Abstract
The serotypes of adeno-associated virus (AAV) have the potential to become important resources for clinical gene therapy. In an effort to compare the role of serotype-specific virion shells on vector transduction, we cloned each of the serotype capsid coding domains into a common vector backbone containing AAV type 2 replication genes. This strategy allowed the packaging of AAV2 inverted terminal repeat vectors into each serotype-specific virions. Each of these helper plasmids (pXR1 through pXR5) efficiently replicated the transgene DNA and expressed helper proteins at nearly equivalent levels. In this study, we observed a correlation between the amount of transgene replication and packaging efficiency. The physical titer of these hybrid vectors ranged between 1.3 × 1011 and 9.8 × 1012/ml (types 1 and 2, respectively). Of the five serotype vectors, only types 2 and 3 were efficiently purified by heparin-Sepharose column chromatography, illustrating the high degree of similarity between these virions. We analyzed vector transduction in reference and mutant Chinese hamster ovary cells deficient in heparan sulfate proteoglycan and saw a correlation between transduction and heparan sulfate binding data. In this analysis, types 1 and 5 were most consistent in transduction efficiency across all cell lines tested. In vivo each serotype was ranked after comparison of transgene levels by using different routes of injection and strains of rodents. Overall, in this analysis, type 1 was superior for efficient transduction of liver and muscle, followed in order by types 5, 3, 2, and 4. Surprisingly, this order changed when vector was introduced into rat retina. Types 5 and 4 were most efficient, followed by type 1. These data established a hierarchy for efficient serotype-specific vector transduction depending on the target tissue. These data also strongly support the need for extending these analyses to additional animal models and human tissue. The development of these helper plasmids should facilitate direct comparisons of serotypes, as well as begin the standardization of production for further clinical development.
The adeno-associated viruses (AAV) are members of the family Parvoviridae and the genus Dependovirus. Serotypes 1 to 4 were originally identified as contaminates of adenovirus preparations (5), whereas type 5 was isolated from a patient’s wart that was human papillomavirus positive. To date, seven molecular clones have been generated representing the serotypes of AAV (2, 8, 9, 30, 34, 39, 46). These clones have provided valuable reagents for studying the molecular biology of serotype-specific infection. Transduction of these viruses naturally results in latent infections, with completion of the life cycle requiring helper functions not associated with AAV viral gene products. As a result, all of these serotypes are classified as nonpathogenic and are believed to share a safety profile similar to the more extensively studied AAV type 2 (AAV2) (5).
The extensive development of AAV2 as a vector has been facilitated by 30 years of studying its biology in vitro. Recombinant AAV2 (rAAV2) has proven to be a suitable gene transfer vector in many different organisms (28, 32). As the number of applications involving gene transfer increases in vitro and in vivo, limitations to efficient rAAV2 transduction have become apparent (3, 11, 18, 37, 41, 46, 49). The natural tropism of any virus, including rAAV2, is a fundamental limitation to efficient gene transfer. With the identification of the AAV2 receptor, the requirements for efficient entry in target cells have become a critical topic of study (40). Efforts have been made to overcome these restrictions by broadening the host range by using either bispecific antibodies to the virion shell (4) or through capsid insertional mutagenesis (13, 45). While these efforts are beginning to bear fruit, utilizing the other serotypes of AAV may yet provide additional resources for making safe efficient gene transfer vectors. To this end, a small number of studies have begun to show the utility of serotype-specific vectors in vitro and in vivo (6, 8, 10, 11, 16, 20, 46, 49). In general, each of these studies uncovered broader cell type specificity with increased gene transfer in vivo. Additional efforts to separate the role of the serotype-specific vector components (cis-acting terminal repeats [TRs] and the trans-acting virion shell) will not only facilitate a comprehensive understanding of efficient vector transduction but will potentially allow the generation of optimized new hybrid AAV vectors (i.e., AAV5 TRs packaged in AAV1 shells).
In an effort to evaluate the role of AAV1 to -5 virion shells on AAV2 transgene transduction, a series of hybrid vectors were designed. Each AAV serotype-specific helper construct contained a high-yield plasmid backbone, the p5 start site mutation ACG (25), removal of the adenovirus terminal repeats (35, 36), and the ability to package transgenes utilizing AAV2 inverted terminal repeats (ITRs). These helper plasmids were characterized in vitro for replication and capsid proteins, ability to replicate vector transgene, total particle number, and purification by heparin-Sepharose chromatography (50). In addition, each hybrid serotype vector was compared for transduction efficiency in both reference and proteoglycan mutant Chinese hamster ovary (CHO) cell lines (40) and in vivo in muscle, liver, and neuronal specific targets.
MATERIALS AND METHODS
Plasmids.
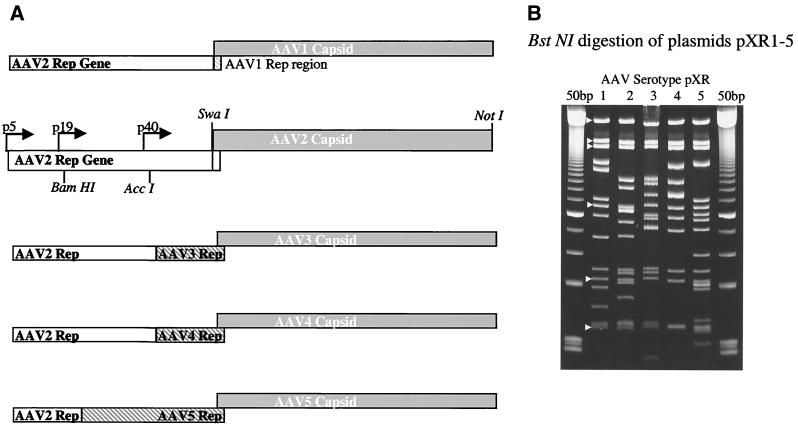
The plasmid pBS+ (Stratagene) was used as the backbone for cloning AAV serotypes 1 to 5 capsid genes. The AAV2 replication gene was subcloned into the plasmid pAAV2Cap (31) by using a XbaI (blunt ended) SwaI digestion of pACG2 (25), and a SmaI/SwaI digestion of pAAV2Cap. This new plasmid, pAAV2rep, was digested with NaeI, blunt ended with mung bean nuclease, and then digested with SwaI, removing the capsid gene and leaving only the replication gene of AAV2. Viral sequences for AAV1, -2, -3, -4, and -5 were cloned from American Type Culture Collection (ATCC) stocks. Primers were designed for each of these serotypes, such that a SwaI site was present before the coding region of Vp1 and a unique NotI site was present after the polyadenylation site. The forward primers were AAV1 and -2 (5′-AAATCAGGTATGGCTGCCGAT-3′), AAV3 (5′-AAATCAGGTATGGCTGCTGAT-3′), AAV4 (5′-AAATCAGGTATGGCTGCTGACGGTTAC-3′), and AAV5 (5′-AAATCAGGTATGGCTTTTGTTGATCAC-3′). The reverse primers were AAV1, -2, -3, and -4 (5′-GCGGCCGCGAGACCAAAGTTCAACTGA-3′) and AAV5 (5′-GCGGCCGCAAGAGGCAGTATTTTACTGA-3′). Pfu polymerase (Stratagene) was used in the PCR to generate the serotype-specific clones with blunt ends. These serotype-specific capsid coding fragments were then cloned into pAAV2rep. A BstNI digestion was used to confirm the presence and orientation of the positive clones (Fig. 1B). Individual clones were chosen after testing for virus production by using the triple transfection technique (26, 47). Clones containing AAV3, -4 and -5 capsid genes and AAV2 replication gene did not yield high-titer viruses from cell lysates or cesium gradients (Table 1). For clones containing the serotype 3, 4, and 5 capsid gene, a portion of the serotype-specific replication gene was substituted for that of AAV2. The AAV3 serotype clone was digested with AccI, as was the original plasmid (nucleotides 1424 to 4355 of the AAV3 sequence, Fig. 1A). The AAV4 serotype clone was digested with AccI and AgeI, as was the original plasmid (nucleotides 1479 to 4488 of the AAV4 sequence, Fig. 1A). The AAV5 serotype clone was digested with BamHI, as was the original plasmid (nucleotides 1071 to 2726 of the AAV5 sequence, Fig. 1A). These clones were designated pXR1, -2, -3, -4, and -5. Plasmid pTR/CMV/GFP (enhanced green fluorescent protein [EGFP] transgene) contains AAV2 ITRs flanking a cytomegalovirus (CMV) immediate-early promoter driven EGFP transgene (16); pXX6-80, a plasmid containing adenovirus genes required for AAV production plasmid, was a gift from X. Xiao, and the plasmid pCB-AAT was provided by Terry Flotte.
FIG. 1.
Construction and characterization of serotype clones. (A) Each serotype capsid domain, generated by PCR, was cloned into the pBS+ AAV2rep plasmid. The serotype-specific capsid insertions (shaded rectangles) were inserted into pBS+ AAV2rep and are listed in order from type 1 to 5. Restriction sites are shown in the AAV2 diagram. Additionally, modifications containing the coding region of the carboxy termini of each serotype’s rep coding domain (gray hatched) were cloned into the constructs as needed. (B) Acrylamide gel of AAV serotypes 1 through 5 digested with BstNI. White arrowheads point to common bands in the backbone and replication gene. The 50-bp ladder (Amersham Pharmacia) flanks the serotype lanes.
TABLE 1.
Comparison of AAV helper constructs containing AAV2-only replication gene or portions of the serotype specific replication genea
| Serotype | Particles/ml (n) | Particles/ml (n) |
|---|---|---|
| rAAV1 | 1.27 × 1011 (10) | ND |
| rAAV2 | 9.79 × 1012 (2) | NA |
| rAAV3 | 9.00 × 108 (1) | 7.16 × 1011 (3) |
| rAAV4 | 2.04 × 1010 (4) | 2.63 × 1011 (2) |
| rAAV5 | 7.64 × 109 (1) | 4.60 × 1011 (5) |
| rAAV5 with AAV5 ITR | 0 | 3.70 × 107 (3) |
ND, not done; NA, not applicable.
Cell culture.
All cell lines (293 human embryonic kidney, HeLa, Cos7, Cos1, CHO K1, and CHO K1 mutant deficient in proteoglycan biosynthesis [pgsA, pgsD, and pgsE]) were originally obtained from the ATCC and were maintained in 5% CO2 saturation at 37°C. 293, HeLa, Cos1, and Cos7 cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (Sigma), and CHO K1 and CHO K1 proteoglycan-deficient lines were cultured in F-12 nutrient mixture (Ham) with 10% fetal bovine serum.
Production of serotype-specific recombinant AAV.
Transfection protocols as previously described (31) were used with the following modifications. At 24 h prior to transfection, 10- to 15-cm-diameter plates were seeded with 1.2 × 107 293 cells and transfected by Superfect (Qiagene) according to manufacturer’s specifications by using the three-plasmid transfection technique (26, 47). Equimolar amounts of each plasmid—75 μg of pTR/CMV/GFP, 150 μg of pXX6-80 (adenovirus helper plasmid), and 75 μg of the serotype-specific plasmids pXR1, -2, -3, -4, or -5—were mixed with 7 ml of DMEM without serum and antibiotics and 1.2 ml of the SuperFect reagent before addition to cells. At 48 h posttransfection, the cells were harvested and virus was isolated as previously described (31). To each milliliter of viral supernatant, 0.59 g of CsCl was added, and 1.38 g of CsCl/ml was added to a final volume of 12 ml. The solution was centrifuged for 36 to 48 h at 37,000 rpm (Sorvall Ultra 80). Peak fractions from the isopycnic gradient were determined by infection of HeLa cells except for AAV4, which was tested on Cos1 cells. A second gradient (Beckman NVT-65 rotor at 65,000 rpm for 4 h) was incorporated for further purity, and peak fractions were determined by infection and dot blot hybridization as described below. Positive fractions were extensively dialyzed against Dulbecco phosphate-buffered saline (PBS) without calcium or magnesium chloride (Gibco-BRL) and supplemented with 10% (wt/vol) d-sorbitol (Sigma).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blots.
Protein samples from transfected human embryonic Kidney cells 293 were lysed as described previously (31), and an aliquot was removed, to which phenylmethylsulfonyl fluoride at 100 μg/ml (Sigma), pepstatin A at 1 μg/ml (Sigma), and leupeptine at 2 μg/ml (Sigma) were added. Protein concentrations were estimated by BCA Protein Assay (Pierce). Equivalent amounts of protein (5 μg/lane) from each serotype cell lysate were fractionated on a 10% denaturing polyacrylamide gel and then transferred to Hybond ECL (Amersham Pharmacia Biotech). Primary monoclonal antibodies 1F11 (22) and B1 (a gift from Jurgen Kleinschmidt) were used to detect AAV Rep and capsid proteins, respectively, as previously described by using SuperSignal West Pico (Pierce) for the detection of secondary antibody conjugate.
Hirt DNA analysis from AAV serotype transfections.
DNA from AAV serotype transfected 293 cells 24 h posttransfection was isolated by using the method described by Hirt (21). Approximately half of each sample was digested with DpnI enzyme, and 2.5 μg of digested and undigested DNA was fractionated on a 0.8% agarose gel. After transfer the blot was probed with a packaged transgene (GFP), the positive signal was visualized by using a phosphorimager, and the intensity of the bands was determined by image quantification.
Heparin column.
After the five serotypes were purified by two rounds of cesium chloride isopycnic centrifugation, dialyzed in 1× PBS (Gibco-BRL) plus 10% sorbitol with three changes, an equal amount of virus was injected into 1 ml of a HiTrap heparin column (Amersham Pharmacia Biotech) by using Amersham Pharmacia Biotech AKTA fast-performance liquid chromatography. A preset program (12 ml of buffer at 137 mM NaCl, 2 mM MgCl2, and 2 mM KCl run over the column at 0.5 ml/min for the flowthrough and wash, followed by a waste wash of 10 ml of the same buffer at 1 ml/min) was implemented before elution. The elution step used a continuous salt gradient from 137 mM NaCl2 to 1 M NaCl2 at a rate of 0.5 ml/min. For the flowthrough, wash, and elution steps, 0.5-ml fractions were collected, with the waste step collected in a single fraction.
Transducing units (TU)/microliter were determined for each fraction by transducing either HeLa (AAV1, -2, -3, and -5) or Cos1 cells (AAV4) in triplicate. The titer of the input virus was also determined so that a percentage of the recovered TU to the total number could be calculated.
Dot blot.
Dot blots were performed essentially as described previously (31). Briefly, 2 to 10 μl of virus from fractions off the cesium gradient, after dialysis, or the heparan column was blotted to GeneScreen Plus by using a dot blot manifold, and then the GeneScreen was UV cross-linked at 60 mJ (UV Stratalinker 1800; Stratagene). Finally, the blot was hybridized for positive signal by using GFP probe.
In vitro transduction assay.
Cell lines were infected with rAAV serotypes by using a volume of virus equivalent to 105 TU. For each datum point at least three experiments were performed, and the average is presented. The titers of the AAV serotypes 1, 2, 3, and 5 were originally determined on HeLa cells, and that of AAV4 was determined on Cos1 cells (reference cell lines). Between 1 × 105 and 1.75 × 105 cells of each cell type were plated in 24-well plates 24 h prior to infection. The volume of each serotype equivalent to 105 TU was added to 100 μl of DMEM that included adenovirus, and this was added to each well dropwise. After 24 h the number of GFP-positive cells was counted. Eight fields/well were counted, and the average number of GFP-positive cells/well was determined; this number was then multiplied by the dilution to obtain the number of TU/microliter.
ELISA analysis of factor IX and α1-antitrypsin.
NOD/Scid, BALB/c, and C57/BL mice were purchased form Jackson Laboratories (Bar Harbor, Maine). Mice were maintained and treated in accordance with the Animal Care and Use Committee of University of North Carolina at Chapel Hill. Portal vein injection, for liver expression, and muscular injection were performed as previously described (6). NOD/Scid mice were each injected with 2 × 1011 particles, and BALB/c mice were each injected with 1010 particles of the rAAV serotypes containing the canine factor IX transgene (6). Canine factor IX antigen was detected by enzyme-linked immunosorbent assay (ELISA) as previously described (H. J. Chao et al., unpublished data). Both BALB/c and C57/BL mice were each injected with 5 × 1010 particles of rAAV serotypes containing the human α1-antitrypsin transgene. A modified double-antibody sandwich ELISA was used for measurement of α1-antitrypsin in biologic fluids (27).
Subretinal injection and in vivo fluorescence imaging.
Subretinal injections were performed via a transcleral transchoroidal approach on wild-type Wistar rats as previously described (33). Briefly, the sclera and the choroid were punctured; a 33-gauge needle was then inserted in a tangential direction under an operating microscope. Then, 3 μl of each of the five hybrid rAAV serotypes (5 × 1011 particles/ml) was delivered into the subretinal space (n = 3, for each serotype). A new method with fundus photography has been developed and performed in order to control the accuracy and reproducibility of subretinal injections (F. Rolling et al., unpublished data).
GFP protein expression in live rats was monitored by fluorescent retinal imaging by using a Canon UVI retinal camera connected to a digital imaging system (Lhedioph Win Software). Retinas were examined at 12, 26, and 46 days postinjection.
RESULTS
Construction of AAV hybrid helper plasmids.
The generation of AAV serotype-specific hybrid helper plasmids utilized a common AAV2 Rep gene pACG2 (25) and the respective capsid coding sequences from each of the five serotypes (Fig. 1A). The ACG mutation of the p5 start site was chosen because this mutation has been shown to improve vector production by reducing rep78/68 while increasing AAV cap expression (25). pACG2 rep sequences were cloned into pAAV2Cap (Stratagene pBS+ backbone previously described [31]), and the capsid gene was removed by SwaI/NsiI digestion. This intermediate plasmid, pAAV2rep, was used for cloning each of the serotype-specific capsid coding sequences (see Materials and Methods for details). The capsid genes of each serotype were PCR amplified from ATCC viral stocks, and the product was cloned into this SwaI blunt-ended NsiI-digested pAAV2rep intermediate. The new hybrid plasmids containing the common AAV2 ACG replication gene and the serotype-specific capsid sequences were further characterized by enzyme digestion and DNA sequencing. BstNI digestion (Fig. 1B) and DNA sequencing (data not shown) determined the correct orientation and nucleotide sequence of the serotype-specific capsid gene. When a number of independent isolates for each serotype-specific helper were compared to traditional type 2 helpers, vector yields varied significantly among these constructs (Table 1). Surprisingly, additions of serotype-specific non-capsid-coding sequences (5′ to the VP1 start site) were required to reduce this variation (see Fig. 1, Table 1, and Materials and Methods for further details). Additional analysis (data not shown) suggests that a Rep-specific capsid interacting domain is required for efficient serotype-specific encapsidation (unpublished data). These constructs were designated pXR1 to -5. Since these modified serotype-specific helper plasmids (as described in Fig. 1) produced vector yields within type 2 range (Table 1), they were further analyzed in detail for AAV Rep and capsid production, as well as for the ability to replicate AAV2 transgenes, as described below.
AAV hybrid helper functions.
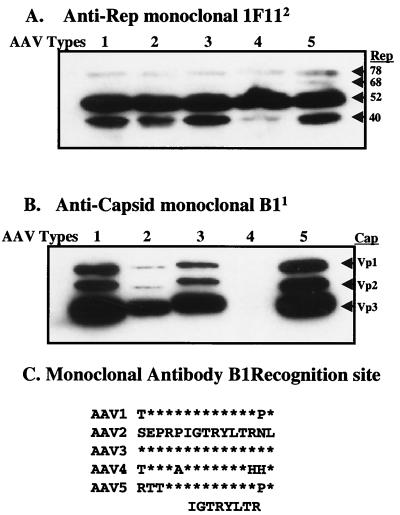
A series of experiments were carried out on the pXR1 to -5 helper plasmids to determine levels of type 2 rep- and serotype-specific capsid expression after transfection in 293 cells. Western blot analysis was used to detect the AAV rep gene products Rep78/68 and Rep52/40 (Fig. 2A) and the three capsid subunits Vp1, -2, and -3 (Fig. 2B) 24 h posttransfection. A comparison of AAV2 Rep proteins in the context of different serotype helper plasmids was carried out by using the monoclonal antibody 1F11 that recognizes each of the four AAV2 replication proteins (22). Previous studies demonstrated that the p5 mutation in the context of the helper plasmid pACG-2 downregulated the expression of Rep78/68, without affecting Rep52/40 (25). The results, shown in Fig. 2A, demonstrate that in each of the serotype-specific helper constructs all four Rep proteins were made at levels equivalent to those described for the original AAV2 helper construct pACG-2 (25). With additional analysis, we observed a lower level of Rep40 from the AAV4 helper construct. The exact reason for this observation remains unknown. The B1 monoclonal antibody (43), which recognizes the amino acid recognition sequence IGTRYLTR in AAV2 structural proteins (44), was used to identify the serotype-specific capsid subunits. This motif is conserved in all serotypes except AAV4 (Fig. 2C), which is evident by the lack of positive signal after Western analysis (Fig. 2B lane 4). For the other helper plasmids, all three capsid proteins were detected (Fig. 2B). Although the data provided constitute a representative example, we consistently observed higher amounts of the structural proteins from serotype 1 compared to the other helper constructs (repeated 10 times [data not shown]). Taken together, the results for replication and capsid protein expression for the five helper plasmids is within the range of AAV helper plasmids currently available for production (14).
FIG. 2.
Western blot of cell lysates from serotype-specific transfections. At 24 h after triple transfection, 5 μg of total protein was loaded into each well. After transfer the blots were incubated with the anti-Rep monoclonal antibody 1F11, with the sizes of the proteins listed on the right side (A), or the anticapsid monoclonal antibody B1, with the capsid subunits listed on the right side (B). The serotype-specific helpers used in the transfection are listed above each blot. (C) B1 recognition site as determined by Wobus et al. (44) is shown at the bottom of the figure. The amino acid sequence from this region for all five serotyes is shown. Asterisks indicate amino acids identical to those of type 2.
Replication and cross packaging of type 2 vectors.
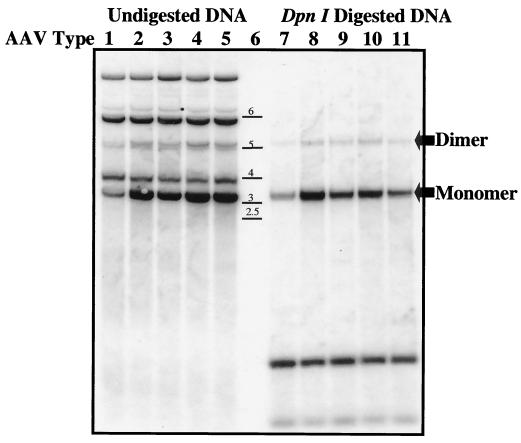
The functional activities of these helper proteins were determined by replication and encapsidation of the rAAV2 GFP template. The five serotype helper plasmids, the rAAV2 transgene pTR/CMV/GFP (packaging 3,400 bases), and the adenovirus helper plasmid (pXX6-80) were triple transfected into 293 cells, and Hirt analysis was carried out 24 h posttransfection. DpnI digestion, followed by Southern blot analysis with GFP gene-specific probe, determined the ratio of input plasmid to newly replicated vector DNA. All serotype-specific helpers replicated the vector template (Fig. 3, DpnI-digested lanes). Although nearly equivalent amounts of input DNA were observed in all lanes of undigested samples, replication of the transgene from XR2, as determined by phosphorimaging, produced the highest levels (Fig. 3, lane 2). Repeated replication analysis demonstrated that, while all other helpers were essentially equal, XR1 consistently generated 40% of XR2 replication levels (Fig. 3 lanes 7 and 8). Packaging efficiencies were determined by dot blot hybridization (Table 1). All helper constructs generated high vector yields within 1011 to 1012 particles/ml. In general, we observed that serotypes 1, 4, and 5 had yields within fourfold of each other after numerous production runs (five times), with serotypes 2 and 3 demonstrating the highest yields (9.8 × 1012 and 7.2 × 1011, respectively [Table 1]). The serotype 1 helper produced the least amount of virus (1.27 × 1011 [Table 1]), a finding consistent with Hirt replication analysis.
FIG. 3.
Hirt assay. Low-molecular-weight DNA was isolated from 293 cells, at 24 h after triple transfected with serotype-specific plasmids. Then, 2.5 μg of undigested (lanes 1 to 5) and DpnI-digested DNA (lanes 7 to 11) from each serotype sample was loaded onto a 1% agarose gel (lane 6, DNA ladder). After transfer the blot was probed with a 735-bp fragment of the GFP gene. Input DNA digested with DpnI reveals the replicating monomer and dimer transgene (arrowed). The lower bands in lanes 7 to 11 are DpnI digestion products of input plasmids.
Heparin column binding profiles of AAV hybrid serotypes.
The use of iodixanol gradient and/or heparin column binding has allowed for the rapid purification of rAAV2 (1, 50). In, addition, these methods allow for purification of vector with better transducing to particle numbers. This purification scheme was applied to the hybrid serotypes and elution profiles determined by GFP transduction as described in Materials and Methods.
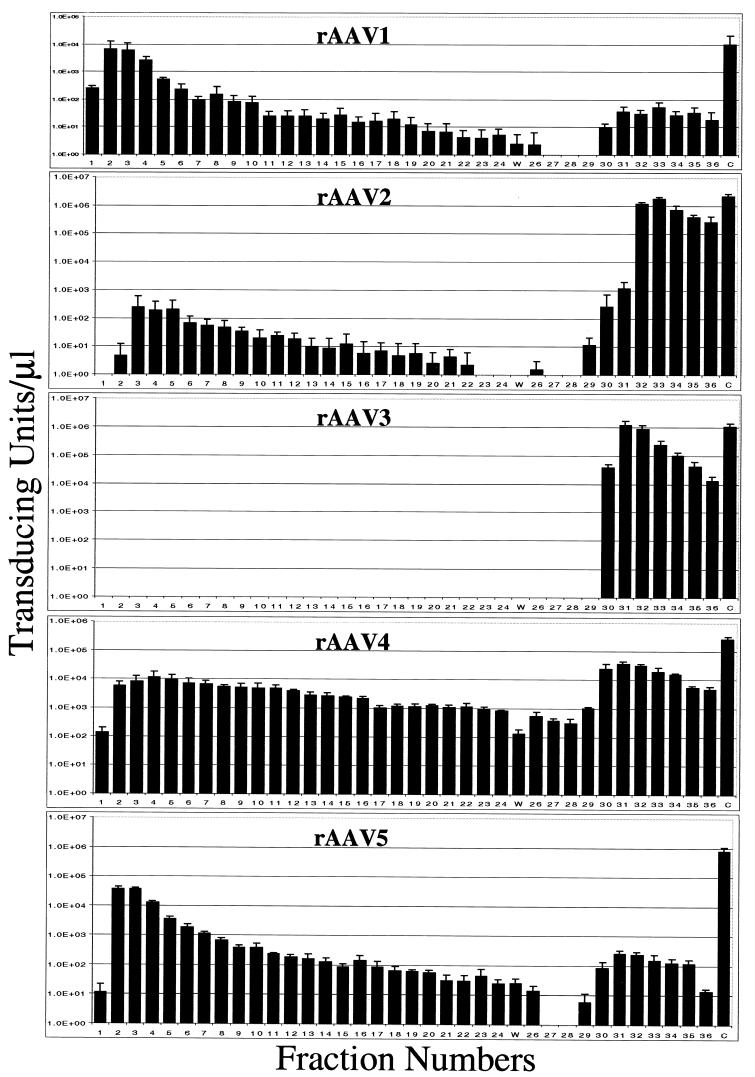
Recombinant AAV2 demonstrated elution profiles as previously published (50), with <1% of starting material recovered in the flowthrough and the majority of the TU being recovered in the elution step (Fig. 4, AAV2). rAAV3 displayed more efficient binding to the column and elution profiles nearly identical to those of type 2. These results suggest that the elution conditions used for rAAV2 purification can be applied to the hybrid rAAV3. Recombinant AAV1 and -5 displayed similar profiles to one another with 60% of type 1 and 80% of type 5 TU recovered in the flowthrough (Fig. 4, AAV1 and -5). The virus eluted from the salt gradient represents only a small portion of the total applied to the column (Fig. 4, AAV1 and -5). These data suggest that the heparin-Sepharose columns are not suitable for the purification of rAAV1 or rAAV5.
FIG. 4.
Transduction efficiency of fractions off a heparin-Sepharose affinity column for rAAV serotypes 1 through 5. The elution conditions were optimized originally for AAV2 (50). Numbered fractions were collected in 0.5-ml volumes, waste (W) was collected in a single 10-ml fraction, and the control (C) was a 1-ml aliquot of virus applied to the column. Infections were done in reference cell lines, with between 1/100 to 20 μl of each fraction. Salt elution began with fraction 26. Each bar represents the average of three separate infections, with the standard deviation indicated by an error bar.
The elution profile of rAAV4 was unique. Less than half of the TU were recovered in the flowthrough, yet the majority of TU were recovered in the elution step (Fig. 4, AAV4). These results suggest that if the salt conditions were altered, virus might be recovered in the elution step or that another ion-exchange column might improve the recovery of AAV serotype 4.
Hybrid vector transduction on rodent, monkey, and human cell lines.
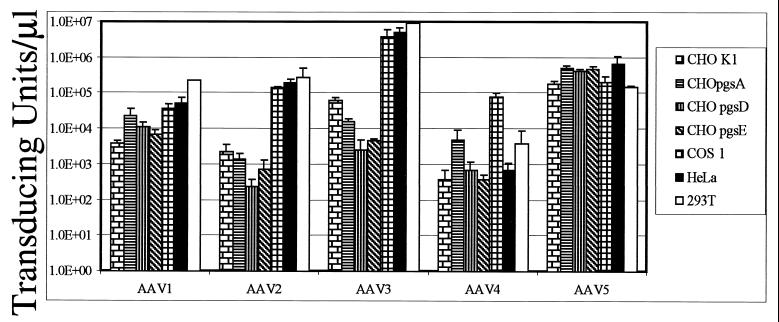
Various cell lines were infected with cesium gradient-purified hybrid vectors in an effort to determine the transducing titer in vitro (Fig. 5). Parental and mutant cell lines defective in heparan sulfate proteoglycan biosynthesis were analyzed for serotype-specific vector transduction since previous studies have demonstrated a role for this cell surface protein in type 2 infection. For rAAV1, a decrease in transduction on CHO pgsD cells, which are heparan sulfate deficient, was not observed. In fact, transduction efficiency for rAAV1 was nearly identical for all cell lines tested (Fig. 5, AAV1). The lack of specific binding to heparin columns and the ability to transduce mutant heparan sulfate cell lines suggest that type 1 entry is distinct from type 2 and yet to be identified. The transduction efficiency for rAAV2 and -3 were similar to what has been previously published (17, 40), with each showing a dependence on cell surface heparan sulfate (Fig. 5, AAV2 and -3) and efficient binding to heparan sulfate columns (Fig. 4, AAV2 and -3). Hybrid AAV5 transduction paralleled that of AAV1 for all cell lines tested. After completion of these studies, evidence identifying sialic acid as a cell surface molecule involved in type 5 transduction was published (23). The presence of sialic acid on the battery of cell lines tested here was not determined, although efficient transduction was observed. Hybrid vector type 4 also transduced all cell lines tested. However, the level of transduction never approached that observed on monkey-derived cell lines (Fig. 5, AAV4). The restriction in the in vitro host range that has been observed with both the hybrid and the traditional type 4 vectors (data not shown) suggests that efficient transduction requires a monkey-specific factor. Overall, these results suggest that, unlike types 2 and 3, AAV1, -4, and -5 do not require heparan sulfate for infection. These observations are in agreement with other studies evaluating the transduction requirements of non-AAV2 serotypes (8, 9, 46) and demonstrate the influence of the serotype-specific virion shells on vector transduction.
FIG. 5.
Transduction efficiency in CHO mutant and reference cell lines. Cells (cell types are specified in the legend on the right) were transduced with ca. 0.3 TU/cell, as determined by the reference cell lines (HeLa cells for AAV serotypes 1, 2, 3, and 5 and Cos1 cells for AAV4). The transducing titers are given for each serotype in each cell line. The particle numbers used in this experiment (per microliter) were as follows: rAAV1, 1.2 × 108; rAAV2, 5.6 × 108; rAAV3, 9.1 × 108; rAAV4, 2.3 × 108; and rAAV5, 5.4 × 108. Each bar represents the average of three separate infections, with the standard deviation indicated by an error bar.
Hierarchy of gene expresssion for AAV hybrid vectors in vivo.
The efficiency of gene expression of both therapeutic and marker transgenes were tested when delivered by each of the five hybrid vectors in vivo. The expression of human α1-antitrypsin (hAAT) and factor IX were tested in outbred and inbred mice, as well as immunodeficient mouse lines (Table 2). The hAAT expression was also examined with respect to the route of injection (i.e., portal vein, intramuscular, and intravenous) (Table 2). ELISA was used to monitor expression of the therapeutic transgenes which consistently demonstrated that AAV1 was superior to the other hybrid vectors tested. This observation was true for all strains of mice, routes of administration, or transgenes tested (Table 2). However, we did observe small differences in the efficiency of expression for the remaining serotypes with respect to each variable. For example, of the five serotypes tested, the rAAV4 vector was the least efficient in the expression of hAAT. In contrast, the rAAV2 vector was the least efficient in the expression of factor IX. These observations point to the importance of evaluating specific transgene and cis-acting regulatory sequences when certain serotype-specific virions for efficient trandsuction are compared.
TABLE 2.
Animal experiments with different serotypes of rAAV
| Genea | Mouse strain (route of injection)b | Score with serotypelc:
|
||||
|---|---|---|---|---|---|---|
| AAV1 | AAV2 | AAV3 | AAV4 | AAV5 | ||
| hAAT | C57BL (i.m.) | +++++ | +++ | ++ | + | ++++ |
| C57BL (p.v.) | +++++ | ++ | +++ | + | ++++ | |
| C57BL (i.v.) | +++++ | ++ | +++ | + | ++++ | |
| BALB/c (i.v.) | +++++ | ++++ | ++ | + | ++++ | |
| dF9 | Scid (i.m.) | +++++ | + | ++++ | +++ | ++++ |
| BALB/c (i.m.) | +++++ | + | ++ | +++ | +++ | |
The transgenes were chicken β-actin promoter with the CMV enhancer-driven hAAT (both C57/BL and BALB/c mice were at 5 × 1010 particles/mouse) and CMV immediate-early promoter-driven dog (d) factor IX (SCID mice at 2 × 1011 particles/mouse and BALB/c mice at 1010 particles/mouse).
For each group of animals, serotype, and route of injection a minimum of three animals were used. i.m., intramuscular injection; p.v., portal vein injection; i.v., intravenous injection.
Scores ranged from the maximum level of protein observed for each set of animals (+++++) to the lowest level of expression in the group (+).
Evaluation of AAV serotypes gene transfer efficiency in the rat retina.
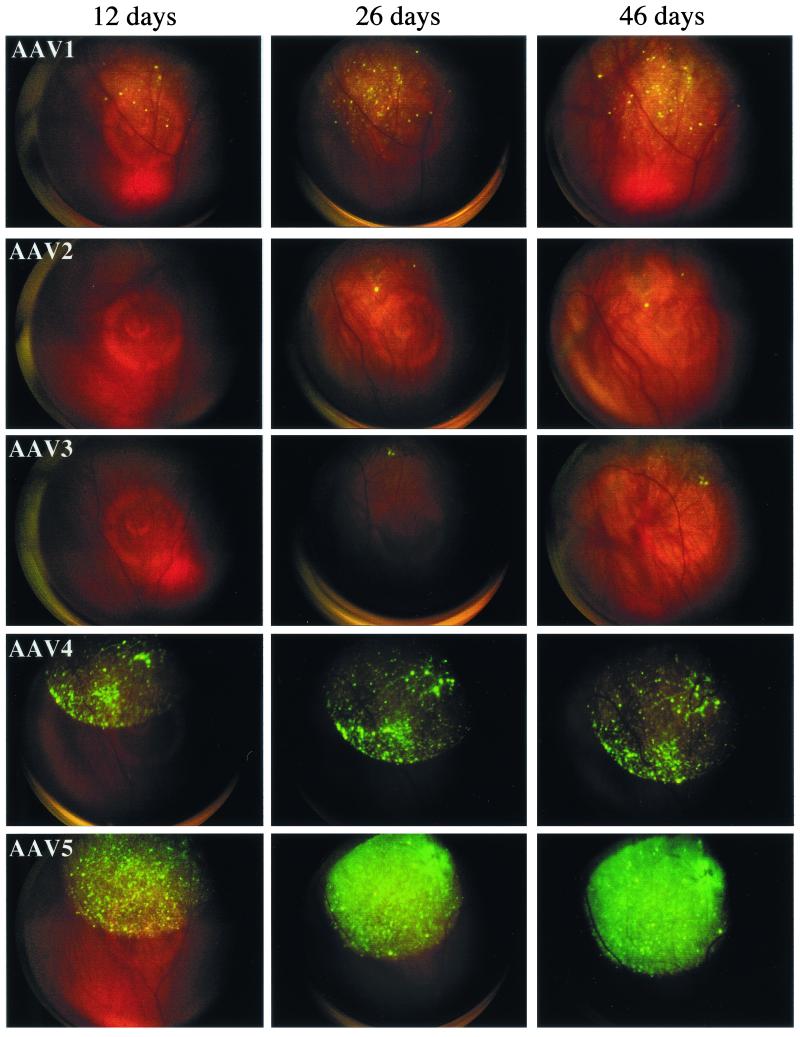
Previous studies have shown type 5 transduction to be more efficient in the brain compared to type 4 or 2 (11). In our study, type 1 vector appeared to be superior to all the others when tested in non-neuronal targets. We extended our hybrid vector analysis to neuronal targets by using rat retina as a substrate and GFP as a reporter. Each of the five serotype vectors was determined for efficient in vivo transduction by using fundus color photography after subretinal injections. At 12 days postinjection, GFP expression could be detected in rAAV5, -4 and -1-injected animals, with type 5 and 4 hybrids displaying the most intense GFP signal. No signal was detected in animals injected with rAAV2 or -3 at this concentration and time point (Fig. 6). At 26 days postinjection, GFP expression increased proportionally for rAAV5, -4, and -1, with types 2 and 3 eventually displaying a small but positive signal (Fig. 6, 26 days). This trend continued for up to 46 days (Fig. 6, 46 days), and these animals remained positive for the duration of the experiment (4 months). We also observed cell type-specific tropism when we used these hybrid vectors in the retina. The exact mechanism and cell types being transduced are being investigated. These results may be significant when an appropriate vector is chosen for a specific disease and also demonstrate, in addition to efficiency, features that may be advantageous when serotype-specific hybrid vectors are used.
FIG. 6.
Subretinal injection and in vivo fluorescence imaging. Subretinal injections were performed via a transcleral transchoroidal approach on wild-type Wistar rats, as previously described (33). Briefly, the sclera and the choroid were punctured, and a 33-gauge needle was then inserted in a tangential direction under an operating microscope. Three microliters of each of the five rAAV serotypes (5 × 1010 particles/ml) was delivered into the subretinal space of rats (n = 3). A new method using fundus photography has been developed and was performed here in order to control the accuracy and reproducibility of subretinal injections (Rolling et al., unpublished). GFP protein expression in live rats was monitored by fluorescent retinal imaging with a Canon UVI retinal camera connected to a digital imaging system (Lhedioph Win Software). Retinas were examined at 12, 26, and 46 days postinjection.
Based on these observations, a different hierarchy of serotype-specific transgene expression was observed in retina compared to non-neuronal tissues. In this setting, serotypes 5 and 4 were superior, followed by types 1, 2, and 3 (Fig. 6). Low to moderate levels of expression from AAV1 was in sharp contrast to data obtained in non-neuronal tissue. However, we cannot distinguish whether these differences are due to properties associated with the virions or simply species differences between rats and mice. Based on AAV2 transduction now established in rodents, dogs, rabbits, and monkeys, it is unlikely that the hybrid vector transduction efficiencies are species related, although additional nonrodent experiments will be required to completely rule out this concern. Taken together, the expression of therapeutic and marker genes from the five hybrid serotypes demonstrates that AAV1 is the superior vehicle for non-neuronal delivery, while in the retina and in the brain (11) AAV5 produces the highest levels of marker gene expression. All in all, these data demonstrate the importance of determining serotype-specific transduction in vivo when AAV is being considered as the delivery system of choice.
DISCUSSION
The serotypes of AAV have the potential to become important resources for clinical gene therapy. In an effort to compare the role of serotype-specific virion shell on vector transduction, we cloned each of the serotype capsid coding domains into a common vector backbone containing AAV2 replication genes. This strategy allowed the packaging of AAV2 ITR vectors into each serotype-specific shell. We reasoned that it would not be uncommon to expect each of the serotype-specific cis-acting terminal repeats to have unique influences on transgene expression. In support of this concept, the potential to alter transgene specific promoter activity with ITR elements both in vitro and in vivo has been observed (12, 15). Therefore, we chose the AAV2 ITRs for our common cis-acting vector sequences since these elements are best understood of the five serotypes. Using type 2 vectors and the hybrid AAV helper strategy described above, we in effect normalized the contribution of cis-acting sequences on vector transduction. We also focused only on AAV serotypes 1 to 5 shells since immunological studies have now determined that AAV1 and -6 are indistinguishable (C. Li et al., unpublished data). In addition to determining the role of the hybrid capsid shell in vector transduction, this approach has facilitated an effort for standardizing AAV serotype-specific vector production. Based on this strategy, similar studies can now be carried out to determine the role of serotype-specific cis-acting ITRs (i.e., type 5 ITRs in AAV1 to -5 shells). The outcome of such studies may determine still more features and combinations of hybrid vectors that enhance or influence efficient transduction. These combinations will be important in determining the best AAV-derived delivery reagent for use in humans.
Our initial analysis of AAV3, -4, and -5 hybrid helper constructs resulted in reduced ability to generate high-titer vectors. Recent studies have demonstrated that the first 232 amino acids of AAV2 rep is required for origin specific binding when the AAV2/Goose parvovirus rep chimera is used (48). With respect to the type 5 hybrid construct, we were able to use as few as the first 250 amino acids of AAV2 rep to efficiently replicate and package rAAV2 ITR containing templates into type 5 virions. These results suggest that sequences downstream of the Rep78/68 DNA-binding domain are required for efficient encapsidation. In support of this concept, previous studies have shown that the small Rep proteins have helicase activity (38) and that this activity is required for efficient packaging of the viral genome into preformed capsids (24). Therefore, we would predict that inclusion of serotype-specific small Rep proteins would be sufficient for efficient encapsidation. Our results are consistent with this hypothesis, because substitution of AAV5 sequences downstream of the BamHI site in pXR5 dramatically increased virus production. Similarly, substitution of AAV3 and AAV4 sequences downstream of the AccI site in pXR3 and pXR4, respectively, improved virus production 10- to 1,000-fold. Because these sequences all lie downstream of the p19 promoter within coding sequences shared between the large Rep proteins and the small Rep proteins, their effect on DNA encapsidation may be due to domain in the small Rep proteins that interacts with the capsids in a sertotype-specific manner (unpublished data). Due to high sequence similarity between type 2 and 1 rep sequences, no additional modification were made to this helper construct. Our vector yields with this hybrid helper were in agreement with that generated from type 1 helper constructs only (data not shown; see also reference 46).
The ability to produce recombinant virus depends on the function of the replication and capsid proteins. Protein analysis demonstrated that nearly equivalent amounts of Rep proteins were generated for all of the serotypes. With respect to capsid proteins, rAAV1 and -5 consistently expressed higher levels (Fig. 2B, lanes 1 and 5). We were unable to assess the AAV4 capsid due to the absence of antibody recognition epitope (Fig. 2B, lane 4, and C). Previous studies have suggested a delicate balance between replication, capsid proteins, and efficient packaging (25, 42). We did not observe a correlation between capsid level and vector yield; instead, the level of virions produced tracked more closely with transgene replication (Fig. 3). This was most obvious with type 1 helper (Fig. 3 and Table 1). Although vector replication appeared to be uniform for helper constructs 2 to 5, type 1 was consistently lower (40% of type 2). These differences were observed in multiple experiments and are difficult to explain since the vector ITR and Rep proteins are all type 2 derived.
Regardless of the vector yields, these new helper constructs allowed for further examination with respect to virion purification and influence of serotype-specific virion shell on transducing titer in vitro and in vivo.
The ability to bind heparan sulfate has changed the method of choice for AAV2 purification from cesium chloride isopycnic centrifugation, which has been shown to damage AAV2, to column chromatography (1, 40, 50). These methods have resulted in lower ratios of particles to transducing numbers and may have application to the other serotypes. Using heparin column purification conditions, we determined that AAV2 and -3 could be efficiently purified by heparin-Sepharose column chromatography with >91% of the TU being recovered (Fig. 4). In contrast, <1% of AAV1 and -5 were recovered during the elution step. Interestingly, with respect to type 1 binding, Wu et al. (45) described two substitution mutants in the AAV2 capsid that resulted in amino acid changes which lost heparan column binding, of which the first was located between amino acids 561 to 565 DEEEI and is conserved in AAV1. The second change (from positions 585 to 588, i.e., RGNR) is SSST in AAV1, suggesting that both or either of these may be responsible for loss of heparan sulfate binding. With respect to AAV5, recent studies have determined that sialic acid serves as a receptor for vector transduction, providing a reason for lack of heparan sulfate binding. AAV4 did not demonstrate any specific binding with the heparan sulfate columns based on elution profiles but appears to interact more strongly with the column matrix, as evidenced by the relatively large recovery of TU in the flowthrough, wash, and waste. Although it does not appear that either AAV1 or -5 will be efficiently purified with the heparin-Sepharose column, if binding conditions for AAV4 were altered it may be possible to purify this serotype by heparin chromatography. Our results suggest the need for a universal purification system, potentially based on ion-exchange chromatography; however, for in vitro and in vivo transduction analysis, we relied on conventional CsCl purification.
Reference cell lines were used to establish the titers of the serotypes. We defined these as cell lines that consistently showed high levels of transduction, ease of counting, good plating efficiency, and consistent susceptibility to helper virus coinfection. HeLa cells were used as the reference cell line for four of the serotypes—AAV1, -2, -3, and -5—and Cos1 cells for AAV4. Using the titers established in the reference cell lines as a guide, we transduced cell lines defective in proteoglycan biosynthesis to determine the vector proteoglycan binding properties (40). Consistent with heparan sulfate column affinity, AAV serotypes 2 and 3 showed a dependence on heparan sulfate proteoglycans for efficient transduction, as previously observed (17, 40). Interestingly, both serotypes showed nearly equivalent changes (>1,000 times) in susceptibility to transduction when the mutant CHO pgsD and HeLa cell line titers were compared (Fig. 5, AAV2 and -3, CHO pgsD versus HeLa). When the mutant to parental CHO lines were evaluated, we observed a 10- to 25-fold change in transduction (Fig. 5, AAV2 and -3, CHO K1 versus CHO pgsD). Based on this analysis, the CHO cell appears to be suboptimum for AAV2 and -3 vector transduction, suggesting that additional step(s) may be required. This is not the case for type 1 and 5 virions. Originally, it was thought that AAV1 bound heparan agarose (31); however, the transduction efficiency in CHO pgsD cells was only fivefold less than the reference cell line and threefold more efficient than the CHO K1 line (Fig. 5). In contrast, AAV4 and AAV5 do not show a dependence on heparan sulfate for transduction (Fig. 5), where the CHO mutant lines were transduced better than or the same as the parental CHO K1 line, respectively. The serotypes show preferences in vitro for transduction of specific cell lines (Fig. 5), so we then assayed these reagents in vivo to further define vector performance.
Based on recent studies, a hierarchy for both qualitative and quantitative expression of AAV serotype vectors is being elucidated (6, 11, 46, 49). In our experiments, we determined that AAV1 was superior in liver and muscle. In contrast, others have reported that AAV2 was more efficient than AAV1 in transducing the liver (46). At present we do not have a simple explanation for this discrepancy, but we have now tested numerous routes of administration and various transgenes that consistently result in superior expression from type 1 vectors in the liver (Table 2). Interestingly, we observe a change in the order of efficient transduction when hybrids were tested in neuronal tissue. Expression from rAAV5 has been shown to be most efficient in the brain compared to types 2 and 4, and our studies have now determined that this is also true for retina (11, 49) (see also Fig. 6). Surprisingly, we observed that type 4 expression in the retina was far more efficient than that of type 1, 2, or 3. This observation appears to be distinct compared to brain and other non-neuronal tissue (Fig. 6 and Table 2) (7, 11, 49). All of these data begin to support the need for additional studies analyzing serotype-specific vector distribution before we can extrapolate conclusions to other animal models or humans. For diseases such as diabetes, cystic fibrosis, muscular dystrophy, Alzheimers, and cancer, etc., the transgene target may become an important component when one is determining which serotype shell to use. Previous analysis of type 2 transduction in animal models translated from rodent, to canine, to nonhuman primate (29) and have now been extended in humans through a phase I safety trial (19). If serotype-specific vectors have similar attributes, then the results of this study strongly suggest that hybrids AAV1 and -5 are superior delivery agents for non-neuronal and neuronal targets, respectively.
The serotypes of AAV have the potential to become powerful tools for safe gene delivery to the target of interest. The ability to better compare and contrast that potential will allow for a more considered choice of delivery reagent for specific gene therapy applications. These plasmids represent the first set of AAV serotype helpers that can be used in this way. The construction of five serotypes in the same genetic backbone, packaging an identical transgene with common serotype ITRs, will now provide researchers the ability to make these comparisons.
Acknowledgments
We acknowledge the technical assistance of Angelique Camp and Karissa Gable in this study. We also thank Doug McCarty for reading the manuscript before its submission and the reviewers for constructive comments. We thank Terry Flotte for AAT and Jurgen Kleinschmidt for providing B1 monoclonal antibody.
This work was supported by NIH grants DK54419 and GM59290.
REFERENCES
- 1.Auricchio, A., M. Hildinger, E. O’Conner, G. Gao, and J. Wilson. 2001. Isolation of highly infectious and pure adeno-associalted virus type 2 vectors with a single-step gravity-flow column. Hum. Gene Ther. 12:71–76. [DOI] [PubMed] [Google Scholar]
- 2.Bantel-Schaal, U., H. Delius, R. Schmidt, and H. zur Hausen. 1999. Human adeno-associated virus type 5 is only distantly related to other known primate helper-dependent parvoviruses. J. Virol. 73:939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74:2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett, J. S., J. Kleinschmidt, R. S. Boucher, and R. J. Samulski. 1999. Targeted adeno-associated virus vector transduction of non-permissive cells mediated by bispecific F(ab′γ)2 antibody. Nat. Biotechnol. 17:181–186. [DOI] [PubMed] [Google Scholar]
- 5.Carter, B. J., and C. A. Laughlin. 1984. Adeno-associated virus defectiveness and the nature of the adenovirus helper function, p.67–152. In K. I. Berns (ed.), The parvoviruses. Plenum Press, New York, N.Y.
- 6.Chao, H., Y. Liu, J. Rabinowitz, C. Li, R. J. Samulski, and C. E. Walsh. 2000. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2:619–623. [DOI] [PubMed] [Google Scholar]
- 7.Chao, H., L. Mao, A. Bruce, and C. Walsh. 2000. Sustained expression of human factor VIII in mice using a parvovirus-based vector. Blood 95:1594–1599. [PubMed] [Google Scholar]
- 8.Chiorini, J., L. Yang, Y. Liu, B. Safer, and R. Kotin. 1999. Cloning and characterization of adeno-associated virus type 5. J. Virol. 73:1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiorini, J. A., L. Yang, Y. Liu, B. Safer, and R. M. Kotin. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 71:6823–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiorini, J. A., B. Zimmermann, L. Yang, R. H. Smith, A. Ahearn, F. Herberg, and R. M. Kotin. 1998. Inhibition of PrKX, a novel protein kinase, and the cyclic AMP-dependent protein kinase PKA by the regulatory proteins of adeno-associated virus type 2. Mol. Cell. Biol. 18:5921–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson, B., C. Stein, J. Heth, I. Martins, M. Kotin, T. Derksen, J. Zabner, A. Ghodsi, and J. Chiorini. 2000. Recombinent adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous sytsem. Proc. Natl. Acad. Sci. USA 97:3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flotte, T. R., S. A. Afione, R. Solow, M. L. Drumm, D. Markakis, W. B. Guggino, P. L. Zeitlin, and B. J. Carter. 1993. Expression of the cystic fibrosis transmembrane conductance regulator from a novel adeno-associated virus promoter. J. Biol. Chem. 268:3781–3790. [PubMed] [Google Scholar]
- 13.Girod, A., M. Ried, C. Wobus, H. Lahm, K. Leike, J. Kleinschmidt, G. Deleage, and M. Hallek. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 5:1052–1056. [DOI] [PubMed] [Google Scholar]
- 14.Haberman, R., G. Kroner-Lux, T. McCown, and R. J. Samulski. 1999. Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration, p.4.17.1–4.17.25. In J. Crawley, C. Gerfen, M. Rogawski, D. Sibley, P. Skolnick, and S. Wray (ed.), Current protocols in neuroscience, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 15.Haberman, R., T. McGown, and R. J. Samulski. 2000. Novel transcriptional regulatory signals in the adeno-associated virus terminal repeat A/D junction element. J. Virol. 74:8732–8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halbert, C. L., E. A. Rutledge, J. M. Allen, D. W. Russell, and A. D. Miller. 2000. Repeat transduction in the mouse lung by using adeno-associated virus vectors with different serotypes. J. Virol. 74:1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handa, A., S. Muramatsu, J. Qiu, H. Mizukami, and K. Brown. 2000. Adeno-associated virus (AAV)-3-based vectors transduce haematopoietic cells not susceptible to transduction with AAV-2-based vectors. J. Gen. Virol. 81:2077–2084. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, J., K. Qing, and A. Srivastava. 2001. Adeno-associated virus type 2-mediated gene transfer: Altered endocytic processing enhances transduction efficiency in murine fibroblasts. J. Virol. 75:4080–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kay, M., C. Manno, M. Ragni, P. Larson, L. Couto, A. McClelland, B. Glader, A. Chew, S. Tai, R. Herzog, V. Arruda, F. Johnson, C. Scallan, E. Skarsgard, A. Flake, and K. High. 2000. Evidence for gene transfer and expression of factor IX in hempophilia B patients treated with an AAV vector. Nat. Genet. 24:257–261. [DOI] [PubMed] [Google Scholar]
- 20.Hildinger, M., A. Auricchio, G. Gao, L. Wang, N. Chirmule, and J. Wilson. 2001. Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J. Virol. 75:6199–6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365–369. [DOI] [PubMed] [Google Scholar]
- 22.Hunter, L. A., and R. J. Samulski. 1992. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J. Virol. 66:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaludov, N., K. Brown, R. Walters, J. Zabner, and J. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King, J., R. Dubielzig, D. Grimm, and J. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, J., R. J. Samulski, and X. Xiao. 1997. Role for highly regulated rep gene expression in adeno-associated virus vector production. J. Virol. 71:5236–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsushita, T., S. Elliger, C. Elliger, G. Podsakoff, L. Villarreal, G. J. Kurtzman, Y. Iwaki, and P. Colosi. 1998. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 5:938–945. [DOI] [PubMed] [Google Scholar]
- 27.Michalski, J., C. McCombs, S. Sheth, M. McCarthy, and R. deShazo. 1985. A modified double antibody sandwich enzyme-linked immunosorbent assay for measurement of alpha-1-antitrypsin in biologic fluids. J. Immunol. Methods 83:101–112. [DOI] [PubMed] [Google Scholar]
- 28.Monahan, P., and R. Samulski. 2000. AAV Vectors: is clinical success on the horizon? Gene Ther. 7:24–30. [DOI] [PubMed] [Google Scholar]
- 29.Monohan, P., and R. J. Samulski. 2000. Adeno-associated virus vectors for gene therapy: more pros than cons? Mol. Med. Today 6:433–440. [DOI] [PubMed] [Google Scholar]
- 30.Muramatsu, S., H. Mizukami, N. Young, and K. Brown. 1996. Nucleotide sequencing and generation of an infectious clone of adeno-accociated virus 3. Virology 221:208–217. [DOI] [PubMed] [Google Scholar]
- 31.Rabinowitz, J., W. Xiao, and R. J. Samulski. 1999. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology 265:274–285. [DOI] [PubMed] [Google Scholar]
- 32.Rabinowitz, J. E., and J. Samulski. 1998. Adeno-associated virus expression systems for gene transfer. Curr. Opin. Biotechnol. 9:470–475. [DOI] [PubMed] [Google Scholar]
- 33.Rolling, F., W. Shen, H. Tabarias, I. Constable, Y. Kanagasingam, C. Barry, and P. Rakoczy. 1999. Evaluation of adeno-associated virus-mediated gene transfer into the rat retina by clinical fluorescence photography. Hum. Gene Ther. 10:641–648. [DOI] [PubMed] [Google Scholar]
- 34.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salvetti, A., S. Oreve, G. Chadeuf, D. Favre, Y. Cherel, P. Champion-Arnaud, J. David-Ameline, and P. Moullier. 1998. Factors influencing recombinant adeno-associated virus production. Hum. Gene Ther. 9:695–706. [DOI] [PubMed] [Google Scholar]
- 36.Samulski, R. J., L.-S. Chang, and T. Shenk. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 63:3822–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samulski, R. J., M. Sally, and N. Muzyczka (ed.). 1999. Adeno-associated viral vectors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Smith, R. H., and R. M. Kotin. 1998. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J. Virol. 72:4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Summerford, C., and R. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associted virus type 2 virions. J. Virol. 72:1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walters, R. W., D. Duan, J. F. Engelhardt, and M. J. Welsh. 2000. Incorporation of adeno-associated virus in a calcium phosphate coprecipitate improves gene transfer to airway epithelia in vitro and in vivo. J. Virol. 74:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weger, S., A. Wistuba, D. Grimm, and J. A. Kleinschmidt. 1997. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J. Virol. 71:8437–8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wobus, C., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao, W., N. Chirmule, S. C. Berta, B. McCullough, G. Gao, and J. M. Wilson. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon, M., D. Smith, P. Ward, F. Medrano, A. Aggarwal, and R. Linden. 2001. Amino-terminal domain exchange redirects origin-specific interactions of adeno-associated virus rep78 in vitro. J. Virol. 75:3230–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zabner, J., M. Seiler, R. Walters, R. M. Kotin, W. Fulgeras, B. L. Davidson, and J. A. Chiorini. 2000. Adeno-associated virus type 5 (AAV5) but not AAV2 binds to the apical surfaces of airway epithelia and facilitates gene transfer. J. Virol. 74:3852–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zolotukhin, S., B. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973–985. [DOI] [PubMed] [Google Scholar]