Abstract
The plant-pathogenic bacterium Xanthomonas campestris pv. vesicatoria possesses a type III secretion (TTS) system which is encoded by the 23-kb hrp (hypersensitive response and pathogenicity) gene cluster. The TTS system is necessary for pathogenicity in susceptible hosts and induction of the hypersensitive response in resistant plants. At the cell surface, the TTS system is associated with an extracellular filamentous structure, the Hrp pilus, which serves as a conduit for the transfer of bacterial proteins into the plant cell cytosol. The major pilus component, the HrpE pilin, is unique to xanthomonads. Previous work showed that HrpE contains two regions: a hypervariable surface-exposed domain, including the N-terminal secretion signal, and a C-terminal polymerization domain. In this study, the evolutionary rate of the hrpE gene was analyzed. Twenty-one alleles were cloned, sequenced, and compared with five known hrpE alleles. The ratio of synonymous (Ks) and nonsynonymous (Ka) substitution rates shows that parts of the HrpE N terminus are subjected to positive selection and the C terminus is subjected to purifying selection. The trade-off between positive and purifying selection at the very-N terminus allowed us to ascertain the amphipathic α-helical nature of the TTS signal. This is the first report of a surface structure from a plant-pathogenic bacterium that evolved under the constraint of positive selection and hints to the evolutionary adaptation of this extracellular appendage to avoid recognition by the plant defense surveillance system.
The type III secretion (TTS) system is a hallmark of many gram-negative bacterial pathogens. This specialized secretion system is responsible for the transport of proteins across the two bacterial membranes, across the host cell plasma membrane, and in some cases across the plant cell wall into the host cell interior. A defect in this system leads to a complete loss of bacterial pathogenicity, as demonstrated for the animal pathogens Yersinia spp., Shigella spp., Salmonella spp., and enteropathogenic and enterohemorrhagic Escherichia coli and for the plant pathogens Ralstonia solanacearum, Pseudomonas syringae, Erwinia spp., and Xanthomonas spp. (10). TTS systems are encoded by approximately 20 genes, 11 of which are conserved between different bacterial species, thus suggesting that they constitute the core of the secretion machinery. While the TTS apparatus is conserved, the secreted substrates differ considerably. The recognition of these proteins by the TTS system is still not well understood. The TTS signal is located in the first 15 to 20 amino acids of the protein and is not conserved on the amino acid level (17, 23). Additionally, the 5′ region of the corresponding mRNA may contribute to recognition by the secretion machinery (1).
Our laboratory studies the plant pathogen Xanthomonas campestris pathovar vesicatoria, which is the causal agent of bacterial spot disease in pepper and tomato plants. The TTS system of X. campestris pv. vesicatoria is encoded by the 23-kb hrp (hypersensitive response and pathogenicity) gene cluster, which is essential to cause disease in susceptible plants as well as to induce the hypersensitive response in resistant plants (9). A key component of the X. campestris pv. vesicatoria pathogenicity machinery is the Hrp pilus, which is thought to serve as a conduit for protein translocation between the bacterium and the plant host cell (30). The Hrp pilus is a filamentous structure that extends up to several micrometers from the bacterial cell surface and is mainly composed of a large number of identical pilin subunits, the 10-kDa HrpE protein, which is unique for the genus Xanthomonas (30). HrpE itself is a TTS substrate and harbors a secretion signal in the first 17 N-terminal amino acids (29). Sequence alignment and secondary structure predictions of five hrpE sequences showed that the very-C terminus is entirely conserved and predominantly α-helical, whereas the N terminus is hypervariable and mostly devoid of regular secondary structure. Reporter protein fusions and pentapeptide insertions demonstrated that HrpE contains two functional domains, a surface-exposed domain, including the TTS signal, and the polymerization domain (29). The sequence hypervariability of the surface-exposed domain prompted us to analyze the evolutionary rate of 26 hrpE sequences, representing Xanthomonas strains belonging to 14 different pathovars. Our results reveal different modes of selection acting on different portions of the protein. The N terminus of HrpE undergoes positive selection, whereas the C terminus is subjected to purifying selection. Since the positively selected parts are exposed at the surface of the assembled Hrp pilus, the plant pathogen Xanthomonas thus generates surface diversity, most probably to evade recognition by the plant perception and defense systems. Furthermore, we propose an amphipathic α-helix as the TTS signal of HrpE which is on one side highly conserved and polar and on the opposite side positively selected and hydrophobic.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The following pathovars/strains of X. campestris were used: alfalfae KS, amoraciae XA, begoniae 077-3382, citri 50E, dieffenbachiae 729, glycines 202, malvacearum G-34, phaeseoli 85-6, vesicatoria 56, vesicatoria 85-10, vignicola 81-30, and vitians 164 (4); amoraciae 3838, amoraciae 5 (48A), raphani 23, and raphani 25 (R. E. Stall, University of Florida, Gainesville); and amoraciae LMG535, amoraciae LMG7383, raphani LMG860, raphani LMG7505, and raphani LMG8134 (Belgium coordinated collections of microorganisms, Brussels, Belgium). Escherichia coli cells were cultivated at 37°C in LB medium, and Xanthomonas strains were grown at 30°C in NYG broth (6) or on NYG 1.5% agar. Antibiotics were added to the medium at the following final concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; rifampin, 100 μg/ml.
Sequence data.
The coding regions of 21 hrpE alleles were amplified using PCR primers derived from conserved regions of hrpD6 (hrpEup, 5′-CAACATCGTCGCGCACATCGC) and hpaB (hrpEdown, 5′-TCGAATCGCGCGCTGCTCAT) located upstream and downstream of the hrpE coding sequence, respectively. For colony PCR, a single colony of each strain was used as the source of template DNA. PCR conditions were an initial 95°C step for 300 s, followed by 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 40 s, and a final step at 72°C for 10 min. PCR products were cloned into pCR 2.1-TOPO and transformed into DH10b (Invitrogen, Carlsbad, CA). Five clones of each hrpE allele were pooled and sequenced using vector-specific primers (TOPO TA cloning kit; Invitrogen).
Additionally, the hrpE nucleotide sequences of X. campestris pv. vesicatoria strain 75-3 (GenBank accession number [AC] AF056246), X. oryzae pv. oryzae strain KACC 10331 (AC AE013598), X. axonopodis pv. glycines strain 8ra (AC AF499777), X. axonopodis pv. citri strain 306 (AC AE011665), and X. campestris pv. campestris strain ATCC 33913 (AC AE012221) were used for bioinformatic analyses.
Sequence analysis.
The nucleotide sequences were translated and aligned using CLUSTAL X (26). The multiple alignment was then manually edited using the BIOEDIT program (11). The amino acid alignments were transposed back to the nucleotide sequence level to gain a codon-based alignment. Mean diversity estimates within sequences were performed with the MEGA 2 software, version 2.1 (15), using the modified Nei-Gojobori method with the Jukes-Cantor corrected distance and the complete-deletion-of-gaps option. Unweighted pair group method with averages (UPGMA) trees were constructed using PAUP 3.1.1 (25). Distances were determined by using the Kimura two-parameter model. Support for nodes was assessed with a bootstrap confidence level using 1,000 replicates.
Calculation of synonymous and nonsynonymous mean pairwise diversities.
The method of Comeron, as implemented in the K-Estimator program, version 6.1 (5), was applied to test whether positive or purifying selection has taken place on hrpE. K-Estimator takes into account the divergence values, number of nucleotides or codons, the transition/transversion substitution ratio (α/β), the amino acid composition, and the G+C content at the third codon position. The confidence intervals for Ka and Ks were obtained by Monte Carlo simulations (5). Synonymous mean pairwise diversity per synonymous site (Ks) and nonsynonymous mean pairwise diversity per nonsynonymous site (Ka) were calculated with the complete-deletion-of-gaps option and a sliding window of 15 nucleotides with a step size of 9 nucleotides.
Nucleotide sequence accession numbers.
The nucleotide sequences for the 21 hrpE alleles were submitted to GenBank (accession numbers DQ286679 to DQ286699).
RESULTS
Rate of synonymous and nonsynonymous substitutions (Ks and Ka).
To analyze the general extent of sequence variation in the genus Xanthomonas, we analyzed five genes from five Xanthomonas strains (Table 1). Generally, a neutral rate test compares the number of synonymous (silent) substitutions per synonymous site (Ks) with the number of nonsynonymous (amino acid-changing) substitutions per nonsynonymous site (Ka) in protein coding sequences. Thus, the ratio Ka/Ks measures the difference between the two rates, in which Ks is often regarded as a value of the underlying mutation rate (19). If an amino acid change is neutral, it will be fixed at the same rate as a synonymous mutation, with Ka/Ks of 1. If the amino acid change is deleterious, purifying selection will reduce its fixation rate, and thus Ka/Ks is <1. However, only if the amino acid change offers a selective advantage will it be fixed at a higher rate than a synonymous mutation, with Ka/Ks of >1 (positive selection) (31). Three of the analyzed genes (hrcS, hrcT, and hrpE) are located in the hrp gene cluster and encode crucial components of the TTS system. For reference, two housekeeping genes, malate dehydrogenase (mdh) and glyceraldehyde-3-phosphate dehydrogenase (gapA), were used. The most notable feature is an unusually high level of amino acid substitutions in the hrpE gene product, which is reflected by the Ka value of 0.278 (Table 1). In contrast, the Ka values for hrcT, hrcS, mdh, and gapA are uniformly low and fall within the narrow range of 0.007 to 0.039. In addition, the Ks value for hrpE is strongly elevated (0.494) in comparison with those of hrcT, hrcS, mdh, and gapA (0.172 to 0.258) (Table 1). The Ka/Ks ratio, which describes the evolutionary rate of a protein coding sequence, is much higher for hrpE than for the other four genes. Although the Ka value does not exceed the Ks value for the full-length coding sequence of hrpE, this high value indicates that parts of hrpE may be subjected to positive selection.
TABLE 1.
Sequence variations of five genes among five Xanthomonas strainsa
| Gene | Size (bp) | Mean pairwise value (102)
|
Ka/Ks ratio | |
|---|---|---|---|---|
| Ks | Ka | |||
| hrpE | 279 | 49.4 ± 6.4 | 27.8 ± 4.3 | 0.560 |
| hrcS | 261 | 25.8 ± 4.8 | 0.80 ± 0.1 | 0.031 |
| hrcT | 828 | 22.2 ± 2.3 | 3.90 ± 0.6 | 0.170 |
| gapAb | 999 | 17.2 ± 1.8 | 0.70 ± 0.2 | 0.040 |
| mdhb | 987 | 18.8 ± 2.0 | 2.50 ± 0.5 | 0.130 |
X. campestris pv. vesicatoria strain 75-3, X. oryzae pv. oryzae strain KACC 10331, X. axonopodis pv. glycines strain 8ra, X. axonopodis pv. citri strain 306, and X. campestris pv. campestris strain ATCC 33913.
X. axonopodis pv. glycines strain 8ra was not included in the analysis.
Comparison of hrpE sequences.
For a more detailed analysis, the hrpE regions of 21 Xanthomonas strains belonging to 12 pathovars were cloned and sequenced. The phylogenetic tree of all 26 hrpE sequences was constructed using the UPGMA method (Fig. 1). Interestingly, the genealogy revealed two major branches (groups I and II).
FIG. 1.
UPGMA tree of hrpE sequences from 26 Xanthomonas strains. Numbers on branches indicate the number of times (percent) that the node was supported by 1,000 replicates of the bootstrap analysis (only values greater than 80% are shown).
Five of 13 hrpE sequences in group I and 3 of 13 hrpE sequences in group II were identical. These siblings were excluded from further analyses. Figure 2 shows an amino acid sequence alignment of the nonredundant HrpE proteins. The last 24 amino acid residues of HrpE are highly conserved in all xanthomonads, suggesting that this region is crucial for HrpE function. Another conserved sequence motif (PxxxSxxxRxxQxxD) is located at the very-N terminus, whereas the region between the two conserved parts is hypervariable. Intriguingly, the conserved N-terminal amino acid pattern which corresponds to the TTS signal is predicted to form an α-helix (30). A helical wheel plot revealed that the conserved polar residues are located on one side of the helix, whereas the opposite side consists of hydrophobic amino acids (Fig. 3). These findings strongly suggest that the HrpE secretion signal is formed by an amphipathic α-helix.
FIG. 2.
Nonredundant amino acid sequence alignment of HrpE. HrpE sequences of group I and II xanthomonads were aligned using CLUSTAL X (26). Residues which are identical in at least 90% of the sequences are shown in white on a black background. Proposed functional domains are indicated on top of the alignment (29).
FIG. 3.
Type III secretion signal of the X. campestris pv. vesicatoria 85-10 HrpE in a helical wheel representation. Conserved amino acids and hydrophobic residues are shaded in gray and black, respectively. The N-terminal proline residue is oriented to the bottom.
Whether or not recombination in the hrpE sequence has occurred was analyzed by assessing the degree of phylogenetic congruence. Based on the amino acid sequence alignment (Fig. 2), the hrpE alleles were split into two halves: a variable region consisting of codons 4 to 50 and the relatively conserved region encompassing codons 51 to 93. Phylogenetic trees were then estimated for each half. No significant differences in topologies of the trees were observed (data not shown), thus indicating that no recombination between hrpE alleles belonging to groups I and II had occurred.
Modes of selection operating on hrpE.
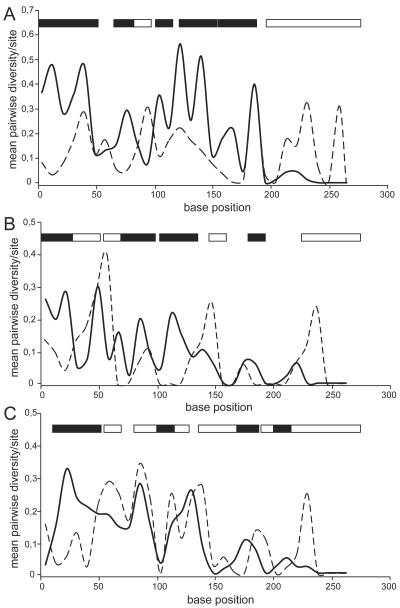
Using the enlarged set of hrpE sequences, we investigated whether certain parts of HrpE show different modes of selection. A sliding window analysis of Ka and Ks values was performed. Since the first amino acid of HrpE from group I and the first four amino acids of HrpE from group II give no substantial information, they were not considered. Windows in which Ka exceeded Ks (one tailed Z-test, P < 0.05) (Fig. 4A) were found in the N-terminal and central regions of the protein. This finding shows that diversifying selection had occurred at these regions. In contrast, Ks exceeded Ka in windows 98 and 215 to 266 (one tailed Z-test, P < 0.05 [Fig. 4A]; the number indicates the central nucleotide of the window), indicating that these regions evolved under purifying selection.
FIG. 4.
Mean pairwise diversity plots of Ks and Ka values. The plots were constructed for windows of 15 nucleotides sliding along the hrpE sequence alignment. The first codon of group I and the first four codons of group II sequences are not included. (A) Comparison of all hrpE sequences. (B) Group I sequence comparison. (C) Group II sequence comparison. Filled boxes indicate the windows (step size of 9 nucleotides) in which nonsynonymous differences significantly exceed synonymous differences (one-tailed Z test, P < 0.05). White boxes indicate positions in the sequence where the reverse condition applies (Ks > Ka; P < 0.05) or sequences which are identical. Solid line, mean Ka; dashed line, mean Ks.
The observation that the hrpE sequences belong to two clearly distinct phylogenetic groups prompted us to analyze whether different modes of selection operated after separation of groups I and II. Individual analyses of group I and II sequences showed that positive selection occurred at the very-N terminus in both groups (group I, 8 to 26; group II, 17 to 44), whereas the C terminus was subjected to purifying selection (group I, 224 to 266; group II, 242 to 266) (Fig. 4B and C). Intriguingly, the intermediate parts of hrpE evolved differently in groups I and II. Most notably, the region between windows 71 and 125 shows strong signatures of positive selection in group I, while in group II this region evolved mainly under purifying constraints.
The positively selected positions are surface exposed.
Certain modes of selection act often highly localized within genes. For many plant resistance proteins, plant chitinases, and the outer membrane porin PorB of Neisseria meningitidis, positive selection has been detected primarily for solvent-exposed amino acids (3, 7, 28). Since solvent-exposed parts of a protein are in general rather hydrophilic and core regions of proteins are more hydrophobic, we performed hydrophobicity plot analyses of groups I and II and compared the localizations of positively selected and hydrophilic regions. As shown in Fig. 5, the HrpE regions which are positively selected in group I or II correspond to hydrophilic regions. Of special interest is the region between amino acids 25 and 35, which is hydrophilic in group I and hydrophobic in group II. This different physico-chemical character is perfectly reflected by different modes of selection, indicating that this region might be surface exposed and therefore positively selected in group I. Notably, for the C-terminal hydrophilic region an indication of positive selection is missing. This finding may be related to the proposed function of this domain in the polymerization process (29).
FIG. 5.
Positively selected parts of HrpE are surface exposed. Mean hydrophobicity plots of group I (dashed line) and group II (solid line) HrpE proteins were calculated over a sliding window of five amino acid residues, using the Kyte and Doolittle hydrophobicity scale (16). Values below 0.4 are considered hydrophilic. Positively selected parts in groups I and II are indicated by open and filled boxes, respectively.
DISCUSSION
Many gram-negative plant pathogens possess a surface-exposed Hrp pilus that mediates the long-distance transport of TTS substrates across the plant cell wall. Although the major pilus subunits of different genera do not share sequence homology, they share several features with each other, such as small size (6 to 11 kDa), predominantly α-helical secondary structure, and sequence hypervariability (14). Interestingly, sequence hypervariability is not only observed between different species but also between pathovars of the same species (12). In this study, we analyzed the sequence variability of the Xanthomonas Hrp pilus subunit HrpE, showed that the observed sequence hypervariability is caused by positive selection, and speculated about the biological significance of hypervariability.
A sliding window analysis of 18 hrpE alleles revealed several regions in the N-terminal two-thirds in which Ka exceeds Ks, thus indicating that these regions are positively selected. In contrast, the far C terminus is almost identical or subjected to purifying selection in all Xanthomonas sequences. Positive selection has been reported as a beacon for proteins involved in genetic conflict and has been documented in genes that encode surface proteins of pathogen (2, 28) and host defense systems, such as the human major histocompatibility complex (13), plant chitinases (3), and plant resistance genes (7). The correlation between positive selection and host-pathogen interactions is particularly strong. For example, a GenBank survey uncovered remarkably few sequences (0.45%) evolving under positive selection, but more than half of these sequences were involved in host-pathogen interactions (8). Only six examples of bacterial surface proteins which have undergone positive selection have been described, but all of them originated from animal pathogens (27). This is the first report of a surface protein from a plant pathogen that is subjected to positive selection. This points to an unexpected analogy between animal and plant pathogenic bacteria in their strategy to avoid recognition by the host.
Despite the lack of an adaptive immune system, plants possess recognition capacities for several microbial compounds often indispensable for the microbial lifestyle. This set of general elicitors are referred to as pathogen-associated molecular patterns (PAMPs) and include the eubacterial flagellin, lipopolysaccharide of gram-negative bacteria, fungal cell wall-derived glucans, chitins, mannans, and surface proteins of microbial pathogens (20). An Hrp pilus of 500 nm in length would consist of approximately 1,000 HrpE subunits. As an abundant key component of the TTS system which is crucial for pathogenicity, HrpE would be a prime candidate to act as a PAMP. However, there is no report of a plant species or cultivar which has evolved a mechanism to detect HrpE or Hrp pilins from other plant pathogens as a PAMP.
Diversification of the Hrp pilus surface by positive selection acting on surface-exposed parts of the protein would make perfect sense in order to conceal the attack by Xanthomonas spp. Such a surface-limited diversification is a well-known strategy of human pathogenic bacteria (2, 28). Indeed, in both phylogenetic groups of Xanthomonas a remarkable correlation between hydrophilic regions and positive selection was found. Notably, only the first four hydrophilic stretches of the HrpE sequence were positively selected, whereas the C-terminal hydrophilic stretch was not. These findings are in perfect agreement with the proposed domain structure of HrpE (29). In this previous work it was found that the N-terminal two-thirds of the protein are tolerant to pentapeptide insertions, whereas the C-terminal third is not. Moreover, we isolated four dominant-negative insertions in the C-terminal third, thus supporting our hypothesis that this domain is conserved due to its function in polymerization. In line with this model, the C-terminal hydrophilic stretch would be hidden within the assembled Hrp pilus, and varying this sequence would give no selective advantage. In conclusion, the N-terminal surface-exposed domain functions as a camouflage suit for the highly conserved assembly domain at the C terminus.
An alternative mechanism to achieve sequence variation is based on a high frequency of recombination, as has been reported for components of the outer membrane or cell surface appendages of mammalian pathogens, such as the outer membrane protein PorB and the pilus protein PilE of N. meningitidis and the flagellin FliC of pathogenic E. coli (2, 22, 28). However, we did not observe discrepancies in the phylogenies between the full-length sequence and the separately analyzed N-terminal and C-terminal halves, indicating that recombination is not responsible for the diversification of hrpE. This finding is supported by the absence of a second copy of hrpE-related sequences in the completely sequenced genomes of five Xanthomonas strains. Interestingly however, the plant pathogens P. syringae and R. solanacearum possess a second copy of their Hrp pilin genes. It has been speculated that these second copies might act as capping or scaffolding proteins (12). Alternatively, one might speculate about a possible role of these extra genes in surface variation.
The very-N terminus of HrpE was shown to serve as a TTS signal (29). Despite 15 years of intense research, and even with the three-dimensional structures of the N termini of TTS substrates at hand, the nature of the TTS signal is still enigmatic (24). The fact that the N termini of these substrates do not share any obvious consensus sequence at the primary sequence level makes it difficult to understand how TTS substrates are recognized and secreted even by heterologous TTS machineries (24). Comparisons of Yersinia TTS substrates indicated that a more general property serves as secretion signal. Yersinial secretion signals consist of an amphipathic series of randomly alternating polar and hydrophobic residues, and even synthetic amphipathic signals consisting of serine and isoleucine were able to mediate secretion of a reporter protein (18). Similar biophysical features were identified in TTS substrates from the plant pathogen P. syringae (21). However, amphipathic N-terminal sequences derived from cytosolic components of the yersinial Yop regulon did not promote secretion of the reporter protein YopE (17). These findings imply that, in addition to the amphipathic character, there must be a sequence dependence on N-terminal secretion signals that is currently not understood.
While most studies on TTS signals have compared the N termini of distantly related or unrelated TTS substrates or have introduced point mutations into the N terminus of TTS substrates (21, 24), we analyzed a set of naturally occurring HrpE variants. From previous work it is known that the HrpE TTS signal is located in the first 17 amino acids (29). Since all HrpE N termini were obtained from pathogens, it is reasonable to assume that they are all functional TTS signals. The trade-off between sequence diversification (to avoid host recognition) and functional conservation (to allow secretion) operating at the HrpE N terminus offered a unique advantage to identify residues and principles that are of crucial importance for function as a TTS signal. The α-helix that would be formed by the amphipathic N-terminal sequence displays a polar and a hydrophobic side. In addition to the conserved amphipathic character of the α-helix, the five amino acid residues at the polar side seem to be critical for function, as indicated by their strong conservation. Probably, these two features are crucial for function of the HrpE TTS signal. To what extent this rule may apply to other TTS substrates remains to be clarified in future studies.
Acknowledgments
We thank Ulla Bonas for generous support of this study, Hannelore Espenhahn for excellent technical assistance, Jens Boch, Daniela Büttner, Sabine Kay, Diana Kühn, and Ute Wahrmund for critical reading of the manuscript, and Martin Beye and Martin Hasselmann for fruitful discussions.
E.W. was supported by a GradFG fellowship from Sachsen-Anhalt.
REFERENCES
- 1.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, T. D., and T. Gojobori. 2004. Strong positive selection and recombination drive the antigenic variation of the PilE protein of the human pathogen Neisseria meningitidis. Genetics 166:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, J. G., A. M. Dean, and T. Mitchell-Olds. 2000. Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA 97:5322-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonas, U., R. Schulte, S. Fenselau, G. V. Minsavage, B. J. Staskawicz, and R. E. Stall. 1991. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant-Microbe Interact. 4:81-88. [Google Scholar]
- 5.Comeron, J. M. 1999. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15:763-764. [DOI] [PubMed] [Google Scholar]
- 6.Daniels, M. J., C. E. Barber, P. C. Turner, M. K. Sawczyc, R. J. W. Byrde, and A. H. Fielding. 1984. Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J. 3:3323-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis, J., P. Dodds, and T. Pryor. 2000. The generation of plant disease resistance gene specificities. Trends Plant Sci. 5:373-379. [DOI] [PubMed] [Google Scholar]
- 8.Endo, T., K. Ikeo, and T. Gojobori. 1996. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 13:685-690. [DOI] [PubMed] [Google Scholar]
- 9.Fenselau, S., I. Balbo, and U. Bonas. 1992. Determinants of pathogenicity in Xanthomonas campestris pv. vesicatoria are related to proteins involved in secretion in bacterial pathogens of animals. Mol. Plant-Microbe Interact. 5:390-396. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999:95-98. [Google Scholar]
- 12.He, S. Y., K. Nomura, and T. S. Whittam. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181-206. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, A. L., and M. Nei. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167-170. [DOI] [PubMed] [Google Scholar]
- 14.Koebnik, R. 2001. The role of bacterial pili in protein and DNA translocation. Trends Microbiol. 9:586-590. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 16.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd, S. A., A. Forsberg, H. Wolf-Watz, and M. S. Francis. 2001. Targeting exported substrates to the Yersinia TTSS: different functions for different signals? Trends Microbiol. 9:367-371. [DOI] [PubMed] [Google Scholar]
- 18.Lloyd, S. A., M. Norman, R. Rosqvist, and H. Wolf-Watz. 2001. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol. Microbiol. 39:520-531. [DOI] [PubMed] [Google Scholar]
- 19.Miyata, T., T. Yasunaga, and T. Nishida. 1980. Nucleotide sequence divergence and functional constraint in mRNA evolution. Proc. Natl. Acad. Sci. USA 77:7328-7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nürnberger, T., F. Brunner, B. Kemmerling, and L. Piater. 2004. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198:249-266. [DOI] [PubMed] [Google Scholar]
- 21.Petnicki-Ocwieja, T., D. J. Schneider, V. C. Tam, S. T. Chancey, L. Shan, Y. Jamir, L. M. Schechter, M. D. Janes, C. R. Buell, X. Tang, A. Collmer, and J. R. Alfano. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 99:7652-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid, S. D., R. K. Selander, and T. S. Whittam. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorg, J. A., N. C. Miller, and O. Schneewind. 2005. Substrate recognition of type III secretion machines -testing the RNA signal hypothesis. Cell. Microbiol. 7:1217-1225. [DOI] [PubMed] [Google Scholar]
- 25.Swofford, D. L. 1993. PAUP: phylogeny analysis using parsimony, version 3.1.1. Illinois Natural History Survey, Champaign, Ill.
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tümmler, B., and P. Cornelis. 2005. Pyoverdine receptor: a case of positive Darwinian selection in Pseudomonas aeruginosa. J. Bacteriol. 187:3289-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urwin, R., E. C. Holmes, A. J. Fox, J. P. Derrick, and M. C. Maiden. 2002. Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol. Biol. Evol. 19:1686-1694. [DOI] [PubMed] [Google Scholar]
- 29.Weber, E., and R. Koebnik. 2005. Domain structure of HrpE, the Hrp pilus subunit of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 187:6175-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber, E., T. Ojanen-Reuhs, E. Huguet, G. Hause, M. Romantschuk, T. K. Korhonen, U. Bonas, and R. Koebnik. 2005. The type III-dependent Hrp Pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J. Bacteriol. 187:2458-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, Z., and J. P. Bielawski. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]