Abstract
We investigated the influence of regulatory and pathogenicity island-associated factors (Hha, RpoS, LuxS, EvgA, RfaH, and tRNA5Leu) on biofilm formation by uropathogenic Escherichia coli (UPEC) strain 536. Only inactivation of rfaH, which encodes a transcriptional antiterminator, resulted in increased initial adhesion and biofilm formation by E. coli 536. rfaH inactivation in nonpathogenic E. coli K-12 isolate MG1655 resulted in the same phenotype. Transcriptome analysis of wild-type strain 536 and an rfaH mutant of this strain revealed that deletion of rfaH correlated with increased expression of flu orthologs. flu encodes antigen 43 (Ag43), which mediates autoaggregation and biofilm formation. We confirmed that deletion of rfaH leads to increased levels of flu and flu-like transcripts in E. coli K-12 and UPEC. Supporting the hypothesis that RfaH represses biofilm formation through reduction of the Ag43 level, the increased-biofilm phenotype of E. coli MG1655rfaH was reversed upon inactivation of flu. Deletion of the two flu orthologs, however, did not modify the behavior of mutant 536rfaH. Our results demonstrate that the strong initial adhesion and biofilm formation capacities of strain MG1655rfaH are mediated by both increased steady-state production of Ag43 and likely increased Ag43 presentation due to null rfaH-dependent lipopolysaccharide depletion. Although the roles of rfaH in the biofilm phenotype are different in UPEC strain 536 and K-12 strain MG1655, this study shows that RfaH, in addition to affecting the expression of bacterial virulence factors, also negatively controls expression and surface presentation of Ag43 and possibly another Ag43-independent factor(s) that mediates cell-cell interactions and biofilm formation.
Sessile bacterial communities embedded in an extracellular matrix are referred to as biofilms. Biofilms generally play an important role in the colonization of niches and in survival in hostile conditions (i.e., interactions of bacteria with eukaryotic hosts). Whereas biofilms in the gastrointestinal tract, which consist of commensals and nonpathogens, may prevent establishment of pathogenic bacteria (71), formation of biofilms by pathogenic bacteria results in increased resistance to antibiotic therapy and activity of the host immune system. This biofilm phenotype is a potential virulence factor involved in the establishment of several bacterial infections (38).
Escherichia coli biofilms are frequently described for catheter-associated or chronic urinary tract infections (UTIs), one of the most common bacterial diseases (16, 28). Uropathogenic E. coli (UPEC) accounts for about 80% of all acute community-acquired UTI cases and a high proportion of nosocomial UTIs (33, 60, 76). The ability of E. coli to form a biofilm may contribute to the colonization of catheter surfaces, which could protect the bacteria from the mechanical flow of urine, host defenses, and antibiotics. UPEC biofilm formation may also be involved in chronic and recurring UTIs, since it has recently been shown that some UPEC strains can form intracellular biofilm-like structures that serve as a reservoir that can be the source of recurrent infections (2, 51). Nevertheless, little is known about the persistence of UPEC in the urinary tract and the establishment of chronic and recurring UTIs. Consequently, a better understanding of the mechanisms involved in E. coli biofilm formation, as well as the relationship between pathogenesis and biofilm formation, is needed.
Among the factors known to contribute to the formation of biofilms by E. coli are flagella, various classes of fimbriae, curli, antigen 43 (Ag43), and the extracellular matrix compounds colanic acid, cellulose, and poly-β-1,6-N-acetyl-d-glucosamine (19, 20, 22, 28, 94, 96). Regulation of gene expression during the different steps of E. coli biofilm formation is complex and not well understood. Surface attachment and microcolony formation seem to respond to signals arising from surface contact and osmolarity sensed via the CpxA/CpxR (6, 29, 49, 66) and EnvZ/OmpR (68, 75) two-component signaling pathways, respectively. The availability of nutrients and other stress signals also play an important role in E. coli biofilm formation. For example, catabolite repression (46, 47) and the alternative sigma factor RpoS (1, 15) have been shown to affect biofilm maturation, as do the regulators H-NS, YaiC, and RcsC (10, 32, 57). Another level of biofilm regulation is exemplified by expression of cell surface-associated structures which undergo phase variation, i.e., the reversible periodic switch between an “ON” phase and an “OFF” phase of gene expression that results in variation of the level of expression in individual cells of a population. Phase variation results from the interplay of different regulatory factors (37, 89, 91, 92).
In this study, the effects of different global regulators and of pathogenicity island I536 (PAI I536)- and PAI II536-encoded factors on the formation of biofilms by UPEC strain 536 were analyzed. The RfaH protein, a transcriptional antiterminator, was identified as a repressor of biofilm formation in E. coli. According to the current model of action, RfaH recognizes an 8-bp DNA motif upstream of RfaH-dependent genes, the ops element, where it can be recruited by the transcription elongation complex. This complex is then able to read through terminator structures within polycistronic operons, thus preventing operon polarity (3, 5). RfaH-affected operons include the operons coding for lipopolysaccharide (LPS) biosynthesis (17), the F factor (79), different capsules (14, 70, 85), hemin uptake receptor (64), and the toxins alpha-hemolysin and cytotoxic necrotizing factor 1 (58, 59). The mechanism underlying biofilm formation in rfaH mutants of uropathogenic E. coli strain 536 and nonpathogenic E. coli K-12 strain MG1655 was then investigated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Uropathogenic E. coli strain 536 (O6:K15:H31) was isolated from a patient with acute pyelonephritis (7). The E. coli strains were routinely grown in Luria-Bertani (LB) medium or M63B1 glucose medium (78) with or without 1.5% Bacto agar (Difco Laboratories, Detroit, MI). When appropriate, kanamycin, ampicillin, and chloramphenicol were added to the growth medium at concentrations of 25 μg/ml, 50 μg/ml, and 30 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli K-12 strains | ||
| SM10λpir | thi-1 thr-1 leuB6 supE44 tonA21 lacY1 recA::RP4-2-Tc::Mu Kmr | 61 |
| SY327 | F−araDΔ(lac pro) argE(Am) recA56 RifrnalAλpir | 61 |
| MG1655 | Wild-type E. coli K-12 | 48 |
| CSH50 oxyR | ara(pro-lac) rpsL thi oxyR::Kmr | 91 |
| TG1 flu | F′[traD36, proAB+lacIqlacZΔM15] supE hsdΔ5 thiΔ(lac-proAB) Δflu::Kmr | Laboratory collection |
| CS2057 | ΔrfaGPSBI::ΩCmr(rfa-1) cps-5::Tn10 thr-1 leuB6 proA argE his thi-1 galK lacY1 trpE mtl-1 xyl ara14 rpsL | 67 |
| MG1655 flu | MG1655, Δflu::cat | 77 |
| MG1655 rfaH | MG1655, ΔrfaH::cat, cat resistance removed by FLP flippase carried by pCP20 | This study |
| MG1655 rfaH flu | P1 vir of Δflu::cat in MG1655 rfaH | This study |
| MG1655 oxyR | P1 vir of oxyR::Kmr in MG1655 | 77 |
| MG1655 oxyR flu | P1 vir of Δflu::cat in MG1655 oxyR | 77 |
| MG1655 rfaH oxyR | P1 vir of oxyR::Kmr in MG1655 rfaH | This study |
| MG1655 rfaH oxyR flu | P1 vir of Δflu::cat in MG1655 rfaH oxyR | This study |
| MG1655 cps | P1 vir of cps-5::Tn10 in MG1655 | This study |
| MG1655 rfa1 | P1 vir of ΔrfaGPSBI::ΩCmr in MG1655 | This study |
| MG1655 cps rfa1 | P1 vir of cps-5::Tn10 in MG1655 cps | This study |
| MG1655 rfa1 flu | P1 vir of Δflu::Kmr in MG1655 rfa1 | This study |
| MG1655 cps rfa1 flu | P1 vir of Δflu::Kmr in MG1655 cps rfa1 | This study |
| Uropathogenic E. coli strains | ||
| 536 | Uropathogenic E. coli isolate (O6:K15:H31), Smr | 7 |
| 536-21 | 536, PAI I536−, PAI II536−, tRNA5Leu− | 9 |
| 536 ORF47V | 536, ΔORF47V::cat, cat resistance removed by FLP flippase carried by pCP20 | This study |
| 536 ORF52III | 536, ΔORF52III::cat, cat resistance removed by FLP flippase carried by pCP20 | This study |
| 536 ORF47V ORF52III | 536, ΔORF52III::cat, ΔORF47V::cat, cat resistance removed by FLP flippase carried by pCP20 | This study |
| 536 rfaH | 536, ΔrfaH::cat, cat resistance removed by FLP flippase carried by pCP20 | This study |
| 536 rfaH-1 | 536rfaH, complemented by insertion of rfaH into lambda attB | This study |
| 536 rfaH ORF47V | 536rfaH, ΔORF47V::cat, cat resistance removed by FLP flippase carried by pCP20 | This study |
| 536 rfaH ORF52III | 536rfaH, ΔORF52III::cat, cat resistance removed by FLP flippase carried by pCP20 | This study |
| 536 rfaH ORF47V ORF52III | 536rfaH, ΔORF52III::cat, ΔORF47V::cat, cat resistance removed by FLP flippase carried by pCP20 | This study |
| 536 hha | 536, hha::cat | 27 |
| 536 evgA | 536, evgA::neo | This study |
| 536 luxS | 536, ΔluxS | This study |
| 536 rpoS | 536, ΔrpoS | This study |
| 536 cpsG | 536, ΔcpsG::cat | This study |
| 536 manB | 536, ΔmanB::cat | This study |
| 536 waaG | 536, ΔwaaG::cat | This study |
| 536 ΔkpsK15 | 536, ΔkpsK15::cat | 83 |
| Plasmids | ||
| pAg43 | Pflu::lacZ transcriptional fusion in pQF50, Apr | 91 |
| pCP20 | ts (replicates at 30°C) plasmid bearing the flp recombinase gene, Cmr Ampr | 12 |
| pKD46 | Arabinose-inducible λ Red recombinase expression plasmid (oriR101, repA101ts, ParaB-gam-bet-exo, Ampr) | 21 |
| pKD3 | Template plasmid for amplification of a cat cassette flanked by FLP recognition sites (FRT) (oriRγ, Ampr Cmr) | 21 |
| pKOBEGA | Arabinose-inducible λ Red recombinase expression plasmid (oriR101, repA101ts, ParaB-gam-bet-exo, Ampr) | 11 |
| pGEM-T Easy | Cloning vector, Ampr | Promega |
| pBluescript-II KS | Cloning vector, Ampr | Stratagene |
| pLDR8 | int gene expression vector, helper plasmid, Kmr | 25 |
| pLDR10 | Cloning vector for integration into λ-attB, Ampr Cmr | 25 |
| pLDR10-1 | Cloning vector for integration of the functional rfaH gene into λ-attB, Ampr Cmr | This study |
Sand column adhesion assays.
Initial attachment of bacterial cells to sea sand-filled columns was performed using the method of Simoni and coworkers (84). Briefly, cells from exponentially growing or overnight cultures grown in M9 glucose medium at 28°C or 37°C were washed and resuspended in phosphate-buffered saline (PBS) (30 ml) at an optical density at 280 nm (OD280) between 0.8 and 1.0 (corresponding roughly to an OD600 of 0.2), which corresponded to a concentration of ca. 108 CFU/ml. The suspension was loaded at a flow rate of 0.5 ml/min onto a 12-cm column filled with 9 g of purified sea sand grains, which had been preequilibrated with PBS. Ten fractions (3 ml each) were collected, and the adhesion efficiency was calculated by determining the ratio of the OD280 of each fraction of the bacterial suspension collected at the column output (A) to the OD280 of the bacterial suspension loaded onto the column (Ao). The average A/Ao ratio for the last six fractions was used to calculate the percentage of attached bacteria by using the formula (1 − A/Ao) × 100. Inspection of the column sand grains with an electron microscope showed that for the most part bacteria attached as single cells in the conditions used in our experiments and that the sand surface was not fully occupied by the bacteria (data not shown), suggesting that cell-cell interactions did not play a major role in attachment to the sand columns.
Determination of physicochemical properties of bacterial strains.
The electrophoretic mobility, a function of the overall surface charge of the bacterial cells, was determined by the method of van Loosdrecht and coworkers (90), using a Doppler electrophoretic light-scattering analyzer (Zeta-Master; Malvern Instruments Ltd., United Kingdom). For measurement, cells grown overnight in M9 glucose medium at 37°C were harvested, washed, and resuspended in PBS (pH 7.0) at a concentration of ca. 5 × 106 CFU/ml. The same suspensions were used to determine bacterial cell hydrophobicity, using the contact angle measurement method described by Jucker et al. (50).
Biofilm formation assay.
Comparisons of biofilm development in microfermentors were performed at least in triplicate as described previously (6, 34). Briefly, overnight cultures were grown in M63B1 medium containing 0.4% glucose at 37°C. Inoculation was performed by dipping the microfermentor removable spatula in a culture containing 108 bacteria/ml for 1 min, followed by rapid rinsing into M63B1 medium; then the spatula was reintroduced into the microfermentor. The medium was pumped through the microfermentors at a constant rate (0.75 ml/min). Images of the biofilm formed on the internal Pyrex spatula were captured after 24 h to 48 h of growth, and the organisms were resuspended in 10 ml of M63B1 minimal medium. The OD600 of each resulting suspension was then determined. This optical density directly reflected the biomass on the spatula.
Microscopy and image analysis.
Biofilm development was recorded with a Nikon Coolpix 950 digital camera. Scanning laser electronic microscopy was performed with biofilms grown in microfermentors on Thermanox slides (Nalgene) fixed on internal removable glass slides as described by Prigent-Combaret et al. (69) at the Laboratoire de Biologie Cellulaire et Microscopie Electronique, UFR Médecine, Tours, France.
Autoaggregation assays.
Autoaggregation assays were performed as described by Roux et al. (77). Cells were grown in LB medium at 37°C for 8 h. Cultures were diluted 1:100 in M63B1 medium containing 0.4% glucose and grown overnight at 37°C (16 to 18 h). The culture OD600 was adjusted to 3 by dilution with M63B1 medium, and 3-ml portions of each culture were transferred to 5-ml tubes. The tubes were incubated without agitation at room temperature. The OD600 of the upper part of the culture in each standing tube was determined every hour for 6 h and after 24 h before images were captured.
Detection of flu expression in E. coli K-12 strains MG1655 and 536 by immunoblot analysis.
Overnight cultures were harvested by centrifugation and washed in 0.9% (wt/vol) NaCl, and the concentrations were standardized to a final OD600 of 1.0 in 75 mM NaCl-0.5 mM Tris (pH 7.4). The passenger domain of Ag43 variants was released from the surface of the cells by heating at 60°C for 20 min. The cells were immediately removed by centrifugation, and the resultant protein in the supernatant was precipitated overnight at 4°C with 10% (vol/vol) trichloroacetic acid. The pellet was dried and resuspended in Tris-EDTA buffer. After the protein concentrations of the samples were adjusted, sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis buffer was added (56) and the samples were boiled for 3 min prior to gel electrophoresis. Separation of the protein samples and detection of the Ag43 passenger domains were carried out as described previously (27). A polyclonal serum raised against the Ag43 passenger domain of strain MG1655 (P. Owen, Dublin, Ireland) was used as the primary serum, and the secondary antibody was horseradish peroxidase-conjugated anti-rabbit immunoglobulin (DAKO, Hamburg, Germany). Chemiluminescence was detected using a Chemi Lux imager (Intas Science Imaging Instruments GmbH, Göttingen, Germany).
Analysis of long-chain LPS expression in E. coli K-12 strains MG1655 and 536 and derivatives of these strains.
LPS was isolated from the E. coli strains used in this study as previously described (36).
DNA techniques.
QIAGEN (Hilden, Germany) products were used to isolate plasmid DNA and to purify DNA fragments or PCR products. For cloning experiments, the Dap Goldstar DNA polymerase (Eurogentec, Seraing, Belgium) was used. Primers were obtained from Sigma-GENOSYS (Taufkirchen, Germany), while restriction enzymes were purchased from New England Biolabs (Frankfurt am Main, Germany). For Southern blot hybridization, DNA was transferred to Nytran Supercharge nylon membranes (Schleicher & Schuell BioSciences, Dassel, Germany). Hybridization with and detection of horseradish peroxidase-labeled probes were performed with the ECL labeling and signal detection system (Amersham Biosciences, Freiburg, Germany). Gene inactivation was carried out by allelic exchange using the suicide vector pCVD442 as described previously (61, 62) or by using lambda Red-mediated recombination of linear DNA fragments as previously described (11, 21, 24). The complete list of primers used for gene inactivation is available on the homepage of the “Enterobacteria” research group of the Institute for Molecular Biology of Infectious Diseases (http://www.uni-wuerzburg.de/infektionsbiologie/imi-start.htm). rfaH, ORF52III, and ORF47V were disrupted by an antibiotic cassette flanked by specific FLP recombinase sites (frt). After interruption of the genes, the antibiotic cassette was subsequently removed by use of plasmid pCP20 expressing the FLP flipase (12). Complementation of strain 536rfaH was achieved by chromosomal insertion of the rfaH gene into the λattB site as described previously (25). For this purpose, a 927-bp DNA fragment that included rfaH together with its flanking up- and downstream regions was amplified by PCR from strain 536 and cloned (BamHI/EcoRI) into pLDR10. The NotI fragment of the resulting plasmid, including the bla and rfaH genes as well as the λattP site, was religated and used for chromosomal insertion into λattB of strain 536rfaH with use of helper plasmid pLDR8. All of the rpoS gene and all of the luxS gene, except their start and stop codons, were completely deleted by allelic exchange using pCVD442-based constructs. hha and evgA were inactivated by insertion of cat and a neo cassette, respectively, via allelic exchange using pCVD442-based constructs. The different constructions were verified by PCR and/or Southern blot hybridization.
RNA techniques.
A small quantity of total RNA was isolated from E. coli using an RNeasy purification kit (QIAGEN). Large quantities of total RNA were extracted from 25-ml portions of bacterial cultures which were immediately mixed with 10 g of crushed ice precooled to −80°C and harvested by centrifugation at 4°C. The cells were resuspended in 0.6 ml of an ice-cold buffer containing 10 mM KCl, 5 mM MgCl2, and 10 mM Tris (pH 7.4) and then immediately added to 0.6 ml of hot lysis buffer (0.4 M NaCl, 40 ml EDTA, 1% sodium dodecyl sulfate) containing 1% β-mercaptoethanol and 200 μl of water-saturated phenol. The mixture was incubated at 90°C for 2 min (13). Cell debris was removed by centrifugation for 10 min at room temperature, and the supernatant was then extracted two or three times with phenol and one or two times with chloroform, precipitated with LiCl overnight, pelleted, washed, and dried. The dried RNA pellet was dissolved in 100 μl sterile diethyl pyrocarbonate-treated water. To obtain DNA-free RNA, the raw RNA was subjected twice to 20 min of DNase treatment (0.5 U/μl of RNase-free DNase I [Roche Diagnostics]), followed by LiCl precipitation.
Genome-wide transcriptional analysis using DNA macroarrays.
According to the results of the initial adhesion tests, the factor(s) involved in increased adherence was expressed in the exponential growth phase. In order to assess differences in the transcription profiles of exponentially growing planktonic cells (M63 medium, 37°C) of wild-type strain 536 and the isogenic rfaH mutant, E. coli K-12 strain MG1655-specific DNA arrays (Panorama E. coli gene arrays; Sigma-Genosys, Cambridge, United Kingdom) were used in combination with the so called E. coli “pathoarray” carrying an assortment of probes specific for many of the known pathogenicity island- or virulence-associated genes of uropathogenic E. coli strain 536, as well as other extraintestinal pathogenic and intestinal pathogenic E. coli strains (26). For each strain, four different RNA preparations from independent cultures were used for cDNA synthesis as follows. DNA-free total RNA was reverse transcribed and radioactively labeled. In short, 30 μg of total RNA was used as the template for cDNA synthesis in a reaction mixture containing 4.8 μg of random hexanucleotide primers (QIAGEN, Hilden, Germany), 30 μCi [α-33P]dATP (1,000 to 3,000 Ci/mmol), 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.5 mM dGTP, 0.5 mM dCTP, 0.5 mM dTTP, 20 U RNase inhibitor (Roche, Mannheim, Germany), and 10 U Omniscript reverse transcriptase (QIAGEN, Hilden, Germany). After incubation for 3 h at 37°C, unincorporated nucleotides were removed with Microspin G 50 spin columns (Amersham Pharmacia) prior to denaturation for 10 min at 90°C. Before hybridization, the DNA macroarrays were rinsed in 2× SSPE and subsequently prehybridized for 3 h at 65°C in 5 ml hybridization solution (5× SSPE, 2% SDS, 1× Denhardt's solution, 100 μg/ml sheared, denatured herring sperm DNA) (1× SSPE is 0.18 M NaCl, 10 mN NaH2PO4, and 1 mM EDTA [pH 7.7]). After addition of the probe denatured in 3 ml of hybridization solution, the arrays were incubated for 12 to 18 h at 65°C. After hybridization, the blots were washed twice with wash solution (0.5× SSPE, 0.2% SDS) for 2 to 3 min at room temperature, followed by three wash steps for 10 min at 65°C. Washed filters were air dried and exposed for 1 or 2 days to a phosphorimager screen (superresolution type) prior to scanning with a Typhoon 8600 variable-mode imager (Molecular Dynamics).
The Panorama E. coli K-12 strain MG1655-specific gene array and the “E. coli Pathoarray” were hybridized in four different experiments using independently generated cDNA probes. The scanned arrays were analyzed with the ArrayVision software (Imaging Research, St. Catharines, Canada), followed by visual inspection. Normalized intensity values for the individual spots were calculated by using the overall-spot-normalization function of ArrayVision. Background values were measured in the four corners of every spot. For identification of genes with statistically significant changes in expression, SAM, a statistical technique for finding significant genes in a set of array experiments, was used (88) (http://www-stat.stanford.edu/∼tibs/SAM/). Genes with statistically significant changes in expression were identified by assimilating a set of gene-specific t tests. Each gene was assigned a score based on its change in expression relative to the standard deviation of repeated measurements for the gene. Genes whose scores exceeded a certain threshold were considered potentially significant. The percentage of such genes identified by chance was the false discovery rate (FDR). The delta value chosen (which corresponded to the FDR) was <10%. The FDR was calculated by determining the ratio of the estimated number of “false significant” genes to the total number of “significant” genes. Genes that were significant according to this analysis and with mRNA levels that were changed at least twofold were considered.
Comparative analysis of flu transcription levels in E. coli wild-type strains and corresponding rfaH mutants by semiquantitative RT-PCR and discrimination of flu orthologs of E. coli strain 536.
For reverse transcription PCR (RT-PCR), 3 μg DNA-free RNA was mixed with 0.5 μg random hexamer primers (QIAGEN, Hilden, Germany) and heated for 10 min at 70°C. After incubation on ice for 5 min, cDNA synthesis was performed with Omniscript reverse transcriptase (QIAGEN, Hilden, Germany) at 37°C. After 3 h of incubation aliquots of the reverse-transcribed RNA were used as cDNA templates for PCRs.
Amplification was performed with Taq polymerase (QIAGEN, Hilden, Germany) under the following conditions: 25 s of denaturation at 92°C, 45 s of annealing at 57°C, 130 s of elongation at 72°C. The primers used for amplification are available in the supplemental material at http://www.uni-wuerzburg.de/infektionsbiologie/imi-start.htm and at http://www.uni-wuerzburg.de/infektionsbiologie/imi-start.htm. To discriminate the transcript levels of the two flu orthologs located on PAI III536 and PAI V536, a 590-bp region of flu was reverse transcribed (primer KM-Ag43_RT) and amplified with primers KM-Ag43_fwd and KM-Ag43_rev, which allowed reverse transcription and amplification of a conserved region of both flu transcript variants. The resulting 590-bp RT-PCR product consisted of a mixture of the two flu ortholog transcripts, which could be distinguished by restriction with SacII or BstEII. There was a SacII restriction site only in the 590-bp RT-PCR product representing the PAI III536-localized flu ortholog, which resulted in 250-bp and 340-bp restriction fragments upon SacII digestion. Conversely, a BstEII restriction site was uniquely present in the flu ortholog on PAI V536, and digestion of the 590-bp RT-PCR product resulted in 200-bp and 390-bp restriction fragments following restriction with BstEII. Consequently, the fraction of the PAI III536- and PAI V536-specific flu RT-PCR products in the pool of the 590-bp RT-PCR product could be assessed by selective digestion with either SacII or BstEII. For this purpose, the RT-PCR products obtained from wild-type strain 536 and its rfaH mutant were column purified (QIAquick spin columns). Aliquots were digested with 10 U of SacII or BstEII for 6 h before separation of the restriction fragments on agarose gels. The signal intensity of the undigested 590-bp RT-PCR product was then considered a measure of the fraction of either the PAI III536- and PAI V536-specific flu RT-PCR product in the common RT-PCR product pool. The primers used in RT-PCR experiments are described in the supplemental material.
RESULTS
Influence of virulence regulators on biofilm formation by uropathogenic E. coli strain 536.
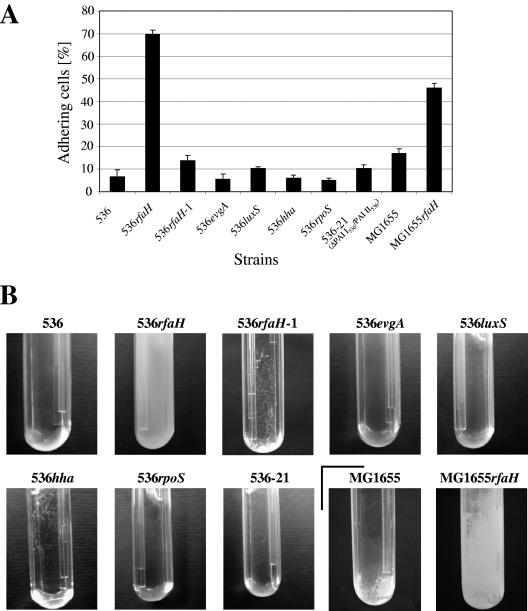
Mutants of UPEC strain 536 lacking the regulatory factors rfaH, evgA, rpoS, hha, and luxS or leuX-encoded tRNA5Leu, as well as two pathogenicity islands (PAI I536 and PAI II536) that encode several virulence-associated factors, including different adhesins (27, 73), were tested to determine their abilities to adhere to solid surfaces. Microbial adhesion to purified sea sand was measured in a column system which specifically examined the initial stages of adhesion (57). Of the mutants examined, only the rfaH mutant displayed strong induction of adhesion. The same phenotype was observed with exponentially growing cells and with cells from the stationary growth phase (Fig. 1A and data not shown). The wild-type adhesion behavior could be restored by complementing strain 536rfaH in trans with a wild-type copy of rfaH, which confirmed that the superadhesion phenotype was dependent on inactivation of the rfaH gene (Fig. 1A). The results of this experiment indicated that inactivation of the rfaH gene results in increased initial adhesion to solid surfaces. This observation is consistent with the strong mature biofilm phenotype displayed by the rfaH 536 null mutant in a flow culture microfermentor system (Fig. 1B).
FIG. 1.
Analysis of the adhesive and biofilm-forming capacities of planktonic cells of uropathogenic E. coli strain 536 and K-12 strain MG1655 and derivatives of these strains. (A) Initial adhesive capacity as measured using a sea sand column system. (B) Mature biofilm development as measured with a flow culture microfermentor system. The results of a representative experiment are shown.
rfaH mutant of a nonpathogenic E. coli K-12 strain also displays an increased-biofilm phenotype.
In uropathogenic E. coli strain 536, RfaH regulates chromosomal core genes shared with many commensal E. coli strains, as well as genes present on UPEC-specific pathogenicity islands. The increased-biofilm phenotype displayed by strain 536rfaH could be due to the effect of RfaH on the expression of PAI-specific genes or the effect of RfaH on the expression of genes belonging to the conserved E. coli core genome or a combination of the two. In order to distinguish between these possibilities, the effect of an rfaH mutation on initial adhesion and biofilm formation by nonpathogenic E. coli K-12 strain MG1655 was examined. The rfaH mutation caused a strong increase in initial adhesion when cells were loaded onto a sand column (Fig. 1A) and also when cells were attached to glass cover slides (10-fold-increased adhesion for the rfaH mutant) (data not shown). As shown in Fig. 1B, an rfaH mutation had a strong positive effect on biofilm formation by K-12 strain MG1655. We also observed that the settlement rates of overnight cultures of both E. coli MG1655 and 536 rfaH mutants were substantially greater than those of their wild-type counterparts (see Fig. S1 in the supplemental material at http://www.uni-wuerzburg.de/infektionsbiologie/imi-start.htm and at http://www.pasteur.fr/recherche/unites/Ggb/supmat.html; data not shown). Since introduction of the rfaH mutation did not significantly affect the growth rate of strain 536 or MG1655 (data not shown), we concluded that the absence of RfaH modified the adhesion properties rather than the growth phenotype of the cells.
These results demonstrated that the biofilm phenotype observed in strain 536 is not restricted to pathogenic E. coli strains and suggested that RfaH affects adhesion by modifying the expression of genes from the core genome shared by pathogenic and nonpathogenic strains of E. coli.
Comparison of transcriptional profiles of strains 536 and 536rfaH by using DNA arrays.
Since the RfaH protein directly affects the expression of several bacterial cell envelope components, we investigated whether the increased-biofilm phenotype of the rfaH mutants could be due to changes in the physicochemical characteristics of the cell surface. The wild-type strains and isogenic rfaH mutants were compared with respect to surface charge and hydrophobicity. No major alterations in either surface charge or hydrophobicity due to rfaH inactivation could be consistently correlated with the different adhesive phenotypes of strains 536rfaH and MG1655rfaH (data not shown). These results, as well as our data showing that the rfaH mutation positively affects the initial adhesion step of biofilm formation, led to the hypothesis that increased adhesion to solid surfaces by the rfaH mutants depends on the production of specific adhesion factors prior to surface adhesion in planktonic cells. In order to identify RfaH-dependent factors that are responsible for the observed enhanced biofilm phenotype, the transcription profiles of strains 536 and 536rfaH were compared. In the absence of an existing UPEC strain 536-specific genome array, we used both an E. coli pathoarray, which allowed us to detect alterations in the levels of transcripts of the majority of the open reading frames (ORFs) present on PAI I536 to PAI V536 of strain 536, and an E. coli K-12-specific core genome array.
The analysis of the transcription profiles showed that expression of only a limited number of genes was significantly altered by more than twofold in the rfaH mutant compared to the wild type (Table 2).
TABLE 2.
Genes found to be significantly induced or repressed in the rfaH mutant of strain 536 compared to the wild-type strain
| Gene | Encoded gene product | Change in expression (fold) |
|---|---|---|
| E. coli K-12-specific DNA array | ||
| b2000 (flu) | Ag43 | 2.5 |
| cpsG | Phosphomannomutase | −2.6 |
| E. coli pathoarray | ||
| ORF52 (PAI III536) | Ag43 ortholog | 3.0 |
| ORF47 (PAI V536) | Ag43 ortholog | 2.2 |
| hlyA | Pore-forming toxin | −3.2 |
| ORF35 (PAI II536) | DNA methyltransferase | −2.3 |
| hlyB | Traffic ATPase required for HlyA secretion | −2.3 |
| hlyD | Inner membrane translocase required for HlyA secretion | −2.2 |
In the E. coli K-12-specific core genome array experiment, we found that the transcript levels of cpsG, encoding a phosphomannomutase involved in colanic acid biosynthesis that is activated by RfaH (86), were reduced by a factor of 2.6 in the rfaH mutant. Surprisingly, transcription of other well-known RfaH-dependent genes, including those coding for LPS biosynthesis, was not found to be significantly repressed. The transcript levels of genes involved in LPS core biosynthesis were reduced only by a factor of 1.3 or less. Nevertheless, using the E. coli K-12-specific array, we observed that the level of the flu (b2000) transcript, which encodes the phase-variable outer membrane protein Ag43, was increased in the 536 rfaH mutant by a factor of 2.5 compared with the wild-type strain.
Interestingly, transcriptome analysis using the E. coli “pathoarray” showed that the transcript levels of five genes localized on PAIs were significantly reduced in the rfaH mutant of strain 536 compared to the wild type by factors greater than 2. These ORFs include the known RfaH-dependent virulence determinant hly (59), which is located on PAI I536 and PAI II536, as well as ORF35 on PAI II536, which encodes a putative DNA methyltransferase. RfaH-dependent K15 capsule-encoding genes (kpsE, kpsF, and kpsS), located on PAI V536, were found to be repressed by the rfaH mutation by a factor of 1.6, as was another previously known RfaH-activated gene, chuA, encoding a hemin receptor. In addition, the transcript levels of two ORFs (ORF52III and ORF47V) were significantly increased in the rfaH mutant. These ORFs are localized on PAI III536 or PAI V536 and code for two similar but not identical Ag43 orthologs. Ag43 variants are usually encoded on variable mobile DNA regions, such as pathogenicity islands or plasmids, and a single strain of pathogenic E. coli may carry multiple copies of the same flu gene or different flu alleles (54, 74, 87). Recent sequence analysis indicated that UPEC strain 536 contains only a single copy of each of the agn43 orthologs ORF52III and ORF47V. This was confirmed by Southern hybridization of genomic DNA of strain 536 with an agn43-specific probe complementary to a conserved part of the passenger domain (see Fig. S2 in the supplemental material). Since the autotransporter protein antigen 43 encoded by these genes is involved in E. coli biofilm formation, its altered expression represented a likely explanation for the increased-biofilm phenotype in rfaH mutant strains.
Different roles for Ag43 in the increased-biofilm phenotype of the rfaH mutants of uropathogenic E. coli strain 536 and nonpathogenic K-12 strain MG1655.
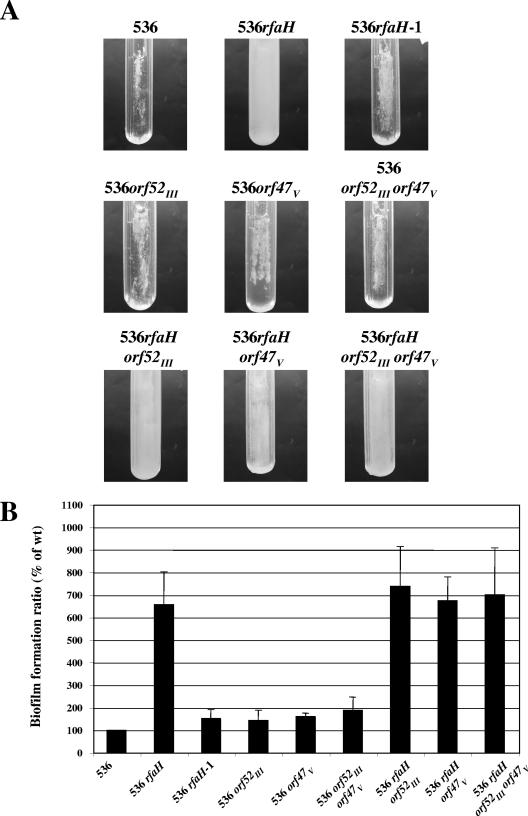
We tested whether Ag43 was involved in the increased-biofilm phenotype of the rfaH mutants. The genes encoding the two flu orthologs (both on PAIs) were deleted in uropathogenic wild-type strain 536 and an isogenic rfaH mutant. Strains carrying the different combination of mutations were tested for the capacity to form biofilms in microfermentors (Fig. 2). The data demonstrated that neither of the flu orthologs was necessary for the increased-biofilm phenotype of strain 536rfaH.
FIG. 2.
Analysis of the importance of different flu orthologs for the RfaH-dependent increased biofilm formation capacity of E. coli strain 536. (A) Biofilm development of strain 536 and its derivatives in microfermentors after 24 h of growth at 37°C in M63B1 glucose medium. The results of a representative experiment are shown. (B) Average results of at least four experiments plotted as a histogram. The error bars indicate standard errors of the means. The level of biofilm formed by wild-type strain 536 (wt) was defined as 100%.
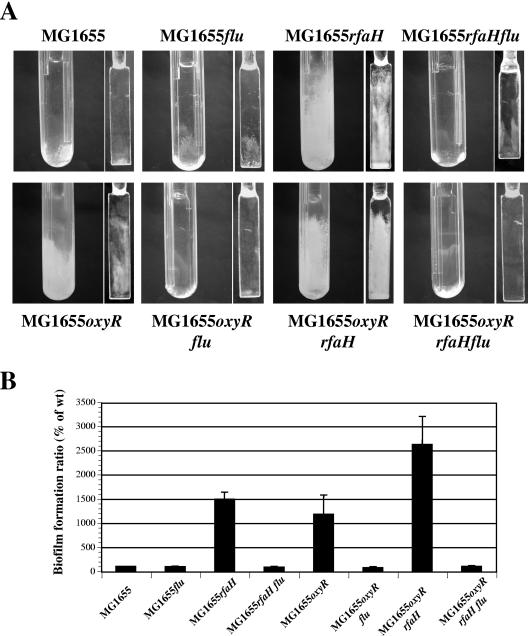
The same experiment was performed in the nonpathogenic K-12 strain background with mutants carrying a disruption of the flu gene. In wild-type strain MG1655, normally a poor biofilm former, a flu mutation had no effect on biofilm formation (Fig. 3). However, in contrast to what was observed for UPEC strain 536, introduction of the flu mutation reversed the increased-biofilm formation phenotype of the rfaH mutant. These data suggest that Ag43 does contribute to the increased-biofilm phenotype observed for the K-12 strain MG1655 rfaH mutant. This situation is reminiscent of the phenotype displayed by an oxyR mutant; in the absence of a functional oxyR gene, the phase-variable flu gene is locked in an “ON” phase, leading to drastically increased flu expression. This observation is depicted in Fig. 3. As shown here, the oxyR mutation increased biofilm formation compared to the wild type. This phenotype was flu dependent since an oxyR flu double mutant displayed poor biofilm formation capacity (Fig. 3). Since all MG1655 derivatives studied had equivalent growth rates (data not shown), these data supported the hypothesis that Ag43 is required for the null rfaH-mediated increased-biofilm phenotype. Moreover, the rfaH and oxyR mutations appeared to have an additive effect both on biofilm formation (Fig. 3) and on autoaggregation phenotypes (see Fig. S1 in the supplemental material) in E. coli K-12.
FIG. 3.
Analysis of the importance of flu for the RfaH-dependent increased biofilm formation capacity of E. coli strain MG1655. (A) Biofilm development of K-12 strain MG1655 and its mutants in microfermentors after 48 h of growth at 37°C in M63B1 glucose medium. The images are representative images of the bottoms of the microfermentors and of the Pyrex spatulas on which biofilms formed. (B) Average results of at least four experiments plotted as a histogram. The error bars indicate standard errors of the means. The level of biofilm formed by wild-type strain MG1655 (wt) was defined as 100%.
Role of RfaH in regulating flu expression.
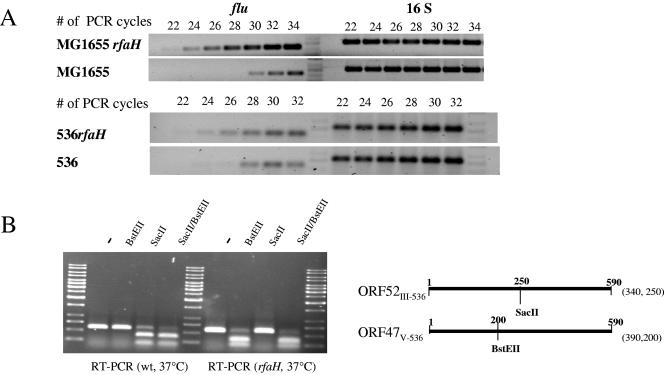
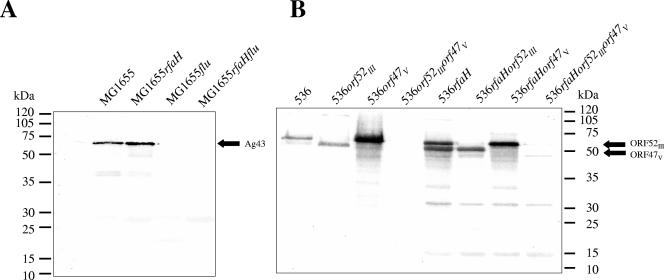
According to a genome-wide transcriptome analysis, flu transcript levels were elevated in the rfaH mutant of E. coli strain 536. Semiquantitative RT-PCR with conserved primers that allowed amplification of fluMG1655, as well as both flu orthologs of strain 536, confirmed that the flu transcript levels in exponentially growing planktonic cells were increased at least fourfold in the rfaH mutant of strain 536 and at least eightfold in strain MG1655rfaH (Fig. 4A). The probe design of the E. coli “pathoarray” did not allow exact discrimination between the PAI III536- and PAI V536-encoded flu orthologs. In order to specifically analyze the effect of rfaH inactivation on the transcript levels of these two genes independently, differences in restriction patterns of the two orthologs were used to distinguish between the transcript levels of ORF47V and ORF52III (Fig. 4B). The results demonstrated that the majority of cDNA generated with general flu-specific primers from total RNA of the wild-type strain was derived from the ORF52III transcript, whereas in the rfaH mutant the majority of cDNA arose from ORF47V transcripts. These data, however, indicate that deletion of rfaH leads to a global increase in flu-like transcripts in the cells.
FIG. 4.
Comparison of flu transcript levels of E. coli wild-type strains and their rfaH derivatives. (A) Comparison of flu transcript levels of uropathogenic E. coli strain 536 and K-12 strain MG1655. The uropathogenic and K-12 wild-type strains, as well as their rfaH mutants, were cultivated at 37°C. The flu transcript levels of exponentially growing, planktonic cells were compared by RT-PCR. 16S RNA levels were used as internal controls of the quantity of RNA used for RT-PCR. (B) Comparison of the flu ortholog transcript levels of uropathogenic E. coli strain 536 and its rfaH mutant. The strains were cultivated at 37°C. The ORF52III-536 and ORF47V-536 transcripts of exponentially growing, planktonic cells were reverse transcribed and amplified by RT-PCR. The cDNA was consecutively digested with SacII and BstEII in order to distinguish between the ORF52III-536- and ORF47V-536-specific cDNA fractions. The resulting restriction fragments were separated on an agarose gel. The localization of the SacII and BstEII restriction sites and the sizes of the resulting cDNA fragments are indicated. wt, wild type.
Additionally, we investigated the possibility that an rfaH mutation could also affect the steady-state level of the Ag43 protein in strains MG1655 and 536. Immunodetection of Ag43 was performed with planktonic cells of wild-type strains MG1655 and 536 and the rfaH mutants of these strains. In planktonic cells of each of the rfaH mutants, the production of Ag43 proteins was increased compared to that in the wild type (Fig. 5 and Table 3), thus corroborating results of the RT-PCR experiments (Fig. 4A). Furthermore, the level of the ORF47V-encoded protein was markedly increased in strain 536rfaH compared to strain 536, whereas the level of the ORF52III-encoded protein was only slightly increased (Fig. 5). Although these results support the hypothesis that there is an overall tendency for a substantial increase in ORF47V expression compared with decreased or more or less constant expression of ORF52III, there is a discrepancy between the transcript and protein levels of ORF52III in the wild-type strain compared to the rfaH mutant of this strain. We assume that posttranscriptional mechanisms that differentially affect the transcript or protein stability could be responsible for the observed differences.
FIG. 5.
Comparison of Ag43 expression in E. coli wild-type strains and their rfaH derivatives. Heat-extracted surface appendages were separated by SDS-polyacrylamide gel electrophoresis. Passenger domains of Ag43 of strain MG1655 (A) or of the Ag43 variants of strain 536 (ORF52III and ORF47V) (B) were detected using a polyclonal antiserum raised against MG1655-specific Ag43. The results of a representative experiment are shown.
TABLE 3.
Increase in Ag43 protein levels in rfaH mutants compared to the corresponding wild-type strains
| Strain background | Relative expression of Ag43 variantsa
|
||
|---|---|---|---|
| Flu | ORF52III | ORF47V | |
| MG1655 (wild type) | 1 | ||
| MG1655rfaH | 2.75 | ||
| 536 (wild type) | 1 | 1 | |
| 536ORF52III | 6.48 | ||
| 536ORF47V | 6.50 | ||
| 536rfaH | 2.26 | 17.02 | |
| 536rfaH ORF52III | 11.94 | ||
| 536rfaH ORF47V | 3.73 | ||
The signal intensity of an Ag43 variant was normalized to the signal intensity of the same Ag43 variant obtained for the wild-type strain.
Role of RfaH in the expression of regulatory factors required for flu transcription.
RfaH has been described as a factor that positively affects gene expression, which is not consistent with the apparent repressor effect of RfaH on the expression of flu and flu-like genes in strains MG1655 and 536. Moreover, the recruitment of RfaH to the transcription elongation complex involves recognition of an upstream binding sequence called the ops element that could not be detected in the regions located upstream of or within the flu genes.
Consequently, we considered the possibility of an indirect role for RfaH through regulation of the expression of other regulatory genes that may repress flu expression. Such an indirect effect could be mediated by an RfaH-dependent modification of the switching frequency of flu transcription. We directly measured the effect of the rfaH mutation on MG1655 flu gene expression switching by using a plasmid-based lacZ fusion that has been shown to provide an accurate estimate of the chromosomal flu ON/OFF promoter switch (91). A colorimetric assay using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) agar plates was performed with the wild-type and rfaH strains. This assay, starting with a colony in either the ON or OFF state, showed that RfaH has no influence on the switching frequency of the flu promoter (data not shown). Additionally, RT-PCR was used to quantify the transcript levels of two known regulators of this switching frequency, oxyR and dam. The RT-PCR experiments were performed with cultures in the exponential and stationary growth phases, as well as with samples of biofilm material. As shown on our website (Fig. S3 in the supplemental material), rfaH inactivation affected neither dam nor oxyR transcript levels in the K-12 and UPEC strains. These results suggest that the role of RfaH in flu gene expression is not mediated either by the regulatory oxyR and dam genes or by a change in the flu ON/OFF promoter switch frequency and may involve another factor. Alternative hypotheses are discussed below.
rfaH mutation induces tight cell packing that is flu independent.
To examine whether the absence of RfaH leads to structural differences in the biofilm formed by E. coli, we performed three independent scanning electron microscopy (SEM) analyses of various derivatives of UPEC strain 536 and K-12 strain MG1655.
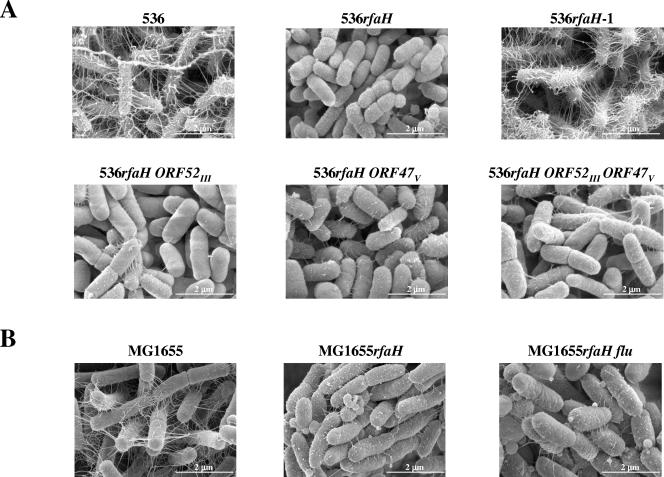
In UPEC strain 536, mutation of the rfaH gene led to tight packing of the bacterial cells within the biofilm, which was not observed with wild-type bacteria. The rfaH mutation also decreased the amount of fibrillar structures, which most likely represented a condensed extracellular matrix due to fixation of biofilm samples and which seemed to connect the bacteria in biofilms formed by wild-type strain 536. Complementation of 536rfaH by a wild-type copy of rfaH restored both of these phenotypes (Fig. 6A). Mutation of the flu orthologs of strain 536 did not appear to modify the compact nature of cells of strain 536rfaH, thus confirming that the presence of the two Ag43-like proteins was dispensable for expression of the biofilm-related null rfaH phenotype.
FIG. 6.
Comparison of biofilm structures formed by strains 536 (A) and MG1655 (B). The phenotypic analysis of the structure of strains 536 and MG1655 and their derivatives was performed in microfermentors with bacteria grown for 48 h at 37°C in M63B1 glucose medium. Thermanox slides were stuck on Pyrex spatulas and removed after biofilm growth for scanning electron microscopy analysis. The images are representative images.
In strain MG1655, the rfaH mutation led to the same phenotype observed in UPEC strain 536 (Fig. 6B), as did an oxyR mutation (data not shown). However, whereas the oxyR phenotype was Ag43 dependent and could be reversed by deletion of the flu gene (data not shown), deletion of flu did not modify the spacing between cells observed with strain MG1655rfaH (Fig. 6B). These results are consistent with the Ag43-independent cellular autoaggregation observed with MG1655rfaH, as opposed to the autoaggregation observed with MG1655oxyR (Fig. S1 in the supplemental material). Therefore, despite the role of Ag43 in the rfaH-dependent biofilm phenotype in E. coli K-12 (Fig. 3), Ag43 is not required for close cell-cell contact in this mutant, which may involve other factors in E. coli strains MG1655 and 536.
Search for alternative mechanisms causing increased biofilm formation by the rfaH mutant of strain 536.
As shown above, up-regulation of flu-like transcripts could not explain the increased biofilm formation ability of the 536rfaH mutant (Fig. 2). This phenotype could be explained by reduction of transcription of putative ORF35 of PAI II536, one of the genes found to be significantly repressed in the absence of RfaH (Table 2). Interestingly, ORF35 of PAI II536 contains a cis-acting upstream ops element. Since ORF35PAI II encodes a putative DNA methyltransferase, a different DNA methylation status due to reduced ORF35PAI II transcript levels in the rfaH mutant may affect the expression of some surface appendages, in analogy to the regulatory role of the Dam methylase in E. coli (31, 43). In order to test whether the reduction of ORF35PAI II transcription could be responsible for the rfaH-dependent biofilm phenotype, ORF35PAI II was deleted in strain 536. However, this mutation had no effect on the ability of strain 536 to form biofilms (data not shown).
Alternatively, it was recently found that capsule could block specific cell-cell or cell-surface interactions. Previous studies demonstrated that LPS and capsule expression are impaired in strain 536rfaH, and our macroarray analysis also indicated that the levels of expression of the phosphomannomutase cpsG gene and K15 capsule genes kpsE, kpsF, and kpsS are reduced in E. coli 536rfaH. Therefore, the reduced capsule and LPS expression caused by an rfaH mutation could allow shorter surface-associated structures to contribute to the initial adherence and/or biofilm formation (39, 65, 80, 83). In order to evaluate whether the lack of LPS or capsule could be involved in the RfaH-dependent biofilm phenotype, we inactivated in wild-type strain 536 the waaG, manB, and cpsG genes, which resulted in elimination of production of long-chain LPS and colanic acid. In addition, we also included a K15 capsule-negative mutant of strain 536 in the biofilm assays. Neither the colanic acid-negative mutant nor the K15 capsule-negative mutant exhibited an increased-biofilm phenotype, indicating that the reduction in expression of the cps and kps determinants could not explain the massive increased-biofilm phenotype of mutant strain 536rfaH (Fig. S4 in the supplemental material). Inactivation of manB, coding for a phosphomannomutase involved in LPS O-side chain biosynthesis, reduced the side chain length of the LPS but did not affect biofilm formation. Furthermore, mutation of waaG, coding for an LPS core glycosyltransferase that resulted in a truncated LPS core structure and a deep rough phenotype, completely abolished biofilm formation by strain 536 without affecting its growth rate in liquid culture (Fig. S4 in the supplemental material), suggesting that an intact LPS core is a major factor for adhesion to abiotic surfaces in E. coli strain 536.
LPS depletion in the rfaH mutant may lead to unmasking of surface adhesins in E. coli MG1655.
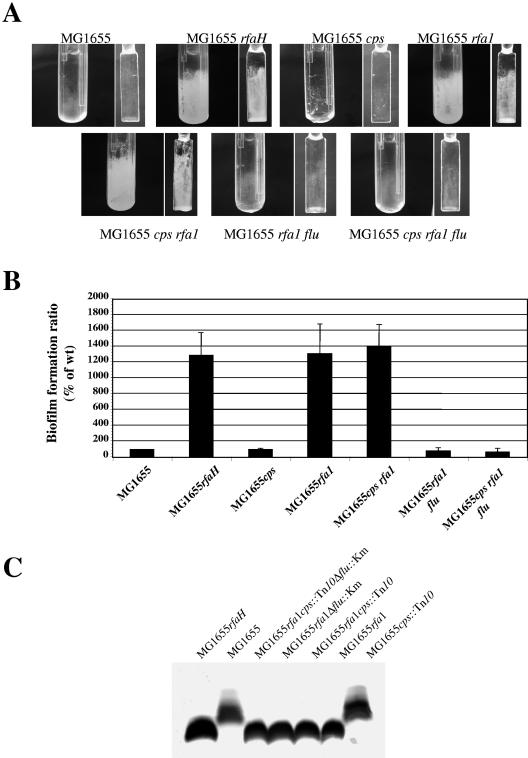
In order to investigate the possible unmasking of a short surface-associated adhesin in the biofilm phenotype observed for the E. coli K-12 rfaH mutant, we estimated the biofilm formation abilities of E. coli K-12 mutants impaired for production of colanic acid and/or semirough LPS, whose synthesis is dependent on RfaH in E. coli K-12. Whereas mutation of colanic acid did not affect biofilm formation by strain MG1655 in a microfermentor, a net increase in biofilm biomass was observed for the strains containing the LPS mutation rfa1 (ΔrfaGPSBI) (Fig. 7). This result suggested that LPS depletion in an rfaH mutant could unmask a short surface-associated adhesin. Based on its role in the biofilm phenotype of an E. coli K-12 rfaH mutant (Fig. 3), Ag43 was an obvious candidate for such a surface adhesin. We therefore introduced a flu mutation into strain MG1655rfa1. As shown in Fig. 7, the introduction of the flu mutation into strain MG1655rfa1 indeed reversed the hyper-biofilm phenotype caused by depletion of long-chain LPS. We also observed that loss of semirough LPS in wild-type strain MG1655 resulted in enhanced autoaggregation (data not shown), a phenotype previously observed upon introduction of the rfaH mutation. However, as in the case of the rfaH-dependent aggregation phenotype (Fig. S1 in the supplemental material), the increased autoaggregation was not dependent on Ag43 (data not shown).
FIG. 7.
Unshielding of Ag43 by LPS depletion causes increasing biofilm formation. (A) Biofilm development for strain MG1655 and its derivatives in microfermentors after 48 h of growth at 37°C in M63B1 glucose medium. The results of a representative experiment are shown. (B) Average results of at least four experiments plotted as a histogram. The error bars indicate standard errors of the means. The level of biofilm formed by wild-type strain MG1655 (wt) was defined as 100%. (C) LPS patterns of the strains.
Taken together, these results suggest that in E. coli K-12, an rfaH mutation leads to strong adhesion and the capacity to form biofilms mediated by an increase in the quantity of Ag43 and possibly by unmasking due to the rfaH-dependent LPS depletion. Our results also support the hypothesis that the rfaH mutation may unmask another unidentified protein(s) responsible for increased cell-cell interactions.
DISCUSSION
It has been proposed that biofilm formation contributes to the virulence of E. coli. To analyze the possible links between biofilms and the expression of virulence-associated genes, we investigated the phenotypic consequences of inactivation of several known regulatory factors that affect E. coli virulence or of deletion of pathogenicity islands I536 and II536, which encode several virulence-associated factors, including different adhesins (27, 73). Inactivation of RfaH leads to an increased-adherence and -biofilm phenotype of uropathogenic E. coli strain 536 and nonpathogenic E. coli K-12 strain MG1655. This suggests that the role of RfaH in the initial adherence and/or cell-cell interactions is a general feature of both pathogenic and nonpathogenic E. coli.
RfaH indirectly regulates the expression of flu and flu orthologs in UPEC strain 536 and K-12 strain MG1655.
To identify factors that are involved in the RfaH-dependent biofilm phenotype, the transcriptomes of the rfaH mutant and wild-type strain 536 were compared. Although the arrays used did not allow whole-genome transcriptional profiling, we nevertheless identified in both uropathogenic E. coli strain 536 and nonpathogenic E. coli K-12 strain MG1655 the flu genes as candidate genes involved in the increased-biofilm phenotype of the rfaH mutants.
In UPEC strain 536, the ORF52III transcript level decreased upon rfaH inactivation, whereas the ORF47V transcript level was markedly increased (Fig. 4B). Similarly, the ORF47III protein level was only slightly increased in the rfaH mutant, whereas the level of the ORF47V protein was increased greatly (Table 3). Owing to the absence of detectable ops elements in the flu upstream regions, it is likely that RfaH does not directly affect flu expression via the ops element and interaction with the RNA polymerase (3, 4). Phase variation may regulate expression of both flu orthologs, although in opposite ways. Phase-variable expression of Ag43 is due to the concerted action of the Dam methylase and the cellular redox sensor OxyR and has been extensively studied in E. coli K-12 (37, 42, 82, 91-93). Although the upstream regions of the two E. coli 536 flu orthologs are not identical, three Dam DNA methylation sites, as well as an OxyR motif, are present upstream of ORF52III and ORF47V. The absence of variation of the Ag43 switch frequency in an rfaH mutant, however, excludes an indirect effect of RfaH on flu expression via modification of dam or oxyR gene expression.
Other possible indirect repression of flu by RfaH may occur via variation of the redox state of OxyR toward its reduced form. The expression of fimbrial organelles containing disulfide bonds could affect the redox status of OxyR and thus Ag43 expression (81, 82). However, since recent indications have shown that phase variation of Ag43 is independent of the oxidation status of OxyR (92) and that the levels of type 1 fimbria expression, as well as S- and P- fimbria expression, are not different in strains 536 and 536rfaH (65), another unknown regulatory mechanism is likely to be involved in the RfaH-mediated flu expression. Hypotheses such as an indirect effect of RfaH on the expression or stability of flu mRNAs or Ag43 proteins have to be considered.
Alternative Ag43-independent mechanisms are involved in the increased biofilm formation by the rfaH mutant of UPEC strain 536.
Ag43 affects colony morphology and mediates autoaggregation, thus contributing to cell-to-cell adhesion and biofilm formation (18, 40, 41, 52, 54). Nevertheless, the flu orthologs may not play a central role in the RfaH-dependent biofilm phenotype of strain 536. It has been found previously that Ag43 enhances biofilm formation primarily by providing efficient cell-to-cell adhesion, which in turn stimulates formation of premature biofilms (53). However, Ag43 has also been shown to be dispensable for biofilm formation as it can be replaced by alternative factors (72). The observation that the difference in biofilm formation between wild-type strain 536 and its rfaH mutant becomes smaller after prolonged incubation (data not shown) also corroborates the findings of Klemm and coworkers (53) that Ag43 expression particularly promotes formation of premature biofilms and that, at later stages, other factors come into play. This factor(s) is probably also responsible for the enhanced cell-cell interactions within the 536rfaH biofilm, as observed by SEM analysis. The larger genome of strain 536 (∼4.94 Mb) compared to that of strain MG1655 (4.63 Mb) underlines the possibility that additional gene products of strain 536 which are absent in strain MG1655 may promote biofilm formation. Other candidate RfaH-regulated factors in strain 536 whose absence could increase biofilm formation, either through modification of the ON-OFF phase switching frequency of surface appendages (ORF35 of PAI II536) or through unmasking of other shorter surface-associated structures (colanic acid, K15 capsule, and long-chain LPS), were tested. None of the corresponding mutations enhanced the capacity of strain 536 to form biofilms. In contrast, the further truncation of the LPS core structure in E. coli strain 536waaG compared to strain 536rfaH severely impaired biofilm formation (see Fig. S4 in the supplemental material at http://www.uni-wuerzburg.de/infektionsbiologie/imistart.htm and at http://www.pasteur.fr/recherche/unites/Ggb/supmat.html). In previous reports workers described LPS as a molecule that is important for adhesion of different pathogenic bacteria, such as Pseudomonas fluorescens, Serratia marcescens, and Klebsiella pneumoniae (23, 45), to abiotic surfaces. The absence of biofilm formation by strain 536waaG is consistent with these reports and suggests that LPS, especially the LPS core, is critical for adhesion of E. coli strain 536 to abiotic surfaces (Fig. S4). Whereas the different extracellular polysaccharides are not responsible for the increased biofilm formation by strain 536rfaH, we cannot rule out the possibility that a surface-associated adhesin is effectively unmasked in 536rfaH by depletion of an unidentified RfaH-regulated E. coli 536 surface component, thus contributing to the increased-biofilm phenotype.
In E. coli MG1655 the rfaH-dependent biofilm phenotype may involve unmasking of Ag43.
In contrast to the situation observed in E. coli 536, the increased Ag43 level is positively correlated with an increased-biofilm phenotype in E. coli K-12 strain MG1655rfaH. Accordingly, the enhanced three-dimensional growth of the biofilm observed in the rfaH mutants could be due to increased cell-cell interactions via Ag43. Loss of semirough LPS in the rfa1 mutant, similar to the loss caused by an rfaH mutation, led to an increased-biofilm phenotype. As in strain MG1655rfaH, this phenotype was reversed by the flu mutation in strain MG1655rfa1. Therefore, as shown for adhesin shielding by expression of the capsule (39, 80), we propose that in the presence of RfaH, the LPS core of K-12 strain MG1655 could physically shield Ag43, preventing its exposure to the cell surface. Modification of the LPS structure may well be used by bacteria to modulate their capacities to form biofilms. Interestingly, in Shigella flexneri, a mechanism that enhances type III secretion, and therefore modulates virulence, has recently been described, and it involves modification of the LPS structure, leading to unmasking of the needle-like secretion structure (95).
Hence, in E. coli K-12, the rfaH mutation could lead to strong adhesion and biofilm formation by two distinct mechanisms, through an increase in the quantity of Ag43 and possibly by unmasking due to LPS core depletion. This hypothesis may account for the macroscopic difference observed between the smooth fragile oxyR mutant biofilms and the granular and more robust biofilms formed by the rfaH mutant (data not shown). The surface exposure and unmasking of a large amount of Ag43 produced in the rfaH oxyR background may explain the additive effect on biofilm formation observed when these two mutations are combined. The different roles of Ag43 proteins in the rfaH biofilm phenotypes of K-12 and UPEC strains suggest that the rfaH mutants of strains produce more biofilm for different reasons. Despite the considerable genetic differences between the two E. coli strains, RfaH affects biofilm formation by different but convergent intermediate pathways in the strains.
rfaH-dependent biofilm phenotype of E. coli K-12 strain MG1655 may also require surface adhesins other than Ag43.
Several lines of evidence presented in this paper support the hypothesis that factors other than Ag43 might also be involved in increased biofilm formation by E. coli strain MG1655. The flu deletion in strain MG1655rfaH did not modify the spacing between cells observed in this strain (Fig. 6B), and Ag43-independent cellular autoaggregation was also observed in strains MG1655rfaH flu and MG1655rfa1 flu (Fig. S1 in the supplemental material; data not shown). This suggests that in strain MG1655rfaH another factor(s) is also induced or, alternatively, unmasked by the RfaH depletion. We recently identified new adhesins in E. coli K-12 by virtue of their sequence homology to Ag43 and the increased abiotic surface adhesion caused by their overexpression (77). Nevertheless, mutation of any of the genes encoding these adhesins, either the type I-encoding gene fimA or the curli-encoding gene csgA, was not able to reverse the increased autoaggregation of the MG1655 rfaH mutant (data not shown).
Relationships between RfaH-mediated virulence and biofilm formation.
There is considerable knowledge regarding virulence-associated factors of E. coli. However, the relationship between pathogenesis and biofilm formation is still poorly understood. In this study we demonstrated that RfaH, besides positively affecting virulence gene expression, plays a role in repressing E. coli biofilm formation.
Since expression of several E. coli virulence-associated genes depends on RfaH, the increased-biofilm phenotype of the nonvirulent rfaH mutant of strain 536 indicates that RfaH-dependent biofilm formation and virulence gene expression (63, 65) are mutually exclusive processes and that biofilm formation may not be regarded as a virulence trait per se.
Recently, the two-component sensor kinase/response regulator hybrid protein RetS of P. aeruginosa has been shown to inversely coordinate the control of genes required for biofilm formation and acute infection in cystic fibrosis patients (35). Whereas the capacity to form a biofilm may be required for other aspects of P. aeruginosa pathogenesis, such as chronic persistence, incompatibility between colonization or infection of the plant vascular system and biofilm formation has been demonstrated in the case of the plant pathogen Xanthomonas campestris. This bacterium alternates between the biofilm and noninfective lifestyle and the fully infective and free-swimming form through cell-cell-signaled dispersal of the biofilm (30). Expression of a type III secretion system of P. aeruginosa and expression of an adenylate cyclase toxin of Bordetella bronchiseptica, both of which are required for acute infection, were shown to inhibit biofilm formation (44, 55).
Consistently, recent data indicate that biofilms of uropathogenic E. coli have to be formed at the right place under appropriate conditions and that this may also promote virulence under certain growth conditions (2, 51). Our findings suggest that derepression of biofilm formation could therefore contribute to the absence of virulence of strain 536rfaH. It has been demonstrated that in Salmonella, rfaH expression itself is affected by the growth phase and the sigma factors RpoN and RpoS (8, 27), which are also known to regulate several virulence determinants in different microorganisms. Hence, modulation of biofilm formation via RfaH could be a mechanism by which bacteria fine-tune their virulence properties, especially in response to environmental signals. This suggests that there are complex regulatory circuits for virulence traits and biofilm formation that have to be analyzed in the future in order to complete our knowledge of factors involved in chronic and recurring UTI.
Acknowledgments
We thank Jean-Claude Lazzaroni for help in constructing the LPS mutant in E. coli K-12 strains, Peter Owen for the gift of an Ag43 antiserum, and Shaynoor Dramsi for help with immunodetection experiments. We also thank Alexander J. B. Zehnder, Jennifer Leeds, Philippe Delepelaire, and Gábor Nagy for helpful discussions and critical reading of the manuscript. We thank Barbara Plaschke, Birgit Schellberg, and Teresa Colangelo for excellent technical assistance and Claude Lebos and Brigitte Arbeille (Laboratoire de Biologie Cellulaire et Microscopie Electronique, Tours, France) for preparation of the SEM micrographs.
The Würzburg group was supported by the DFG (SFB479, TP A1), the “Fonds der Chemischen Industrie,” and the “Bayerische Forschungsstiftung.” J.-M.G. and C.B. were supported by the Institut Pasteur, as well as by CNRS and Fondation BNP Paribas grants.
REFERENCES
- 1.Adams, J. L., and R. J. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. [DOI] [PubMed] [Google Scholar]
- 3.Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109:193-203. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, M. J., C. Hughes, and V. Koronakis. 2000. In vitro recruitment of the RfaH regulatory protein into a specialised transcription complex, directed by the nucleic acid ops element. Mol. Gen. Genet. 262:1052-1059. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 6.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 7.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 152:1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bittner, M., S. Saldias, F. Altamirano, M. A. Valvano, and I. Contreras. 2004. RpoS and RpoN are involved in the growth-dependent regulation of rfaH transcription and O antigen expression in Salmonella enterica serovar Typhi. Microb. Pathog. 36:19-24. [DOI] [PubMed] [Google Scholar]
- 9.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschäpe, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brombacher, E., C. Dorel, A. J. Zehnder, and P. Landini. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847-2857. [DOI] [PubMed] [Google Scholar]
- 11.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 13.Chuang, S. E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke, B. R., R. Pearce, and I. S. Roberts. 1999. Genetic organization of the Escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J. Bacteriol. 181:2279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corona-Izquierdo, F. P., and J. Membrillo-Hernandez. 2002. A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS Microbiol. Lett. 211:105-110. [DOI] [PubMed] [Google Scholar]
- 16.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 17.Creeger, E. S., T. Schulte, and L. I. Rothfield. 1984. Regulation of membrane glycosyltransferases by the sfrB and rfaH genes of Escherichia coli and Salmonella typhimurium. J. Biol. Chem. 259:3064-3069. [PubMed] [Google Scholar]
- 18.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 19.Danese, P. N., L. A. Pratt, and R. Kolter. 2001. Biofilm formation as a developmental process. Methods Enzymol. 336:19-26. [DOI] [PubMed] [Google Scholar]
- 20.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davey, M. E., and A. G. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lima Pimenta, A., P. Di Martino, E. Le Bouder, C. Hulen, and M. A. Blight. 2003. In vitro identification of two adherence factors required for in vivo virulence of Pseudomonas fluorescens. Microbes Infect. 5:1177-1187. [DOI] [PubMed] [Google Scholar]
- 24.Derbise, A., B. Lesic, D. Dacheux, J. M. Ghigo, and E. Carniel. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38:113-116. [DOI] [PubMed] [Google Scholar]
- 25.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration in the lambda attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-24. [DOI] [PubMed] [Google Scholar]
- 26.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobrindt, U., L. Emödy, I. Gentschev, W. Goebel, and J. Hacker. 2002. Efficient expression of the alpha-haemolysin determinant in the uropathogenic Escherichia coli strain 536 requires the leuX-encoded tRNA(5)(Leu). Mol. Genet. Genomics 267:370-379. [DOI] [PubMed] [Google Scholar]
- 28.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 30.Dow, J. M., L. Crossman, K. Findlay, Y. Q. He, J. X. Feng, and J. L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 100:10995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Labany, S., B. K. Sohanpal, M. Lahooti, R. Akerman, and I. C. Blomfield. 2003. Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol. Microbiol. 49:1109-1118. [DOI] [PubMed] [Google Scholar]
- 32.Ferrieres, L., and D. J. Clarke. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665-1682. [DOI] [PubMed] [Google Scholar]
- 33.Foxman, B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S-13S. [DOI] [PubMed] [Google Scholar]
- 34.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 35.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745-754. [DOI] [PubMed] [Google Scholar]
- 36.Grozdanov, L., U. Zähringer, G. Blum-Oehler, L. Brade, A. Henne, Y. A. Knirel, U. Schombel, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, E. T. Rietschel, and U. Dobrindt. 2002. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J. Bacteriol. 184:5912-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haagmans, W., and M. van der Woude. 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35:877-887. [DOI] [PubMed] [Google Scholar]
- 38.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 39.Hanna, A., M. Berg, V. Stout, and A. Razatos. 2003. Role of capsular colanic acid in adhesion of uropathogenic Escherichia coli. Appl. Environ. Microbiol. 69:4474-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasman, H., M. A. Schembri, and P. Klemm. 2000. Antigen 43 and type 1 fimbriae determine colony morphology of Escherichia coli K-12. J. Bacteriol. 182:1089-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henderson, I. R., M. Meehan, and P. Owen. 1997. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol. Lett. 149:115-120. [DOI] [PubMed] [Google Scholar]
- 42.Henderson, I. R., and P. Owen. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernday, A. D., B. A. Braaten, G. Broitman-Maduro, P. Engelberts, and D. A. Low. 2004. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16:537-547. [DOI] [PubMed] [Google Scholar]
- 44.Irie, Y., S. Mattoo, and M. H. Yuk. 2004. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J. Bacteriol. 186:5692-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izquierdo, L., N. Abitiu, N. Coderch, B. Hita, S. Merino, R. Gavin, J. M. Tomas, and M. Regue. 2002. The inner-core lipopolysaccharide biosynthetic waaE gene: function and genetic distribution among some Enterobacteriaceae. Microbiology 148:3485-3496. [DOI] [PubMed] [Google Scholar]
- 46.Jackson, D. W., J. W. Simecka, and T. Romeo. 2002. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jucker, B. A., H. Harms, S. J. Hug, and A. J. B. Zehnder. 1997. Adsorption of bacterial surface polysaccharides on mineral oxides is mediated by hydrogen bonds. Colloids Surf. B Biointerfaces 9:331-343. [Google Scholar]
- 51.Justice, S. S., C. Hung, J. A. Theriot, D. A. Fletcher, G. G. Anderson, M. J. Footer, and S. J. Hultgren. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 101:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kjaergaard, K., M. A. Schembri, H. Hasman, and P. Klemm. 2000. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J. Bacteriol. 182:4789-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 54.Klemm, P., L. Hjerrild, M. Gjermansen, and M. A. Schembri. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51:283-296. [DOI] [PubMed] [Google Scholar]
- 55.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 57.Landini, P., and A. J. Zehnder. 2002. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J. Bacteriol. 184:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landraud, L., M. Gibert, M. R. Popoff, P. Boquet, and M. Gauthier. 2003. Expression of cnf1 by Escherichia coli J96 involves a large upstream DNA region including the hlyCABD operon, and is regulated by the RfaH protein. Mol. Microbiol. 47:1653-1667. [DOI] [PubMed] [Google Scholar]
- 59.Leeds, J. A., and R. A. Welch. 1996. RfaH enhances elongation of Escherichia coli hlyCABD mRNA. J. Bacteriol. 178:1850-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leone, M., F. Garnier, M. Avidan, and C. Martin. 2004. Catheter-associated urinary tract infections in intensive care units. Microbes Infect. 6:1026-1032. [DOI] [PubMed] [Google Scholar]
- 61.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mobley, H. L., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol. Microbiol. 10:143-155. [DOI] [PubMed] [Google Scholar]
- 63.Nagy, G., U. Dobrindt, J. Hacker, and L. Emödy. 2004. Oral immunization with an rfaH mutant elicits protection against salmonellosis in mice. Infect. Immun. 72:4297-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagy, G., U. Dobrindt, M. Kupfer, L. Emödy, H. Karch, and J. Hacker. 2001. Expression of hemin receptor molecule ChuA is influenced by RfaH in uropathogenic Escherichia coli strain 536. Infect. Immun. 69:1924-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagy, G., U. Dobrindt, G. Schneider, A. S. Khan, J. Hacker, and L. Emödy. 2002. Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect. Immun. 70:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 70.Rahn, A., and C. Whitfield. 2003. Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol. Microbiol. 47:1045-1060. [DOI] [PubMed] [Google Scholar]
- 71.Reid, G., J. Howard, and B. S. Gan. 2001. Can bacterial interference prevent infection? Trends Microbiol. 9:424-428. [DOI] [PubMed] [Google Scholar]
- 72.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 73.Ritter, A., G. Blum, L. Emödy, M. Kerenyi, A. Bock, B. Neuhierl, W. Rabsch, F. Scheutz, and J. Hacker. 1995. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol. Microbiol. 17:109-121. [DOI] [PubMed] [Google Scholar]
- 74.Roche, A., J. McFadden, and P. Owen. 2001. Antigen 43, the major phase-variable protein of the Escherichia coli outer membrane, can exist as a family of proteins encoded by multiple alleles. Microbiology 147:161-169. [DOI] [PubMed] [Google Scholar]
- 75.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 76.Ronald, A. 2002. The etiology of urinary tract infection: traditional and emerging pathogens. Am. J. Med. 113(Suppl. 1A):14S-19S. [DOI] [PubMed] [Google Scholar]
- 77.Roux, A., C. Beloin, and J. M. Ghigo. 2005. Combined inactivation/expression strategy to study gene function in physiological conditions: application to the identification of new adhesins in Escherichia coli. J. Bacteriol. 187:1001-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 79.Sanderson, K. E., and B. A. Stocker. 1981. Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J. Bacteriol. 146:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 186:1249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schembri, M. A., L. Hjerrild, M. Gjermansen, and P. Klemm. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J. Bacteriol. 185:2236-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schembri, M. A., and P. Klemm. 2001. Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J. 20:3074-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schneider, G., U. Dobrindt, H. Brüggemann, G. Nagy, B. Janke, G. Blum-Oehler, C. Buchrieser, G. Gottschalk, L. Emödy, and J. Hacker. 2004. The pathogenicity island-associated K15 capsule determinant exhibits a novel genetic structure and correlates with virulence in uropathogenic Escherichia coli strain 536. Infect. Immun. 72:5993-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simoni, S. F., H. Harms, T. N. P. Bosma, and A. J. B. Zehnder. 1998. Population heterogeneity affects transport of bacteria through sand columns at low flow rates. Environ. Sci. Technol. 32:2100-2105. [Google Scholar]
- 85.Stevens, M. P., B. R. Clarke, and I. S. Roberts. 1997. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol. Microbiol. 24:1001-1012. [DOI] [PubMed] [Google Scholar]
- 86.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Torres, A. G., N. T. Perna, V. Burland, A. Ruknudin, F. R. Blattner, and J. B. Kaper. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol. Microbiol. 45:951-966. [DOI] [PubMed] [Google Scholar]
- 88.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van der Woude, M. W., and A. J. Bäumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Loosdrecht, M. C. M., W. Norde, J. Lyklema, and A. J. B. Zehnder. 1990. Hydrophobic and electrostatic parameters in bacterial adhesion. Aquat. Sci. 52:103-114. [Google Scholar]
- 91.Waldron, D. E., P. Owen, and C. J. Dorman. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44:509-520. [DOI] [PubMed] [Google Scholar]
- 92.Wallecha, A., J. Correnti, V. Munster, and M. van der Woude. 2003. Phase variation of Ag43 is independent of the oxidation state of OxyR. J. Bacteriol. 185:2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wallecha, A., V. Munster, J. Correnti, T. Chan, and M. van der Woude. 2002. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184:3338-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313-1317. [DOI] [PubMed] [Google Scholar]
- 96.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]