for helicase function (11). Interestingly, LSU helicases with mutations in the first residue of this motif rarely caused dominant negative growth defects and frequently were able to support growth. Drs1 and Mtr4 were exceptions, in that mutations in this domain were unable to support growth in the absence of the wild-type protein and were dominant negative. In addition, our results are supported by Rocak et al., who also found that a similar mutation, S228A, in motif III of Has1 was not essential for growth, although this mutation inhibited the helicase function in vitro (41). In contrast, mutation of the third position of motif III, T230A, was essential for Has1 function, suggesting that this motif is indeed important in vivo (41). In some cases, mutations made in RNA helicases required for other RNA processes (Prp2, Prp22, Prp16, Sub2) in the conserved serine did cause dominant negative growth defects in a subset of proteins (24, 38, 47, 57). However, mutations made in motif III in the SSU RNA helicases also infrequently caused dominant negative growth defects (only Dhr2) and usually complemented depletion of the wild-type protein (22a). Collectively, our results suggest that the invariant first residue of motif III is not absolutely required for helicase function.
Helicase interactions.
We used coimmunoprecipitation experiments to determine if RNA helicases required for similar pre-rRNA processing steps interact. Our results are in agreement with and expand upon previous results from TAP tag purifications, synthetic lethality screens, and yeast two-hybrid experiments (8, 10, 14, 22, 26). For example, Daugeron and Lindner found synthetic lethal interactions between yeast with mutations made in Dbp7, Dbp6, and Dbp9 but not Spb4, suggesting that these three helicases function at the same pre-rRNA processing steps (10). Similarly, our results show that Dbp6 and Dbp9 coimmunoprecipitate Mak5, another RNA helicase whose depletion causes similar pre-rRNA processing defects. In addition to the synthetic lethal interactions, TAP tag purifications by Gavin et al. (22) frequently revealed RNA helicases associated with other proteins required for LSU biogenesis. For example, in the Ssf1 purification, Dbp10, Dbp9, Drs1, Has1, and Mak5 were identified. Here, we show that Dbp10 and Dbp9 are able to immunoprecipitate Mak5; Mak5 immunoprecipitates Has1 in agreement with those results. Similarly, during the Nop2 purification, Dbp10, Drs1, Has1, Mak5, and Spb4 were identified. Here, we show that Dbp10 immunoprecipitates Spb4, Dbp10 and Mak5 immunoprecipitate Has1, and Dbp10 immunoprecipitates Mak5. Collectively, based upon our results and those previously published, we found that RNA helicases (Dbp6, Dbp7, Dbp9, Mak5, Has1, and Drs1) that are required for the earliest processing steps are all tightly associated. Similarly, proteins required for later processing steps (Dbp10, Mtr4, and Spb4) are also able to coimmunoprecipitate. In addition, a subset of RNA helicases that are associated for the early processing steps are able to coimmunoprecipitate other RNA helicases that function later during processing. For example, Dbp10 is able to immunoprecipitate Mak5 and Has1. These results are consistent with those from Gavin et al., who frequently found Dbp9, Has1, Dbp10, Drs1, Mak5 in complexes after TAP purification (22). Finally, our results are also consistent with those of Jansen et al., who used Bayesian networks to predict protein-protein interactions and reported that Dbp3 could interact with Dbp7 and Mak5 (26).
We determined that growth in yeast depleted of the two nonessential helicases involved in LSU biogenesis, Dbp3 and Dbp7, was worse than when either one was depleted individually. Previously, Daugeron and Linder also observed reduced growth when Dbp7 was genomically disrupted (10). However, in contrast to these results, we found that depletion of Dbp7 synthetically enhanced the growth defects observed by Dbp3 deletion (10). Our results suggest that Dbp7 and Dbp3 are involved in related events during LSU biogenesis.
Our results suggest that although the helicase motifs are highly conserved, they do not function identically in vivo among all the LSU helicases. Since mutations made in many of the conserved motifs are required for function, these results suggest that many of these proteins may have diverse biochemical properties. Unfortunately, we were unable to use these mutants to more precisely pinpoint the helicases substrates. In the future, it will be important to identify the substrates of these putative RNA helicases to better understand their function in pre-rRNA processing. Since depletion of many of these helicases results in identical pre-rRNA processing defects, understanding each helicase's unique function may elucidate why there are so many required for ribosome biogenesis.
Acknowledgments
We thank Neal Janson, Tim “Paul” Stone, Robin Evans, Veronica Segarra, Jennifer Holtzman, Kaury Eisenman, and Amy Schoenfeld for their contributions to the project. We thank Alan Tartakoff for giving us the pGM435L MTR4 plasmid.
K.A.B. was supported by a predoctoral fellowship from the National Institutes of Health (NIH) (GM67564) and by a research service award (GM07499) from the NIH National Institute of General Medical Sciences (NIGMS). S.G. was supported by Anna Fuller and Leslie H. Warner Cancer Research Fellowships. This work was supported by the National Science Foundation (grant MCB-0110403 to S.J.B.).
REFERENCES
- 1.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belli, G., E. Gari, M. Aldea, and E. Herrero. 1998. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14:1127-1138. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, K. A., and S. J. Baserga. 2004. The small subunit processome is required for cell cycle progression at G1. Mol. Biol. Cell 15:5038-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, K. A., J. E. Gallagher, B. M. Mitchell, S. Granneman, and S. J. Baserga. 2004. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell 3:1619-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burger, F., M. C. Daugeron, and P. Linder. 2000. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 28:2315-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 7.Cordin, O., N. K. Tanner, M. Doere, P. Linder, and J. Banroques. 2004. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 23:2478-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daugeron, M. C., D. Kressler, and P. Linder. 2001. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA 7:1317-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daugeron, M. C., and P. Linder. 2001. Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis. Nucleic Acids Res. 29:1144-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daugeron, M. C., and P. Linder. 1998. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA 4:566-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 12.de la Cruz, J., D. Kressler, M. Rojo, D. Tollervey, and P. Linder. 1998. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 4:1268-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Cruz, J., D. Kressler, D. Tollervey, and P. Linder. 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 17:1128-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Cruz, J., T. Lacombe, O. Deloche, P. Linder, and D. Kressler. 2004. The putative RNA helicase Dbp6p functionally interacts with Rpl3p, Nop8p and the novel trans-acting factor Rsa3p during biogenesis of 60S ribosomal subunits in Saccharomyces cerevisiae. Genetics 166:1687-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delagoutte, E., and P. H. von Hippel. 2002. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: structures and properties of isolated helicases. Q. Rev. Biophys. 35:431-478. [DOI] [PubMed] [Google Scholar]
- 16.Delagoutte, E., and P. H. von Hippel. 2003. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Q. Rev. Biophys. 36:1-69. [DOI] [PubMed] [Google Scholar]
- 17.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emery, B., J. de la Cruz, S. Rocak, O. Deloche, and P. Linder. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 52:141-158. [DOI] [PubMed] [Google Scholar]
- 19.Fairman, M. E., P. A. Maroney, W. Wang, H. A. Bowers, P. Gollnick, T. W. Nilsen, and E. Jankowsky. 2004. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science 304:730-734. [DOI] [PubMed] [Google Scholar]
- 20.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 21.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 22.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 22a.Granneman, S., K. A. Bernstein, F. Bleichert, and S. J. Baserga. 2006. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol. Cell. Biol. 26:1183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herskowitz, I. 1987. Functional inactivation of genes by dominant negative mutations. Nature 329:219-222. [DOI] [PubMed] [Google Scholar]
- 24.Hotz, H. R., and B. Schwer. 1998. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics 149:807-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291:121-125. [DOI] [PubMed] [Google Scholar]
- 26.Jansen, R., H. Yu, D. Greenbaum, Y. Kluger, N. J. Krogan, S. Chung, A. Emili, M. Snyder, J. F. Greenblatt, and M. Gerstein. 2003. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science 302:449-453. [DOI] [PubMed] [Google Scholar]
- 27.Kikuma, T., M. Ohtus, T. Utsugi, S. Koga, K. Okuhara, T. Eki, F. Fujimori, and Y. Murakami. 2004. Dbp9p, a member of the DEAD box protein family, exhibits DNA helicase activity. J. Biol. Chem. 279:20692-20698. [DOI] [PubMed] [Google Scholar]
- 28.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1998. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1855-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857-860. [DOI] [PubMed] [Google Scholar]
- 30.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713-724. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S. J., and S. J. Baserga. 1999. Imp3p and Imp4p: two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19:5441-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang, S., M. Hitomi, Y. H. Hu, Y. Liu, and A. M. Tartakoff. 1996. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol. Cell. Biol. 16:5139-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 34.Martin, A., S. Schneider, and B. Schwer. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277:17743-17750. [DOI] [PubMed] [Google Scholar]
- 35.Milkereit, P., H. Kühn, N. Gas, and H. Tschochner. 2003. The pre-ribosomal network. Nucleic Acids Res. 31:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh, J. Y., and J. Kim. 1999. ATP hydrolysis activity of the DEAD box protein Rok1p is required for in vivo ROK1 function. Nucleic Acids Res. 27:2753-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pause, A., and N. Sonenberg. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11:2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plumpton, M., M. McGarvey, and J. D. Beggs. 1994. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 13:879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 40.Ripmaster, T. L., G. P. Vaughn, and J. L. Woolford, Jr. 1992. A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl. Acad. Sci. USA 89:11131-11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocak, S., B. Emery, N. K. Tanner, and P. Linder. 2005. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 33:999-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5:232-241. [DOI] [PubMed] [Google Scholar]
- 43.Rozen, F., J. Pelletier, H. Trachsel, and N. Sonenberg. 1989. A lysine substitution in the ATP-binding site of eucaryotic initiation factor 4A abrogates nucleotide-binding activity. Mol. Cell. Biol. 9:4061-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid, S. R., and P. Linder. 1991. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol. 11:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwer, B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8:113-116. [DOI] [PubMed] [Google Scholar]
- 46.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwer, B., and T. Meszaros. 2000. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 19:6582-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverman, E., G. Edwalds-Gilbert, and R. J. Lin. 2003. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1-16. [DOI] [PubMed] [Google Scholar]
- 49.Sung, P., and S. A. Stratton. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983-27986. [DOI] [PubMed] [Google Scholar]
- 50.Tanner, N. K., O. Cordin, J. Banroques, M. Doere, and P. Linder. 2003. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11:127-138. [DOI] [PubMed] [Google Scholar]
- 51.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251-262. [DOI] [PubMed] [Google Scholar]
- 52.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weaver, P. L., C. Sun, and T. H. Chang. 1997. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol. Cell. Biol. 17:1354-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wehner, K. A., and S. J. Baserga. 2002. The sigma 70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9:329-339. [DOI] [PubMed] [Google Scholar]
- 56.Zagulski, M., D. Kressler, A. M. Becam, J. Rytka, and C. J. Herbert. 2003. Mak5p, which is required for the maintenance of the M1 dsRNA virus, is encoded by the yeast ORF YBR142w and is involved in the biogenesis of the 60S subunit of the ribosome. Mol. Genet. Genomics 270:216-224. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, M., and M. R. Green. 2001. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev. 15:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]