Abstract
Endogenous retroviral sequences in the pig genome (PERV) represent a potential infectious risk in xenotransplantation. All known infectious PERV have been asssigned to the PERV γ1 family, consisting of the subfamilies A, B, and C. The aim of the study was the concise examination of PERV γ by the analysis of the retroviral pro-pol sequences. The analysis of 52 pro-pol clones amplified in this study revealed eight PERV γ families. In addition to four already-described families (γ1, γ4, γ5, γ6), four novel families (γ7, γ8, γ9, γ10) were identified. Quantitative analysis of the novel PERV γ sequences in selected breeds revealed variations in the endogenous retroviral load. Open reading frames (ORF) in the amplified proviral fragment were only found for PERV γ1. In addition, novel ORF-containing PERV γ1 clones consisting of hybrid sequences were revealed. Sequence comparison from published full-length PERV γ1 clones of the PERV subfamilies A, B, and C resulted in a lack of strict correlation of the classification of pro-pol and env. The results indicated the occurrence of causative recombination events between retroviral genomes. Thus, our study on PERV γ provides new data for the evaluation and selection of pigs intended to be used in xenotransplantation.
Xenotransplantation of functional pig cells, tissues, and organs is discussed as a possible solution for the compensation of the shortage of human donor organs. Prerequisites are the solution of the severe host-versus-graft reactions and the prevention of the cross-species transfer of pathogens via the transplant. Use of specific-pathogen-free animals focuses the potential risk of infection to the porcine endogenous retroviral sequences (PERV) (2).
Endogenous retroviruses (ERV) are copies of exogenous retroviral genomes integrated into the host genome. They are transmitted vertically to the offspring. ERV have been found in multiple copy numbers in all vertebrates examined (6). Most ERV are defective due to deleterious mutations (24). Whereas retrospective examinations of patients exposed to living porcine tissues have not yielded any indications for the infection of the recipients with PERV (10, 21), the potential risk of cross-species infection of PERV has been shown in in vitro and in vivo experiments (4, 8, 28). Xenotransplantation of tissues derived from pigs which are genetically altered for the aim of the reduction of the serious host-versus-graft reactions in humans (7, 14) has been suggested to enhance the risk of infection with PERV (25, 30).
Referring to the envelope (env) sequences, ERV are classified into the retroviral β (B- or D-type) and γ (C-type) genera (29). PERV A, B, and C, which have been observed to be infectious in vitro, are highly homologous in their gag and pro-pol retroviral genes and, therefore, have been classified as PERV γ1 (22). Significant differences in the env gene explain their different host tropisms (16, 26). All pig breeds examined contain PERV A and B with a copy number ranging from 10 to 23 and from 7 to 12, respectively. PERV C is only present in some of the breeds with 8 to 15 copies. Differences in the proviral load have been observed in the genome of different animals, and the correlation of an increased proviral load in highly inbred pig breeds has been discussed previously (1, 5, 13, 15-17, 23). Although most of the copies of PERV A and B are defective, several full-length functional PERV A and B from genomic pig loci have been sequenced (11, 19). Recently, additional PERV γ clones, including defective sequences as well as mutant full-length copies (PERV E), have been described (17, 22).
Here we precisely examined the PERV γ pro-pol nucleotide sequences. We identified the lack of the strict correlation of the classification of pro-pol and env in already-described PERV γ1 sequences as well as novel γ1 pro-pol clones harboring hybrid sequences and an open reading frame (ORF).
Amplification and analysis of the PERV sequences.
Previous reports have shown a high PERV load in Landrace pigs (16, 17); therefore, the search for additional PERV γ families was carried out with genomic DNA of this breed. pro-pol sequences were amplified by PCR with six pairs of degenerate primers (two 5′ primers [9, 27] and three 3′ primers [9]) and an annealing temperature of 38°C. The 0.5- to 1.2-kb fragments were separated in agarose gels, isolated, and cloned into the pGEM-T Easy vector (Promega, Madison, Wis.). Thirty independent clones were sequenced from the amplification products of each of the six primer pairs. Sequence analysis was carried out with the Amersham DNA sequencing kit (Amersham Pharmacia Biotech, Vienna, Austria) and the ABI PRISM 377 automated DNA sequencer. The clones were sequenced bidirectionally with additional primers annealing within the cloned sequences. Analysis by BLAST search of the GenBank DNA database revealed that 53 of the 180 clones were of retroviral origin, which showed a length of 0.7 to 1.0 kb. Fifty-two clones were classified as PERV γ, one clone as PERV β, and no clone was grouped as a spumavirus. From the 52 PERV γ clones amplified in this study, several 0.9-kb clones showed identical sequences, thereby indicating a low rate of polymerase errors in the PCR process as previously described (15).
Phylogenetic analysis.
By using Gene Jockey II (Biosoft, Cambridge, United Kingdom), further comparison of the complete amplified pro-pol fragment of the 53 clones revealed nine families (eight PERV γ families and a single clone belonging to a novel PERV β family) with 1 to 21 members. Amplified clones showing more than 90% identity were pooled to families. Nomenclature of the families was carried out according to Patience et al. (22). In addition to four already-described PERV γ families (γ1, γ4, γ5, γ6), four novel families (γ7, γ8, γ9, γ10) were found. Thirty-eight of the 52 PERV γ clones were assigned to two families, γ1 (n = 21) and γ6 (n = 17).
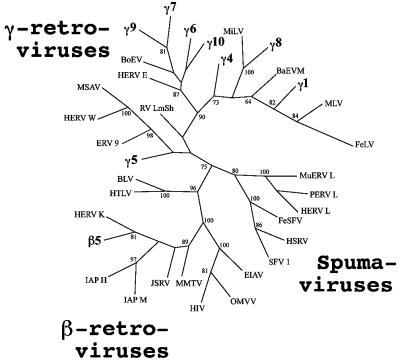
The phylogenetic examination of the PERV sequences was carried out both for the DNA and for the protein sequences. Partial and full-length sequences were prepared in SeqApp (http://ftp.bio.indiana.edu/soft/molbio/seqapp/) and were aligned by ClustalW (12), with manual adaptation. Most-parsimony, neighbor-joining, and maximum-likelihood phylogenetic trees were generated by using PAUP* (http://www.lms.si.edu/PAUP) and PHYLIP (http://evolution.genetics.washington.edu/phylip.html). The least defective clone was chosen as the representative of the family. Due to the differing sequences in the 5′ region of the pro-pol fragments, 510 nucleotides (nt) of the 3′ end and the C-terminal 170 amino acids, respectively, were used according to Herniou et al. (9). Figure 1 shows the tree obtained with the nucleotide sequences by using the neighbor-joining method. The phylogenetic relationship of the sequences was confirmed by analogous results which were generated with the different data sets and the various algorithms used (data not shown).
FIG. 1.
Neighbor-joining tree of retroviruses produced by PHYLIP. The analysis was done with the 510-nt 3′ fragment of the amplified pro-pol sequences and a data set of 1,000 bootstrap replicates. Percent bootstrap values higher than 60% are shown. The PERV sequences obtained in this study (γ1, γ4 to γ10, and β5) are depicted in bold. The viruses and GenBank accession numbers of the sequences (in parentheses) used in the phylogenetic tree are as follows: BaEVM (M16550), baboon ERV; BLV (AF257515), bovine leukemia virus; BoEV (X99924), bovine ERV; EIAV (AF247394), equine infectious anemia virus; ERV 9 (X57147), human ERV 9; FeLV (AF052723), feline leukemia virus; FeSFV (U78765), feline syncytial virus; HERV E (M10976), human ERV E; HERV K (M14123); HERV L (X89211); HERV W (AF135487); HIV (K03455), human immunodeficiency virus; HSRV (U21247), human spumaretrovirus; (L03561), human T-cell lymphotropic virus; IAP H (M10134), syrian hamster intracisternal A-particle; IAP M (M17551), murine intracisternal A-particle; JSRV (M80216), Jaagsiekte sheep retrovirus; MiLV (X99931), mink ERV; MLV (J01998), murine leukemia virus; MMTV (M15122), mouse mammary tumor virus; MSAV (AF009668), multiple sclerosis-associated retrovirus; MuERV L (Y12713), murine ERV L; OMVV (NC_001511), ovine lentivirus; PERV L (AJ233661); RVLmSh (Y07810), lemon shark ERV; and SFV 1 (X54482), simian foamy virus 1.
PERV γ1 GenBank sequences.
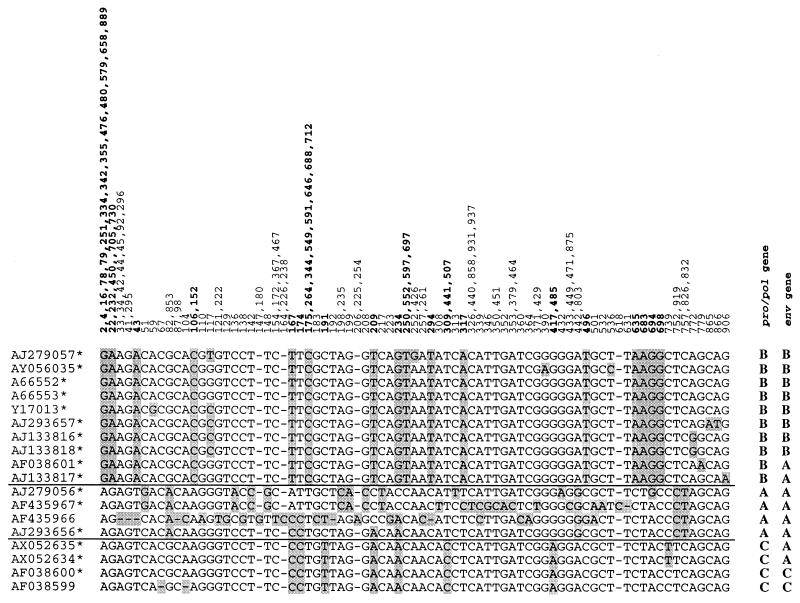
The intensively investigated mouse leukemia virus-related PERV γ1 family includes the three subfamilies PERV A, B, and C, which are defined by their env gene (1, 16). For the classification of the γ1 pro-pol sequences amplified in this study, first-step GenBank sequences of full-length PERV γ1 clones were compared. Designation of the clones according to their env gene as PERV A, B, and C was carried out by sequence comparison to Y12238, Y12239, and AF038600, respectively. Classification into pro-pol subfamilies was done according to identities in the polymorphic nucleotide positions of the aligned sequences (Fig. 2). Fourteen of 18 examined 928-nt pro-pol fragments strictly correlated to their env gene, whereas two PERV B (AF038601 and AJ133817) and two PERV C (AX052634 and AX052635) pro-pol sequences correlated with the PERV A env gene. The PERV A pro-pol sequences (AF435966, AF435967, AJ279056, and AJ293656) shared only a few common polymorphic nucleotides and showed an increased sequence polymorphism (Fig. 2). Further analysis of five published 928-nt pro-pol sequences which lack the sequence information for their corresponding env gene resulted in classification as γ1B (U77599 and X99933) and γ1C (AF033259, AX052636, and U77600). One additional sequence (AF274705) harboring a stop codon showed 40-, 42-, and 48-nt mismatches to γ1A (AJ293656), γ1B (A66552), and γ1C (AF038600), respectively, and therefore remained unclassified (data not shown).
FIG. 2.
Alignment of the 928-nt pro-pol fragments from published full-length PERV γ1 clones. Designation of the env gene to PERV A, B, and C subfamilies was carried out by sequence comparison to Y12238, Y12239, and AF038600, respectively (in the column labeled env gene). The classification of the pro-pol fragment to γ1A, γ1B, and γ1C is shown in the column labeled pro-pol gene. The polymorphic nucleotides (n = 144; 15.4%) are depicted with the numbers of their positions in the alignment of the data set shown at the top. The 51 nucleotide positions whose sequence invariably determined the pro-pol subfamilies γ1B and γ1C are shown in bold. Mutant nucleotides are depicted in shaded boxes. pro-pol fragments harboring an ORF are indicated with an asterisk.
PERV γ1 clones amplified in this study.
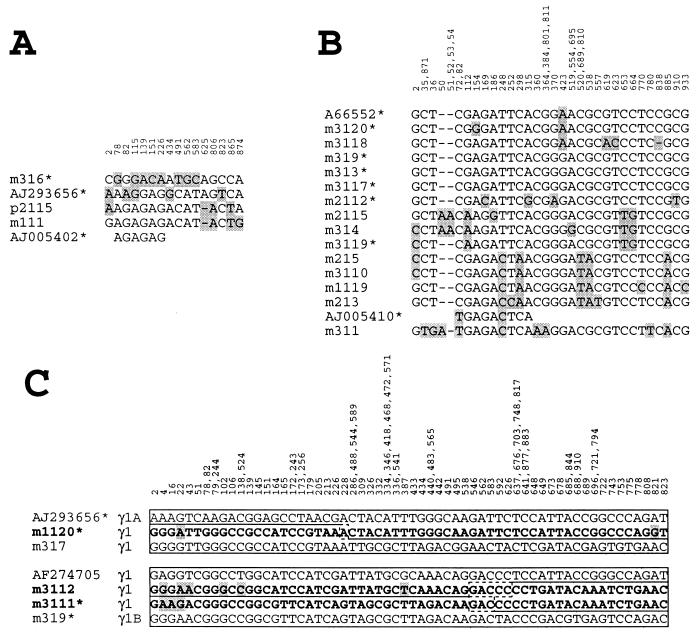
The classification of the 21 PERV γ1 clones amplified in this study was carried out by sequence comparison of the clones to all published PERV γ1 sequences. The pro-pol GenBank sequences showing the highest identity were grouped with the respective clones according to identities in the polymorphic nucleotide positions (Fig. 3). Within the examined 928-nt pro-pol fragment, 3 clones were classified as γ1A (Fig. 3A) and 14 clones were assigned to γ1B (Fig. 3B).
FIG. 3.
Alignment of the 21 PERV γ1 pro-pol clones amplified in this study. The amplified clones assigned to γ1A (A) and γ1B (B) and the published GenBank sequences which showed the highest identity are shown. The polymorphic nucleotides (n = 16 [1.7%] and n = 44 [4.7%] for γ1A and γ1B, respectively) of the amplified 928-nt pro-pol sequence are depicted, with the numbers of their positions in the alignment of the respective data set shown at the top. Mutant nucleotides are depicted in shaded boxes. Fragments harboring an ORF are indicated with an asterisk. Sequence A66552 represents seven GenBank sequences which are highly homologous in the pro-pol region and are identical at nt A423 (A66553, AF038601, AF147808, AJ133817, AJ279057, and U77599). (C) Sequence comparison of the clones m317, m1120, m3111, and m3112 to other PERV γ1 sequences amplified in this study or taken from GenBank. The three hybrid clones m1120, m3111, and m3112 are depicted in bold letters. Homologous regions are shown in boxes, and polymorphic nucleotides of the hybrid sequences are shown in shaded boxes.
Four PERV γ1 clones (m317, m1120, m3111, and m3112) did not match to the published PERV γ1 sequences (Fig. 3C). Clone m317 (AF511091) showed differences with γ1A (AJ293656), γ1B (A66552), and γ1C (AF038600) in 60, 29, and 61 nt, respectively. Clones m1120 (AF511090), m3111 (AF511096), and m3112 (AF511097) showed hybrid sequences with high homologies to various pro-pol sequences for different regions. Specifically, clone m1120 harboring an ORF was identical to m317 in its 5′ region until nt 285 and was identical to AJ293656 downstream of nt 226, with the exception of one A/G nucleotide exchange each. The two clones m3111 and m3112 showed sequence identity to each other 3′ of nt 539, whereas high homologies were found in their 5′ regions to m319 and to AF274705, respectively. The overlapping region of the clones m3111 and m3112 (nt 539 to 928) showed no identity to other PERV γ1 sequences.
In total, sequence analysis revealed nine clones harboring an ORF in the examined pro-pol sequence which were derived from at least six independent loci. Five of these loci were previously undescribed. In detail, PERV γ1A included the ORF containing clone m316 (AF511088). Three PERV γ1B clones (m319, m313, and m3117) were identical (AF511092), and clone m2112 (AF511095) was closely related. Clone m3119 (AF511098) maintaining an ORF showed relationship to the mutant sequences m314 and m2115. Clone m3120 differed only slightly from several GenBank sequences represented by A66552. In addition, the hybrid clones m1120 (AF511090) and m3111 (AF511096) harbored an ORF.
Determination of the approximate copy number of the PERV γ families was carried by Southern blot analysis (BamHI) with six pig breeds (Duroc, Landrace, Large White, Mangaliza, Pietrain, and Turopolja). Ten micrograms of DNA from three individuals per breed was examined. A specific probe hybridizing to ryr1 was used as a quantitative loading control (31). Probes specific for the different PERV γ families were amplified by PCR. Genomic mouse DNA was used as a negative control. PERV clones, which were found to be the closest relatives by phylogenetic analysis, were diluted in genomic mouse DNA and were used to monitor cross-hybridization. For the estimation of the copy number, the signal strength was compared to the signals of the probe used diluted in genomic mouse DNA. For PERV γ1, the results were consistent with the data already described (Table 1) (16, 17).
TABLE 1.
Approximate copy numbers of PERV γ sequences per haploid genome in different pig breeds determined by using Southern blot analysis
| PERV family | Copy no. in the following pig breeds:
|
|||||
|---|---|---|---|---|---|---|
| Duroc | Landrace | Large White | Mangaliza | Pietrain | Turopolja | |
| γ1 | 50 | 50 | 50 | 50 | 50 | 50 |
| γ4 | 10 | 10 | 10 | 10 | 10 | 10 |
| γ5 | 2 | 2 | 2 | 2 | 2 | 2 |
| γ6a | 50 | 50 | 50 | 50 | 50 | 25 |
| γ7a | 10 | 5 | 5 | 5 | 10 | 2 |
| γ8 | 1 | 1 | 1 | 1 | 1 | 1 |
| γ9 | 0 | 5 | 5 | 0 | 0 | 0 |
| γ10 | 2 | 2 | 2 | 2 | 2 | 0 |
Copy numbers were observed to differ within the breeds.
PERV γ4 and γ5.
Mutant members have been previously described for both PERV γ4 and γ5 families (22). Eight amplified clones were representatives of PERV γ4. Multiple frameshift and premature stop codon mutations were observed in the clones. About 10 copies per haploid genome were determined for PERV γ4 (Table 1). One clone belonging to PERV γ5 was observed to harbor multiple missense mutations. The sequence was assigned to a branch with human ERV-9, human endogenous retrovirus W (HERV-W), and MSAV (Fig. 1) and was present in a low copy number in all pig breeds examined (Table 1). The amplified clones of PERV γ4 and γ5 showed high identity to the published sequences AF274708 and AF274709, respectively.
PERV γ6.
Mutant full-length PERV E members have been observed in the pig genome (17). Seventeen of our clones corresponded to this family, which was named PERV γ6. Compared to clones of PERV γ1, the PERV γ6 clones showed a higher sequence diversity. Phylogenetic analysis was done as for PERV γ1 and revealed at least 11 novel loci (data not shown). None of the clones was found to have an ORF. Analogous to the PERV γ1 hybrid sequences, comparison of the 945-nt clones m116 (AF511100) and m2110 (AF511101) revealed sequence identity 3′ of nt 480, whereas the 5′ region showed differences in 26 nt (data not shown). The copy number of PERV γ6 sequences estimated for different pig breeds (Table 1) was similar to that described in previously published studies (17).
Novel PERV families (γ7 to γ10 and β5).
Six clones of endogenous retroviral sequences were found not to match to previously described families. Therefore, they represented PERV γ7, consisting of two independent clones, and PERV γ8, γ9, γ10, and β5. None of the clones was found to have an ORF. Southern blot analysis revealed low copy numbers for the novel PERV γ families. In addition, breed-specific differences were detected (Table 1).
The extensive search for new PERV γ copies carried out by the identification and phylogenetic analysis of more than 50 independent pro-pol sequences resulted in previously undescribed PERV families as well as novel clones harboring an ORF. Occurrence of several independent 0.9-kb PCR clones in PERV γ1B, γ4, and γ6 showing the identical sequence indicated a low incidence for misincorporation of nucleotides during the PCR procedure as previously described (15). The copy numbers of the different PERV γ families observed in Landrace pigs (Table 1) correlated well with the number of clones which were amplified in this study from Landrace genomic DNA and subsequently were assigned to the respective families. Previous studies have suggested the correlation of an increased PERV load in highly inbred breeds (13, 17). However, all breeds examined showed a high proviral load for PERV γ1 and γ6. Recently, breed-specific differences in the chromosomal integration sites of PERV have been described (15) which were confirmed by our results. This indicates that the use of preselected pigs in subsequent breeding concepts might lead to the reduction of the genomic load of putative infectious PERV.
Within the examined 928-nt pro-pol region, five novel PERV γ1 loci harboring an ORF (one γ1A, two γ1B, and two hybrid clones) were identified. This suggests that additional functional PERV γ1 proviruses that have not yet been described may exist in the pig genome. In contrast, in addition to the published mutant full-length PERV E (17), 17 defective clones representing at least 11 novel loci were found for PERV γ6. Taking the estimated copy number into account, these results indicate a low incidence of the presence of intact PERV γ6 genomes.
After in vitro coculture experiments with porcine cells, human-tropic replication-competent particles with hybrid sequences have been observed in human cells which have not been found in the genome of the donor pigs (20). This suggests that coexpression and recombination events of distinct PERV might lead to the production of recombined virus particles with unknown consequences on the potential risk of infection. In this study we showed the presence of such chimeric PERV γ1 sequences in the pig genome, where the classification of pro-pol and env did not correlate within the subfamilies. In humans, chimerism between the reverse transcriptase and transmembrane domains has been found both for endogenous retroviruses and infectious retroviruses (3). Recombination events within the diploid retroviral genome have been described to cause this phenomenon (18). We also found hybrid PERV pro-pol sequences within a given animal. This was confirmed by the observation of hybrid PERV pro-pol sequences (5′ region including nt 307) in the published clones AF435967 and AJ279056 (Fig. 2). In addition, intragenic recombinants between PERV γ1A and γ1B have previously been described for the env gene (15).
Thus, the results of our study indicate the presence of sequences deriving from recombination events between distinct PERV γ1 sequences in the pig genome. The potential risk of infection can therefore not be ruled out for mutant PERV loci.
Nucleotide sequence accession numbers.
The nucleotide sequence data of the representatives of the PERV families have been submitted to GenBank for PERV γ1 (AF511088-AF511099), γ6 (AF511100 to AF511110), γ7 (AF511111), γ8 (AF511112), γ9 (AF511113), γ10 (AF511114), and β5 (AF511115).
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckmann, J. P., G. Brem, F. W. Eigler, W. Günzburg, C. Hammer, W. Müller-Ruchholtz, E. M. Neumann-Held, and H. L. Schreiber. 2000. Xenotransplantation von Zellen, Geweben oder Organen. Springer Verlag, Berlin, Germany.
- 3.Benit, L., P. Dessen, and T. Heidmann. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 75:11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boneva, R. S., T. M. Folks, and L. E. Chapman. 2001. Infectious disease issues in xenotransplantation. Clin. Microbiol. Rev. 14:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, S., C. Arnauld, and A. Jestin. 2000. Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J. Virol. 74:8575-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 7.Dai, Y., T. D. Vaught, J. Boone, S. H. Chen, C. J. Phelps, S. Ball, J. A. Monahan, P. M. Jobst, K. J. McCreath, A. E. Lamborn, J. L. Cowell-Lucero, K. D. Wells, A. Colman, I. A. Polejaeva, and D. L. Ayares. 2002. Targeted disruption of the α1,3-galactosyltransferase gene in cloned pigs. Nat. Biotechnol. 20:251-255. [DOI] [PubMed] [Google Scholar]
- 8.Deng, Y. M., B. E. Tuch, and W. D. Rawlinson. 2000. Transmission of porcine endogenous retroviruses in severe combined immunodeficient mice xenotransplanted with fetal porcine pancreatic cells. Transplantation 70:1010-1016. [DOI] [PubMed] [Google Scholar]
- 9.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herring, C., D. A. Cunningham, A. J. Whittam, X. M. Fernandez-Suarez, and G. A. Langford. 2001. Monitoring xenotransplant recipients for infection by PERV. Clin. Biochem. 34:23-27. [DOI] [PubMed] [Google Scholar]
- 11.Herring, C., G. Quinn, R. Bower, N. Parsons, N. A. Logan, A. Brawley, K. Elsome, A. Whittam, X. M. Fernandez-Suarez, D. Cunningham, D. Onions, G. Langford, and L. Scobie. 2001. Mapping full-length porcine endogenous retroviruses in a large white pig. J. Virol. 75:12252-12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 13.Jin, H., Y. Inoshima, D. Wu, A. Morooka, and H. Sentsui. 2000. Expression of porcine endogenous retrovirus in peripheral blood leukocytes from ten different breeds. Transplant. Infect. Dis. 2:11-14. [DOI] [PubMed] [Google Scholar]
- 14.Lai, L., D. Kolber-Simonds, K. W. Park, H. T. Cheong, J. L. Greenstein, G. S. Im, M. Samuel, A. Bonk, A. Rieke, B. N. Day, C. N. Murphy, D. B. Carter, R. J. Hawley, and R. S. Prather. 2002. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295:1089-1092. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J. H., G. C. Webb, R. D. Allen, and C. Moran. 2002. Characterizing and mapping porcine endogenous retroviruses in Westran pigs. J. Virol. 76:5548-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 17.Mang, R., J. Maas, X. Chen, J. Goudsmit, and A. C. van Der Kuyl. 2001. Identification of a novel type C porcine endogenous retrovirus: evidence that copy number of endogenous retroviruses increases during host inbreeding. J. Gen. Virol. 82:1829-1834. [DOI] [PubMed] [Google Scholar]
- 18.Negroni, M., and H. Buc. 2001. Mechanisms of retroviral recombination. Annu. Rev. Genet. 35:275-302. [DOI] [PubMed] [Google Scholar]
- 19.Niebert, M., C. Rogel-Gaillard, P. Chardon, and R. R. Tonjes. 2002. Characterization of chromosomally assigned replication-competent gamma porcine endogenous retroviruses derived from a large white pig and expression in human cells. J. Virol. 76:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oldmixon, B. A., J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H. J. Schuurman, and C. Patience. 2002. Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J. Virol. 76:3045-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paradis, K., G. Langford, Z. Long, W. Heneine, P. Sandstrom, W. M. Switzer, L. E. Chapman, C. Lockey, D. Onions, and E. Otto. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. The XEN 111 Study Group. Science 285:1236-1241. [DOI] [PubMed] [Google Scholar]
- 22.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogel-Gaillard, C., N. Bourgeaux, A. Billault, M. Vaiman, and P. Chardon. 1999. Construction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements. Cytogenet. Cell. Genet. 85:205-211. [DOI] [PubMed] [Google Scholar]
- 24.Stoye, J. P. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11:R914-R916. [DOI] [PubMed] [Google Scholar]
- 25.Takefman, D. M., G. T. Spear, M. Saifuddin, and C. A. Wilson. 2002. Human CD59 incorporation into porcine endogenous retrovirus particles: implications for the use of transgenic pigs for xenotransplantation. J. Virol. 76:1999-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tristem, M., P. Kabat, L. Lieberman, S. Linde, A. Karpas, and F. Hill. 1996. Characterization of a novel murine leukemia virus-related subgroup within mammals. J. Virol. 70:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Laan, L. J., C. Lockey, B. C. Griffeth, F. S. Frasier, C. A. Wilson, D. E. Onions, B. J. Hering, Z. Long, E. Otto, B. E. Torbett, and D. R. Salomon. 2000. Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature 407:90-94. [DOI] [PubMed] [Google Scholar]
- 29.Van Regenmortel, M. H., C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 30.Weiss, R. A. 1998. Transgenic pigs and virus adaptation. Nature 391:327-328. [DOI] [PubMed] [Google Scholar]
- 31.Zinovieva, N., D. Vasicek, B. Aigner, M. Müller, and G. Brem. 1996. Single tube allele specific (STAS) PCR for direct determination of the mutation in the porcine ryanodine receptor gene associated with malignant hyperthermia. Anim. Biotechnol. 7:173-177. [Google Scholar]