Abstract
Previously, we reported that human immunodeficiency virus type 1 (HIV-1) recombines approximately two to three times per genome per replication cycle, an extremely high rate of recombination given the relatively small genome size of HIV-1. However, a recombination hot spot involving sequence of nonretroviral origin was identified in the vector system utilized, raising the possibility that this hot spot skewed the rate of recombination, and the rate of recombination observed was an overestimation. To address this issue, an HIV-1-derived vector system was used to examine the rate of recombination between autologous HIV-1 sequences after restricting replication to a single cycle in the absence of this hot spot. Viral DNA and RNA were analyzed by a combination of the heteroduplex tracking assay, restriction enzyme analysis, DNA sequencing, and reverse transcription-PCR. The results indicate that HIV-1 undergoes recombination at a minimum rate of 2.8 crossovers per genome per cycle. Again, this is a very high rate given the small size of the HIV-1 genome. The results also suggested that there might be local hot spots of recombination at different locations throughout the genome since 13 of the 33 strand transfers identified by DNA sequencing shared the same site of recombination with one or two other clones. Furthermore, identification of crossover segments also allowed examination of mutations at the point of recombination, since it has been predicted from some studies of cell-free systems that mutations may occur with a frequency of 30 to 50% at crossover junctions. However, DNA sequence analysis of crossover junctions indicated that homologous recombination during viral replication was not particularly mutagenic, indicating that there are other factors or conditions not yet reproduced in cell-free systems which contribute to fidelity during retroviral recombination.
The human immunodeficiency virus type 1 (HIV-1) genome in nature is characterized by its rapid evolution, which permits the virus to escape immune surveillance and to develop drug-resistant variants as well as making it difficult to produce an effective vaccine. HIV-1 can diversify in at least two ways. First, mutations, including point mutations, deletions, and insertions, can be introduced into the genome during viral DNA synthesis by the viral reverse transcriptase (RT) (12, 22, 36, 50) owing in part to its lack of DNA proofreading activity (4). Second, sequence diversity can also be obtained via recombination. HIV-1, like all other retroviruses, is diploid, containing two genomic RNA molecules per virion. Therefore, cells infected with two different strains of HIV-1 might produce heterozygous virions, providing an opportunity for recombination to occur during reverse transcription. Previous studies have shown that such a process does occur in tissue culture systems (18, 55, 57), and the occurrence of HIV-1 recombination in nature is borne out by the identification of genomes that are recombinants between different HIV-1 subtypes (25, 28, 38, 45, 46). Some of these recombinant viruses have become fixed in the human population and are referred to as circulating recombinant forms (21, 25, 49), and in at least a few cases circulating recombinant forms have become the predominant strain in specific geographic areas of infection (6, 13, 23-25, 29, 30, 32). Thus, it appears that recombination has an important influence upon HIV-1 population dynamics throughout the world.
Retroviral recombination was initially identified in avian retroviruses (20, 51) and appears to be a common feature of retroviral replication in general (7, 54, 56). To examine the rates and mechanism of retroviral recombination, a system based upon spleen necrosis virus (SNV), isolated from birds, was established (2, 16, 17). This was a cell culture based system in which replication of vector virus was restricted to a single cycle, thereby yielding insight into the process of recombination (2, 16, 17). One of the interesting findings from this work was that homologous recombination seemed to occur in a distinct subpopulation exhibiting “high negative interference” (10, 16). To explain this phenomenon, the hypothesis was posed that either the subpopulation is predisposed to recombination or once recombination occurs it becomes more prone to recombination (2, 16).
A link between mutation and recombination has been established in cell-free systems. Several groups using cell-free systems have reported an increased mutation rate due to recombination (37, 39, 40, 55). In one case, mutations occurred approximately 30 to 50% of the time at the point of strand transfer using purified HIV-1 RT (37, 55). Furthermore, base misincorporations by RT have been shown to enhance strand transfer in vitro (11, 35). However, it remains to be tested whether recombination is mutagenic during HIV-1 replication in intact cells.
Previously, we reported that an HIV-1 vector recombines at a rate of two to three times per genome per replication cycle over 7,300 bases of HIV-1 autologous sequence (18). These results were obtained using two defective vectors based upon HIV-1 strains HXB2 and BCSG3 (14, 34). Recombination events were examined across the entire HIV-1 vector genome employing a heteroduplex tracking assay (HTA). The recombination rate obtained was similar to the rate found at the end of the viral genome during minus-strand DNA synthesis, indicating that most recombination events occurred during minus-strand DNA synthesis. A nonviral recombination hot spot was found, suggesting that sequence context can play a role in HIV-1 recombination. It is possible that the presence of a nonviral hot spot affected the overall recombination rate in the previous report. To address this contingency, recombination was examined in the absence of this nonviral hot spot using vectors based upon HIV-1 strains HXB2 and NL4-3 (1, 34). These vectors were used in conjunction with an inducible envelope-producing cell line to study HIV-1 recombination in a single cycle of retroviral replication between autologous HIV-1 sequences (58). Recombination events occurred at a minimum rate of 2.4 crossovers per vector genome per cycle. When extrapolated to the full-length HIV-1 genome, this would yield a minimum rate of 2.8 crossovers per genome per cycle. Furthermore, the proviral DNA sequence was examined at crossover points, and it was found that mutations do not occur at the high frequencies observed in cell-free systems, indicating that additional factors and/or conditions, which have not yet been reproduced in cell-free systems, affect fidelity during HIV-1 recombination. Finally, there appear to be local hot spots of recombination.
MATERIALS AND METHODS
Plasmids.
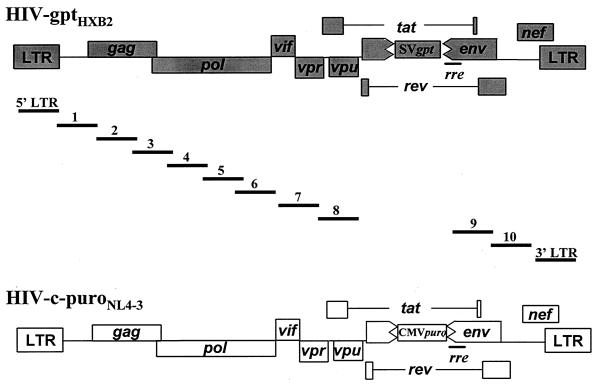
HIV-gptHXB2 (Fig. 1) and pNL4-3 were obtained from the AIDS Research and Reference Reagent Program (1, 34). To construct pHIV-c-puroNL4-3 (Fig. 1), a 1.2-kbp deletion was made in the envelope gene of pNL4-3 from the NdeI site at nucleotide 6400 to the BglII site at nucleotide 7611. The deletion was replaced with a 1.4-kbp NruI-to-XbaI fragment from pcPuro, which contains the cytomegalovirus immediate-early promoter and puromycin resistance gene coding sequence.
FIG. 1.
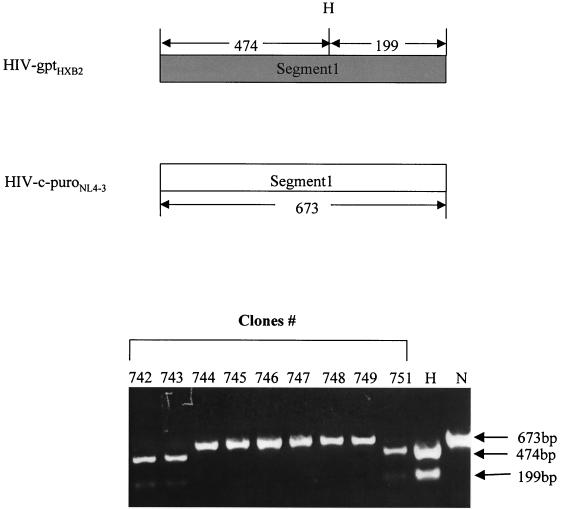
HIV-1 vectors and segments of provirus DNA amplified to study HIV-1 recombination rate. HIV-c-puroNL4-3 is based upon HIV-1 strain NL4-3. HIV-gptHXB2 is based upon HIV-1 strain HXB2. CMVpuro represents the puro gene expressed from the cytomegalovirus immediate-early promoter. SVgpt represents the gpt gene under control of the simian virus 40 early gene promoter. The 12 different overlapping segments amplified are depicted by the bars with numbers or LTR designations above them. The coordinates of the amplified segments are as follows: 5′LTR, 53 to 799; 1, 611 to 1283; 2, 1261 to 2049; 3, 2028 to 2826; 4, 2807 to 3663; 5, 3547 to 4398; 6, 4271 to 5066; 7, 4932 to 5797; 8, 5778 to 6390; 9, 7643 to 8454; 10, 8336 to 9156; 3′LTR, 8966 to 9716 (sequence coordinates are according to the HXB2 provirus sequence).
pcPuroΔAS and pcGPTΔAE were used to make internal control (IC) RNA for RT-PCR (see Fig. 6). pcPuroΔAS was constructed by deleting a 105-bp AccI-to-SacII fragment from pcPuro, followed by cloning the BamHI and NotI fragment, with the deletion, into pcDNA3.1(+) (Invitrogen). pcGPTΔAE was constructed from pSV2gpt by deleting a 107-bp AgeI-to-EcoRV fragment, followed by cloning the BglII and NotI fragment, with the deletion, into pcDNA3.1(+). All maps are available upon request.
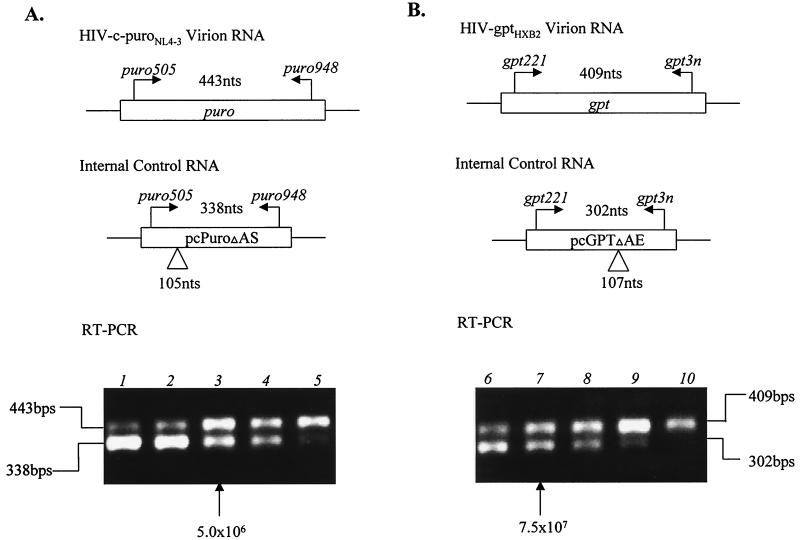
FIG. 6.
RT-PCR to determine relative levels of HIV-c-puroNL4-3 and HIV-gptHXB2 RNAs. (A) Quantitation of HIV-c-puroNL4-3 virion RNA. The primers used to amplify the puro-specific sequence are indicated in Materials and Methods. The primers are also specific for the puro control RNA, which has an internal deletion of 105 bases to distinguish it from the signal obtained from the viral RNA. Viral RNA (625 ng) was mixed with different dilutions of the puro control RNA ranging from 107 to 106 molecules (1.0 × 107, 7.5 × 106, 5 × 106, 2.5 × 106, and 1.0 × 106 in lanes 1 to 5, respectively). As can be seen, the signal from the viral RNA is about the same as the signal obtained with 5 × 106 molecules of control RNA. (B) RT-PCR for quantitation of HIV-gptHXB2 RNA. The gpt-specific segment to be amplified and the control RNA with an internal deletion are indicated. Again, 625 ng of virion RNA was added to each sample with the amount of control RNA ranging from 108 to 107 molecules (1.0 × 108, 7.5 × 107, 5 × 107, 2.5 × 107, and 1.0 × 106 in lanes 6 to 10, respectively). As is evident, equivalent signals are obtained when 7.5 × 107 molecules of control RNA are added.
Cell culture.
293T cells were grown in Dulbecco's modified Eagle's minimum essential medium (DMEM) supplemented with 10% fetal bovine serum.HeLaT4 cells were grown in DMEM supplemented with 10% fetal bovine serum and hygromycin (0.1 mg/ml). The tetracycline-inducible cell line 69TIRevEnv was grown in DMEM supplemented with 10% fetal bovine serum, G418 (0.2 mg/ml), hygromycin (0.1 mg/ml), and tetracycline (2 μg/ml) (58).
To create inducible producer cell clones (58), HIV-gptHXB2 and HIV-c-puroNL4-3 were separately cotransfected with an amphotropic murine leukemia virus env expression vector, pEnvAm (27), into 293T cells. Pseudotyped HIV-gptHXB2 vector virus produced from 293T cells was then used to inoculate 69TIRevEnv cells, which express HIV-1 Env under tetracycline regulation (58); this was followed by selection with GPT (xanthine [250 μg/ml], hypoxanthine [15 μg/ml], mycophenolic acid [7 μg/ml]), which was in turn followed by isolation of cell clones. The cell clones were subsequently inoculated with pseudotyped HIV-c-puroNL4-3 vector virus produced from 293T cells; this was followed by selection with puromycin (1 μg/ml), again followed by isolation of cell clones.
Transfection and infection.
Transfections were performed by using a modified calcium phosphate precipitation method (15). To perform infections, 2 × 105 HeLaT4 target cells were treated with Polybrene (50 μg/ml) for 30 min. Treated cells were subsequently inoculated with 10-fold serial dilutions of filtered (pore size, 0.45 μm) virus stocks in 0.3 ml of medium and incubated for 2 h, with occasional rocking. Alternatively, infection was carried out by premixing virus dilutions with Polybrene (8 μg/ml). One milliliter of the virus-Polybrene mixture was used to inoculate 2.5 × 105 target cells, which were incubated for approximately 6 h at 37°C in 5% CO2. Twenty-four hours after infection, the cells were selected for resistance to either GPT or puromycin (1 μg/ml).
PCR and RT-PCR.
Genomic DNA was isolated from infected cell clones using the DNeasy tissue kit (QIAGEN Inc.). Proviral DNA was amplified from this genomic DNA by either conventional or touchdown PCR using Taq DNA polymerase (Promega). The sequences of the 12 pairs of primers used to amplify different segments of proviral DNA are listed in Table 1. Viral RNA was isolated from the cell supernatant using the NucleoSpin virus kit (Clontech) following the manufacturer's protocol. IC RNA was transcribed in vitro by T7 RNA polymerase (Life Technologies) from pcPuroΔAS and pcGPTΔAE, respectively. The concentrations of both the viral and IC RNAs were determined by spectrophotometry. The cDNA was amplified using the SuperScript first-strand synthesis system kit for RT-PCR (Invitrogen Life Technology) following the manufacturer's protocol. More specifically, 625 ng of viral RNA was mixed with different dilutions of IC RNA as indicated (see Fig. 6) ranging from 106 to 108 molecules. Two picomoles of the antisense gene specific primers (puro948 and gpt3n for puro and gpt, respectively) was added along with deoxynucleoside triphosphates at a concentration of 1 mM in a total volume of 25 μl. The samples were incubated at 65°C for 5 min and immediately transferred to 50°C. As prescribed by the manufacturer, 25 μl of an RT reaction mixture, prewarmed at 50°C, was then added, and this was followed by the addition of 1 μl of SuperScript II RT (50 U) and incubation at 50°C for 50 min. The RT reaction was terminated by heating at 70°C for 15 min. One microliter of RNase H (2 U) was then added to the reaction mixture, and the mixture was incubated at 37°C for 20 min. Two microliters of the first-strand reaction was used as a template to perform touchdown PCR using Platinum Taq DNA polymerase (Life Technologies). Gene-specific primer pair puro505 and puro948 and primer pair gpt 221 and gpt3n were used in the touchdown PCR to amplify puro and gpt segments, respectively. The sequences of these primers were, respectively, 5′-CCGCGTTCGCCGACTACC-3′, 5′-GCTCGTAGAAGGGAGGTTG3′, 5′-GACATGTTGCAGATCCATGC-3′, and 5′-ACGAATACGACGCCCATATC-3′. The touchdown PCR protocol entailed incubating the DNA mixture at 95°C for 5 min, followed by 20 cycles at 95°C for 30 s, 63°C for 30 s with a decrease of 0.5°C per cycle, and 72°C for 40 s. Then, 20 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 40 s were carried out, ending with a 10-min extension at 72°C.
TABLE 1.
Primers used in amplification of proviruses
| Segment | Primer sequence | Positiona | Orientation |
|---|---|---|---|
| 5′LTR | 5′-CTACCACACACAAGGCTACT-3′ | 53-72 | 5′ |
| 5′LTR | 5′-CTCGCACCCATCTCTCTCCTT-3′ | 779-799 | 3′ |
| 1 | 5′-AGTCAGTGTGGAAAATCTCT-3′ | 611-632 | 5′ |
| 1 | 5′-GCTGAAAGCCTTCTCTTCTACTA-3′ | 1261-1283 | 3′ |
| 2 | 5′-TAGTAGAAGAGAAGGCTTTCAGC-3′ | 1261-1283 | 5′ |
| 2 | 5′-GTCCTTCCTTTCCACATTTCC-3′ | 2028-2049 | 3′ |
| 3 | 5′-TGGAAATGTGGAAAGGAAGGAC-3′ | 2028-2049 | 5′ |
| 3 | 5′-CCTAATTGAACTTCCCAGAA-3′ | 2807-2826 | 3′ |
| 4 | 5′-TTCTGGGAAGTTCAATTAGG-3′ | 2807-2826 | 5′ |
| 4 | 5′-ACTGCCTCTGTTAATTGTTT-3′ | 3644-3663 | 3′ |
| 5 | 5′-GCAAGGCCAATGGACATATCAA-3′ | 3547-3568 | 5′ |
| 5 | 5′-CTACAGTCTACTTGTCCATGCA-3′ | 4377-4398 | 3′ |
| 6 | 5′-GATGGAATAGATAAGGCCCAAGA-3′ | 4271-4290 | 5′ |
| 6 | 5′-CATCCTGTCTACTTGCCACACA-3′ | 5047-5066 | 3′ |
| 7 | 5′-GGAAAGGACCAGCAAAGCT-3′ | 4932-4940 | 5′ |
| 7 | 5′-CTGCTATGTCGACACCCAAT-3′ | 5778-5797 | 3′ |
| 8 | 5′-ATTGGGTGTCGACATAGCAG-3′ | 5778-5797 | 5′ |
| 8 | 5′-GATGCACAAAATAGAGTGGTGGT-3′ | 6368-6390 | 3′ |
| 9 | 5′-GATATGAGGGACAATTGGAGAA-3′ | 7643-7664 | 5′ |
| 9 | 5′-CTGTCTCTGTCTCTCTCTCCA-3′ | 8434-8454 | 3′ |
| 10 | 5′-GTGAATAGAGTTAGGCAGGGAT-3′ | 8336-8357 | 5′ |
| 10 | 5′-AGTAGCCTTGTGTGTGGTAG-3′ | 9137-9156 | 3′ |
| 3′LTR | 5′-GCTAGAAGCACAAGAGGAGGA-3′ | 8966-8986 | 5′ |
| 3′LTR | 5′-CTAGAGATTTTCCACACTGACT-3′ | 9695-9716 | 3′ |
Primer position according to the HXB2 provirus sequence numbering.
HTA.
HTA is described elsewhere (18, 59). Briefly, a 32P-labeled, single-stranded probe was prepared from HIV-c-puroNL4-3 plasmid DNA by asymmetrical PCR utilizing the primer pairs listed in Table 1. The ratio of primer concentrations used to amplify the single-stranded probe was either 1:100 (final concentration ratio, 2 nM:200 nM) or 1:200 (final concentration ratio, 1 nM:200 nM). The reaction was carried out in a mixture of deoxynucleoside triphosphates (each at 0.1 mM), 2 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.02% Nonidet P-40, 2 U of Taq polymerase (Promega), and 10 μCi of [32P]dCTP. A 2-μl aliquot of 32P-labeled single-stranded probe was mixed with 8 to 10 μl of symmetric PCR product amplified with the same primers from the genomic DNA of target cell clones and 1.5 μl of 10× annealing buffer (1 M NaCl, 100 mM Tris-HCl [pH 7.8], 20 mM EDTA). The DNA mixtures were then denatured at 98°C for 2 min and annealed by decreasing the temperature 1°C every 10 s, and this was followed by incubation at 22°C for 10 min and storage on ice. Homoduplex and heteroduplex DNAs were differentiated by electrophoresis in a 6% nondenaturing polyacrylamide gel, dried onto 3MM Whatman paper, and visualized by autoradiography.
DNA sequencing.
QIAGEN-purified PCR products were sequenced directly using the corresponding primer pairs (Table 1). Alternatively, purified PCR products were cloned by using the pGEM-T Easy Vector system (Promega) and sequenced using T7 and SP6 primers.
Calculation of the composition of virion RNA.
Because recombination cannot be scored from homozygous virions, the number of proviruses assayed should be corrected to exclude the proviruses from homozygous virions. According to the Hardy-Weinberg equation (47), G2 + 2GP + P2 = 1, where G = titer of GPT-resistant virus/(titer of GPT-resistant virus + titer of puromycin-resistant virus) and P = titer of puro-resistant virus/(titer of GPT-resistant virus + titer of puromycin-resistant virus), G2 represents the fraction of homozygous HIV-gptHXB2 virions, 2GP represents the fraction of heterozygous virions, and P2 represents the fraction of homozygous HIV-c-puroNL4-3 virions. In this experimental system, the titer of puromycin-resistant virus (1.5 × 102 CFU/ml [average of two experiments]) was 20-fold lower than the titer of GPT-resistant virus (3.0 × 103 CFU/ml [average of two experiments]), while virion RNA levels of HIV-c-puroNL4-3 were approximately 15-fold less than that of HIV-gptHXB2 (see Results). That is, G is 20 (15 for RNA level) and P is 1 (1 for RNA level), so the percentage of heterozygous virions among all virions conferring puromycin resistance is represented by the following equation: GP/(P2 + GP) = 95%. Therefore, of the 16 puro progeny clones examined, it is anticipated that proviruses from 15 clones should have resulted from heterozygous virions, assuming random copackaging.
RESULTS
Generation of heterozygous virions.
Two HIV-1 env-defective vectors were used, in combination with an HIV-1 env producer cell line (58), to study recombination in a single cycle of viral replication. These two vectors, HIV-gptHXB2 and HIV-c-puroNL4-3, are based upon HIV-1 strains HXB2 and NL4-3, respectively (Fig. 1) (1, 34). Strains HXB2 and NL4-3 are approximately 97% homologous, providing sufficient heterogeneity for the study of recombination. Note that strain NL4-3 was substituted for BCSG3 to examine if the rate remains high even with a different strain and to utilize a strain that a larger number of laboratories employ on a more regular basis.
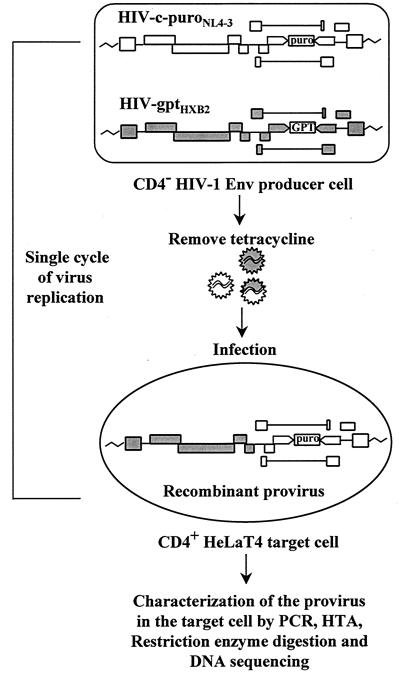
The protocol used for studying recombination is shown in Fig. 2. A producer cell clone containing a single copy of each HIV-gptHXB2 and HIV-c-puroNL4-3 was established (see Materials and Methods). These producer cells contain HIV-1 env under the control of an inducible tetracycline promoter (58). To induce virus production, tetracycline was withdrawn and the supernatant containing infectious vector virus was used to infect CD4-positive HeLaT4 target cells, which was followed by selection for puromycin resistance. In this system, viral replication is restricted to a single cycle because the producer cells are CD4 negative, so that they cannot be reinfected, and the target cells lack HIV-1 Env, such that new virus cannot be produced. Since the titer of puromycin-resistant virus was approximately 20-fold lower than the titer of GPT-resistant virus, puromycin-resistant target cell clones were most likely infected by heterozygous virions and were therefore analyzed for recombination (see above, “Calculation of the composition of virion RNA”). To rule out the possibility of double infection, these cell clones were tested for their sensitivity to GPT. Only those clones that were GPT sensitive and puromycin resistant were utilized for further analysis.
FIG. 2.
Protocol for study of HIV-1 recombination. Individual cell clones containing a single copy of both HIV-gptHXB2 and HIV-c-puroNL4-3 were established. Upon induction by removal of tetracycline, vector virus was harvested and used to inoculate CD4-positive HeLaT4 target cells, which was followed by selection and isolation of target cell clones. Going from a provirus in a producer cell to a provirus in a target cell comprised a single cycle of virus replication. Proviruses in each target cell clone were further analyzed by PCR, the HTA, restriction enzyme digestion, and DNA sequencing in order to identify recombinants and crossover points.
HIV-1 recombination occurs at a high rate.
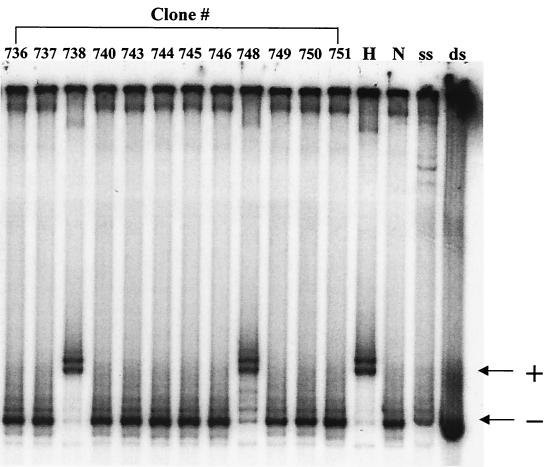
Genomic DNA was extracted from 16 selected target clones, and 12 different segments across the proviral genomes were amplified by PCR using primer pairs common to both HXB2 and NL4-3 strains (Fig. 1; Table 1). These proviral segments were first analyzed for recombination using the HTA (18, 59). HTA can differentiate sequence heterogeneity based upon the electrophoretic mobility differences between homoduplex and heteroduplex DNAs. A typical HTA gel is shown in Fig. 3 for segment 8 (Fig. 1) in which there is approximately a 1% sequence difference between the strains HXB2 and NL4-3. In this assay, a 32P-labeled single-stranded DNA probe, corresponding to NL4-3 nucleotides 5778 to 6390, was prepared from HIV-c-puroNL4-3 plasmid DNA by asymmetric PCR (Fig. 3, lane ss). The same primer pair was also utilized in a conventional PCR to amplify proviral DNA from different progeny cell clones and from HIV-gptHXB2 and HIV-c-puroNL4-3 plasmids as controls. After the probe was annealed to the amplified DNA, homoduplex and heteroduplex DNAs were distinguished via electrophoresis on a nondenaturing polyacrylamide gel. A heteroduplex was formed when the NL4-3-templated probe was annealed to a PCR product derived from HIV-gptHXB2 plasmid DNA (Fig. 3, lane H), whereas a homoduplex was formed when the probe was annealed to a PCR product derived from HIV-c-puroNL4-3 (Fig. 3, lane N). DNA fragments from progeny cell clones 738 and 748 formed heteroduplexes when annealed to the HIV-c-puroNL4-3 probe, indicating they are recombinant. On the other hand, DNA fragments from clones 736, 737, 740, 743 to 746, and 749 to 751 formed homoduplexes when annealed to the probe, suggesting that no sequence change or a mismatch of less than 1% had occurred.
FIG. 3.
Representative example of an HTA. Radioactively labeled single-stranded probe was prepared from HIV-c-puroNL4-3 plasmid DNA by asymmetric PCR using the segment 8 primer pair (lane ss). Radioactively labeled double-stranded segment was amplified at the same time as a control (lane ds). Segment 8 is 612 bp in length. The sequence difference between the two strains within this segment is approximately 1%. The probe was annealed with PCR products amplified from NL-c-puroNL4-3 (lane N, homoduplex control), HIV-gptHXB2 (lane H, heteroduplex control), or genomic DNA from different target cell clones (clones 736 to 738, 740, 743 to 746, and 748 to 751). Clones 738 and 748 are shifted compared to the band for HIV-c-puroNL4-3 DNA and were therefore scored as recombinants. Symbols: +, band shifted due to heteroduplex formation; −, nonshift homoduplex band.
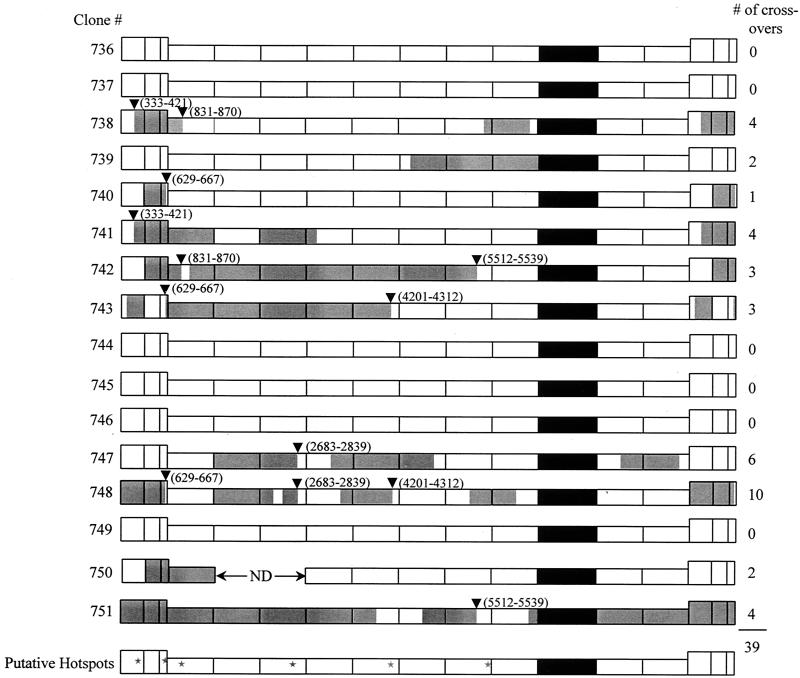
Although the sequence heterogeneity between the HXB2 and NL4-3 strains of HIV-1 is approximately 3% throughout the viral genome, the differences within each separate segment vary between 0.8 and 4.8%. The size of the fragments ranged from 612 to 868 bp. The successful detection of recombinants by HTA was dependent on both the sequence divergence and the size of the segments. Under our conditions, crossover events within some segments could not be detected. Because of this limitation, enzyme digestion and DNA sequencing were also used to screen for recombination events. Figure 4 shows a representative restriction enzyme digestion for segment 1 (Fig. 1). DNA fragments corresponding to HIV-1 strains HXB2 and NL4-3 nucleotides 611 to 1283 were amplified from the proviral DNA. Samples derived from HIV-gptHXB2 and HIV-c-puroNL4-3 plasmid DNA were also prepared as controls. HindIII was used to digest the purified fragments. Bands of 474 and 199 bp indicate that the fragment originates from HIV-gptHXB2, while a 673-bp band indicates that the fragment originates from HIV-c-puroNL4-3. PCR-amplified DNA from clones 742, 743, and 751 produced the 474- and 199-bp bands, indicating that they are of HIV-gptHXB2 origin. PCR products from clones 744 to749 inclusive produced a 673-bp band, indicating that they are of HIV-c-puroNL4-3 origin. In addition, out of a total of 192 segments (12 segments × 16 clones), 121 segments were sequenced. Combining HTA, restriction enzyme digestion, and sequencing results, it was determined that 10 out of 16 proviral clones had undergone a total of 39 recombination events (Fig. 5). The number of crossovers per clone ranged from 0 to 10. The overall recombination rate was 2.4 crossovers per vector genome per replication cycle (39 crossovers/16 clones per cycle). Given that the target size examined in the vector sequence was 7,964 bases (using RNA coordinates) and the size of the full-length genome is 9,181 bases, extrapolating to the full-length genome, the rate is 2.8 crossovers per genome per cycle (9,181/7,964 × 2.4 = 2.8).
FIG. 4.
Representative example of a restriction enzyme digestion to identify crossover points. At the top of the figure, the diagram depicts the anticipated sizes of amplified segment 1 from HIV-gptHXB2 and HIV-c-puroNL4-3, respectively, as well as the predicted HindIII digestion patterns. Purified proviral and plasmid DNA segments were digested with HindIII. Bands of 474 and 199 indicate this segment of the clone is from HIV-gptHXB2 (lane H) while a 673-bp band indicates it is from HIV-c-puroNL4-3 (lane N). The bottom of the figure illustrates the digestion pattern of amplified segment 1 from nine progeny target cell clones, 742 to 749 and 751. H indicates control DNA amplified from HIV-gptHXB2 plasmid, and N indicates control DNA amplified from HIV-c-puroNL4-3 plasmid. Clones 742, 743, and 751 display 474- and 199-bp bands, indicating they are of HIV-gptHXB2 origin. Clones 745 to 749 display a 673-bp band, indicating they are of HIV-c-puroNL4-3 origin.
FIG. 5.
Recombination distribution throughout the full-length proviral genomic DNA. Ten out of 16 clones had undergone a total of 39 recombination events. Thirty-three out of the 39 recombinant sites were sequenced. Six putative hot spots are indicated by inverted triangles and summarized with stars. The coordinates of the putative hot spots are as follows: 333 to 421, 629 to 667, 831 to 870, 2683 to 2839, 4201 to 4312, and 5512 to 5539 (sequence coordinates are according to the HXB2 provirus sequence). White is of HIV-c-puroNL4-3 origin and grey is of HIV-gptHXB2 origin. The marker gene cassette is depicted in black.
HIV-gptHXB2 and HIV-c-puroNL4-3 RNA levels in virion preparations.
The results just described suggest that there were two populations of virions, one that averaged around four crossovers per replication cycle (10 of 16 proviruses) and one that had not undergone any recombination (6 of 16 proviruses) (Fig. 5). These results are reminiscent of high negative interference, which was previously described for the avian retrovirus SNV (2, 16). This assumes that the puromycin-resistant proviruses were progeny of heterozygous virions (see Materials and Methods), which is presumed because the titer of GPT-resistant virus is 20-fold higher than the titer of puromycin-resistant virus, indicating that the HIV-gptHXB2 virion RNA is in approximately 20-fold excess compared to the HIV-c-puroNL4-3 virion RNA. However, it is possible that the penetrances of the GPT- and puromycin-resistant phenotypes are not linearly proportional to the respective virion RNA levels. To address this issue, RT-PCR was performed to measure directly the relative levels of HIV-gptHXB2 and HIV-c-puroNL4-3 RNA in the virus preparations. Virion RNA was isolated from producer cell supernatant using the NucleoSpin virus kit according to the manufacturer's protocol. To differentiate between the two viral RNAs, primers were prepared specific for gpt and puro, respectively (Fig. 6) As ICs, puro- and gpt-specific RNAs with internal deletions permitting them to be distinguished from the virion RNAs were prepared in vitro with T7 RNA polymerase and were added to each sample in various quantities (Fig. 6). The level of each specific virion RNA could then be quantified when the signal was equivalent to that from a known amount of IC RNA. As can be ascertained from Fig. 6, 625 ng of virion RNA contained approximately 7.5 × 107 molecules of HIV-gptHXB2 RNA and 5 × 106 molecules of HIV-c-puroNL4-3 RNA. Thus, the amount of HIV-gptHXB2 RNA is approximately 15-fold greater than that of HIV-c-puroNL4-3 RNA, which correlates well with the differences in titer. However, these results do not rule out the possibilities that copackaging of virions was not random and that the six proviruses not scoring as recombinants are progeny of homozygous virions. This point is addressed in more detail in the Discussion.
Compilation of crossover data and identification of putative hot spots.
The HTA, restriction digestion, and sequencing results were compiled for each proviral clone. The distribution of recombination events across the HIV-1 genome is illustrated in Fig. 5. Note that of the 39 crossovers identified, the DNA sequences of 33 have been determined. Of these, 13 share crossover points with other proviral progeny (Fig. 5). For example, at coordinates 629 to 667, three proviruses share the same transfer point (Fig. 5). At coordinates 333 to 421, 831 to 870, 2683 to 2839, 4201 to 4312, and 5521 to 5539, two proviruses have crossed over at the same small segment (Fig. 5). This suggests that there might be local hot spots spread throughout the genome.
HIV-1 recombination is not highly mutagenic.
The crossover junctions of the recombinant proviral clones were screened for mutations using DNA sequencing. A crossover junction is defined as the sequence between two sites of strain heterogeneity, one of NL4-3 origin and the other of HXB2 origin, within a proviral segment. The length of the junctions examined ranged from 4 to 173 nucleotides. Sequence information obtained for several proviral clones described in previous reports was also included in this analysis (18, 57). These aforementioned clones had been the result of recombination between HIV-1 strains HXB2 and BCSG3 (14, 34). A total of 4,010 nucleotides spanning 69 crossover events within 36 different progeny clones were analyzed for mutations. Recombination junctions in several different regions of the genome were examined. Two point mutations were found in the recombination junctions analyzed (Table 2). This indicates that HIV-1 recombination is not very mutagenic.
TABLE 2.
Analysis of recombination junctions
| Strains analyzed | Genome location | No. of progeny cell clones analyzed | No. of cross-over events | No. of bases analyzed |
|---|---|---|---|---|
| HXB2 + NL4-3 | Varied | 9 | 33 | 2,160b |
| HXB2 + BCSG3 | LTR | 11 | 12 | 238c |
| HXB2 + BCSG3 | 5′ end pol | 18a | 24 | 1,612 |
| Total | 36a | 69 | 4,010 |
Two clones analyzed in the LTR were also analyzed at the 5′ end of pol.
One point mutation observed (C→A).
One point mutation observed (G→A).
DISCUSSION
Previously, we reported that HIV-1 undergoes a high rate of recombination (18). The vectors used in this earlier report contained nonretroviral sequence from the early region of simian virus 40, which introduced an 89-base palindrome that acted as a recombination hot spot, presumably by forming a stable hairpin structure and promoting significant additional pausing. The specific vector crossover rate was 3.0, which was corrected by excluding the crossovers at the exogenous hot spot to 2.1. It should be noted that in the previous study the HIV-1 target sequence was 7,300 bases, so extrapolating to the full-length genome of 9,181 bases, the recombination rates were 3.8 (including the exogenous hot spot) and 2.6 (factoring out the hot spot). There still remained the possibility that even the corrected crossover rate of 2.6 might be artificially high since it had been previously suggested that once recombination occurs it predisposes the virus to further recombination (see below). The present study was undertaken to rule out the possibility that the earlier recombination rate was artificially high because of the exogenous hot spot. The results from the experiments described here demonstrate that in the absence of the nonretroviral recombination hot spot, the rate of HIV-1 recombination between autologous, homologous sequences remained very high. The minimum rate extrapolated to the entire genome was 2.8 crossovers per cycle of replication. Thus, the corrected rate (2.6) obtained in the previous study did not seem to have been biased by the exogenous hot spot. The rate obtained here confirms that given the small size of the HIV-1 viral genome, HIV-1 replication represents the most actively recombinogenic system related to mammals described to date.
In the proviruses analyzed, there appeared to be two populations, one that underwent recombination and one that did not. This was also observed previously for the simple retrovirus, SNV, and the hypothesis was posed that this phenomenon is an example of high negative interference, which maintains that the recombinant population is either predisposed to recombination or becomes more prone to recombination after the first initiating crossover (2, 16). An alternate explanation is that the HIV-1 nonrecombinants were progeny of homodimeric virions. Evidence in support of high negative interference in the case described here is twofold. First, the titer of GPT-resistant virus was 20-fold higher than the titer of puromycin-resistant virus, so it was anticipated that the majority of the puromycin-resistant proviruses should be progeny of heterodimeric virions, assuming random copackaging and equivalent phenotypic penetrance of the GPT- and puromycin-resistant phenotypes. RT-PCR was employed to examine the relative amounts of each packaged viral RNA. The results support the contention that the viral titers accurately reflected the overall viral RNA levels since it was found that the amount of HIV-gptHXB2 RNA was approximately 15-fold greater than the amount of HIV-c-puroNL4-3 RNA, correlating well with the 20-fold differences in titers and suggesting that there is equivalent penetrance of the GPT- and puromycin-resistant phenotypes (Fig. 6). Second, one of the clones, 748 (Fig. 5), underwent 10 crossovers, which, given that the average crossover rate in the identified recombinant population is 4, seems to be quite high and be consistent with the concept that recombination predisposes the reverse transcription complex to further recombination. However, at this juncture, we cannot formally rule out the possibility that RNA copackaging was not random. First, we could not directly measure it. Second, there are sequence differences between the two strains, and these differences might affect copackaging. However, sequences known to be important for packaging and dimer formation such as the six-base palindrome (GCGCGC) in the SL1 loop (8, 9), as well as the rest of the SL1 loop and the SL3 and SL4 stem-loops, are identical between the strains through database searches (GenBank accession number K03455 and M19921 for HXB2 and NL4-3, respectively) and our sequencing results. Thus, further investigation about dimerization and copackaging is needed to clarify this issue. Lastly, it was surprising that of the six putative nonrecombinants, none had undergone an intermolecular strand transfer during minus-strand strong-stop DNA transfer. This can be ascertained by examining the viral long terminal repeats (LTRs). If U3, R, and U5 are of the same viral origin, the primer strand transfers were intramolecular, whereas if U3 differs in strain origin from R and U5, the transfers were intermolecular. Given our previous results (57) and those of others (53) in which it was found that intermolecular strand transfers occurred 50% of the time during HIV-1 strong-stop DNA transfer, we would have anticipated to see approximately three intermolecular strand transfers, yet none were observed (Fig. 5). Although it should be noted that for the simpler retrovirus SNV, most nonrecombinants only underwent intramolecular minus-strand primer transfers. Nevertheless, in light of the possibility that the putative nonrecombinant population might be progeny of homozygous virions, it can be concluded that the 2.8 crossovers per genome per replication cycle is a minimum rate of recombination.
Thirteen of the 33 sequenced crossover segments shared the same crossover region with at least one other proviral clone. In the case of the segment 629 to 667, three clones had undergone recombination within this portion of the genome (Fig. 5). The segment is 38 bp, so assuming random strand transfer, the expected frequency of a single crossover occurring, given that the target sequence analyzed was 7,964 bp, is 18.6% (39 crossovers ×38 bp/7,964 bp), and the expected frequency of three crossovers is 0.64% ([39 × 38 bp/7,964 bp]3). Therefore, at this locus alone, it appears likely that there is a hot spot for recombination. Another way to look at this is by comparing the crossover frequency over the six segments, where more than one crossover occurred, to the frequency over all of the target sequences. More than one recombination transpired over a total of 459 bp, whereas the entire target sequence was 7,964 bp. Thirteen crossovers occurred within the segments comprising the 553 bp regions, yielding a frequency of 28.3 × 10−3 crossovers/bp (13 crossovers/459 bp), which is 5.8-fold greater than the average frequency of 4.9 × 10−3 crossovers/bp (39 crossovers/7,964 bp) over all of the target sequence. This is suggestive of relative hot spots within the six segments comprising the 459bp. The existence of recombinational hot spots is not surprising given that they have been described in cell-free systems (31), and it has been previously reported that a hot spot exists in the dimer initiation sequence of the HIV-1 5′-untranslated region (3). Moreover, in our own case, an exogenous sequence proved to be a very effective hot spot, presumably owing to its palindromic nature and concomitant secondary structure (3, 18). The influence of primary sequence and RNA secondary structure can be studied as more local hot spots are catalogued.
Identification and characterization of recombination junctions also enabled us to address the prediction that HIV-1 recombination is mutagenic. It was previously reported using cell-free systems that HIV-1 incorporates additional nucleotides about 50% of the time beyond the 5′ end of both RNA and DNA template ends (37, 39). It has also been shown using another cell-free system that mutations occur frequently at strand transfer junctions (40). A separate study demonstrated that mutations occurred 30% of the time at one frequently utilized strand transfer point in vitro (55). In this study, 69 crossover junctions were examined by direct DNA sequencing, which encompassed a total of 4,010 bp. Two point mutations were observed, one C-to-A substitution in segment 10 (Fig. 1; Table 2) and one G-to-A substitution in the U3 region of the LTR (Table 2). From these data it is not possible to determine whether the mutations occurred as a result of recombination or due to unrelated mutations by RT. HIV-1 mutation rates have been measured and found to be approximately 3 × 10−5 to 8 × 10−5/bp/replication cycle (26, 33). Two mutations were found for 4,010 bp sequenced, which represents a rate of 5.0 × 10−4/bp/replication cycle. Although this suggests that the mutation rate might be approximately sixfold higher during recombination, with such a small sample of mutations one cannot be certain. However, even if one assumes that the mutation rate is sixfold higher during recombination, it is certainly much lower than that predicted in previous cell-free studies (5, 19, 41-44, 52), since in this study only two mutations occurred at 69 crossover junctions.
In summary, the results presented here support the idea that HIV-1 recombines at an extremely high rate of at least 2.8 crossovers during each cycle of replication, making this the most recombinogenic process observed in any mammalian related system described so far. Moreover, recombination hot spots might exist in a number of segments throughout the HIV-1 genome. Furthermore, HIV-1 RT seems to switch template in a relatively precise manner, suggesting that factors or conditions that contribute to fidelity during recombination are missing in at least some cell-free systems. These findings provide further evidence that the reason retroviruses are diploid is to provide a recombination partner (48), and that recombination is both an integral aspect of the HIV-1 replication process and a possible target for chemotherapeutic intervention.
Acknowledgments
J.Z. and A.E.J. contributed equally to this study.
We thank Martin Adelson, Chiann-Chyi Chen, Sayandip Mukherjee, Annmarie Pacchia, and Amariliz Rivera for discussions and helpful comments on the manuscript.
This work was supported by grants CA50777, AI43886, and AI34834 from the National Institutes of Health.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. A., R. J. Teufel II, P. D. Yin, and W. S. Hu. 1998. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J. Virol. 72:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakrishnan, M., P. J. Fay, and R. A. Bambara. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482-36492. [DOI] [PubMed] [Google Scholar]
- 4.Battula, N., and L. A. Loeb. 1976. On the fidelity of DNA replication. Lack of exodeoxyribonuclease activity and error-correcting function in avian myeloblastosis virus DNA polymerase. J. Biol. Chem. 251:982-986. [PubMed] [Google Scholar]
- 5.Bebenek, K., J. Abbotts, J. D. Roberts, S. H. Wilson, and T. A. Kunkel. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948-16956. [PubMed] [Google Scholar]
- 6.Carr, J. K., B. T. Foley, T. Leitner, M. Salminen, B. Korber, and F. McCutchan. 1998. Reference sequences representing the principal genetic diversity of HIV-1 in the pandemic, p. III-10-III-19. In B. Korber, C. L. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. W. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 7.Clavel, F., M. D. Hoggan, R. L. Willey, K. Strebel, M. A. Martin, and R. Repaske. 1989. Genetic recombination of human immunodeficiency virus. J. Virol. 63:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clever, J. L., M. L. Wong, and T. G. Parslow. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin, J. M. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42:1-26. [DOI] [PubMed] [Google Scholar]
- 11.Diaz, L., and J. J. DeStefano. 1996. Strand transfer is enhanced by mismatched nucleotides at the 3′ primer terminus: a possible link between HIV reverse transcriptase fidelity and recombination. Nucleic Acids Res. 24:3086-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dougherty, J. P., and H. M. Temin. 1988. Determination of the rate of base pair substitution and insertion mutations in retrovirus replication. J. Virol. 62:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, F., D. L. Robertson, S. G. Morrison, H. Hui, S. Craig, J. Decker, Fultz, P. N., M. Girard, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, S. K., P. N. Fultz, E. Keddie, M. S. Saag, P. M. Sharp, B. H. Hahn, and G. M. Shaw. 1993. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology 194:858-864. [DOI] [PubMed] [Google Scholar]
- 15.Gorman, C. 1985. High efficiency gene transfer into mammalian cells, p. 143-190. In D. M. Glover (ed.), DNA cloning. IRL Press, Oxford, United Kingdom.
- 16.Hu, W. S., E. H. Bowman, K. A. Delviks, and V. K. Pathak. 1997. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J. Virol. 71:6028-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 18.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji, J. P., and L. A. Loeb. 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31:954-958. [DOI] [PubMed] [Google Scholar]
- 20.Kawai, S., and H. Hanafusa. 1972. Genetic recombination with avian tumor virus. Virology 49:37-44. [DOI] [PubMed] [Google Scholar]
- 21.Koulinska, I. N., T. Ndung'u, D. Mwakagile, G. Msamanga, C. Kagoma, W. Fawzi, M. Essex, and B. Renjifo. 2001. A new human immunodeficiency virus type 1 circulating recombinant form from Tanzania. AIDS Res. Hum. Retrovir. 17:423-431. [DOI] [PubMed] [Google Scholar]
- 22.Kulpa, D., R. Topping, and A. Telesnitsky. 1997. Determination of the site of first strand transfer during Moloney murine leukemia virus reverse transcription and identification of strand transfer-associated reverse transcriptase errors. EMBO J. 16:856-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liitsola, K., M. Ristola, P. Holmstrom, M. Salminen, H. Brummer-Korvenkontio, S. Simola, J. Suni, and P. Leinikki. 2000. An outbreak of the circulating recombinant form AECM240 HIV-1 in the Finnish injection drug user population. AIDS 14:2613-2615. [DOI] [PubMed] [Google Scholar]
- 24.Liitsola, K., I. Tashkinova, T. Laukkanen, G. Korovina, T. Smolskaja, O. Momot, N. Mashkilleyson, S. Chaplinskas, H. Brummer-Korvenkontio, J. Vanhatalo, P. Leinikki, and M. O. Salminen. 1998. HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad. AIDS 12:1907-1919. [DOI] [PubMed] [Google Scholar]
- 25.Malim, M. H., and M. Emerman. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469-472. [DOI] [PubMed] [Google Scholar]
- 26.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markowitz, D., S. Goff, and A. Bank. 1988. Construction and use of a safe and efficient amphotropic packaging cell line. Virology 167:400-406. [PubMed] [Google Scholar]
- 28.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14(Suppl. 3):S31-S44. [PubMed] [Google Scholar]
- 29.McCutchan, F. E., J. K. Carr, M. Bajani, E. Sanders-Buell, T. O. Harry, T. C. Stoeckli, K. E. Robbins, W. Gashau, A. Nasidi, W. Janssens, and M. L. Kalish. 1999. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology 254:226-234. [DOI] [PubMed] [Google Scholar]
- 30.Montavon, C., F. Bibollet-Ruche, D. Robertson, B. Koumare, C. Mulanga, E. Esu-Williams, C. Toure, S. Mboup, E. Saman, E. Delaporte, and M. Peeters. 1999. The identification of a complex A/G/I/J recombinant HIV type 1 virus in various West African countries. AIDS Res. Hum. Retrovir. 15:1707-1712. [DOI] [PubMed] [Google Scholar]
- 31.Moumen, A., L. Polomack, B. Roques, H. Buc, and M. Negroni. 2001. The HIV-1 repeated sequence R as a robust hot-spot for copy-choice recombination. Nucleic Acids Res. 29:3814-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasioulas, G., D. Paraskevis, E. Magiorkinis, M. Theodoridou, and A. Hatzakis. 1999. Molecular analysis of the full-length genome of HIV type 1 subtype I: evidence of A/G/I recombination. AIDS Res. Hum. Retrovir. 15:745-758. [DOI] [PubMed] [Google Scholar]
- 33.O'Neil, P. K., G. Sun, H. Yu, Y. Ron, J. P. Dougherty, and B. D. Preston. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J. Biol. Chem., in press. [DOI] [PubMed]
- 34.Page, K. A., N. R. Landau, and D. R. Littman. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 64:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palaniappan, C., M. Wisniewski, W. Wu, P. J. Fay, and R. A. Bambara. 1996. Misincorporation by HIV-1 reverse transcriptase promotes recombination via strand transfer synthesis. J. Biol. Chem. 271:22331-22338. [DOI] [PubMed] [Google Scholar]
- 36.Parthasarathi, S., A. Varela-Echavarria, Y. Ron, B. D. Preston, and J. P. Dougherty. 1995. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J. Virol. 69:7991-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, P. H., and B. D. Preston. 1994. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl. Acad. Sci. USA 91:549-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peeters, M., and P. M. Sharp. 2000. Genetic diversity of HIV-1: the moving target. AIDS 14(Suppl. 3):S129-S140. [PubMed] [Google Scholar]
- 39.Peliska, J. A., and S. J. Benkovic. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112-1118. [DOI] [PubMed] [Google Scholar]
- 40.Peliska, J. A., and S. J. Benkovic. 1994. Fidelity of in vitro DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Biochemistry 33:3890-3895. [DOI] [PubMed] [Google Scholar]
- 41.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 42.Ricchetti, M., and H. Buc. 1990. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 9:1583-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, J. D., B. D. Preston, L. A. Johnston, A. Soni, L. A. Loeb, and T. A. Kunkel. 1989. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol. Cell. Biol. 9:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robertson, D. L., B. H. Hahn, and P. M. Sharp. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40:249-259. [DOI] [PubMed] [Google Scholar]
- 46.Sharp, P. M., E. Bailes, D. L. Robertson, F. Gao, and B. H. Hahn. 1999. Origins and evolution of AIDS viruses. Biol. Bull. 196:338-342. [DOI] [PubMed] [Google Scholar]
- 47.Strickberger, M. W. 1976. Genetics. Macmillan Publishing Co. Inc., New York, N.Y.
- 48.Temin, H. M. 1993. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc. Natl. Acad. Sci. USA 90:6900-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tscherning-Casper, C., G. Dolcini, P. Mauclere, E. M. Fenyo, F. Barre-Sinoussi, J. Albert, E. Menu, et al. 2000. Evidence of the existence of a new circulating recombinant form of HIV type 1 subtype A/J in Cameroon. AIDS Res. Hum. Retrovir. 16:1313-1318. [DOI] [PubMed] [Google Scholar]
- 50.Varela-Echavarria, A., C. M. Prorock, Y. Ron, and J. P. Dougherty. 1993. High rate of genetic rearrangement during replication of a Moloney murine leukemia virus-based vector. J. Virol. 67:6357-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogt, P. K. 1971. Genetically stable reassortment of markers during mixed infection with avian tumor viruses. Virology 46:947-952. [DOI] [PubMed] [Google Scholar]
- 52.Weber, J., and F. Grosse. 1989. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural DNA. Nucleic Acids Res. 17:1379-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilhelm, M., M. Boutabout, T. Heyman, and F. X. Wilhelm. 1999. Reverse transcription of the yeast Ty1 retrotransposon: the mode of first strand transfer is either intermolecular or intramolecular. J. Mol. Biol. 288:505-510. [DOI] [PubMed] [Google Scholar]
- 54.Wong, P. K., and J. A. McCarter. 1973. Genetic studies of temperature-sensitive mutants of Moloney-murine leukemia virus. Virology 53:319-326. [DOI] [PubMed] [Google Scholar]
- 55.Wu, W., B. M. Blumberg, P. J. Fay, and R. A. Bambara. 1995. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J. Biol. Chem. 270:325-332. [DOI] [PubMed] [Google Scholar]
- 56.Wyke, J. A., J. G. Bell, and J. A. Beamand. 1975. Genetic recombination among temperature-sensitive mutants of Rous sarcoma virus. Cold Spring Harb. Symp. Quant. Biol. 39:897-905. [DOI] [PubMed] [Google Scholar]
- 57.Yu, H., A. E. Jetzt, Y. Ron, B. D. Preston, and J. P. Dougherty. 1998. The nature of human immunodeficiency virus type 1 strand transfers. J. Biol. Chem. 273:28384-28391. [DOI] [PubMed] [Google Scholar]
- 58.Yu, H., A. B. Rabson, M. Kaul, Y. Ron, and J. P. Dougherty. 1996. Inducible human immunodeficiency virus type 1 packaging cell lines. J. Virol. 70:4530-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, Cooper, D. A., and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]