Abstract
To evaluate host range differences between two different strains of feline leukemia virus subgroup B (FeLV-B), we compared the binding and infectivity patterns of retrovirus vectors bearing either FeLV-B-90Z or FeLV-B-GA envelopes. We report here that the ability of these envelopes to utilize different Pit1 orthologs is mediated primarily by the receptor binding domain; however, in the case of FeLV-B-90Z, the C terminus also contributes to the recognition of certain Pit1 orthologs.
Feline leukemia virus (FeLV) is a naturally occurring gammaretrovirus of domestic cats often associated with degenerative diseases of the hematopoietic system, immunodeficiency, and neoplasia (13). The surface glycoproteins (SU) of FeLV are of interest because not only do these viral envelope proteins mediate attachment and subsequent infection of target cells, but as a result, they are critical determinants of disease specificity as well. Receptor recognition by gammaretrovirus envelope proteins has been attributed to the N-terminal half of SU, which contains two variable regions termed VRA and VRB (4-6, 29). This region is collectively known as the receptor binding domain (RBD) and is separated from the C-terminal half of SU by a conserved proline-rich region found in all gammaretrovirus envelope proteins. Work with murine retrovirus envelopes indicates that the C-terminal domain of SU may play a role in fusion activation and perhaps other postbinding events (23, 37). In the case of subgroup B FeLV envelopes, sequences in the C-terminal half of SU have been shown previously to be important in influencing their ability to use Pit2 as a receptor (1, 37).
FeLV is categorized into four subgroups (FeLV-A, -B, -C, and -T) (19, 27, 32, 36). The FeLV-B viruses represent a heterogeneous population of FeLVs that evolved by recombination of exogenous FeLV-A with different portions of endogenous feline retroviral envelope gene sequences (enFeLV) (11, 30, 34). FeLV-A is thought to be the transmissible form of the virus (14, 17, 18); its receptor remains to be identified. FeLV-A infects primarily feline cells, whereas FeLV-B isolates also infect many heterologous (nonfeline) cells (19, 27). Acquisition of endogenous FeLV sequences in SU results in the ability of FeLV-B to infect a variety of cells by using Pit1 as its receptor (42) and, in the case of feline cells, the feline Pit2 ortholog as well (1, 35, 42).
Pit1, the receptor for FeLV-B as well as gibbon ape leukemia virus (GALV), woolly monkey virus (WMV), and the 10A1 murine leukemia virus (MuLV), normally functions as a type III phosphate transporter in human cells and shares approximately 62% amino acid identity with Pit2, another transmembrane protein with a similar transporter function. Pit2 serves as the receptor for amphotropic MuLV (A-MuLV) and 10A1 MuLV (26, 28, 42, 48). Both Pit1 and Pit2 are ubiquitously expressed in a wide variety of cell types, resulting in the broad cellular tropism exhibited by retroviruses such as GALV and A-MuLV that co-opt these proteins to serve as their viral receptors. FeLV-B and GALV are classified as members of the same gammaretrovirus family. Despite their common receptor usage, GALV and FeLV-B have distinct host ranges (35, 42).
Although the FeLV-B and GALV envelope glycoproteins utilize Pit1 (28, 42), they lack significant homology in the N-terminal region of SU (9, 11). FeLV-B isolates vary in the amount of feline endogenous envelope sequences that they contain and in single residue differences scattered throughout the rest of their SU regions. Two representative strains of FeLV-B are 90Z (FeLV-B-90Z) (6), which contains enFeLV throughout SU, and Gardner-Arnstein (FeLV-B-GA) (47), which contains enFeLV only in the N-terminal two-thirds of SU, with the C-terminal one-third comprised of FeLV-A sequence. Both FeLV-B-90Z and FeLV-B-GA virus strains utilize Pit1 to infect human cells (6, 42).
The primary host range and receptor determinants of FeLV-B map to VRA and VRB in the SU region of the viral envelope (1, 6, 37). For example, an arginine in place of glutamine at position 73 within VRA was shown elsewhere to be necessary for viral entry into cells via the human Pit2 receptor (37). These recent studies also identified the C terminus of SU as being an important determinant for the use of human Pit2 by certain FeLV-Bs (37). While the FeLV-B envelope determinants required for human Pit1 and Pit2 receptor usage have been characterized in some detail, the influence of these determinants on FeLV-B host range remains largely unresolved. Studies addressing these determinants may provide insight into how specific regions in the SU of the envelope confer host range and specific recognition of various Pit1 orthologs. Initial observations in our laboratory indicate that FeLV-B-90Z and FeLV-B-GA enveloped vectors exhibit distinct infectivity patterns, not only among different species but also among tissue types within the same species. We therefore sought to examine the specific regions and sequences within the FeLV-B envelope that modulate FeLV-B-90Z and FeLV-B-GA cellular tropism.
Rabbit SIRC cells are susceptible to FeLV-B vectors.
In previous host range studies, rabbit SIRC cells (ATCC CCL60) were shown to express a functional Pit1 ortholog, as these cells were susceptible to infection by GALV and GALV pseudovirions (35, 42, 45). Interestingly, in the same studies SIRC cells were found to be resistant to FeLV-B pseudovirions and this resistance was attributed to either a nonfunctional receptor for FeLV-B or a virus-specific postreceptor block in replication (42). In our lab, we have found that SIRC cells are susceptible to FeLV-B-90Z pseudotyped retrovirus vectors. In order to determine if this discrepancy in SIRC cell susceptibility was FeLV-B strain specific, retrovirus vector stocks of FeLV-B-90Z and FeLV-B-GA carrying a selectable marker (lacZ) were prepared as described elsewhere (37, 43). Target cells were seeded into 12-well dishes at 4 × 104 cells per well. The following day, the medium was removed and viral supernatant containing Polybrene (10 μg/ml) was added to each well. Twenty-four hours later the medium was changed and cells were cultured for an additional 24 to 48 h before analysis for expression of β-galactosidase by histochemical staining with X-Gal (5-bromo-4 chloro-3-indolyl-β-d-galactopyranoside) (46).
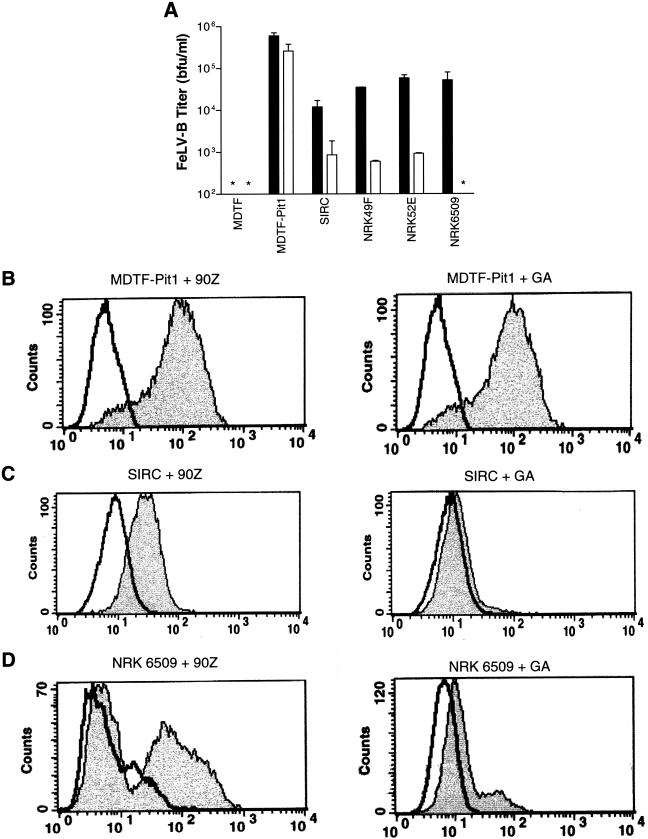
MDTF cells expressing Pit1 (MDTF-Pit1) were used as positive controls for infection, as the murine orthologs of Pit1 and Pit2 are not functional FeLV-B receptors (10). Expression of human Pit1 renders MDTF cells susceptible to infection by both FeLV-B-90Z and FeLV-B-GA (37). We found that FeLV-B-90Z enveloped vectors infect SIRC cells, albeit one-half log less efficiently than they infect MDTF-Pit1 cells (Fig. 1). Vectors bearing FeLV-B-90ZRBD envelopes, which contain the 90Z RBD with the C-terminal region of the SU replaced with the corresponding sequence from FeLV-A, also infect SIRC cells at levels nearly identical to those of control cells (data not shown). In contrast, FeLV-B-GA enveloped vectors infect SIRC cells but at significantly reduced titers (2.5-log reduction) compared to those of FeLV-B-GA infection of MDTF-Pit1 cells. In addition, SIRC cells are significantly less susceptible to infection by FeLV-B-GA vectors (8.6 × 102 blue focus units [bfu]/ml) than by FeLV-B-90Z enveloped vectors (1.2 × 104 bfu/ml) and FeLV-B-90ZRBD (2.2 × 105 bfu/ml; data not shown). Of note, titers that we obtained from control MDTF-Pit1 cells with either FeLV-B-90Z or FeLV-B-GA vectors were 3 logs higher than titers reported previously (Fig. 1 and Table 1 in reference 42). The 3-log difference in titers observed in our study is at the lower threshold of sensitivity in the previous studies, which failed to discriminate between inefficient infection and resistance. Titers obtained with FeLV-B pseudovirions (42) are generally lower than those obtained with retroviral vectors. Finally, defined molecular clones of the FeLV-B-90Z and FeLV-B-GA envelopes were used to produce the retrovirus vectors reported herein, whereas the composition of the FeLV-B envelope used by Takeuchi et al. (42) to produce the pseudovirions was unclear. Based on these results, it is likely that an isolate other than FeLV-B-90Z was used in the previous studies.
FIG. 1.
Susceptibility of various cell lines to infection by FeLV-B correlates with cell surface binding. (A) Cells were infected with retrovirus vectors packaging a genome containing the lacZ gene and bearing either FeLV-B-90Z (solid bars) or FeLV-B-GA (open bars) envelopes. Titers are expressed as the mean numbers of bfu per milliliter ± the standard errors of the means and are based on the results of experiments done in triplicate; asterisks indicate a titer of <10 bfu/ml. (B to D) Histograms from flow cytometric analysis of cells stained with fluorescein-conjugated monoclonal antibody HA.11, recognizing soluble HA-tagged FeLV-B-90Z and FeLV-B-GA SU. For each panel, the x axis represents fluorescence intensity (log scale) and the y axis represents cell number. Areas under bold black lines represent negative control cells which were incubated with medium only, followed by addition of fluorescein-conjugated HA.11 antibody. Gray-shaded areas correspond to MDTF-Pit1 (B), SIRC (C), or NRK 6509 (D) cells exposed to either FeLV-B-90Z SU or FeLV-B-GA SU.
TABLE 1.
Infection of cells with FeLV-B-90Z- and FeLV-B-GA-derived vectors
The inability of FeLV-B-GA vectors to efficiently infect SIRC cells could be attributed to either a binding or a postbinding block. To determine if the block to FeLV-B-GA infection of SIRC cells was at the level of binding, a fluorescence-activated cell sorting (FACS) binding assay was used to measure FeLV-B-90Z, FeLV-B-90ZRBD, and FeLV-B-GA SU binding to various target cells (12). Soluble forms of these envelopes fused to tandem C-terminal hemagglutinin (HA) epitope tags (22) were incubated with MDTF, MDTF-Pit1, and SIRC cells. As shown previously, both FeLV-B-90Z and FeLV-B-GA SU bound efficiently to MDTF-Pit1 cells (Fig. 1B) but did not bind to MDTF cells (data not shown; 22, 37). A significant shift in fluorescence intensity occurred when SIRC cells were incubated with either FeLV-B-90Z or FeLV-B-90ZRBD SU, indicating efficient binding, correlating with the ability of this vector to infect SIRC cells (Fig. 1A and C; data not shown). In contrast, FeLV-B-GA SU did not efficiently bind to SIRC cells (Fig. 1C), indicating that the decreased titer of FeLV-B-GA enveloped vectors on SIRC cells correlates with inefficient binding of this SU to the rabbit ortholog of Pit1 present on SIRC cells. Therefore, the inability of FeLV-B pseudotypes to infect SIRC cells does not appear to be due to nonfunctional receptors or a postbinding block to infection as had been previously proposed (42). In addition, use of vectors bearing FeLV-B-GA envelopes resulted in a significant reduction in titer compared to that with FeLV-B-90Z, showing that decreased FeLV-B-GA binding correlates with the decreased titer observed with FeLV-B-GA compared to FeLV-B-90Z. As stated above FeLV-B-GA differs from FeLV-B-90Z in its RBD as well as in the composition of its C-terminal region of the SU, which derives from FeLV-A, whereas the C-terminal region of FeLV-B-90Z derives entirely from enFeLV sequences. To determine whether differences in RBD or C-terminal regions of the SU were responsible for the decreased binding and titer seen with FeLV-B-GA compared with those of FeLV-B-90Z, we examined the ability of FeLV-B-90ZRBD enveloped vectors to bind and infect SIRC cells (data not shown). The RBDs of FeLV-B-90ZRBD and FeLV-B-90Z are identical; however, similarly to FeLV-B-GA, the C terminus of FeLV-B-90ZRBD is derived from FeLV-A. FeLV-B-90ZRBD can bind and infect SIRC cells with the same efficiency as that of FeLV-B-90Z (data not shown), suggesting that, in the case of SIRC cells, the RBD, and not the C-terminal FeLV-A sequence in the SU, is the main factor influencing the expanded host range observed for FeLV-B-90Z enveloped vectors.
NRK cells exhibit differences in susceptibility to FeLV-B-90Z and FeLV-B-GA enveloped vectors.
It has also been previously reported that rat cells, notably normal rat kidney (NRK) cells, are resistant to infection by FeLV-B (41, 42). Resistance of NRK cells to FeLV-B was ascribed to a difference in the amino acid sequence of region A between the rat and human orthologs of Pit1 (41). Other rat cells, such as rat XC cells, were reported elsewhere to be weakly susceptible to FeLV-B (42). This difference in susceptibility of rat XC cells to FeLV-B was attributed to several possibilities, including sequence variation in the rat XC and NRK Pit1 orthologs (8, 16, 44), differential receptor expression levels, or increased efficiency of infection at a step other than receptor binding (41). It was unclear from these prior studies which FeLV-B isolate was used, nor was the derivation of NRK cells known.
We compared the susceptibilities of NRK cells derived from rat fibroblasts (NRK 49F, ATCC CRL 1570), rat epithelium (NRK 52E, ATCC CRL 1571), and normal rat kidney (NRK 6509, ATCC CRL 6509) to FeLV-B-90Z or FeLV-B-GA enveloped retrovirus vectors by β-galactosidase histochemical assay. The results show that NRK 49F and 52E cells are susceptible to infection by FeLV-B-90Z vectors; however, titers were 1 log lower than those for control MDTF-Pit1 cells (Fig. 1A). In contrast, infection of these cells by FeLV-B-GA enveloped vectors resulted in titers 2.5-log-fold lower than those for MDTF-Pit1. NRK 6509 cells were found to be resistant to infection by FeLV-B-GA enveloped vectors, but titers were equivalent to those for NRK 49F and 52E cells when exposed to FeLV-B-90Z vectors (Fig. 1A). These data suggest that various NRK cell lines derived from different tissue types, but from the same species, can vary in their susceptibility to FeLV-B infection. Of the three NRK cell lines tested, only NRK 6509 cells were resistant to FeLV-B-GA enveloped particles (Fig. 1A). Different susceptibilities of NRK cells to FeLV-B enveloped vectors may be due to variations between Pit1 orthologs expressed in each rat tissue type. This is not without precedent; for example, Chinese hamster cell lines E36 and CHOK1 exhibit dramatically different susceptibilities to infection by GALV vectors. GALV vectors efficiently infect the Chinese hamster lung fibroblast cell line E36; however, CHOK1 cells derived from hamster ovary are significantly less susceptible to infection (10). Cloning of the E36 and CHOK1 Pit1 revealed differences between these receptor orthologs, despite their being derived from the same hamster species (7, 39, 46). Thus, differences in the susceptibility of NRK cells to FeLV-B-90Z and FeLV-B-GA enveloped vectors may be due to differences in Pit1 orthologs unique to each of the cell lines derived from different rat tissue types.
To investigate whether the block to FeLV-B-GA infection of NRK 6509 cells is at the binding or postbinding stage of infection, we performed FACS binding assays and compared the efficiency of binding on NRK cells to that on MDTF-Pit1 cells. Binding analysis revealed that FeLV-B-90Z SU efficiently binds NRK 6509 cells but that binding of FeLV-B-GA SU is reduced (Fig. 1D), suggesting that host range differences between FeLV-B-90Z and FeLV-B-GA for NRK 6509 cells are likely mediated at the level of receptor binding. NRK 6509 cells exhibit a bimodal shift in fluorescence intensity; one population of NRK 6509 cells efficiently binds FeLV-B-90Z SU, and to a lesser extent FeLV-B-GA, while the other does not. This type of heterogeneity has been observed previously (12) and may result from intrinsic properties of the receptor of efficiently binding FeLV-B-GA SU. The combined results of these studies indicate that the reduced binding of FeLV-B-90Z and FeLV-B-GA SU to NRK 6509 cells correlates with a reduced titer compared to that for MDTF-Pit1 cells. The composition of the FeLV-B envelope may also serve to restrict infectivity of NRK 6509 cells, while permitting entry into other NRK cells. The block to FeLV-B-GA infection appears to be mediated at the level of receptor binding, since the reduced level of binding observed fails to permit infection by vectors bearing this envelope (Fig. 1D). A similar block to infection has been observed elsewhere for GALV and WMV, which is related to GALV and also uses Pit1 as a receptor (43). These viruses have similar host ranges with at least one exception: E36 cells are not susceptible to infection by WMV but are susceptible to infection by GALV (43). As seems to be the case with FeLV-B-90Z and FeLV-B-GA, it was shown previously that amino acid differences between the WMV and GALV envelope proteins are responsible for the block to infection of E36 cells and that the block occurs at the level of receptor binding (43).
Another example of different viral isolates from the same receptor group having different host ranges has been observed with Friend MuLV (F-MuLV) and ecotropic PVC-211 MuLV, a neuropathic variant of F-MuLV. PVC-211 MuLV can infect rat brain capillary endothelial cells (BCECs) and CHOK1 hamster cells, both of which are resistant to F-MuLV and other ecotropic MuLVs (25). Host range determinants have been localized to subtle changes in the PVC-211 SU protein (25). These findings are significant because changes in the viral envelope glycoprotein result in the ability of a virus to utilize different orthologs of the ecotropic receptor not recognized by other viruses in the same receptor group. Differences in host range correlate with the distinct pathogenic properties exhibited by PVC-211 and F-MuLV. For example, PVC-211 MuLV causes rapidly progressive neurodegenerative disease when injected into neonatal rats and mice; in contrast, F-MuLV is nonneuropathogenic (15, 21, 24). The FeLV-B-90Z and FeLV-B-GA isolates were obtained from cats in various disease states following infection with FeLV-A, but it remains unclear how differences between the FeLV-B-90Z and FeLV-B-GA envelopes contribute to disease status.
Mink cells are susceptible to infection by vectors bearing FeLV-B-90ZRBD chimeric envelopes.
Another mammalian cell line that has been reported to be resistant to FeLV-B is the mink fibroblast cell line CCL 64 (42). We exposed mink cells to vectors bearing either FeLV-B-90Z or FeLV-B-GA envelopes and confirmed that they were not susceptible to infection by either vector (Table 1). It has been previously reported that the receptor utilization properties of FeLV-B-90Z can be expanded by substituting sequences derived from FeLV-A-61E for the 216 C-terminal residues of FeLV-B-90Z SU; this chimeric envelope is designated FeLV-B-90ZRBD (Table 1) (6, 37). Vectors bearing this chimeric envelope can infect cells by using human and feline Pit1 or Pit2 as receptors (1, 37). We therefore exposed mink cells to various FeLV vectors including those bearing the chimeric FeLV-B-90ZRBD envelope and found that mink cells were susceptible only to vectors bearing FeLV-B-90ZRBD envelopes (Table 1). Therefore, by replacing the C-terminal region of FeLV-B-90Z with the corresponding region of FeLV-A, we expanded the host range of FeLV-B-90Z to include mink cells.
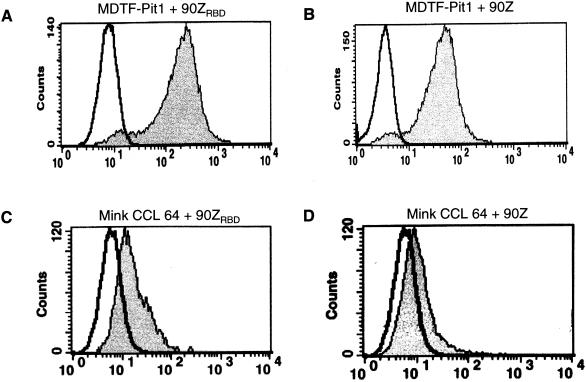
To determine at which stage the block to infection of mink cells by FeLV-B-90Z occurs, we performed binding assays with FeLV-B-90Z or FeLV-B-90ZRBD soluble SU. Both FeLV-B-90Z and FeLV-B-90ZRBD SU efficiently bind MDTF-Pit1 cells (Fig. 2A and B). We observed a shift in fluorescence intensity when mink cells were incubated with supernatant containing soluble FeLV-B-90ZRBD SU (Fig. 2C). In contrast, FeLV-B-90Z SU showed reduced binding (Fig. 2D). The overall reduction in binding of FeLV-B-90ZRBD SU to mink cells correlates with reduced titer compared to those for MDTF-Pit1 and MDTF-Pit2 cells (Table 1). Interestingly, while there was evidence of FeLV-B-90Z binding to mink cells, it appears that this level of binding was below the threshold necessary for infection, suggesting that FeLV-B-90Z vectors fail to infect mink cells because of reduced binding of the viral envelope to the mink Pit1 ortholog compared to MDTF-Pit1. Taken together, these findings show that binding and infectivity data with FeLV-B-90ZRBD envelopes indicate that replacing enFeLV sequences in the C-terminal half of SU with those derived from FeLV-A renders FeLV-B-90Z capable of infecting mink cells. Additionally, our results suggest that, in a manner similar to that described by Lavillette et al. (23), a favorable interaction between the C-terminal region and the RBD is required for the expanded host range of FeLV-B-90ZRBD enveloped vectors, as comparable FeLV-A sequences in FeLV-B-GA or FeLV-B-GARBD do not expand the host range of these vectors.
FIG. 2.
Histograms of flow cytometric analyses showing binding of FeLV-B-90ZRBD or FeLV-B-90Z SU to cells. Binding was detected by staining cells with fluorescein-conjugated monoclonal antibody HA.11, which recognizes soluble HA-tagged SU. The x axis represents fluorescence intensity (log scale), and the y axis represents cell number. Areas under bold black lines represent negative control cells which were exposed to medium only, followed by incubation with fluorescein-conjugated HA.11 antibody. Gray-shaded areas correspond to MDTF-Pit1 or mink CCL 64 cells binding to either FeLV-B-90ZRBD SU (A and C) or FeLV-B-90Z SU (B and D).
As mentioned above, FeLV-B-GA vectors bearing envelopes wherein an arginine is substituted in place of glutamine at position 73 within VRA have been shown to expand receptor utilization properties. FeLV-B-GARBD-73Q→R, like FeLV-B-90ZRBD, can use both Pit1 and Pit2 as receptors (37). Interestingly, the substitution of arginine at position 73 of GA creates a potential mammalian consensus heparin-binding domain (HBD) in the VRA of GA. This HBD, represented by XBBXBX, where X is any residue and B is a basic residue, is also present in FeLV-B-90Z (e.g., MRRWRQ). Recently it has been proposed for the PVC-211 MuLV that the expanded host range of this virus relative to those of other ecotropic viruses may be a result of a single residue change in PVC-211 that creates a novel HBD. The ability of PVC-211 to infect rat BCECs correlates with the enhanced affinity of this virus for heparin and suggests that the ability of PVC-211 to infect cells not susceptible to other ecotropic viruses may be due to its ability to interact with heparin-like molecules on the surface of rat BCECs (20).
To determine whether the presence of a potential HBD in the VRA of FeLV-B is associated with the ability of FeLV-B-90ZRBD to infect mink cells, we assessed the susceptibility of mink cells to vectors bearing FeLV-B-GARBD-73Q→R envelopes. FeLV-B-GARBD-73Q→R enveloped vectors failed to infect mink cells, even though these vectors are capable of utilizing Pit2, in addition to Pit1, as a receptor (Table 1). Thus, neither the presence of a potential HBD in the VRA region nor the ability of FeLV-B-GARBD-73Q→R to use Pit2 as a receptor correlates with the expanded host range observed with FeLV-B-90ZRBD compared to those of FeLV-B-GARBD vectors.
FeLV-B-90ZRBD enveloped vectors use the mink Pit1 ortholog to infect cells.
It was previously demonstrated that some engineered forms of the FeLV-B viral envelope such as FeLV-B-90ZRBD and FeLV-B-GARBD-73Q→R can use human and feline orthologs of Pit2 to infect cells (1, 6, 37). Vectors bearing FeLV-B-90ZRBD envelopes are capable of infecting a wider range of mammalian cells, including mink, than are FeLV-B-GARBD vectors (Table 1; data not shown). However, FeLV-B-GARBD-73Q→R enveloped vectors failed to infect mink cells, even though these vectors are capable of utilizing Pit2, in addition to Pit1, as a receptor (Table 1). Thus, the ability to use Pit2 does not directly correlate with the expanded host range observed with FeLV-B-derived vectors.
To determine which Pit-mediated entry pathway is utilized by FeLV-B-90ZRBD enveloped vectors to infect mink cells, we performed interference assays with mink cells chronically infected with either GALV or A-MuLV. This interference assay is based on the principle that cells infected with GALV are resistant to infection by other viruses that use Pit1; however, Pit2 remains available as a viral receptor. Likewise, cells infected with A-MuLV 4070A cannot be superinfected by viruses that use Pit2 but retain susceptibility to viruses that use Pit1. Exceptions to this pattern include hamster E36 cells and a Japanese feral mouse (Mus musculus molossinus) kidney cell line (MMMol) (10, 31, 33), where analysis of the interference patterns of GALV and A-MuLV indicates that GALV can use either the Pit1 or Pit2 orthologs expressed in E36 and MMMol cells. We wanted to determine whether FeLV-B-90ZRBD entry into mink cells is restricted to only Pit1 or whether the mink Pit2 ortholog also functions as a receptor for FeLV-B-derived particles. When we exposed mink cells productively infected with GALV (mink-GALV) to GALV-SEATO vectors, we found that they could not be superinfected with these vectors. They were, however, susceptible to A-MuLV 4070A retroviral particles, as expected (Table 2). Reciprocal interference was also observed with mink-A-MuLV cells (Table 2). Results of the interference assay showed that mink-GALV cells are resistant to FeLV-B-90ZRBD superinfection (Table 2), indicating that FeLV-B-90ZRBD vectors use the mink Pit1 ortholog. Mink cells persistently infected with A-MuLV remain susceptible to infection by FeLV-B-90ZRBD enveloped vectors. Therefore, the mink Pit1 ortholog, and not the Pit2 ortholog, functions as a receptor for this chimeric envelope when Pit2 utilization is blocked (Table 2).
TABLE 2.
Interference assays of FeLV-B-90ZRBD, GALV-SEATO, and A-MuLV 4070A enveloped vectors
| Cell line | % Infectivity of pseudotypea
|
||
|---|---|---|---|
| 90ZRBD | GALV-SEATO | A-MuLV 4070A | |
| Mink | 100.0 ± 1.7 | 100.0 ± 6.0 | 100.0 ± 2.4 |
| Mink-GALV | 0 ± 0 | 0 ± 0 | 45.5 ± 2.0 |
| Mink-A-MuLV | 39.7 ± 2.9 | 50.9 ± 5.9 | 0 ± 0 |
Values represent titers of vectors in cells infected with various viruses as a percentage of titers in control uninfected mink cells (mean ± range; n = 3).
Earlier work mapped specific residues in VRA and VRB of FeLV-B that interact with discrete domains of human and rat Pit1 orthologs that are critical for receptor recognition and function (38, 40). Results that we obtained with FeLV-B-90ZRBD vectors indicate that envelope sequences that regulate Pit1 ortholog usage are not contained exclusively in the N terminus of SU but include sequences in the C terminus as well. The C terminus of SU has been implicated elsewhere in postbinding stages of viral entry (2, 3, 23, 37). We found that C-terminal sequences significantly influence the ability of the FeLV-B envelope to bind Pit1 receptor orthologs such as that of mink (Fig. 2). In general, we observed a correlation between reduced binding and reduced titer (Fig. 2; Table 1). These data suggest that C-terminal SU sequences are an important host range determinant that mediates binding of the FeLV-B envelope protein to receptors on target cells.
In summary, we have demonstrated that vectors bearing FeLV-B-90Z envelopes efficiently infect a wider range of mammalian cells than do FeLV-B-GA enveloped vectors. Host range differences are mediated at the level of receptor binding and are dependent on the specific composition of the FeLV-B SU. C-terminal domains outside of RBD in the SU influence the host range and infectivity of FeLV-B-90Z-derived vectors at the level of receptor binding. Our demonstration that FeLV-B vectors exhibit distinct infectivity and binding patterns on various NRK cell lines provides a cautionary note on making assumptions about tropism from a single cell line of a given species. Finally, we have shown that FeLV-B-90ZRBD enveloped vectors can utilize the mink Pit1 ortholog, whereas mink cells are resistant to FeLV-B-GA vectors, indicating that, in the case of FeLV-B-90ZRBD, the C-terminal region in addition to RBD is responsible for the expanded host range of this envelope. Work is currently under way to map the residues in the FeLV-B-90Z SU that confer mink cell susceptibility on FeLV-B enveloped vectors and to characterize the mink Pit1 ortholog.
Acknowledgments
We thank Howard Mostowski (CBER, FDA) for FACS binding analysis. We are grateful to Adam Lauring and Maria Anderson for providing the SU constructs and helpful advice. We thank Karen B. Farrell and Jill Russ for critical comments on the manuscript and Harshita Satija for technical assistance.
Support is provided to J.O. by Public Health Service grant CA51080 from the National Cancer Institute.
REFERENCES
- 1.Anderson, M., A. S. Lauring, S. Robertson, C. Dirks, and J. Overbaugh. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J. Virol. 75:10563-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, A. L., and J. M. Cunningham. 2001. Receptor binding transforms the surface subunit of the mammalian C-type retrovirus envelope protein from an inhibitor to an activator of fusion. J. Virol. 75:9096-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, A. L., R. A. Davey, and J. M. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini, J.-L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battini, J.-L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boomer, S., M. Eiden, C. Burns, and J. Overbaugh. 1997. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J. Virol. 71:8116-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudry, G. J., C. Schmitz, Y.-T. Ting, K. B. Farrell, C. J. Petropoulos, Y. S. Lie, and M. V. Eiden. 1999. Molecular characterization of the gibbon ape leukemia virus (GALV) receptor homologs (HaPIT1 and HaPIT2), present in CHOK1 cells, reveals the basis for their failure to function as GALV receptors. J. Virol. 73:2916-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. 1996. Retroviridae: the viruses and their replication, p. 1767-1848. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 9.Delassus, S., P. Sonigo, and S. Wain-Hobson. 1989. Genetic organization of gibbon ape leukemia virus. Virology 173:205-213. [DOI] [PubMed] [Google Scholar]
- 10.Eglitis, M. A., M. V. Eiden, and C. A. Wilson. 1993. Gibbon ape leukemia virus and the amphotropic murine leukemia virus 4070A exhibit an unusual interference pattern on E36 Chinese hamster cells. J. Virol. 67:5472-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elder, J. H., and J. I. Mullins. 1983. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J. Virol. 46:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell, K. B., J. L. Russ, R. K. Murthy, and M. V. Eiden. 2002. Reassessing the role of region A in Pit1-mediated viral entry. J. Virol. 76:7683-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy, W. D., Jr. 1993. Feline oncoretroviruses, p. 109-180. In J. A. Levy (ed.), The Retroviridae, vol. 2. Plenum Press, New York, N.Y.
- 14.Hardy, W. D., Jr., P. W. Hess, E. G. MacEwen, A. J. McClelland, E. E. Zuckerman, M. Essex, S. M. Cotter, and O. Jarrett. 1976. Biology of feline leukemia virus in the natural environment. Cancer Res. 36:582-588. [PubMed] [Google Scholar]
- 15.Hoffman, P. M., E. F. Cimino, D. S. Robbins, R. D. Broadwell, J. M. Powers, and S. K. Ruscetti. 1992. Cellular tropism and localization in the rodent nervous system of a neuropathogenic variant of Friend murine leukemia virus. Lab. Investig. 67:314-321. [PubMed] [Google Scholar]
- 16.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 17.Jarrett, O., and P. H. Russell. 1978. Differential growth and transmission in cats of feline leukemia viruses of subgroups A and B. Int. J. Cancer 21:466-472. [DOI] [PubMed] [Google Scholar]
- 18.Jarrett, O. H., W. D. Hardy, Jr., M. C. Golder, and D. Hay. 1978. The frequency of occurrence of feline leukemia virus subgroups in cats. Int. J. Cancer 21:334-337. [DOI] [PubMed] [Google Scholar]
- 19.Jarrett, O. H., M. Laird, and D. Hay. 1973. Determinants of host range of feline leukemia viruses. J. Gen. Virol. 20:169-175. [DOI] [PubMed] [Google Scholar]
- 20.Jinno-Oue, A., M. Oue, and S. K. Ruscetti. 2001. A unique heparin-binding domain in the envelope protein of the neuropathogenic PVC-211 murine leukemia virus may contribute to its brain capillary endothelial cell tropism. J. Virol. 75:12439-12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kai, K., and T. Furuta. 1984. Isolation of paralysis-inducing murine leukemia viruses from Friend virus passaged in rats. J. Virol. 50:970-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauring, A. S., M. M. Anderson, and J. Overbaugh. 2001. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 75:8888-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavillette, D., B. Boson, S. J. Russell, and F.-L. Cosset. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J. Virol. 75:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda, M., C. A. Hanson, N. V. Dugger, D. S. Robbins, S. G. Wilt, S. K. Ruscetti, and P. M. Hoffman. 1997. Capillary endothelial cell tropism of PVC-211 murine leukemia virus and its application for gene transduction. J. Virol. 71:6168-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, M., M. Masuda, C. A. Hanson, P. H. Hoffman, and S. K. Ruscetti. 1996. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J. Virol. 70:8534-8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, D. G., R. H. Edwards, and A. D. Miller. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 91:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moser, M., C. C. Burns, S. Boomer, and J. Overbaugh. 1998. The host range and interference properties of two closely related feline variants suggest that they use distinct receptors. Virology 242:366-377. [DOI] [PubMed] [Google Scholar]
- 28.O'Hara, B., S. V. Johann, H. P. Klinger, D. G. Blair, H. Rubinson, K. J. Dunne, P. Sass, S. M. Vitek, and T. Robins. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1:119-127. [PubMed] [Google Scholar]
- 29.Ott, D., and A. Rein. 1992. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J. Virol. 66:4632-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overbaugh, J., P. R. Donahue, S. L. Quackenbush, E. A. Hoover, and J. I. Mullins. 1988. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science 239:906-910. [DOI] [PubMed] [Google Scholar]
- 31.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarma, P. S., T. Log, D. Jain, P. R. Hill, and R. J. Huebner. 1975. Differential host range of viruses of feline leukemia-sarcoma complex. Virology 64:438-446. [DOI] [PubMed] [Google Scholar]
- 33.Schneiderman, R. D., K. B. Farrell, C. A. Wilson, and M. V. Eiden. 1996. The Japanese feral mouse PiT1 and PiT2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J. Virol. 70:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheets, R. L., R. Pandey, V. Klement, C. K. Grant, and P. Roy-Burman. 1992. Biologically selected recombinants between feline leukemia virus (FeLV) subgroup A and an endogenous FeLV element. Virology 190:849-855. [DOI] [PubMed] [Google Scholar]
- 35.Sommerfelt, M. A., and R. A. Weiss. 1990. Receptor interference groups of 20 retroviruses plating on human cells. Virology 176:58-69. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, M. A., M. Warnock, A. Wheeler, N. Wilkie, J. I. Mullins, D. E. Onions, and J. C. Neil. 1986. Nucleotide sequence of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinatorial origin of subgroup B viruses. J. Virol. 58:825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugai, J., M. V. Eiden, M. M. Anderson, N. Van Hoeven, C. D. Meiering, and J. Overbaugh. 2001. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 and Pit2. J. Virol. 75:6841-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tailor, C. S., and D. Kabat. 1997. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J. Virol. 71:9383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tailor, C. S., A. Nouri, and D. Kabat. 2000. Cellular and species resistance to murine amphotrophic, gibbon ape, and feline subgroup C leukemia viruses is strongly influenced by receptor expression levels and by receptor masking mechanisms. J. Virol. 74:9797-9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tailor, C. S., A. Nouri, and D. Kabat. 2000. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J. Virol. 74:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tailor, C. S., Y. Takeuchi, B. O'Hara, S. V. Johann, R. A. Weiss, and M. K. Collins. 1993. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J. Virol. 67:6737-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi, Y., R. G. Vile, G. Simpson, B. O'Hara, M. K. L. Collins, and R. A. Weiss. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ting, Y.-T., C. A. Wilson, K. B. Farrell, G. J. Chaudry, and M. V. Eiden. 1998. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite their bearing functional receptors. J. Virol. 72:9453-9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss, R. A., and C. Tailor. 1995. Retrovirus receptors. Cell 82:531-533. [DOI] [PubMed] [Google Scholar]
- 45.Weiss, R. A., and A. L. Wong. 1977. Phenotypic mixing between avian and mammalian RNA tumor viruses. I. Envelope pseudotypes of Rous sarcoma virus. Virology 76:826-834. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, C. A., K. B. Farrell, and M. V. Eiden. 1994. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cell. J. Virol. 68:7697-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wunsch, M., A. S. Schultz, W. Koch, R. Friedrich, and G. Hunsmann. 1983. Sequence analysis of Gardner-Arnstein feline leukemia virus envelope gene reveals common structural properties of mammalian retroviral envelope genes. EMBO J. 2:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeijl, M. V., S. V. Johann, E. Cross, J. Cunningham, R. Eddy, T. B. Shows, and B. O'Hara. 1994. An amphotropic virus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA 91:1168-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]