Abstract
Bacterial chromosome partitioning and cell division are tightly connected cellular processes. We show here that the Caulobacter crescentus FtsK protein localizes to the division plane, where it mediates multiple functions involved in chromosome segregation and cytokinesis. The first 258 amino acids of the N terminus are necessary and sufficient for targeting the protein to the division plane. Furthermore, the FtsK N terminus is required to either assemble or maintain FtsZ rings at the division plane. The FtsK C terminus is essential in Caulobacter and is involved in maintaining accurate chromosome partitioning. In addition, the C-terminal region of FtsK is required for the localization of the topoisomerase IV ParC subunit to the replisome to facilitate chromosomal decatenation prior to cell division. These results suggest that the interdependence between chromosome partitioning and cell division in Caulobacter is mediated, in part, by the FtsK protein.
Bacterial cell division is a complex process that requires the coordination of many mechanisms that ensure proper partitioning of the sister chromosomes to the daughter cells. The segregation of the chromosomal terminus poses a major topological problem, as replication of circular chromosomes leaves two catenated circular chromosomes during the final stages of replication. This intermolecular linkage must be resolved by topoisomerase IV (Topo IV) before cell division (18, 25). The importance of Topo IV activity in the cell is evident in Topo IV mutants, where aberrant cell division yields long anucleate filaments and cell death in Escherichia coli and Salmonella enterica serovar Typhimurium (1, 18, 25). Similar abnormal DNA partitioning and cell division are also observed in Topo IV mutants of Caulobacter crescentus (23, 32, 33). In addition, there is evidence suggesting a role for Topo IV in overall nucleoid organization, as Topo IV mutants cause cells to lose polar positioning of their origins of replication (32).
In E. coli, DNA replication is often still ongoing even as cell division inititates. Lau et al. (19) found that immediately before cell divison, the two terminal regions are frequently localized asymmetrically in one of the daughter cell compartments, causing DNA to become trapped in the division plane of the cell. FtsK, an ATP-dependent DNA translocase, has been reported to be responsible for clearing the division site of chromosomal DNA prior to cell division (4, 9, 21, 38). Similarly, the Bacillus subtilis SpoIIIE protein, belonging to the FtsK family, is required during the sporulation process to translocate the sister chromosomes from the mother cell compartment through the asymmetric division septum and into the smaller forespore compartment (3, 27, 28). E. coli FtsK and B. subtilis SpoIIIE are bifunctional proteins with an N-terminal transmembrane domain and a C-terminal ATPase domain. In E. coli, the two domains are linked by a proline- and glutamine-rich linker region. The N-terminal region has been shown to be responsible for targeting the FtsK protein to the division plane in E. coli and has also been shown to be sufficient to support cell division (10, 37). In contrast, the C-terminal domain belongs to the AAA family of ATPases and is involved in chromosome segregation (21, 38). Furthermore, the C-terminal domain of FtsK (FtsKC) operates as a link between chromosome segregation and cell division. It is known to activate both the decatenation activity of Topo IV and the XerCD recombination reaction that in E. coli resolves chromosomal dimers (2, 5, 7, 12). FtsKC has also recently been shown to be a bidirectional motor in vivo, its directionality dependent upon short asymmetric DNA sequences (24).
In Caulobacter crescentus, DNA replication occurs once and only once per cell division cycle and the replication origin is always positioned at a cell pole (16). Each cell division is asymmetric, producing two distinct cell types: a motile swarmer cell and a stalked cell. In the swarmer cell, which is unable to initiate DNA replication, the origin of replication is located at the flagellated pole and the terminus is located near the opposite end of the cell (15). DNA replication initiates when the swarmer cell differentiates into a stalked cell. As soon as replication is initiated, a copy of the replicated origin moves rapidly to the pole opposite the stalk in what appears to be an active process mediated by the actin homologue, MreB (14, 30), whereas the terminus is gradually displaced to the division plane during the S phase (15).
Here, we have demonstrated that the FtsK protein in Caulobacter plays a major role during the final stages of the cell cycle. Using a gfp fusion to the ftsK gene, we found that FtsK is localized to the division plane just before cell division in predivisional cells and it remains at the newly generated cell pole of each progeny cell following division for a period of time. The C terminus of FtsK is essential for viability, and in cells depleted for the FtsK C terminus, approximately 15 to 20% of the cells had defects in terminal segregation. Furthermore, the localization of the Topo IV ParC replisome component (32), responsible for decatenation at the division plane, is dependent on the presence of the FtsK C terminus. The first 258 amino acids of the N terminus are necessary and sufficient for targeting the FtsK protein to the division plane, where it is required to either assemble or maintain FtsZ rings. Thus, the bifunctional FtsK protein mediates an interdependence between chromosome partitioning and cell division in Caulobacter.
MATERIALS AND METHODS
Strains and growth conditions.
The Caulobacter crescentus parC and parE temperature-sensitive strains were derived from strain CB15 (33) (Table 1). All other Caulobacter strains were derived from CB15N. Synchronization was performed as described previously (13). All strains were grown at 28°C or 37°C in M2-glucose minimal medium (M2G) or peptone yeast extract (PYE) (11). When indicated, strains were induced with 0.3% xylose or 0.5 mM Na-vanillate (pH 7.5). Transductions were performed using ΦCr30 (11).
TABLE 1.
Table of C. crescentus strains
| Strain | Relevant genetic marker(s) or featuresa | Source or reference |
|---|---|---|
| LS4203 | CB15N (NA1000) xylX::ftsK; ftsK::ΩaacC4 | This study |
| LS4200 | CB15N (NA1000) xylX::ftsK-gfp | This study |
| LS4204 | CB15N (NA1000) xylX::ftsK1-258-gfp | This study |
| LS4205 | CB15N (NA1000) xylX::ftsK1-333-gfp | This study |
| LS4206 | CB15N (NA1000) xylX::ftsK334-819-gfp | This study |
| LS4215 | CB15N (NA1000) xylX::ftsK; ftsKΔaa334-806 | This study |
| LS4216 | CB15N (NA1000) xylX::ftsK334-819-gfp; ftsKΔaa334-806 | This study |
| LS4202 | CB15N (NA1000) ftsK::Pxyl-ftsK; ftsKΔaa327-820 | This study |
| LS4209 | CB15 parC ts (divF310); xylX::ftsK-gfp | This study |
| LS4208 | CB15 parE ts (divC307); xylX::ftsK-gfp | This study |
| LS4207 | CB15N (NA1000) vanA::parC-gfp; ftsK::Pxyl-ftsK; ftsKΔaa327-820 | This study |
| LS4210 | CB15N (NA1000) dnaB-yfp; ftsK::Pxyl-ftsK; ftsKΔaa327-820 | This study |
| LS4211 | CB15N (NA1000) holC-yfp; ftsK::Pxyl-ftsK; ftsKΔaa327-820 | This study |
| LS4212 | CB15N (NA1000) vanA::ftsZ-yfp; xylX::ftsK; ftsKΔaa334-806 | This study |
| LS4213 | CB15N (NA1000) vanA::ftsZ-yfp; pMR20-Pxyl::ftsK | This study |
| LS4214 | CB15N (NA1000) vanA::ftsZ-yfp; xylX::ftsK; ftsK::ΩaacC4 | This study |
| LS4201 | CB15N (NA1000) pMR20-Pxyl::ftsK | This study |
ts, temperature sensitive.
The ftsK gene was amplified by using primers FtsK-13 (5′-CATATGCGACGGTTGAGTTCGGAGAC-3′) and FtsK-14 (5′-ATGAAGCCGCGTTCAGGTTG-3′) with CB15N chromosomal DNA as template. The resulting product was cloned into the pCR-Blunt II-TOPO vector (Invitrogen) to make pSCW569.
Strain LS4239 was constructed as follows. The NdeI- and EcoRI-digested fragment from pSCW569 and the NheI-NdeI Pxyl-containing fragment from pXGFP4 (gift from M. R. K. Alley, Anacor Pharmaceuticals, Palo Alto, CA) were cloned into the SpeI- and BamHI-digested integration vector pXGFP7C1 (M. R. K. Alley). The resulting plasmid, pSCW642, was integrated into the chromosome of CB15N at the xylX locus by a single crossover event.
Strain LS4203 was constructed as follows. The 5′ ftsK flanking region was amplified using primers FtsK-38 (5′-TATAGAATTCGGACGTTTGGGAAACCAGA-3′) and FtsK-39 (5′-TATAGGATCCAAGGAGCTCCGTCGTGGA-3′) with CB15N chromosomal DNA as template. The 3′ flanking region of the ftsK gene was amplified using primers FtsK-11 (5′-TATAGGATCCGGCAAGCGGGAGATTTTG-3′) and FtsK-12 (5′-GGACTAGTCAGGTTCAGCGGCGTCTT-3′) with CB15N chromosomal DNA as template. The EcoRI- and BamHI-digested and BamHI- and SpeI-digested PCR products were cloned into the allele exchange vector pNPTS138 (M. R. K. Alley). The resulting plasmid (pSCW755) was then digested with BamHI, and an apramycin resistance cassette from pHP45ΩaacC4 (6) was then cloned into that site between the upstream and downstream sequences, resulting in plasmid pSCW761. Plasmid pSCW761 was integrated into the chromosome of CB15N at the ftsK locus by a single crossover event. Clones containing tandem copies of the wild-type and disrupted ftsK genes were selected, and this locus was then transduced into LS4239. Subsequently, excision of the integrated pNPTS138 derivative was selected for by growing the resulting strain on PYE plates containing 3% sucrose and 0.3% xylose. Colonies were then screened by PCR for correct recombinants.
Strain LS4200 was constructed as follows. The ftsK gene was amplified using pSCW569 as template, using primers FtsK-6 (5′-GGAATTCCATATGGCGCGAGCCGCGCGGCGATCCA-3′) and FtsK-7 (5′-TATAGAATTCGAGCGGCGGCGTCGGCGGGGCCAAAAT-3′). The NdeI- and EcoRI-digested PCR products were cloned into the integration vector pXGFP4, containing ∼2,300 bp of the xylX locus and the gfp fragment from pEGFP-N2 (Clontech). The resulting plasmid (pSCW658) was integrated into the chromosome of CB15N at the xylX locus by a single crossover event to produce strain LS4200.
Strains LS4204, LS4205, and LS4206 were constructed as follows. For all three strains, the truncated ftsK genes were amplified using pSCW569 as template. For LS4202, primers FtsK-6 and FtsK-42 (5′-TATAGAATTCGGCCGCGATCGGGGTTTCGT-3′) were used for amplification. For LS4205, primers FtsK-6 and FtsK-43 (5′-TATAGAATTCCTTGGACTTGGCCAGCAT-3′) were used for amplification. For LS4206, primers FtsK-44 (5′-GGAATTCCATATGCCGCGCTCGTCCGAGGTGGA-3′) and FtsK-7 were used for amplification. The NdeI- and EcoRI-digested PCR products were cloned into the integration vector pXGFP4. The resulting plasmids, pSCW804, pSCW807, and pSCW796, respectively, were integrated into the chromosome of CB15N at the xylX locus by a single crossover event.
Strain LS4215 was constructed as follows. The 5′ ftsK C-terminal flanking region was amplified using primers FtsK-9 (5′-TATAGAATTCGCCATCTGGCAGCTAGAAAA-3′) and FtsK-10 (5′-TATAGGATCCCTTGGACTTGGCCAGCAT-3′) with CB15N chromosomal DNA as template. The 3′ ftsK C-terminal flanking region was amplified using primers FtsK-11 and FtsK-12 with chromosomal DNA as template. The EcoRI- and BamHI-digested and BamHI- and SpeI-digested PCR products were cloned into the allele exchange vector pNPTS138. The resulting plasmid (pSCW583) was integrated into the chromosome of CB15N at the ftsK locus by a single crossover event. Clones containing tandem copies of the wild-type and disrupted ftsK genes were selected, and this locus was then transduced into LS4239. Excision of the integrated pNPTS138 plasmid was selected for by growing the resulting strain on PYE plates containing 3% sucrose and 0.3% xylose. Colonies were then screened by PCR for correct recombinants.
Strain LS4202 was constructed as follows. The genomic region encoding the N5004 terminus of FtsK was amplified using pSCW569 as template, using primers FtsK-6 and FtsK-22 (5′-TATAGGATCCCTACAGTTCGGGCAGCTGGAA-3′). The NdeI- and BamHI-digested PCR products were cloned into the integration vector pXGFP5 (M. R. K. Alley), containing ∼200 bp of the xylX locus and the gfp fragment from pEGFP-N2 (Clontech). The resulting plasmid (pSCW620) was integrated into the chromosome of CB15N at the ftsK locus by a single crossover event.
Strain LS4207 was constructed as follows. pSCW449 (32) was digested with NdeI and HpaI. The fragment containing parC-gfp was gel purified and cloned into the NdeI- and EcoRV-digested integration vector pMT355 (M. Thanbichler, unpublished). The resulting plasmid (pSCW818) was integrated into the chromosome of CB15N at the vanA locus by a single crossover event. The vanA::parC-gfp fusion was then transduced into LS4202.
Strain LS4201 was constructed as follows. pSCW569 was digested with NdeI and HindIII. The fragment containing the ftsK gene was gel purified and, along with the NheI-NdeI Pxyl fragment from pXGFP4, cloned into the SpeI- and HindIII-digested low-copy-number plasmid pMR20. The resulting plasmid (pSCW599) was transformed into CB15N.
Strains LS4212, LS4213, and LS4214 were constructed as follows. The vanA::ftsZ-yfp fusion from strain MT196 (M. Thanbichler, unpublished) was transduced into LS4215, LS4201, and LS4203, respectively.
Live cell microscopy.
Cells were imaged as described previously (17) with the following modifications. Cells were immobilized on a thin layer of agarose containing M2G or M2G including either 0.3% xylose or 0.5 mM Na-vanillate when appropriate.
FISH.
The protocol for fluorescence in situ hybridization (FISH) was as described previously (15).
RESULTS
FtsK is an essential cell division protein in Caulobacter.
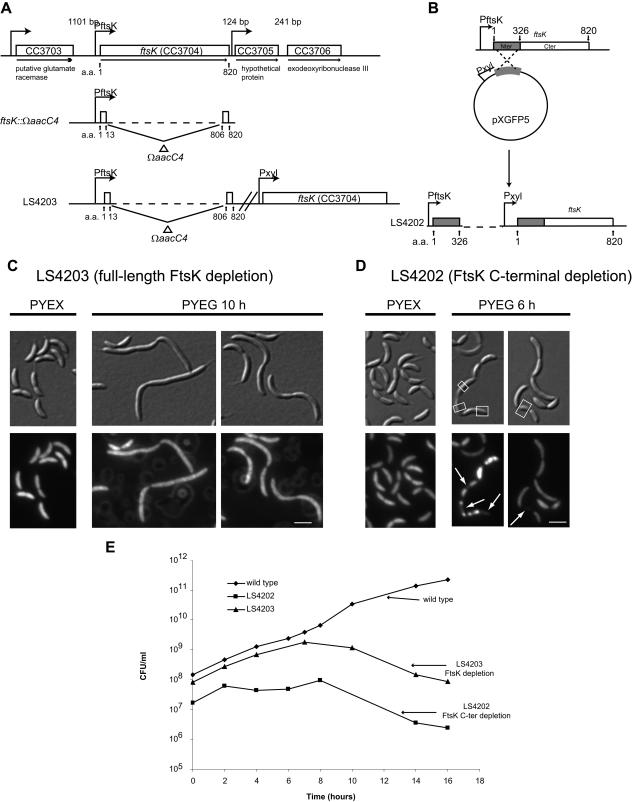
A chromosomal deletion of the ftsK gene was constructed by replacing the majority of the coding region with a gene cassette that confers resistance to apramycin (ΩaacC4), resulting in a null allele of ftsK (ftsK::ΩaacC4) (Fig. 1A). In the same cell, the full-length ftsK gene was integrated at the chromosomal xylX locus, placing ftsK expression under the control of a xylose-inducible promoter in strain LS4203. The ftsK::ΩaacC4 strain was viable only in the presence of the wild-type ftsK gene. When strain LS4203 was grown in the presence of PYE supplemented with 0.2% glucose (PYEG), expression of ftsK was repressed. FtsK depletion by growth of LS4203 in PYEG for 10 h resulted in the accumulation of smooth filamentous cells, suggesting that FtsK is necessary for cell division (Fig. 1C). In addition, cultures of LS4203 exhibited a decrease in CFU after approximately 7 to 10 h of growth in PYEG (Fig. 1E). These results indicate that FtsK is essential. To visualize the DNA content within the cell, strain LS4203 was stained with 4′, 6-diamidino-2-phenylindole (DAPI). Similar to the wild type, DNA was dispersed throughout the entire cell even after FtsK depletion by 10 h of growth in PYEG (Fig. 1C). Flow cytometry analysis of LS4203 after FtsK depletion by growth in PYEG showed that the filamentous cells continue to initiate DNA replication even while being blocked in cell division (data not shown).
FIG. 1.
Construction and phenotype of full-length and C-terminal depletion strains of FtsK. (A) Schematic representation of ftsK and surrounding genes on the chromosome. Horizontal arrows indicate the direction of transcription. Perpendicular arrows indicate putative promoters. Numbered arrows point to relevant amino acid residues. Lengths of intergenic regions in base pairs are indicated above. A deletion of ftsK, ftsK::ΩaacC4, was constructed by replacing amino acids (a.a.) 14 to 805 with the ΩaacC4 cassette. In the same cell, the full-length ftsK gene was integrated at the chromosomal xylX locus, creating strain LS4203. (B) Schematic diagram of the construction of an FtsK C-terminal depletion strain, LS4202. The first 978 base pairs (first 326 amino acids) of ftsK (N terminus [Nter]), indicated in gray, were put under the control of a xylose-inducible promoter and ligated to the integration vector pXGFP5 to yield pSCW620. pSCW620 was then transformed into the wild-type CB15N strain. A single recombination event generated a full-length copy of ftsK under the control of Pxyl and the first 326 codons under the control of the native PftsK promoter. Cter, C terminus. (C and D) Nomarski differential interference contrast (DIC) and fluorescent images of cells treated with DAPI. In panel C, strain LS4203 was grown in rich medium with 0.3% xylose (PYEX) to induce expression of the wild-type copy of ftsK. Cells were then washed and grown in PYEG for 10 h to deplete full-length FtsK. In panel D, strain LS4202 was grown in PYEX to induce expression of wild-type FtsK. Cells were then washed and grown in PYEG for 6 h to deplete full-length FtsK, leaving only the synthesis of the FtsK N terminus. Arrows indicate DNA-free areas resulting from a DNA-partitioning defect. White boxes indicate regions that lack DNA. White scale bars, 2 μm. (E) CFU for the wild type, LS4202, and LS4203 that were grown in rich medium with 0.3% xylose (PYEX) to log phase and then were washed and grown in the presence of 0.2% glucose (PYEG).
C terminus of FtsK is essential and is involved in chromosome partitioning.
To explore the role of the FtsK C-terminal region, we attempted to construct an in-frame deletion of the C terminus of FtsK (aa 334 to 806) utilizing the counterselectable sacB gene system. It was not possible to delete the DNA region encoding the C terminus of FtsK unless a complementing copy of the full-length ftsK gene was present at the chromosomal xylX locus. Thus, the C-terminal domain of FtsK is essential in Caulobacter.
To study the function of the FtsK C-terminal region, we constructed a strain in which the DNA region encoding the N terminus of FtsK (aa 1 to 326) was under the control of the endogenous ftsK promoter at its native locus and the full-length ftsK gene was under Pxyl control and integrated in the same cell (LS4202) downstream (Fig. 1B). Thus, when LS4202 is grown in the presence of glucose, full-length FtsK is depleted from the cell and only the region encoding the N-terminal domain of ftsK is expressed. Heretofore, we refer to growth of LS4202 in the presence of glucose as the depletion of the C terminus of FtsK, for simplicity.
When strain LS4202 was grown in PYEG for 6 hours, cells exhibited a late-stage cell division defect, resulting in the formation of chains of cells (Fig. 1D), and we observed a decrease in CFU (Fig. 1E), confirming that the FtsK C-terminal domain is essential for viability. Flow cytometry analysis showed that cells of strain LS4202 are able to continue to initiate DNA replication even though they are unable to complete cell division (data not shown).
To determine if the FtsK C terminus has a role in chromosome partitioning, we grew strain LS4202 in the presence of glucose for 6 h and stained DNA with DAPI (Fig. 1D). Approximately 15 to 20% of the cells had an obvious partitioning defect, where DNA was unevenly distributed throughout the cell or chains of cells. In a low percentage of cells, DAPI staining was entirely missing from some cell compartments (Fig. 1D).
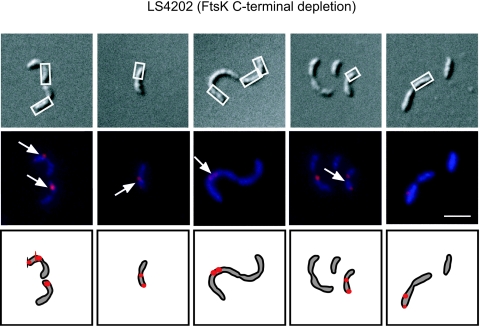
To further characterize the partitioning defect, we performed FISH with probes to the terminus region of the chromosome. When strain LS4202 was grown in the presence of glucose for 6 h to deplete the FtsK C-terminal region, 15 to 20% of cells had defects in terminal distribution. Chains of cells accumulated foci in one cell compartment and had other compartments devoid of termini, thus demonstrating an inability to completely partition chromosomes among daughter cells in the absence of the FtsK C-terminal domain (Fig. 2). In contrast, cells of strain LS4202 grown in the presence of xylose for 6 h, and thus induced for wild-type ftsK expression, did not display a partitioning defect.
FIG. 2.
Terminal regions are mislocalized in strain LS4202. Strain LS4202 was grown in PYEX to induce expression of wild-type FtsK. Cells were then washed and grown in PYEG for 6 h to deplete full-length FtsK, leaving only the synthesis of the FtsK N terminus. FISH was then performed with probes to the terminal region of the chromosome. (Top row) DIC images. White boxes indicate cell compartments that lack termini. (Middle row) Overlay of DAPI staining (pseudocolored blue) and Cy5 terminal probe (pseudocolored red). Arrows indicate cell compartments that contain one or more terminal foci. (Bottom row) Schematic diagram of terminus localization. Termini are shown as red dots, and DAPI staining is shown in gray. White scale bar, 2 μm.
FtsK is localized to the division plane.
The cell cycle-regulated expression of ftsK peaks at 90 min in a 135-min cell cycle (20). To determine the cellular position of FtsK in synchronized cultures, we constructed a gfp fusion to the 3′ end of the full-length ftsK gene and placed it under the control of a xylose-inducible promoter at the chromosomal xylX locus. The wild-type copy of the ftsK gene is still present at its normal chromosomal site, creating the merodiploid strain LS4200.
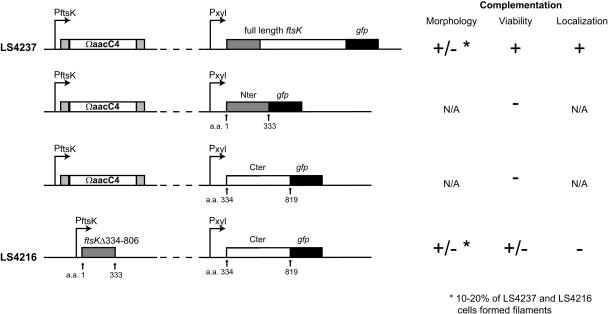
To determine if the FtsK-green fluorescent protein (GFP) fusion is functional, the ftsK::ΩaacC4-null allele was transduced into LS4200, replacing the wild-type copy of the ftsK gene while leaving Pxyl::ftsK-gfp at the xylose locus to create strain LS4237 (see Fig. 5). When strain LS4237 was grown in the presence of xylose, inducing expression of ftsK-gfp, localization of the FtsK-GFP protein was similar to that of the wild type (data not shown). The FtsK-GFP fusion protein alone complemented the FtsK deletion phenotype.
FIG. 5.
FtsK N-terminal (Nter) and C-terminal (Cter) domains, fused to GFP, are able to complement in trans. Schematic diagrams of complementation constructs are shown on the left. Complementation of morphology, viability, and localization are indicated with a +/−, +, −, or N/A (not applicable).
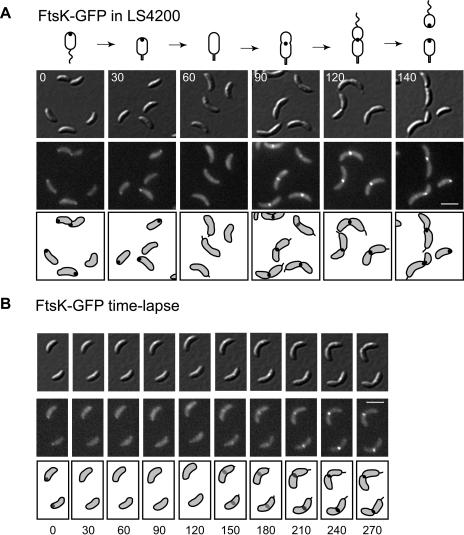
Strain LS4200 was incubated in the presence of 0.3% xylose to induce expression of ftsK-gfp. Swarmer cells were isolated and then allowed to progress synchronously through the cell cycle (Fig. 3A). At the swarmer cell stage (0 min), FtsK-GFP was observed at a cell pole, where it remained during the swarmer-to-stalked-cell transition (30 min). At the stalked-cell stage (60 min), FtsK-GFP disperses and then reappears at the division plane in early and late predivisional cells (90 and 120 min, respectively). As cells divide, FtsK-GFP stays at the new poles in both the progeny swarmer and stalked cells (140 min).
FIG. 3.
FtsK localization as a function of the cell cycle. (A) Intracellular localization of FtsK-GFP in LS4200 as a function of the cell cycle. LS4200 was grown in minimal medium in the presence of 0.3% xylose for 2 h to induce expression of ftsK-gfp. LS4200 swarmer cells were isolated from the FtsK-induced culture and allowed to progress synchronously through the cell cycle. Images were collected at the indicated times (in minutes). A schematic of the Caulobacter cell cycle showing the dynamic localization of FtsK is shown above. (B) Time lapse microscopy of FtsK-GFP in LS4200. Cells were grown in minimal medium with 0.3% xylose to induce expression of FtsK-GFP. Swarmer cells were isolated and placed on an agarose pad containing xylose. Images of the same cells were acquired every 30 min as the cells progressed through the 300-min cell cycle. For panels A and B: top rows, DIC images; middle rows, GFP fluorescence; bottom rows, schematic diagrams of FtsK-GFP fluorescence in gray. White scale bars, 2 μm.
To ascertain the cellular location of polar FtsK-GFP, we followed individual cells as they progressed through the cell cycle (Fig. 3B). Swarmer cells were isolated, immobilized on agarose pads, and imaged every 30 min during the cell cycle using the fluorescence microscope. Swarmer cells exhibit a focus of FtsK-GFP at the pole opposite the flagellum, based on time lapse imaging, which disperses as the cells move through the swarmer-to-stalked-cell transition and then reappears at the division plane in predivisional cells (Fig. 3B).
N terminus of FtsK is necessary and sufficient for localization to the division plane.
To identify the region of the FtsK protein responsible for targeting FtsK to the division plane, we constructed GFP fusions to two N-terminal fragments of FtsK, comprising the first 258 and 333 amino acids, respectively, both of which include the four predicted N-terminal transmembrane domains in Caulobacter. In addition, we constructed a GFP fusion to a C-terminal fragment of FtsK (amino acids 334 to 819) (Fig. 4A). The gfp fusions were integrated at the chromosomal xylX locus, placing expression of the gfp fusions under the control of a xylose-inducible promoter while the wild-type copy of ftsK was retained at its endogenous chromosomal site. When strains LS4204 and LS4205 were grown for 2 h in the presence of 0.3% xylose to induce the expression of ftsK1-258-gfp and ftsK1-333-gfp, respectively, both N-terminal FtsK fragments were able to localize to the division plane (Fig. 4B). We cannot exclude the possibility that the N-terminal GFP fusions interacted with the wild-type FtsK protein present at the division plane. However, when strain LS4206 was induced for expression of ftsK334-819-gfp, the protein did not localize (Fig. 4B), suggesting that at least the first 258 amino acids of the N-terminal domain of FtsK are necessary and sufficient for targeting the FtsK protein to the division plane.
FIG. 4.
N terminus of FtsK is necessary and sufficient for localization to the division plane. (A) Schematic representation of full-length FtsK and FtsK truncations. Numbered arrows point to relevant amino acid residues. The transmembrane domain (TM) is shown in dark gray. Transmembrane segments are shown as black bars. GFP was fused to the C termini of fragments comprising amino acids 1 to 258 (LS4204), 1 to 333 (LS4205), and 334 to 819 (LS4206). In these strains, the ftsK-gfp fusions were placed under the control of the Pxyl promoter by ectopic integration into the xylX locus while the wild-type ftsK gene was still present. Localization of the fusion proteins to the division plane is indicated with “+,” while no localization of the strains is indicated with “−.” (B) Localization of the fusion proteins produced by strains LS4204, LS4205, and LS4206. Each strain was grown in rich medium (PYE) and then transferred to rich medium with 0.3% xylose (PYEX) for 2 h to induce expression of the fusion proteins. Top, DIC images; middle, GFP fluorescence; bottom, schematic diagrams of GFP fluorescence in gray. White scale bar, 2 μm.
Separated N- and C-terminal regions of FtsK fused to GFP can function in trans to complement an FtsK deletion.
To determine if individual N-terminal domains and C-terminal domains, on the same chromosome but not contiguous, can complement an ftsK-null allele, we first examined the functionality of either the FtsK N-terminal region (aa 1 to 333) or the C-terminal region (aa 334 to 819) fused to GFP in an ftsK deletion background (Fig. 5). Neither the N-terminal GFP fusion in an ftsK deletion background nor the C-terminal GFP fusion in an ftsK deletion was able to complement the loss of wild-type FtsK. We then transduced the Pxyl::ftsK334-819-gfp allele into strain LS4215. Thus, we produced a strain (LS4216) in which Pxyl::ftsK334-819-gfp replaces Pxyl::ftsK at the xylX locus while the 3′ (C-terminal) in-frame deletion of ftsK remains at the endogenous ftsK locus (Fig. 5). Thus, when LS4216 is grown in glucose, only the N-terminal domain of ftsK is produced, but when it is grown in xylose, the C-terminal domain fused to GFP is also produced. There were no colonies when selection for transductants was performed on medium without xylose. However, in the presence of xylose, the FtsK C-terminal domain fused to GFP and the FtsK N-terminal domain are produced in the same cell, but not from contiguous genes. Under these conditions, cells were able to grow, albeit somewhat slower. Furthermore, as in the case of strain LS4237 with an intact ftsK gene fused to gfp, approximately 15 to 20% of the cells of strain LS4216 were slightly elongated or filamentous. These results suggest that the N- and C-terminal domains of FtsK can function in trans to complement the growth phenotype of the FtsK deletion. Because FtsK334-819-GFP did not localize in strain LS4216, complementation of the growth phenotype occurred when the N-terminal region was localized to the division plane, but the C-terminal region was not localized.
FtsK C terminus is necessary for localizing the topoisomerase IV subunit, ParC, to the replisome.
We have previously shown that the Topo IV ParC subunit colocalizes with the DnaB helicase component of the replisome throughout the Caulobacter cell cycle (32). Both ParC-GFP and DnaB-YFP are dispersed in swarmer cells, where DNA replication is normally repressed (8). At the swarmer-to-stalked-cell transition, when DNA replication is initiated, ParC-GFP and DnaB-YFP localize to the stalked-cell pole coincident with the assembly of the replisome at the origin of replication (32). As DNA replication progresses, ParC-GFP and DnaB-YFP foci move with the replisome from the stalked pole toward the division plane in predivisional cells. Finally, in late predivisional cells, when DNA replication is completed and the replisome disassembles, both ParC-GFP and DnaB-YFP foci disperse and then reappear at the stalked-cell pole, where the replisome is again assembled and DNA replication is initiated (32).
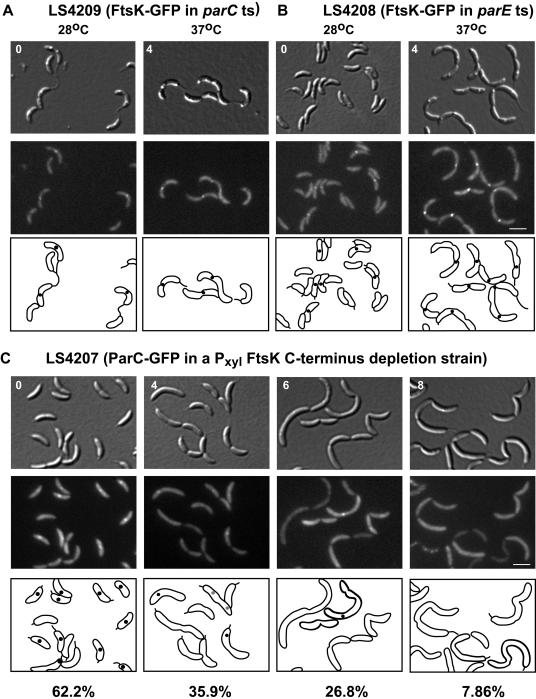
Because both ParC (32) and FtsK (Fig. 3) appear at the division plane prior to cell separation, we asked if their cellular positioning is interdependent. Accordingly, we transduced the Pxyl::ftsK-gfp allele into a temperature-sensitive mutant of parC (LS4209). Upon shifting of the parC temperature-sensitive strain to the nonpermissive temperature (37°C), chains of cells were formed, indicating a late-stage cell separation defect (Fig. 6A). When strain LS4209 was incubated with 0.3% xylose for 2 h to induce expression of ftsK-gfp at the permissive temperature (28°C), cells showed the wild-type FtsK localization pattern. When cells were shifted to the nonpermissive temperature for 4 h, FtsK-GFP maintained the localization pattern observed in wild-type cells and in LS4209 at 28°C (Fig. 6A). Thus, FtsK is able to localize to the division plane independent of the presence of active ParC. The same experiment was repeated with a temperature-sensitive mutant of the other topo IV subunit, parE (LS4208). Again, FtsK localized to the division plane independent of the presence of ParE (Fig. 6B), which we showed previously is required for ParC localization (32).
FIG. 6.
ParC requires FtsK to localize to the replisome. Mixed cultures of LS4209 (A) and LS4208 (B) were grown in minimal medium at the permissive temperature (28°C) (0 h) to log phase and then shifted to the nonpermissive temperature (37°C) for 4 h. Cells were incubated with 0.3% xylose for 2 h before each time point to induce expression of ftsK-gfp. Top, DIC images; middle, GFP fluorescence; bottom, schematic diagrams of FtsK-GFP fluorescence in black. White scale bar, 2 μm. (C) A mixed culture of LS4207 was grown in rich medium with 0.3% xylose (PYEX) to log phase and then depleted for the FtsK C terminus by growth in rich medium with 0.2% glucose (PYEG) for the indicated number of hours. Before each time point, cells were also incubated with 0.5 mM Na-vanillate (pH 7.5) for 1 h to induce expression of parC-gfp. Top, DIC images; middle, GFP fluorescence; bottom, schematic diagrams of ParC-GFP fluorescence in black. The percentage of cells with ParC-GFP foci in strain LS4207 after depletion of the FtsK C terminus for the indicated number of hours is indicated below the images (n > 140 cells). ts, temperature sensitive.
To determine if ParC can localize to the division plane in the absence of the FtsK C-terminal region, we constructed a strain in which only the FtsK N-terminal domain is expressed upon growth in the absence of xylose and which contains an inducible parC-gfp allele. To generate this strain, parC-gfp was integrated at the chromosomal vanA locus, placing parC-gfp under the control of a vanillate-inducible promoter (M. Thanbichler, unpublished). The vanA::parC-gfp construct was then transduced into strain LS4202, resulting in a strain (LS4207) in which the full-length FtsK was depleted by growth in PYEG. In this strain, the presence of the localized FtsK N-terminal domain was maintained and the expression of parC-gfp was induced by vanillate. Effectively, this strain was depleted of the FtsK C-terminal domain. When LS4207 was grown in PYE containing xylose (PYEX) (thus expressing wild-type ftsK) and induced with 0.5 mM Na-vanillate for 1 h, approximately 62.2% of cells showed ParC-GFP foci (Fig. 6C, 0). Upon incubating strain LS4207 in PYEG to deplete the FtsK C terminus, the percentage of cells with ParC-GFP foci decreased to 35.9% after 4 h. ParC-GFP foci decreased to 26.8% 6 h after incubation in PYEG and to 7.86% after 8 h (Fig. 6C). Thus, the localization of the FtsK N-terminal domain to the division plane was not sufficient for the division plane localization of ParC-GFP. Furthermore, this experiment suggested that the C-terminal domain of FtsK may be necessary for ParC-GFP foci to localize to the replisome or that the loss of ParC-GFP foci could be due to a general loss of replisome localization. However, we observed that two other components of the replisome, DnaB-YFP and HolC-YFP, shown previously to form foci at the assembled replisome (32), were able to localize independent of FtsK C-terminal function (data not shown). Therefore, the localization of ParC to the division plane requires the FtsK C-terminal domain.
FtsK is required to maintain FtsZ rings.
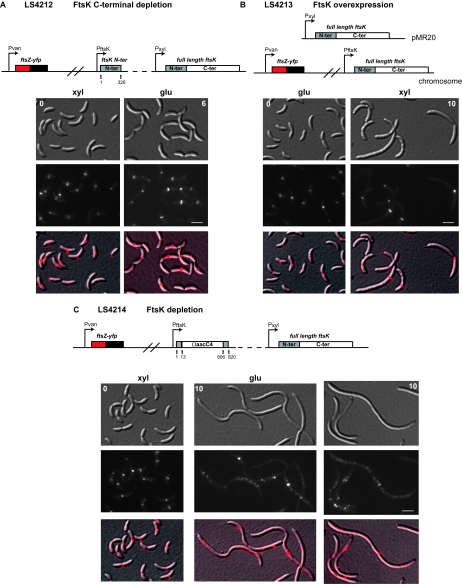
Because FtsK appears to be involved in both chromosome segregation and cell division, we examined the effect of FtsK on FtsZ ring formation. To accomplish this, we observed FtsZ-YFP ring formation in three different ftsK mutants: (i) an FtsK C-terminal depletion strain (LS4212), (ii) an FtsK overexpression strain (LS4213), and (iii) an FtsK full-length depletion strain (LS4214).
Strain LS4212 contained an ftsK C-terminal deletion under the control of the ftsK promoter, the full-length ftsK gene under the control of the Pxyl promoter, and an ftsZ-yfp fusion integrated at the chromosomal vanA locus such that its expression was controlled by a vanillate-inducible promoter. When this strain was grown in medium containing glucose and vanillate, the FtsK C terminus was depleted and FtsZ-YFP was induced. When it was grown in the presence of xylose and induced for expression of ftsZ-yfp with 0.5 mM vanillate for 1 h, we observed a wild-type pattern of FtsZ rings at the division plane. Similarly, upon depleting the FtsK C terminus after 6 h in glucose-containing medium, we observe that the wild-type pattern of FtsZ-YFP localization was maintained (Fig. 7A). These results suggest that the C terminus of FtsK is not involved in the assembly of FtsZ rings at the division plane.
FIG. 7.
FtsK is required to maintain FtsZ rings. (A) A culture of LS4212 was grown in PYEX to log phase (0 h). Cells were then washed and grown in PYEG for 6 h to deplete the C terminus (C-ter) of FtsK. xyl, xylose; glu, glucose. (B) A culture of the FtsK overexpression strain LS4213 was grown in PYEG to log phase (0 h) and then induced with 0.3% xylose for 10 h to induce expression of ftsK. Before each time point in panels A to C, cells were also incubated in 0.5 mM Na-vanillate (pH 7.5) for 1 h to induce expression of ftsZ-yfp. (C) A culture of LS4214 was grown in PYEX to log phase (0 h). Cells were then washed and grown in PYEG for 10 h to deplete them of FtsK. White scale bar, 2 μm. For panels A to C, schematic diagrams of each construct are shown above the images. Top, DIC images; middle, YFP fluorescence; bottom, overlay of DIC and YFP fluorescence. FtsZ-YFP is pseudocolored red.
We also examined the cellular position of FtsZ in an FtsK overexpression strain. This strain contains wild-type ftsK at its chromosomal locus, Pxyl::ftsK on the pMR20 low-copy-number plasmid, and the Pvan::ftsZ-yfp allele integrated at the chromosomal vanA locus, creating strain LS4213 (Fig. 7B). Upon overexpression of FtsK with 0.3% xylose for 10 h, we observed that cells become filamentous and smooth, indicating a block in cell division (Fig. 7B). The FtsK overexpression strain appeared to retain FtsZ-YFP rings at the old division sites (Fig. 7B).
To determine the effect on FtsZ ring formation of a full-length FtsK depletion, we constructed a strain such that Pvan::ftsZ-yfp was integrated at the chromosomal vanA locus, Pxyl::ftsK was integrated at the chromosomal xylX locus, and a null allele of ftsK (ftsK::ΩaacC4) replaced the wild-type copy of ftsK, creating strain LS4214. Before the depletion of FtsK, FtsZ-YFP displayed Z rings at the division plane. However, upon depletion of FtsK for 10 h, we observed the formation of long filaments in which there was a random distribution of multiple diffuse FtsZ rings (Fig. 7C). Cumulatively, the experiments shown in Fig. 7 suggest that the N terminus of FtsK is involved in assembling or maintaining FtsZ rings at the division plane.
DISCUSSION
The final stage of DNA replication is a critical juncture in the cell cycle during which multiple events must occur in a defined temporal order and in a precise position in the cell. These events include the decatenation of newly replicated chromosomes by Topo IV so that chromosome segregation and subsequent cell division can occur (1, 25). Because formation of the division septum is initiated while DNA replication is still in progress, chromosomal termini are frequently found in only one of the two daughter cell compartments, trapping DNA in the dividing septum (19). The FtsK (SpoIIIE) protein has been shown in both B. subtilis (3, 35) and E. coli (21, 38) to play a role in the completion of chromosome movement into the daughter cells. Here, we have investigated the role of the FtsK protein in Caulobacter and shown it to play a critical role in both chromosome segregation and cell division.
The Caulobacter FtsK protein was found to be dynamically localized to the division plane during the cell cycle and to remain transiently positioned at the new cell poles following cell division. In both E. coli (31, 37) and B. subtilis (36), FtsK is present at the division plane but is not retained at the new poles generated by cell division. Multiple proteins involved in cytokinesis are localized to the division plane in E. coli, B. subtilis, and C. crescentus (22, 26, 34), suggesting a role for FtsK in this process or in the final steps of chromosome replication.
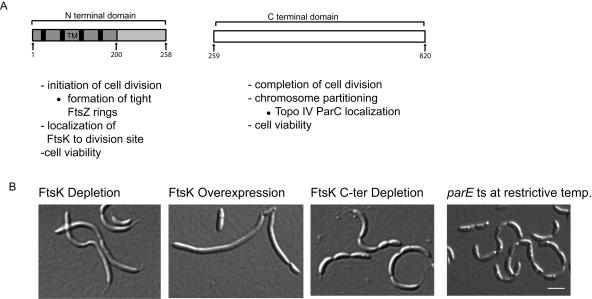
We have shown here that the first 258 amino acids of the FtsK N terminus are sufficient to target the protein to the division plane; the FtsK C terminus was unable to localize in the cell. These results suggest that the N-terminal domain of FtsK is necessary and sufficient for targeting the protein to the division plane (Fig. 8A). A fusion of GFP to the C terminus of the full-length FtsK protein restored, albeit at a slower growth rate, the viability and localization pattern of an ftsK deletion strain, although 10 to 20% of the cells were filamentous. We found that a distal C-terminal FtsK334-819-GFP fragment was able to complement, in trans, a strain containing the N-terminal domain but depleted of the C-terminal domain (Fig. 5). Since the C-terminal fragment in this strain was unable to localize to the division plane and yet viability was restored, we conclude that the cellular position of the C terminus is not a critical determinant of cell viability. However, we cannot rule out the possibility that only a few molecules of FtsK are sufficient for function and these would not be detected through the background fluorescence.
FIG. 8.
Summary of the functions of the FtsK N- and C-terminal domains. (A) Schematic diagram of the N- and C-terminal domains with their separate functions listed below. (B) DIC images comparing the full-length FtsK depletion strain (LS4203), the FtsK overexpression strain (LS4201), the FtsK C-terminal depletion strain (LS4202), and the parE temperature-sensitive (ts) strain (LS4208) with the indicated time (in hours) of depletion, overexpression, or incubation at the restrictive temperature shown. TM, transmembrane domain.
Cells depleted of FtsK formed long, smooth filaments prior to cell death, indicating that FtsK is required for cell division (Fig. 8A and B). When only the C-terminal domain of FtsK was depleted, cells formed bead-like chains of cells, similar to that exhibited by the temperature-sensitive mutants of the Topo IV subunits ParC and ParE at the restrictive temperature (32, 33) (Fig. 8A and B). The similarity in phenotype may be caused by the lack of DNA clearance from the division plane in both cases, creating a block in cell division. This phenotype is also seen in Caulobacter SMC deletion mutants, where chromosomes exhibit abnormal segregation with a consequent block in cell division (15). The C-terminal domain of FtsK is essential in Caulobacter, which is in direct contrast to what has been observed in E. coli (21, 38) (Fig. 8A). Chromosome segregation may be more tightly coordinated with cell division in Caulobacter, as seen in the temperature-sensitive Topo IV mutants and the Δsmc strain. Therefore, without the FtsK C-terminal region, abnormal chromosome segregation causes a cell division block, producing a cell separation defect (Fig. 8A and B).
We have previously shown that the Topo IV subunit, ParC, colocalizes with the DnaB helicase, a component of the replisome during the cell cycle (32). ParC, DnaB, and FtsK all localize to the division plane at the final stages of DNA replication. In E. coli, the C-terminal domain of FtsK was shown to interact with ParC and stimulate its decatenation activity (12). Here, we have shown that although FtsK is not dependent on ParC for its localization to the division plane, the C-terminal domain of FtsK is required for the formation of ParC foci, presumably indicative of its interaction with the replisome (Fig. 8A). It is possible that the FtsK C terminus acts indirectly by serving as an activator of an adapter protein linking ParC to the replisome. Another possibility is that free FtsK in the cytoplasm may serve as a direct link between ParC and components of the replisome complex.
The smooth filaments formed in the absence of FtsK reflect the cells' inability to form functional FtsZ rings (Fig. 8A and B), although numerous diffuse FtsZ foci are found dispersed along the long filamentous cells. However, in the FtsK C-terminal depletion strain, FtsZ rings are formed at constriction sites and at potential division sites. Because the N-terminal domain is still present in this strain and is able to localize normally to the incipient division plane, the localized FtsK N terminus may be sufficient to recruit downstream effectors of cell division to the division plane, thus allowing the cell to constrict at division sites. However, chromosome segregation is disrupted when the C terminus is depleted, causing cells to block at the cell separation stage.
Overexpression of FtsK causes the formation of smooth filaments (Fig. 8B). It is possible that the excess copies of FtsK in the cell may form complexes with downstream effectors of cell division in the cytoplasm. This may disrupt the normal stoichiometry of cell division effectors interacting with the Z ring at the division plane. Thus, productive division fails to occur.
Surprisingly, FtsZ-GFP did not form rings in the full-length FtsK depletion mutant. Instead, FtsZ was delocalized and diffuse, suggesting that the N-terminal domain of FtsK is required to maintain FtsZ in tight rings. The FtsK N terminus may regulate FtsZ directly by acting as a structural component necessary to maintain FtsZ ring formation or indirectly by binding to and controlling a structural regulator of FtsZ ring formation. The appearance of diffuse nucleoids and multiple FtsZ bands or spirals has been observed in E. coli upon general inhibition of transcription (29). Sun and Margolin (29) suggested that blocking general transcription suppresses nucleoid occlusion such that FtsZ is then able to form on top of nucleoids, albeit as multiple nonfunctional bands or spirals. Because nucleoid occlusion is not known to function in Caulobacter, it is possible that the FtsK N terminus instead acts as an activator of a Caulobacter-specific mechanism, preventing FtsZ ring formation in an incorrect position within the cell. Therefore, in the full-length FtsK depletion mutant, such a mechanism is suppressed and FtsZ becomes diffuse throughout the cell. Taken together, these results demonstrate that the multifunctional FtsK protein contributes to the interdependence of chromosome segregation and cell division.
Acknowledgments
We thank Joseph Chen, Antonio Iniesta, and Martin Thanbichler for critical reading of the manuscript. We also thank M.T. for use of his Pvan promoter and FtsZ-YFP constructs.
This work was supported by NIH grant GM51426 and DOE grant DE-FG03-O1ER63219. S.C.W. was supported by a predoctoral fellowship from the National Science Foundation.
REFERENCES
- 1.Adams, D. E., E. M. Shekhtman, E. L. Zechiedrich, M. B. Schmid, and N. R. Cozzarelli. 1992. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell 71:277-288. [DOI] [PubMed] [Google Scholar]
- 2.Aussel, L., F. X. Barre, M. Aroyo, A. Stasiak, A. Z. Stasiak, and D. Sherratt. 2002. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108:195-205. [DOI] [PubMed] [Google Scholar]
- 3.Bath, J., L. J. Wu, J. Errington, and J. C. Wang. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995-997. [DOI] [PubMed] [Google Scholar]
- 4.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigot, S., J. Corre, J. M. Louarn, F. Cornet, and F. X. Barre. 2004. FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol. Microbiol. 54:876-886. [DOI] [PubMed] [Google Scholar]
- 6.Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 7.Boyle, D. S., D. Grant, G. C. Draper, and W. D. Donachie. 2000. All major regions of FtsK are required for resolution of chromosome dimers. J. Bacteriol. 182:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degnen, S. T., and A. Newton. 1972. Chromosome replication during development in Caulobacter crescentus. J. Mol. Biol. 64:671-680. [DOI] [PubMed] [Google Scholar]
- 9.Diez, A. A., A. Farewell, U. Nannmark, and T. Nystrom. 1997. A mutation in the ftsK gene of Escherichia coli affects cell-cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J. Bacteriol. 179:5878-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draper, G. C., N. McLennan, K. Begg, M. Masters, and W. D. Donachie. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 180:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 12.Espeli, O., C. Lee, and K. J. Marians. 2003. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 278:44639-44644. [DOI] [PubMed] [Google Scholar]
- 13.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, R. B., and L. Shapiro. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. USA 96:10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, R. B., S. C. Wang, and L. Shapiro. 2002. Dynamic localization of proteins and DNA during a bacterial cell cycle. Nat. Rev. Mol. Cell Biol. 3:167-176. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, R. B., S. C. Wang, and L. Shapiro. 2001. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 20:4952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, J., Y. Nishimura, R. Imamura, H. Niki, S. Hiraga, and H. Suzuki. 1990. New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 19.Lau, I. F., S. R. Filipe, B. Soballe, O. A. Okstad, F. X. Barre, and D. J. Sherratt. 2003. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 49:731-743. [DOI] [PubMed] [Google Scholar]
- 20.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 21.Liu, G., G. C. Draper, and W. D. Donachie. 1998. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol. Microbiol. 29:893-903. [DOI] [PubMed] [Google Scholar]
- 22.Martin, M. E., M. J. Trimble, and Y. V. Brun. 2004. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol. Microbiol. 54:60-74. [DOI] [PubMed] [Google Scholar]
- 23.Ohta, N., A. J. Ninfa, A. Allaire, L. Kulick, and A. Newton. 1997. Identification, characterization, and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J. Bacteriol. 179:2169-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pease, P. J., O. Levy, G. J. Cost, J. Gore, J. L. Ptacin, D. Sherratt, C. Bustamante, and N. R. Cozzarelli. 2005. Sequence-directed DNA translocation by purified FtsK. Science 307:586-590. [DOI] [PubMed] [Google Scholar]
- 25.Peng, H., and K. J. Marians. 1993. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl. Acad. Sci. USA 90:8571-8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quardokus, E. M., N. Din, and Y. V. Brun. 2001. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol. Microbiol. 39:949-959. [DOI] [PubMed] [Google Scholar]
- 27.Sharp, M. D., and K. Pogliano. 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295:137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharpe, M. E., and J. Errington. 1995. Postseptational chromosome partitioning in bacteria. Proc. Natl. Acad. Sci. USA 92:8630-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, Q., and W. Margolin. 2004. Effects of perturbing nucleoid structure on nucleoid occlusion-mediated toporegulation of FtsZ ring assembly. J. Bacteriol. 186:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viollier, P. H., M. Thanbichler, P. T. McGrath, L. West, M. Meewan, H. H. McAdams, and L. Shapiro. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. USA 101:9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, L., and J. Lutkenhaus. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol. 29:731-740. [DOI] [PubMed] [Google Scholar]
- 32.Wang, S. C., and L. Shapiro. 2004. The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins. Proc. Natl. Acad. Sci. USA 101:9251-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward, D., and A. Newton. 1997. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 26:897-910. [DOI] [PubMed] [Google Scholar]
- 34.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 35.Wu, L. J., and J. Errington. 1994. Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]
- 36.Wu, L. J., and J. Errington. 1997. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 16:2161-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, X. C., A. H. Tran, Q. Sun, and W. Margolin. 1998. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J. Bacteriol. 180:1296-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, X. C., E. K. Weihe, and W. Margolin. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol. 180:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]