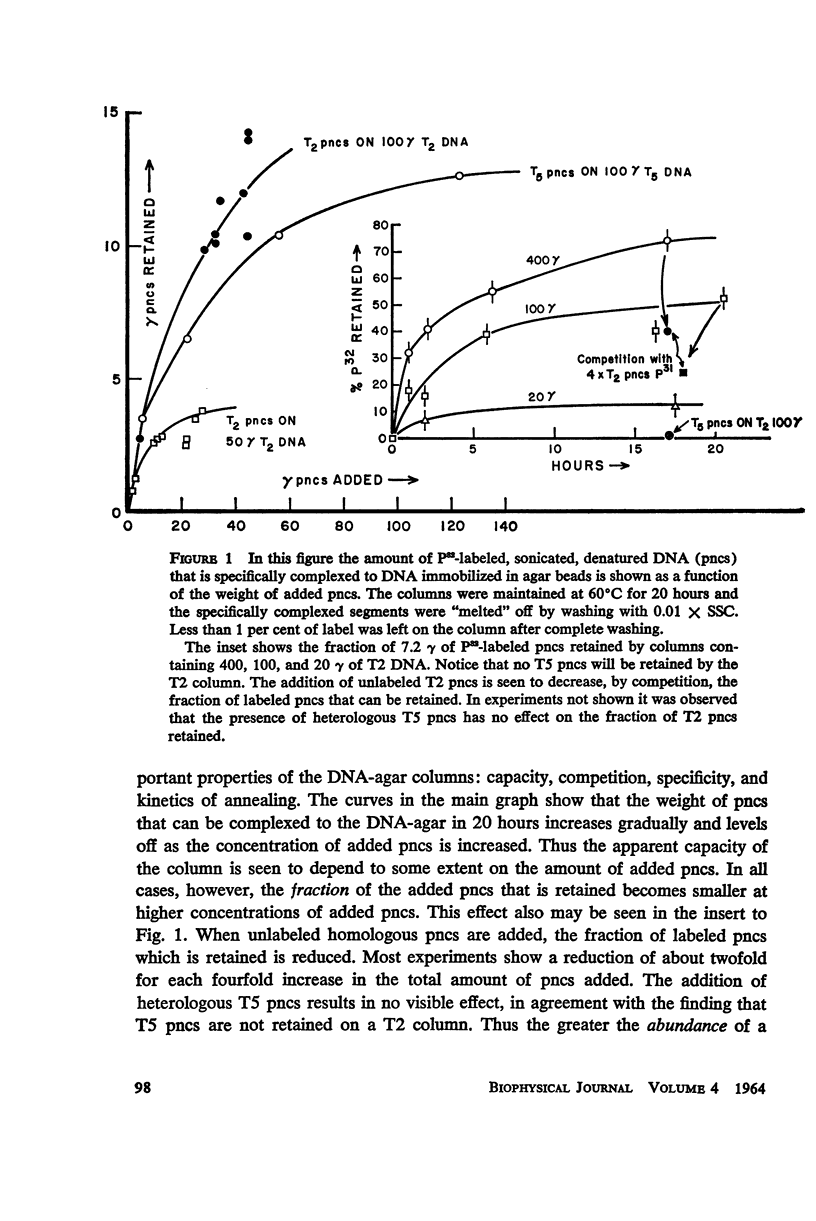
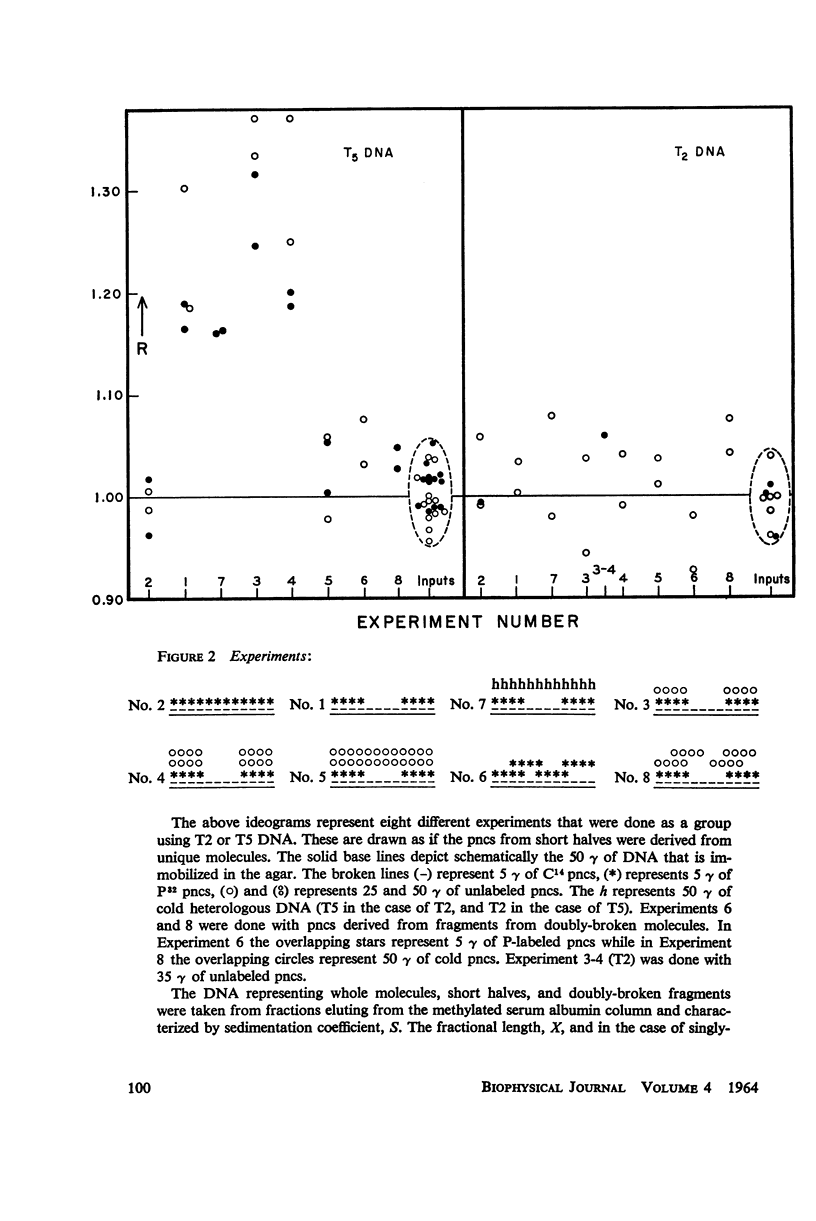
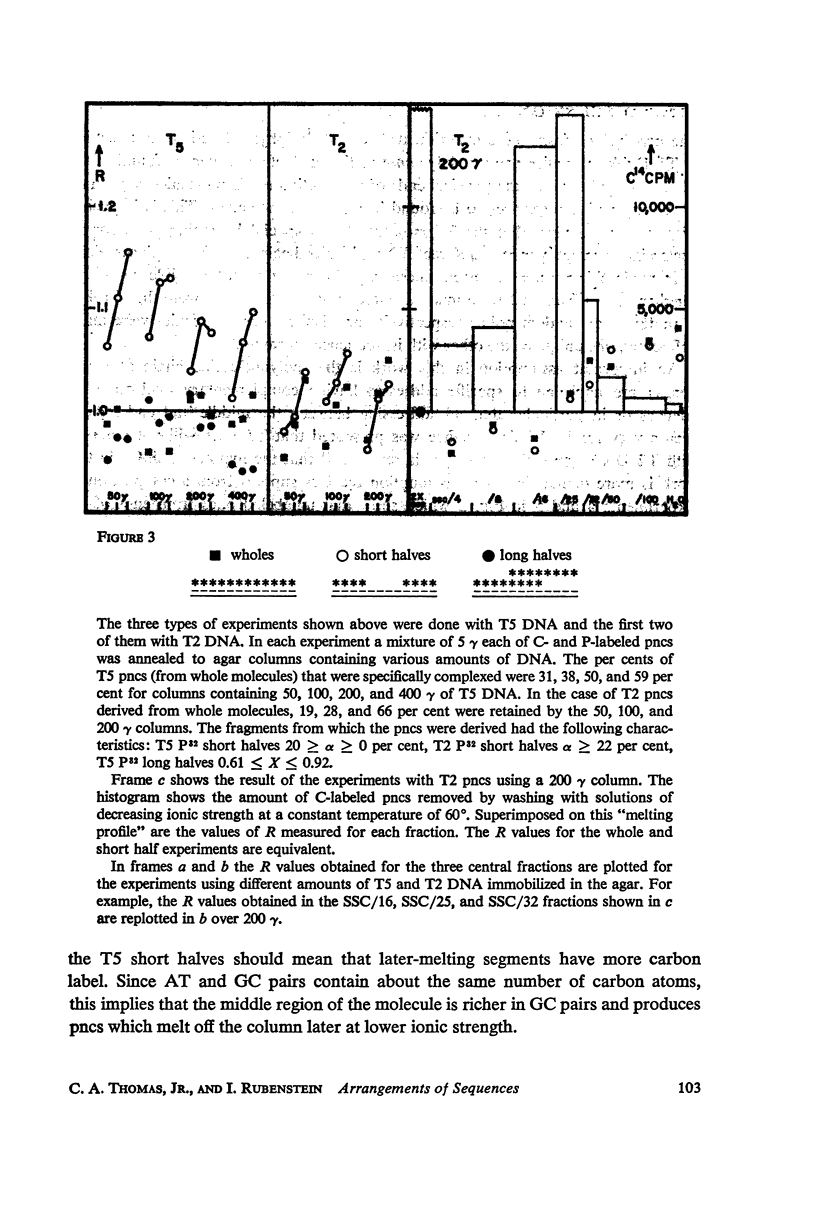
Abstract
The genetic map of T4 (and T2) bacteriophage is circular but the DNA molecule that is liberated by phenol extraction is a linear duplex of polynucleotide chains. If the genetic map is related to the physical structure of the DNA molecule, the problem arises as to how a linear molecule can give rise to a circular map. An explanation can be made on the basis that the bacteriophage liberate molecules which have nucleotide sequences which are circular permutations of each other. Thus, markers which are most distant on one molecules are closest together on another. To test this hypothesis, the middles of T2 and T5 DNA molecules were mechanically deleted and the absence of certain nucleotide sequences was tested by “renaturation” or “reannealing” experiments using columns containing denatured DNA immobilized in agar beads. The results indicate that when the middles are deleted from the T5 DNA molecule, some special sequences are removed; whereas, when the middles are deleted from the T2 DNA molecule, no special group of sequences is removed. This would indicate that T2 molecules begin at different points in their nucleotide sequence, while T5 molecules all begin at the same point. It is likely that this permutation of sequences of T2(T4) molecules is related to the circularity of their genetic map.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BEER M. Electron microscopy of unbroken DNA molecules. J Mol Biol. 1961 Jun;3:263–266. doi: 10.1016/s0022-2836(61)80067-3. [DOI] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. A relative molecular weight series derived from the nucleic acid of bacteriophage T2. J Mol Biol. 1961 Aug;3:458–472. doi: 10.1016/s0022-2836(61)80058-2. [DOI] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAIRNS J. An estimate of the length of the DNA molecule of T2 bacteriophage by autoradiography. J Mol Biol. 1961 Dec;3:756–761. doi: 10.1016/s0022-2836(61)80080-6. [DOI] [PubMed] [Google Scholar]
- DAVISON P. F., FREIFELDER D., HEDE R., LEVINTHAL C. The structural unity of the DNA of T2 bacteriophage. Proc Natl Acad Sci U S A. 1961 Aug;47:1123–1129. doi: 10.1073/pnas.47.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty P., McGill B. B., Rice S. A. THE PROPERTIES OF SONIC FRAGMENTS OF DEOXYRIBOSE NUCLEIC ACID. Proc Natl Acad Sci U S A. 1958 May;44(5):432–438. doi: 10.1073/pnas.44.5.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKEL F. R. An unusual DNA extracted from bacteria infected with phage T2. Proc Natl Acad Sci U S A. 1963 Mar 15;49:366–372. doi: 10.1073/pnas.49.3.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERSHEY A. D., GOLDBERG E., BURGI E., INGRAHAM L. Local denaturation of DNA by shearing forces and by heat. J Mol Biol. 1963 Mar;6:230–243. doi: 10.1016/s0022-2836(63)80072-8. [DOI] [PubMed] [Google Scholar]
- Hershey A. D., Burgi E., Ingraham L. Sedimentation Coefficient and Fragility under Hydrodynamic Shear as Measures of Molecular Weight of the DNA of Phage T5. Biophys J. 1962 Nov;2(6):423–431. doi: 10.1016/s0006-3495(62)86865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER A. D., HOGNESS D. S. The transformation of Escherichia coli with deoxyribonucleic acid isolated from bacteriophage lambda-dg. J Mol Biol. 1960 Dec;2:392–415. doi: 10.1016/s0022-2836(60)80050-2. [DOI] [PubMed] [Google Scholar]
- KAISER A. D. The production of phage chromosome fragments and their capacity for genetic transfer. J Mol Biol. 1962 Apr;4:275–287. doi: 10.1016/s0022-2836(62)80005-9. [DOI] [PubMed] [Google Scholar]
- KLEINSCHMIDT A. K., LANG D., JACHERTS D., ZAHN R. K. [Preparation and length measurements of the total desoxyribonucleic acid content of T2 bacteriophages]. Biochim Biophys Acta. 1962 Dec 31;61:857–864. [PubMed] [Google Scholar]
- LEVINTHAL C., DAVISON P. F. Degradation of deoxyribonucleic acid under hydrodynamic shearing forces. J Mol Biol. 1961 Oct;3:674–683. doi: 10.1016/s0022-2836(61)80030-2. [DOI] [PubMed] [Google Scholar]
- LURIA S. E. Genetics of bacteriophage. Annu Rev Microbiol. 1962;16:205–240. doi: 10.1146/annurev.mi.16.100162.001225. [DOI] [PubMed] [Google Scholar]
- MANDELL J. D., HERSHEY A. D. A fractionating column for analysis of nucleic acids. Anal Biochem. 1960 Jun;1:66–77. doi: 10.1016/0003-2697(60)90020-8. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Thermal renaturation of deoxyribonucleic acids. J Mol Biol. 1961 Oct;3:585–594. doi: 10.1016/s0022-2836(61)80023-5. [DOI] [PubMed] [Google Scholar]
- MESELSON M., WEIGLE J. J. Chromosome brekage accompanying genetic recombination in bacteriophage. Proc Natl Acad Sci U S A. 1961 Jun 15;47:857–868. doi: 10.1073/pnas.47.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G, Bruce V. Linkage of Genetic Markers in Phages T2 and T4. Genetics. 1960 Sep;45(9):1289–1296. doi: 10.1093/genetics/45.9.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]


