Abstract
Papillomaviruses possess small DNA genomes that encode five early (E) proteins. Transient DNA replication requires activities of the E1 and E2 proteins and a DNA segment containing their binding sites. The E6 and E7 proteins of cancer-associated human papillomavirus (HPV) transform cells in culture. Recent reports have shown that E6 and E7 are necessary for episomal maintenance of HPV in primary keratinocytes. The functions of E6 necessary for viral replication have not been determined, and to address this question we used a recently developed transfection system based on HPV31. To utilize a series of HPV16 E6 mutations, HPV31 E6 was replaced by its HPV16 counterpart. This chimeric genome was competent for both transient and stable replication in keratinocytes. Four HPV16 E6 mutations that do not stimulate p53 degradation were unable to support stable viral replication, suggesting this activity may be necessary for episomal maintenance. E7 has also been shown to be essential for episomal maintenance of the HPV31 genome. A point mutation in the Rb binding motif of HPV E7 has been reported to render HPV31 unable to stably replicate. Interestingly, HPV31 genomes harboring two of the three p53 degradation-defective E6 mutations combined with this E7 mutation were maintained as replicating episomes. These findings imply that the balance between E6 and E7 functions in infected cells is critical for episomal maintenance of high-risk HPV genomes. This model will be useful to dissect the activities of E6 and E7 necessary for viral DNA replication.
Papillomaviruses are small, double-stranded DNA viruses that infect the skin of animals and humans and are maintained for extended periods of time as extrachromosomal episomes (29). High-risk human papillomaviruses (HPV) infect the mucosal epithelia and are the causative agents of cervical cancer (53). The cancer-promoting activity of these viruses has been attributed largely to their ability to manipulate cell-cycle-related proteins (52). The most extensively studied of these interactions have been those between papillomavirus E6 and the tumor suppressor protein p53 (43, 51) and between papillomavirus E7 and the cell cycle regulatory protein Rb (10, 35). High-risk HPV E6, in association with the ubiquitin ligase E6AP, targets p53 for proteasome-mediated degradation (42). E7 binds to Rb and causes release of the E2F protein, thereby inducing cell cycle progression (23). E6 and E7 have also been shown to bind and modify the action of numerous other cellular factors. These interactions are less understood and may function in manipulating the cell cycle or other viral activities. E6 has been shown to promote the degradation of the tumor suppressor protein hDLG (38) and the Rap-GAP-like protein E6TP1 (16) and to interact with ERC-55 (E6BP) (7), paxillin (48, 49), protein kinase N (15), hMCM-7 (26), AP-1 (6), Bak (45), IRF-3 (39), Tyk2 (30), GPS2 (9), and ADA3 (28). Recently it was reported that E6 activates the cellular telomerase protein by increasing transcription of the catalytic subunit hTERT (19, 37, 50). Telomerase activation by E6 appears to correlate with its ability to immortalize human mammary epithelial cells (27). In addition to binding Rb, E7 has been shown to interact with Rb-regulated factors, such as the E2F/cyclin A complex (1) and cyclin E (33) and with p21Cip1 (14, 23, 40).
Mutational analyses of HPV16 E6 have begun to address the biological significance of interactions between E6 and specific cellular proteins. HPV E6 mutations have been described that abrogate interaction with p53, E6AP, E6BP, hDLG, and telomerase (7, 17, 18, 25, 27, 31, 32, 46). The significance of these biochemical associations has usually been tested for inhibition of p53-mediated growth arrest and apoptosis and primary cell immortalization. The relationship of E6-mediated p53 inactivation and its other interactions in HPV replication are largely unknown. A recent report showed that HPV E6 is necessary for the long-term maintenance of HPV31 genomes as episomes in keratinocytes (47). While an extensive analysis was not performed, an HPV31 genome harboring a p53-binding-defective E6 was incapable of long-term episomal maintenance. An HPV16 genome with inactivation of the E7 gene was not maintained in primary keratinocytes but was replication competent in an immortal keratinocyte cell line (11, 47). Efficient Rb binding function of HPV31 E7 was reported to be necessary for viral replication in human foreskin keratinocytes (HFKs) (11, 47).
We have begun to examine in detail the activities of E6 necessary for stable HPV genome maintenance by using previously characterized HPV16 E6 mutations, with initial focus on its p53 interaction. Our results demonstrate that E6-mediated p53 degradation is necessary only in the context of high-risk E7, and therefore it may be related to maintaining a balance between p53 and Rb within the cell. These data imply that E6 has an additional role in the productive viral life cycle distinct from induction of p53 degradation.
MATERIALS AND METHODS
Plasmids.
pBR322-HPV31 was provided by Frattini et al. (13). The HPV31 genome was released from pBR322 by digestion with EcoRI. The genome was then cloned into the EcoRI site of pSG5 (Stratagene) to create pSG5-HPV31. This clone was used as a template for PCR-directed cloning of HPV16 E6 into HPV31.
HPV31 E6 was replaced with HPV16 E6 by creating restriction sites at the N- and C-termini of both genes. After restriction digestion and cloning of HPV16 E6 in place of HPV31 E6, another series of PCR steps were performed to remove the cloned restriction sites. The resulting clone, pSG5-HPV31/16, contains the entire HPV31 genome with the exception of the E6 gene derived from HPV16.
HPV16 E6 mutations were cloned into the context of pSG5-HPV31/16 either by repeating the above protocol with mutant HPV16 E6 DNA as a template for PCR or by site-directed PCR. Clones containing the following HPV16 E6 mutations were constructed: F2L, F2V, 8S/9A/10T, Y54H, C63R, L110Q, and Δ118-122. In addition, a mutation was introduced into the HPV31 E7 gene at amino acid 21 to create pSG5-HPV31/16E7D21G. The following double E6/E7 mutant genomes were created: F2L-D21G, F2V-D21G, 8S/9A/10T-D21G, C63R-D21G, and Δ118-122-D21G. All mutant genomes were confirmed by DNA sequencing.
Transfection of HFKs.
HFKs were grown on mitomycin C-treated mouse J2 3T3 fibroblasts (treated for 2 to 4 h with 8 ng of mitomycin C/ml) in E medium (47). HFKs were plated onto 60-mm-diameter dishes at 106 cells per dish. Three to 4 h later cells were transfected by using 3 μl of Fugene (Roche) per 1 μg of DNA. Ten micrograms of each pSG5-HPV31 or pSG5HPV31/16 genome was digested with EcoRI to release the viral genome. Genomes were gel purified and then unimolecularly ligated by using T4 DNA ligase. The entire precipitated ligation (3 μg of DNA) was cotransfected with 1 μg of pCDNA3 (neo resistance) or 1 μg of pSVNeo into the HFKs. One day posttransfection cells were split into two 100-mm-diameter dishes containing mitomycin C-treated J2 3T3 cells and were serially passaged for stable replication assays.
Transient replication assays.
C33a cells were transfected by using calcium phosphate with 5 μg of each HPV genome. Four to 5 days posttransfection low-molecular-weight DNA was isolated from each 100-mm-diameter dish by Hirt extraction (20). The DNA was digested with DpnI to remove residual methylated input DNA and with EcoRI to linearize the viral genomes. The samples were transferred to a nylon membrane prehybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.5% sodium dodecyl sulfate (SDS), 5× Denhardt's reagent, and 0.1 mg of denatured salmon sperm DNA/ml for 1 h at 68°C. A 32P-labeled HPV31-specific probe was made by using the Prime-a-Gene labeling system (Promega). The labeled probe was denatured and added to hybridization solution (6× SSC, 0.5% SDS, 0.1 mg of salmon sperm DNA/ml) and incubated with the membrane overnight at 68°C. The membrane was washed twice with 2× SSC plus 0.5% SDS, once with 2× SSC plus 0.1% SDS, once at 60°C with 0.1× SSC plus 0.5% SDS, and finally with 0.1× SSC. Hybridizing species were visualized by using the Personal Molecular Imager FX (Bio-Rad).
Stable episomal maintenance assays.
HFKs were transfected as described above. One day posttransfection cells were split into 100-mm-diameter dishes in selection medium containing 150 μg of G418/ml. Cells were maintained in this medium for 2 weeks, at which point G418-resistant colonies were pooled and plated onto 60-mm-diameter dishes. Pooled populations were expanded and split once at 1:3. After cells reached 80 to 90% confluence, Hirt extracts were made. In some cases, in addition to Hirt extracts the cellular DNA pellet was analyzed for the presence of HPV DNA as follows: after precipitation of low-molecular-weight DNA from cells, the pellet was resuspended in lysis buffer (400 mM Tris-HCl [pH 7.4], 10 mM EDTA, 50 μg of RNase A/ml, 50 μg of proteinase K/ml, 0.2% SDS) and incubated at 37°C overnight. DNA was sheared by passage through an 18-gauge needle. The DNA was then extracted with phenol:chloroform and was ethanol precipitated. After either method of extraction, DNA was resuspended in Tris-EDTA and digested with EcoRI and DpnI and a Southern blot was performed as described above.
RESULTS
The HPV31/16 hybrid genome is maintained as a stable episome in primary keratinocytes.
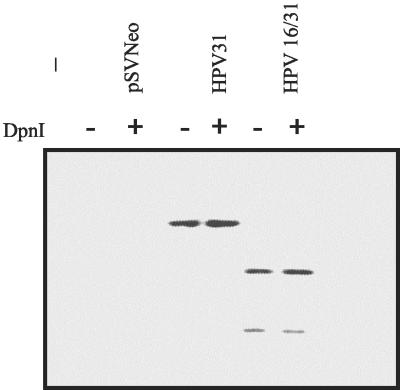
To take advantage of a set of previously characterized HPV16 E6 mutations for the study of viral replication, we first replaced HPV31 E6 with HPV16 E6 in the context of the HPV31 genome by using a PCR-based approach. The HPV16 and -31 E6 proteins are 65% identical and 77% similar (Fig. 1), so we predicted that the hybrid HPV31 genome would replicate in HFKs and then, if successful, the HPV16 E6 mutations could be evaluated in this model. We transfected primary HFKs with ligated HPV31 and HPV31/16 E6 genomes and assayed for genome replication after serial passage during 4 to 5 weeks in culture. The low-molecular-weight Hirt extracts were digested with DpnI to remove input DNA produced by bacteria, EcoRI to linearize the genomes, and BspEI to distinguish the HPV16 E6 gene. We had previously engineered this site within HPV16 E6, and it is not present in HPV31 (8). The wild-type and chimeric HPV31 genomes were detected by Southern blot, indicating that HPV16 E6 could replace its HPV31 counterpart in this assay (Fig. 2). In addition, the primary HFKs transfected with the hybrid genome became immortal, as did the wild-type HPV31-containing cells. Control cells transfected with only the pCDNA3 vector senesced approximately 5 to 6 weeks posttransfection.
FIG. 1.
Alignment of the E6 proteins from HPV31 and HPV16. Conserved residues are listed between the two protein sequences. Pluses indicate similar amino acids. Mutations of the amino acids within the E6 protein, designated in bold type, were created in the context of the hybrid HPV31/16 genome. Underlined residues are deleted in Δ118-122. Numbers listed below the alignment correspond to the position in HPV16 E6 with the second methionine of the open reading frame as amino acid number one.
FIG. 2.
Stable maintenance of HPV31 and the hybrid plasmid HPV31/16 in primary human foreskin keratinocyte cells 4 weeks posttransfection. The Hirt-extracted DNAs were digested with EcoRI and BspEI (a site engineered into HPV16 E6) with or without DpnI and analyzed by Southern blot.
Several HPV16 E6 mutations were cloned into the context of this hybrid genome. The phenotypes of these mutations have been described previously (8, 31). F2L, C63R, and Δ118-122 have decreased ability to bind p53 in vitro but facilitate its degradation in vivo. F2V and Y54H are not able to bind or degrade p53 in vitro or in vivo. However, these mutant proteins are able to bind E6AP and can immortalize mammary epithelial cells (31). L110Q is defective for p53 binding and degradation and does not bind to E6AP or E6BP; however, it maintains the ability to immortalize mammary epithelial cells. The 8S/9A/10T mutation in HPV16 E6 renders E6 unable to degrade p53 (25) but maintains the ability to bind to E6AP and to activate telomerase (19). The HPV16 E6 mutations used in this study and their reported phenotypes are listed in Table 1.
TABLE 1.
Summary of HPV16 E6 mutant characterizationa
| HPV16 gene | E6AP binding | P53 (37°C) degradation | P53 in early passage MECsb | Stable replication |
|---|---|---|---|---|
| Wild type | +++ | +++ | Low | Yes |
| F2L | +++ | ++ | NDc | Yes |
| F2V | +++ | − | Normal | No |
| Y54H | +++ | − | Normal | No |
| C63R | ++ | + | Normal | Yes |
| L110Q | − | − | Normal | No |
| Δ118-122 | + | +/− | Low | No |
| 8S9A10T | + | − | Normal | No |
The first three columns refer to previously published data for in vitro binding between E6 and E6AP, in vitro degradation of P53 by E6, and level of p53 proteins in mammary epithelial cells expressing wild-type or mutant HPV16 E6, respectively (17, 20, 33). The fourth column summarizes the results from replication assays in this report.
MECs, human mammary epithelial cells.
ND, not done.
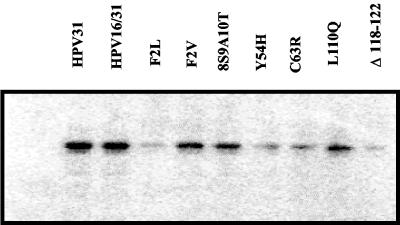
Previous studies have shown that only the E1 and E2 proteins are required for transient replication of papillomavirus genomes. Thus, we expected that mutation of E6 would not affect transient replication of our genomes. However, past experiments have revealed that mutations in E6 can affect splicing of the E1 and E2 transcripts and thereby reduce viral replication (22, 47). Therefore, we tested the mutant genomes for their ability to replicate transiently in C33a cells. As shown in Fig. 3, all were competent for transient replication. This indicated that the E1 and E2 proteins were expressed in each of the mutant genomes. This assay was performed three times, and in one experiment the Δ118-122 genome showed very low levels of replicated DNA, while in the other two assays the replicated genomes were readily detectable. We suspect that this deletion mutation may reduce levels of E1 or E2.
FIG. 3.
Transient replication of HPV31 and HPV31/16 mutant genomes. C33a cells were transfected with each of the indicated plasmids. Four days posttransfection, plasmid DNAs were isolated by Hirt extraction and were digested with DpnI and EcoRI and analyzed by Southern blot.
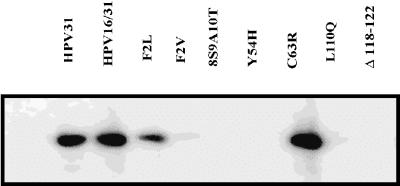
After establishing that these mutant genomes were capable of transient replication, the goal was to determine whether these E6 mutations would support long-term episomal maintenance in HFKs. We cotransfected each mutant genome along with a plasmid conferring neomycin resistance. The epithelial cells were maintained in selective medium on 3T3 feeder layers and were assayed for the presence of episomal viral DNA 4 to 5 weeks posttransfection. DpnI-resistant HPV genomes harboring the F2L, C63R, and T149V mutations were observed on Southern blots (Fig. 4). These mutant E6 proteins bind and degrade p53. In contrast, genomes harboring the F2V, 8S/9A/10T, Y54H, L110Q, and Δ118-122 mutations were not maintained. With the exception of Δ118-122, these E6 mutants are defective for p53 degradation. The failure of the 8S/9A/10T mutant genome to replicate suggests that induction of telomerase by E6 is not sufficient for long-term episomal maintenance. Interestingly, Δ118-122 has been reported to be unable to induce telomerase activation (25), suggesting that this E6 activity may be necessary for HPV genome maintenance in keratinocytes. Southern blotting of genomic DNA from these transfected HFKs after 4 to 5 weeks did not reveal integrated segments of HPV31. Taken together, these data implied that p53 degradation may be necessary for stable episomal maintenance of high-risk papillomavirus genomes. However, the inability of the Δ118-122 genome to be maintained in this assay indicates that p53 degradation is not sufficient for long-term episomal maintenance of HPV genomes and infers that another function of E6, perhaps telomerase activation, may be required.
FIG. 4.
Stable maintenance of HPV episomes in primary HFKs. Primary HFKs were cotransfected with pSVNeo and the viral DNAs. Cells were transferred to keratinocyte medium containing G418 and were grown on a feeder layer of mitomycin-treated J2 3T3 cells for 4 weeks. Hirt extracts DNAs were digested with EcoRI and DpnI and were analyzed by Southern blot.
Coordinate requirement for E6 and E7 activities in replication.
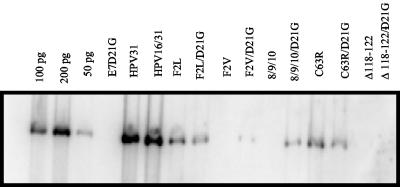
The observation that E6-mediated p53 degradation was necessary for persistent HPV genome replication was not anticipated. While these results might reflect a requirement for p53 degradation in cultured cells, another explanation was more likely. We hypothesized that the requirement for p53 degradation by high-risk HPVs might be dependent on the concomitant expression of high-risk HPV31 E7. Recent studies have demonstrated the requirement of functional E7 protein for the long-term maintenance of high-risk papillomavirus genomes in primary keratinocytes (11, 47). High-risk HPV E7 binds and degrades the tumor suppressor Rb (5, 24). Expression of high-risk HPV E7 in primary human keratinocytes has been shown to induce apoptosis, which can be reduced by coexpression of E6 (44). Low-risk HPV E7 has a much lower affinity for Rb and has not been reported to induce its proteolysis. Mutation of high-risk E7 in the Rb-binding motif (aspartic acid [D] at amino acid 21 in HPV31) reduced the affinity for Rb to levels similar to those for low-risk E7 (36). HPV31 genomes harboring this mutation were not maintained as stable episomes in long-term maintenance assays, suggesting a requirement for Rb inactivation (47). Alternatively, it was possible that the p53 degradation activity of E6 might only be necessary when the cells also expressed high-risk E7. To test this hypothesis, we engineered the mutation of aspartic acid at position 21 in HPV31 E7 to glycine (D21G). Consistent with the previous report, the HPV31/16 genomes harboring the E7 D21G mutation were not maintained as stable episomes. The E6 mutations were then transferred into the D21G genome. We first tested these double-mutant genomes for the ability to support transient replication, which would indicate that they produced functional levels of E1 and E2 proteins, and as expected, we found DpnI-resistant linear genomes on Southern blot (data not shown). We subsequently transfected HFKs with the double-mutant genomes and tested for replicated viral DNA after 4 to 5 weeks (Fig. 5). Interestingly, the p53 degradation-defective E6 mutants F2V and 8S9A10T supported episomal replication in the context of E7 D21G, contrasting with their inability to be maintained in the background of wild-type E7. These results imply that p53 degradation by E6 is necessary for viral replication in the presence of strong Rb inactivation by E7. However, the E6 Δ118-122/E7D21G genome was not detectable after 4 to 5 weeks in culture, suggesting a necessary and permissive function of E6 was inactivated. E6 F2L and C63R were replication competent in the context of wild-type E7 and in combination with E7 D21G. However, the E7 D21G mutation itself does not support episomal maintenance, yet these E6 mutations restored replication competence. Paradoxically, these data suggest that an activity of high-risk E6 may also restrict replication of HPV genomes expressing low-risk type E7 proteins.
FIG. 5.
Stable maintenance of HPV31 and HPV16/31 mutant genomes. Primary foreskin keratinocytes were transfected with 5 μg of each genome. Four weeks posttransfection, plasmid DNA was isolated and digested with EcoRI and DpnI and analyzed by Southern blot. Genomes designated D21G contain the HPV31 E7 mutation aspartic acid to glycine.
DISCUSSION
Recently it has become possible to transfect HPV genomes into primary keratinocytes, which replicate as autonomous plasmids, providing a biologically relevant model to test the functions of HPV proteins necessary for virus replication (12, 34, 47). Genomes from high-risk HPVs associated with cervical cancer have shown the most success in this culture system, perhaps because of their ability to immortalize the primary cells. To explore the activities of E6 necessary for viral episomal maintenance, we used a modified HPV31 genome containing HPV16 E6. E6 has been reported to interact with multiple cellular factors, the majority of which were identified by using HPV16. Furthermore, many mutations in HPV16 E6 have been generated and characterized, while only a few have been described for HPV31.
We first demonstrated that HPV16 E6 could functionally replace its counterpart in the HPV31 genome. Previously characterized E6 mutations were then transferred to the viral genome to examine the roles of E6AP and p53 and induction of telomerase in viral replication. Genomes harboring p53 degradation-defective mutations were not maintained as stable episomes in primary keratinocytes, while those that retained p53 degradation activity were maintained. Genomes carrying E6 F2V and Y54H, which bind E6AP but are unable to degrade p53, were not maintained, indicating that E6AP interaction alone is not sufficient for long-term episomal replication of HPV genomes. These data imply that E6-mediated p53 inactivation is necessary for the long-term episomal maintenance of high-risk papillomavirus genomes.
Our finding that p53 degradation was necessary for long-term episomal maintenance of high-risk papillomavirus genomes was surprising. Since the E6 proteins of low-risk anogenital and cutaneous HPVs do not bind p53, we anticipated p53 degradation may solely be relevant to HPV oncogenicity. We considered that in the context of the HPV31 genome, expression of the high-risk E7 protein might be deleterious to the cells in the absence of p53 inactivation. It has been reported that high-risk HPV E7 can activate a p53 response leading to apoptosis (2, 3). When E6 was present, it prevented E7-induced keratinocyte death (11, 44). Furthermore, while it has been suggested that Rb inactivation by E7 may be necessary for HPV replication (47), we hypothesized that the requirement for efficient Rb inactivation may be obviated in the context of a p53 degradation E6 mutant. Mutation of amino acid 21 within the high-risk HPV E7 protein reduced its ability to bind Rb to levels similar to those of the low-risk HPV E7 proteins (36). This E7 mutation alone did not allow maintenance of the viral DNA. We found that HPV genomes harboring mutations in E7 D21G in the context of two p53 degradation-defective E6 mutants were maintained as stable episomes for 4 to 5 weeks. Thus, the p53 degradation activity of high-risk E6 is necessary for stable maintenance of high-risk HPV genomes as a mechanism for balancing E7 function.
HPV genomes harboring the E7 D21G mutation but expressing wild-type HPV16 or −31 E6 (47) were not maintained as stable episomes. This suggests that expression of low-risk E7 in the context of wild-type E6 may prohibit episomal maintenance of HPV genomes. It is not clear what E6 activity might result in a growth disadvantage in the presence of E7. It is likely not due to p53 inactivation, as E6 C63R and F2L retain this activity and were replication competent with both wild-type and D21G E7. Interestingly, it has been reported that HPV16 genomes defective for E7 expression were maintained as stable episomes after transfection into an immortal keratinocyte cell line but not in primary keratinocytes (11). It will be interesting to examine the HPV31 E7 D21G mutant genomes in these immortal cells. It is possible that E7 activity is specifically required in primary cells and is not necessary for the maintenance of high-risk HPV genomes after immortalization occurs.
The Δ118-122 E6 protein induces efficient p53 degradation in vivo, yet genomes harboring this mutation with either wild-type or mutant E7 were not maintained as stable episomes. This implies that a function of E6 distinct from p53 degradation may be required. While one group reported that Δ118-122 did not activate telomerase (25), another laboratory found elevated telomerase activity in human mammary epithelial cells immortalized by Δ118-122 E6 (16). Because this mutant genome also showed reduced transient replication, it may express decreased levels of E1 or E2. Analysis of additional E6 mutations will be necessary to determine the relevance of telomerase activation for the maintenance of high-risk HPV genomes as stable episomes.
The major differences between high-risk HPV E6 and E7 proteins and their low-risk counterparts are their abilities to bind to the tumor suppressor proteins p53 and Rb (21, 36, 41). A number of additional interactions have been reported for the high-risk E6 and E7 proteins. The functions of E6 and E7, independent of their abilities to inactivate p53 and Rb that are required for stable episomal maintenance, remain to be determined. The observation that HPV31 genomes containing E6 mutations unable to degrade p53 and an E7 mutation with reduced Rb binding provides a model to explore the activities of E6 and E7 necessary for genome replication that are shared among all HPVs. Because the complete viral replication program requires a differentiated environment, it will be interesting to conduct future experiments using organotypic models to further analyze the balance between E6 and E7 functions permissive for viral DNA replication in differentiating keratinocytes.
Acknowledgments
We thank L. Laimins and J. Thomas for HPV constructs and for teaching us the methods of transfecting HPV genome into human keratinocytes.
This work was supported by National Cancer Institute grant R01 CA73558 to E.J.A.
REFERENCES
- 1.Arroyo, M., S. Bagchi, and P. Raychaudhuri. 1993. Association of the human papillomavirus type 16 E7 protein with the S-phase-specific E2F-cyclin A complex. Mol. Cell. Biol. 13:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 3.Bates, S., and K. H. Vousden. 1996. p53 in signaling checkpoint arrest or apoptosis. Curr. Opin. Genet. Dev. 6:12-18. [DOI] [PubMed] [Google Scholar]
- 4.Bohl, J., K. Das, B. Dasgupta, and S. B. Vande Pol. 2000. Competitive binding to a charged leucine motif represses transformation by a papillomavirus E6 oncoprotein. Virology 271:163-170. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 6.Chen, J. J., Y. H. Hong, E. Rustamzadeh, J. D. Baleja, and E. J. Androphy. 1998. Identification of an alpha helical motif sufficient for association with papillomavirus E6. J. Biol. Chem. 273:13537-13544. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269:529-531. [DOI] [PubMed] [Google Scholar]
- 8.Dalal, S., Q. Gao, E. J. Androphy, and V. Band. 1996. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J. Virol. 70:683-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degenhardt, Y. Y., and S. J. Silverstein. 2001. Gps2, a protein partner for human papillomavirus E6 proteins. J. Virol. 75:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 11.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, C. A. Sattler, and P. F. Lambert. 1999. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology 262:344-354. [DOI] [PubMed] [Google Scholar]
- 13.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funk, J. O., S. Waga, J. B. Harry, E. Espling, B. Stillman, and D. A. Galloway. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, Q., A. Kumar, S. Srinivasan, L. Singh, H. Mukai, Y. Ono, D. E. Wazer, and V. Band. 2000. PKN binds and phosphorylates human papillomavirus E6 oncoprotein. J. Biol. Chem. 275:14824-14830. [DOI] [PubMed] [Google Scholar]
- 16.Gao, Q., L. Singh, A. Kumar, S. Srinivasan, D. E. Wazer, and V. Band. 2001. Human papillomavirus type 16 E6-induced degradation of E6TP1 correlates with its ability to immortalize human mammary epithelial cells. J. Virol. 75:4459-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardiol, D., and L. Banks. 1998. Comparison of human papillomavirus type 18 (HPV-18) E6-mediated degradation of p53 in vitro and in vivo reveals significant differences based on p53 structure and cell type but little difference with respect to mutants of HPV-18 E6. J. Gen. Virol. 79:1963-1970. [DOI] [PubMed] [Google Scholar]
- 18.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 19.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 75:7198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 21.Howley, P. M., K. Munger, H. Romanczuk, M. Scheffner, and J. M. Huibregtse. 1991. Cellular targets of the oncoproteins encoded by the cancer associated human papillomaviruses. Princess Takamatsu Symp. 22:239-248. [PubMed] [Google Scholar]
- 22.Hubert, W. G., and L. A. Laimins. 2002. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 76:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, D. L., R. M. Alani, and K. Münger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, D. L., D. A. Thompson, and K. Munger. 1997. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology 239:97-107. [DOI] [PubMed] [Google Scholar]
- 25.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 26.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, A., Y. Zhao, G. Meng, M. Zeng, S. Srinivasan, L. M. Delmolino, Q. Gao, G. Dimri, G. F. Weber, D. E. Wazer, H. Band, and V. Band. 2002. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol. Cell. Biol. 22:5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law, M.-F., D. R. Lowy, I. Dvoretzky, and P. M. Howley. 1981. Mouse cells transformed by bovine papillomavirus contain only extrachromosonal viral DNA sequences. Proc. Natl. Acad. Sci. USA 78:2727-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, S., S. Labrecque, M. C. Gauzzi, A. R. Cuddihy, A. H. Wong, S. Pellegrini, G. J. Matlashewski, and A. E. Koromilas. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727-5737. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., J. J. Chen, Q. Gao, S. Dalal, Y. Hong, C. P. Mansur, V. Band, and E. J. Androphy. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 73:7297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansur, C. P., B. Marcus, S. Dalal, and E. J. Androphy. 1995. The domain of p53 required for binding HPV16 E6 is separable from the degradation domain. Oncogene 10:457-465. [PubMed] [Google Scholar]
- 33.McIntyre, M. C., M. N. Ruesch, and L. A. Laimins. 1996. Human papillomavirus E7 oncoproteins bind a single form of cyclin E in a complex with cdk2 and p107. Virology 215:73-82. [DOI] [PubMed] [Google Scholar]
- 34.Meyers, C., T. J. Mayer, and M. A. Ozbun. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J. Virol. 71:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munger, K., C. L. Yee, W. C. Phelps, J. A. Pietenpol, H. L. Moses, and P. M. Howley. 1991. Biochemical and biological differences between E7 oncoproteins of the high- and low-risk human papillomavirus types are determined by amino-terminal sequences. J. Virol. 65:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh, S. T., S. Kyo, and L. A. Laimins. 2001. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 75:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pim, D., M. Thomas, R. Javier, D. Gardiol, and L. Banks. 2000. HPV E6 targeted degradation of the discs large protein: evidence for the involvement of a novel ubiquitin ligase. Oncogene 19:719-725. [DOI] [PubMed] [Google Scholar]
- 39.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruesch, M. N., and L. A. Laimins. 1997. Initiation of DNA synthesis by human papillomavirus E7 oncoproteins is resistant to p21-mediated inhibition of cyclin E-cdk2 activity. J. Virol. 71:5570-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sang, B. C., and M. S. Barbosa. 1992. Single amino acid substitutions in “low-risk” human papillomavirus (HPV) type 6 E7 protein enhance features characteristic of the “high-risk” HPV E7 oncoproteins. Proc. Natl. Acad. Sci. USA 89:8063-8067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheffner, M., J. M. Huibregtse, and P. M. Howley. 1994. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc. Natl. Acad. Sci. USA 91:8797-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 44.Stöppler, H., M. C. Stöppler, E. Johnson, C. M. Simbulan-Rosenthal, M. E. Smulson, S. Iyer, D. S. Rosenthal, and R. Schlegel. 1998. The E7 protein of human papillomavirus type 16 sensitizes primary human keratinocytes to apoptosis. Oncogene 17:1207-1214. [DOI] [PubMed] [Google Scholar]
- 45.Storey, A., M. Thomas, A. Kalita, C. Harwood, D. Gardiol, F. Mantovani, J. Breuer, I. M. Leigh, G. Matlashewski, and L. Banks. 1998. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature 393:229-234. [DOI] [PubMed] [Google Scholar]
- 46.Talis, A. L., J. M. Huibregtse, and P. M. Howley. 1998. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 273:6439-6445. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 96:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong, X. A., and P. M. Howley. 1997. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 94:4412-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vande Pol, S., M. C. Brown, and C. E. Turner. 1998. Association of Bovine Papillomavirus Type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene 16:43-52. [DOI] [PubMed] [Google Scholar]
- 50.Veldman, T., I. Horikawa, J. C. Barrett, and R. Schlegel. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 75:4467-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werness, B. A., K. Munger, and P. M. Howley. 1991. Role of the human papillomavirus oncoproteins in transformation and carcinogenic progression. Imp. Advan. Onc. 1991:3-18. [PubMed] [Google Scholar]
- 52.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]
- 53.zur Hausen, H. 1999. Papillomaviruses in human cancers. Proc. Assoc. Am. Physicians 111:581-587. [DOI] [PubMed] [Google Scholar]