Abstract
Retroviruses package two copies of genomic RNA into viral particles. During the minus-sense DNA synthesis step of reverse transcription, the nascent DNA can transfer multiple times between the two copies of the genome, resulting in recombination. The mechanism for this process is similar to the process of obligate strand transfers mediated by the repeat and primer binding site sequences. The location at which the DNA 3′ terminus completely transfers to the second RNA strand defines the point of crossover. Previous work in vitro demonstrated that reverse transcriptase pausing has a significant impact on the location of the crossover, with a proportion of complete transfer events occurring very close to pause sites. The role of pausing in vivo, however, is not clearly understood. By employing a murine leukemia virus-based single-cycle infection assay, strong pausing was shown to increase the probability of recombination, as reflected in the reconstitution of green fluorescent protein expression. The infection assay results were directly correlated with the presence of strong pause sites in reverse transcriptase primer extension assays in vitro. Conversely, when pausing was diminished in vitro, without changing the sequence of the RNA template involved in recombination, there was a significant reduction in recombination in vivo. Together, these data demonstrate that reverse transcriptase pausing, as observed in vitro, directly correlates with recombination during minus-sense DNA synthesis in vivo.
Retroviruses exhibit a high frequency of homologous recombination (7, 29, 35, 36, 37, 43, 58, 59, 61, 65). Human immunodeficiency virus type 1 (HIV-1) recombination occurs at a rate of approximately 2.4 × 10−4/bp per replication cycle or higher while the direct repeat deletion rate for murine leukemia virus (MLV) was reported at 1.09 × 10−5/bp per replication cycle (29, 46, 50). The rate of nonhomologous recombination is much lower but is still mediated by short homologous sequences (62, 64). During one round of reverse transcription, multiple crossover events often appear to occur at a significantly higher rate than mathematically anticipated, suggesting that a second crossover is not an independent event (negative interference) (2, 3). There are conflicting reports concerning this phenomenon in HIV-1; early work suggested the existence of high negative interference (29), while a more recent report obtained the expected distribution of multiple crossover events (51).
Recombination of two different retrovirus strains relies on coinfection of a host cell by two genetically distinct viruses and copackaging of one genomic RNA from each integrated provirus into a single virion (26, 27, 55). The heterozygous virion then infects a new target cell where a chimeric recombinant provirus can be generated during reverse transcription. Recently, copackaging of heterozygous virions was demonstrated to be strongly affected by the identity of the dimerization initiation sequence. Genomes with different dimerization initiation sequences were packaged together at a lower frequency than those with identical palindromes, resulting in an apparent decrease in the rate of recombination in the former case (12). Homozygous genomes can also recombine when polymerization switches from one genome to the other during reverse transcription, but this does not necessarily change the genotype of the progeny virus.
Nearly all retroviral recombination occurs during minus-sense DNA synthesis, when reverse transcriptase (RT) utilizes the genome RNA as a template for polymerization (3, 63). The mechanism by which two RNA genomes recombine was first described as “copy choice” by Coffin (13). Variations of this model have been formulated to incorporate factors that influence various stages of the process, namely, RT processivity and RNase H activity (8-10, 17-19, 23, 28, 45, 48, 49, 52, 53, 57, 60), RNA secondary structure (1, 2, 4, 5, 20, 38, 40-42, 66), and the viral nucleocapsid protein (NC) (16, 30, 44, 52, 54, 66). Overall, the recombination process is very similar to the obligate strand transfers mediated by the repeat and primer binding site (PBS) sequences during reverse transcription (14). As RT synthesizes DNA on one RNA template (the “donor” template), the RNase H activity hydrolyzes the template RNA behind the polymerase, allowing the nascent DNA to base pair with a second RNA template (the “acceptor” template). Branch migration then “zippers” together the DNA and acceptor template RNA. The position on the acceptor template where the DNA 3′ terminus ultimately anneals defines the crossover point, and the annealing appears to be facilitated by single-stranded regions of the acceptor RNA, such as loops and bulges, that are potentially more accessible for base pairing (20, 22, 29).
One version of this recombination mechanism was coined the “dynamic” copy choice model. A mutational analysis of the MLV RT demonstrated that a decrease in polymerase activity increased the rate of homologous recombination while a decrease in RNase H activity decreased recombination in vivo (28, 57), suggesting that a balance exists between RNase H activity and DNA synthesis, which determines the overall rate of recombination. Once the nascent DNA starts to anneal to the acceptor RNA template, extension of the nascent DNA continues until branch migration forces complete DNA 3′ terminus transfer.
A major factor affecting the rate of continued RT-mediated DNA synthesis is the presence of pause sites. Pausing during synthesis on RNA templates has been attributed to stretches of homopolymers G and C and to duplex RNA structures (30, 31, 34, 47, 56, 60). In vitro, such pause sites are associated with an overall increase in strand transfer (52, 53, 67), but more recent work has demonstrated that pausing has a greater influence on the point of crossover. When RT encounters a pause site, the location of the 3′ terminus transfer to an acceptor template clusters near the pause position (15, 33). If pausing is minimal, however, transfer still occurs but with a more random crossover point distribution (15). Similarly, pausing on RNA templates in vitro was correlated with a preferred region of viral recombination in vivo; however, the effects of removing the pause sites were not determined (21).
Pausing within the duplex region of stem-loop structures has been shown to diminish when base pairing contiguity was disrupted by the introduction of single unpaired nucleotides or mismatches without changing the template sequence (33, 34). This observation provides a unique opportunity to determine if pausing or lack of pausing affects strand transfer in vitro and retroviral recombination in vivo. By employing a variation of a previously characterized Moloney MLV (M-MLV)-based recombination assay (66), recombination in the presence or absence of predicted strong pause sites was monitored with the reconstitution of green fluorescent protein (GFP) expression. Comparing in vitro primer extension assays on RNA segments derived from the proviral constructs with the proportion of GFP-expressing, MLV-infected target cells provides the first direct evidence that the rate of homologous recombination directly corresponds to the extent of pausing during reverse transcription.
MATERIALS AND METHODS
Materials and reagents.
Restriction enzymes, T4 DNA ligase, and Vent DNA polymerase were purchased from New England Biolabs. T4 polynucleotide kinase and proteinase K were purchased from Invitrogen. M-MLV RT was purchased from U.S. Biochemicals. Plasmid mutagenesis was performed using the QuikChange site-directed mutagenesis kit from Stratagene. DNA oligonucleotides were synthesized by Operon or QIAGEN. [γ-32P]ATP was from PerkinElmer Life Sciences. Reagents for RNA solutions were from Ambion. The neomycin analogue G418 was from Sigma or Mediatech Inc. Dulbecco's modified Eagle's medium (DMEM), trypsin, and fetal bovine serum (FBS) were purchased from Gibco. PGEM9Zf(−) and the T7 RiboMax transcription kit were from Promega. The pMP-1 plasmid was a very generous gift from Vinay Pathak (NIH HIV Drug Resistance Program). Cloning disks, methotrexate, aminopterin, 50× hypoxanthine-aminopterin-thymidine, and 50× hypoxanthine-thymidine medium supplements were from Sigma.
Plasmids and retroviral vectors.
The parent vector for all retroviral constructs is the M-MLV-based pMP-1 (11). New proviral constructs were made as follows. Using site-directed mutagenesis, an EcoRI site and SalI site were introduced into pMP-1 between the GFP initiating ATG and the rest of the GFP coding sequence, producing pMPes, which has the initiating ATG, followed by 5′ GAATTCTTCGGTCGAC 3′ and the remaining GFP sequence. pMPes was digested with EcoRI and SalI and ligated to PLf (5′ AATTCTTCCTCGAGGCGATCGATCTGG3′) and PLr (5′ TCGACCAGATCGATCGCCTCGAGGAAG 3′) that had been previously annealed to each other. This generated pMPmcs (for multi-cloning site), which contains the restriction sites EcoRI, XhoI, PvuI, ClaI, and SalI between the GFP initiation codon and the remaining GFP coding sequence (5′ to 3′). The oligonucleotides HAf and HAr (5′ CCGGTTACCCTTACGACGTGCCCGACTACGCGC 3′ and 5′ TCGAGCGCGTAGTCGGGCACGTCGTAAGGGTAA 3′, respectively) and PSf and PSr (5′ AATTCGGTCTAACTAGGGAGACCCACA 3′ and 5′ [Phosph]CCGGTGTGGGTCTCCCTAGTTAGACCG 3′, respectively) were ligated to each other, generating a duplex fragment with EcoRI and XhoI complementary overhangs. This PS/HA duplex was then ligated to EcoRI-XhoI-cut pMPmcs to generate pMPph. pMPph was then digested with XhoI and SalI and ligated to create an in-frame N-terminal fusion to the GFP coding sequence. Separately, pMPph was transformed into Escherichia coli strain GM48, a most generous gift from Joyce Karlinsey of K. Hughes' lab (University of Washington). This was necessary, since the ClaI site is known to be resistant to cleavage when grown in strains capable of methylation. pMPph purified from GM48 was digested with ClaI and XhoI. The final product was then ligated with previously annealed oligonucleotides STOPf and STOPr (5′ TCGAGTAGATAACTGATTAAGACTGACATAACGTGACAT 3′ and 5′ CGATGTCACGTTATGTCAGTCTTAATCAGTTATCTAC 3′, respectively) creating pNoDR (no direct repeat). E. coli strain GM48-derived pNoDR was digested with SalI and ClaI and gel purified. The heat-annealed oligonucleotides TARhpF and TARhpR (5′ CGATGGGTCTCTCTGGTTAGACCAGATCTGAGCCTGGGAGCTCAGATCTGGTCTAAC TAGGGAGACCCACA 3′ and 5′ CCGGTGTGGGTCTCCCTAGTTAGACCAGATCTGAGCTCCCAGGCTCAGATCTGGTCTAACCAGAGAGACCCA T 3′, respectively) were ligated to previously annealed HAp (5′ [Phosph]CCGGTTACCCTTACGACGTGCCCGACTACGCGC 3′) and HAr. The resulting ligation product was subsequently ligated to ClaI-SalI-cut pNoDR, generating pHPnb (hairpin, no-bulge). pHPnb was used as template for site-directed mutagenesis to create pHP-1b (one bulge) and pHP-2b (two bulges) (primer sequences available on request). All plasmids were sequenced in the region of the inserted sequences for final verification.
In vitro RNA expression vectors and RNA synthesis.
Primers that flank the inserted sequences described above were used for PCR amplification of proviral vector templates. The PCR products were digested with EcoRI and HindIII and ligated into EcoRI-HindIII-cut pGEM9Zf(−) (Promega), creating pIVNOSTOP, pIVNoDR, pIVHPnb, pIVHP-1b, and pIVHP-2b. The ligated fragment was oriented such that positive-sense RNA could be produced using the T7 promoter. All constructs were verified by sequencing. The pGEM9Zf(−)-derived vectors were digested with HindIII, and RNA was synthesized using the RiboMax transcription kit (Promega) with T7 RNA polymerase. Full-length RNA product was purified on a 5% denaturing polyacrylamide gel.
Primer extension assays.
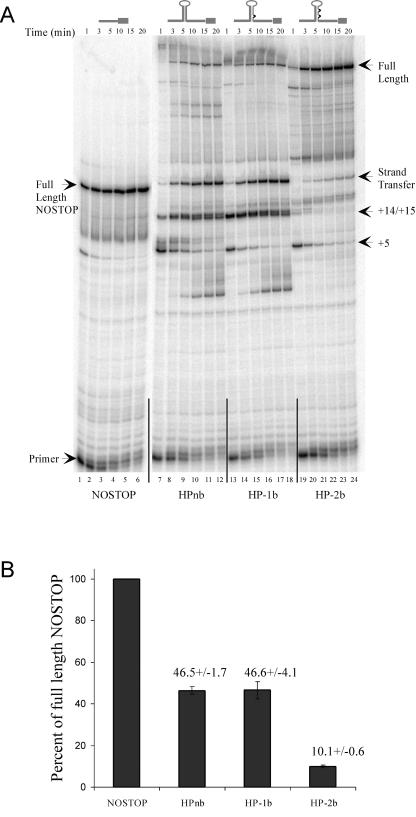
Ten picomoles of the DNA primer 40extMP (5′ TCACATCGCCATCCAGTTCCACGAGAATTGGGACCACGCC 3′) was 5′ end labeled with polynucleotide kinase in the presence of [γ-32P]ATP at 37°C in a total reaction volume of 20 μl. Approximately 0.5 pmol labeled primer was annealed to 1 pmol of the RNA templates described above by heating to 85°C for 2 min and cooling to 37°C at a rate of 0.02°C/s using a Hybaid Thermocycler (Savant) in the presence of 71 mM Tris (pH 8.0) and 71 mM KCl. Dithiothreitol, MgCl2, and 10 pmol of M-MLV RT (U.S. Biochemicals) were added, and the mixture was further incubated at 37°C for 5 min. Primer extension was initiated by the addition of deoxynucleoside triphosphates. In a total volume of 20 μl, the final concentrations were as follows: 50 mM Tris (pH 8.0), 50 mM KCl, 10 mM dithiothreitol, 5 mM MgCl2, 100 μM (each) deoxynucleoside triphosphates, 25 nM primer, 50 nM template, and 500 nM M-MLV RT. At each time point, 3-μl aliquots were removed and transferred to 12 μl of 96% formamide-20 mM EDTA. Denaturing polyacrylamide gel electrophoresis was used to separate the reaction products. Gels were dried and exposed to a PhosphorImager screen, which in turn was scanned using a STORM PhosphorImager. To verify the identity of in vitro strand transfer product (see Fig. 4), the putative transfer products for HPnb and HP-1b and the full-length NOSTOP product were gel purified and PCR amplified using the primers 40extMP (see above) and SText [TAACCTAATCTGAAAGTAGGGAGCTCGTCGACGAATTC] and the PCR products were sequenced. It should be noted that a PCR product could be generated only if strand transfer had occurred, since the EcoRI site (in boldface) to which SText anneals is not present if the band in the gel resulted from pausing during sequential extension. The sequences of the PCR products clearly identified the bands in the HPnb and HP-1b experiments as the strand transfer product (data not shown).
FIG. 4.
(A) An 8% denaturing polyacrylamide gel of an in vitro primer extension time course using 5′ end-labeled 40extMP (see Materials and Methods) DNA primer. Labeled primer was annealed to in vitro-synthesized RNA templates NOSTOP (lanes 1 to 6; 160 full-length nucleotides), HPnb (lanes 7 to 12; 301 full-length nucleotides), HP-1b (lanes 13 to 18; 300 full-length nucleotides), and HP-2b (lanes 19 to 24; 299 full-length nucleotides). Above each set of lanes are time points and a schematic of the RNA template structure. Pause sites within the stem-loop structures are labeled on the right and numbered in relation to the first base pair at the base of the stem, which is denoted +1. (B) Analysis of the proportion of strand transfer product generated during primer extension of HPnb, HP-1b, and HP-2b relative the amount of full-length product synthesized on the NOSTOP template. For a given experiment, the proportion of signal corresponding to the full-length extension on the positive control NOSTOP template at the 20-min time point was determined and defined as 100% strand transfer. The amount of strand transfer product at 20-min time points for HPnb, HP-1b, and HP-2b was then calculated and normalized to the NOSTOP full-length template. Values are results from three independent experiments.
Generation of virus producing cell lines.
Clone PG13 was a generous gift from V. Pathak (NIH HIV Drug Resistance Program). Selection for packaging function was performed as outlined in the ATCC protocol (http://www.atcc.org). To generate stable virus-producing lines, PG13 cells were transfected with 5 μg of each proviral construct using the calcium phosphate method (25). Cells were trypsinized at 24 h and serially diluted into 100-mm dishes with selective medium. Selection was maintained for 9 days before isolating clones. Titer was determined for each clone by infecting D17 cells and counting G418-resistant colonies (28).
Expression of proviral vectors in D17 cells.
D17 cells are a dog osteosarcoma-derived cell line susceptible to gibbon ape leukemia virus (GALV)-mediated MLV infection. D17 cells were transfected with 5 μg of each plasmid by the calcium phosphate method. G418-resistant cells were selected by growing the cells in DMEM supplemented with 6% FBS and 400 μg/ml G418 (active) for 10 days. Fluorescence microscopy and imaging were performed using a Nikon TS-100 inverted fluorescence microscope with Q-capture software generously provided by M. Lagunoff (University of Washington).
Infection of D17 with MLV produced by PG13 clones for flow cytometry analysis.
For each virus strain, duplicate 60-mm dishes were seeded with 5 × 105 D17 cells and grown overnight. The next day, the medium was changed to DMEM supplemented with 6% FBS and 4 μg/ml polybrene and virus was added from each clone to two separate 60-mm plates at an approximate multiplicity of infection [MOI] of 2 × 10−3 CFU/cell per plate. As a negative control, 100 μl of supernatant collected from mock-transfected PG13 cells grown in selective medium for 2 days (before die-off) was added to a separate plate. After 24 h, each 60-mm plate was split into two 100-mm dishes and selected for 9 to 12 days. On the final day of selection, colonies from duplicate 100-mm plates were pooled and the numbers of GFP+ and GFP− cells were determined using the Influx flow cytometer from Cytopeia. Flow data were analyzed with Summit 3.1 software from DakoCytomation.
RESULTS
Design of vectors used to compare pausing in vitro with recombination in vivo.
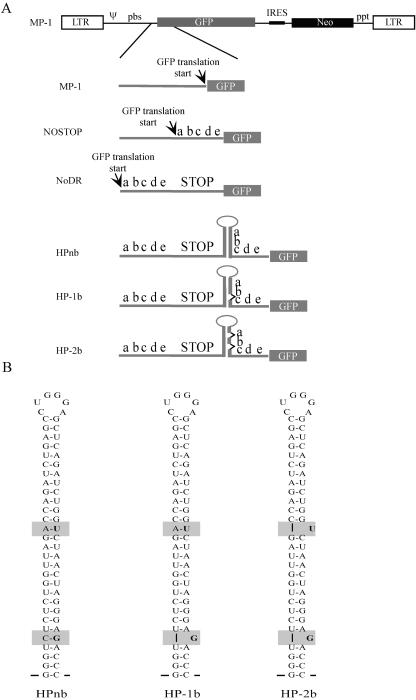
M-MLV proviral vectors used in this study are shown schematically in Fig. 1. The parent plasmid was pMP-1, which has been described previously (11). This vector possesses all of the cis genomic sequences necessary for RNA packaging and reverse transcription. The gag/pol/env coding region has been replaced with the GFP coding sequence, followed by the neomycin resistance gene (neo), and this construct can be transfected into an MLV packaging line that provides the essential components for virus production in trans (Fig. 1A, MP-1). After reverse transcription and integration, the GFP and neo sequences yield a single transcript controlled by the viral long terminal repeat promoter. Translation of the neo gene relies on an internal ribosomal entry site sequence.
FIG. 1.
Schematic diagram of proviral constructs. (A) Constructs used for generating virus from PG13 packaging lines. At the top is the general provirus structure of the unmodified MP-1 vector. LTR, long terminal repeat; Ψ, packaging sequence; Neo, neomycin resistance gene; ppt, polypurine tract. Below the structure are expanded schematics of the region containing the GFP translation start site. The letters abcde represent the direct repeat sequence described in Materials and Methods. STOP corresponds to a sequence with stop codons in all three reading frames. Unpaired nucleotides in HP-1b and HP-2b are represented by sideways carats between “a” and “b” and “b” and “c” along the stem-loop. (B) Schematic diagram of the stem-loops introduced into constructs HPnb, HP-1b, and HP-2b. The sequence is derived from the HIV-1 TAR sequence. Gray boxes indicate modified regions, and the specific nucleotides are in boldface. Note that the sequence involved in the recombination process does not change.
To determine the effects of pausing and displacement synthesis during reverse transcription on M-MLV recombination, the translation of the GFP was modified in the following way. First, a sequence encoding a 22-amino-acid N-terminal fusion with GFP (Fig. 1A, labeled “abcde”; see also Materials and Methods) was inserted in frame between the GFP initiating ATG and the remainder of the coding sequence (Fig. 1A, NOSTOP). Next, a sequence containing all three stop codons in all three reading frames (STOP sequence) was inserted downstream of the N-terminal fusion sequence (Fig. 1A, NoDR). A third construct was engineered with a stable hairpin derived from the HIV-1 transactivation response element (TAR) sequence that has a stem of 29 contiguous base pairs (33). As the schematic shows, this construct has direct repeats (abcde) that flank the STOP sequence (Fig. 1A, HPnb; Fig. 1B). Importantly, the first 19 nucleotides (abc) of the proximal direct repeat (in relation to the direction of reverse transcription of the RNA) are base paired in the stem-loop structure. The fourth and fifth constructs (Fig. 1A, HP-1b and HP-2b, respectively) contain one and two bulges in the stem of the hairpin, respectively (Fig. 1B). For HPnb and HP-1b, the insertion of the hairpin and the deletion of 1 nucleotide create a frameshift between the initiating ATG and the GFP coding region. HP-2b is in frame; however, translation machinery must bypass an additional stop codon (there are three before it in the STOP sequence) to reach the GFP coding sequence. Overall, the length of the direct repeat is 61 nucleotides in each construct, separated by 80 nucleotides. Of the 61 nucleotides in the proximal direct repeat, 21 are involved in the hairpin region. It should be noted that the identity of the direct repeat remains the same in each of the hairpin constructs (HPnb, HP-1b, and HP-2b).
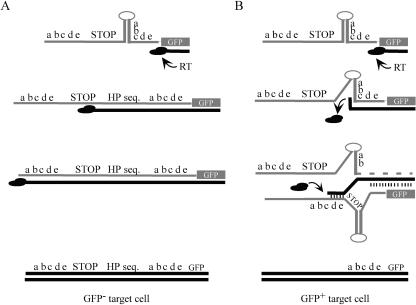
Figure 2 schematically outlines the recombination process predicted to occur during reverse transcription of the packaged MLV genome derived from the provirus vectors described in Fig. 1. Previous work has shown that during synthesis through stem-loop structures by RT, the polymerase pauses at specific positions on the proximal strand. This pausing is relieved if unpaired nucleotides interrupt hairpin-stem base pair contiguity (33). As RT synthesizes minus-strand DNA, it replicates the proximal direct repeat first. If RT is able to replicate through the first direct repeat, the hairpin, the STOP sequence, and the second direct repeat, then the resulting integrated provirus will produce a transcript in which the GFP would not be translatable because of the presence of the stop codons and, as mentioned above, in some cases, a frame shift (Fig. 2A). If pausing occurs, however, polymerization through the various sequences may be delayed long enough for the process of strand transfer (recombination) to occur in which the 3′ terminus of the nascent DNA switches from the original RNA template (donor template) to the second packaged template (acceptor template) (Fig. 2B). This crossover could take place at the proximal or distal repeat of the acceptor strand during intermolecular recombination. Alternatively, although not depicted in Fig. 2B, the 3′ terminus could transfer to the distal repeat of the same donor template (intramoleculer recombination). Previous work indicated that the overall rate of recombination would not be affected by which of the repeats is utilized for the transfer (29). If complete DNA 3′ terminus transfer occurs intermolecularly at the proximal repeat of the acceptor template, then RT would still replicate the hairpin and STOP sequence and the infected cell would not express GFP. If complete transfer occurs at the distal repeat on either template, however, the region containing the hairpin and STOP sequence would loop out and be deleted. This process would restore the proper GFP reading frame and remove all stop codons between the initiating ATG and the GFP coding region. The expressed protein would be GFP with a 22-amino-acid N-terminal fusion as in the NOSTOP construct, and infected cells in which this process occurred would be expected to exhibit GFP fluorescence.
FIG. 2.
Schematic diagram depicting two possible outcomes of synthesis through stem-loop structures during reverse transcription. Gray indicates RNA, and black indicates DNA; short vertical lines indicate annealing. (A) Synthesis through a stem-loop without pausing. RT polymerizes through the structure and replicates the STOP and distal direct repeat sequences. The resulting provirus (bottom) will not express GFP due to the presence of the stop codons. (B) Synthesis through a stem-loop with pausing. RT dissociates at the pause site (“c”), and RNase H degrades the donor template RNA (dashed line) behind the DNA 3′ terminus. Base pairing and branch migration transfer the nascent DNA to an acceptor template such that RT resumes synthesis at the distal direct repeat. The stem-loop and STOP sequences are deleted, and the resulting provirus translates GFP with a 22-amino acid N-terminal fusion (abcde).
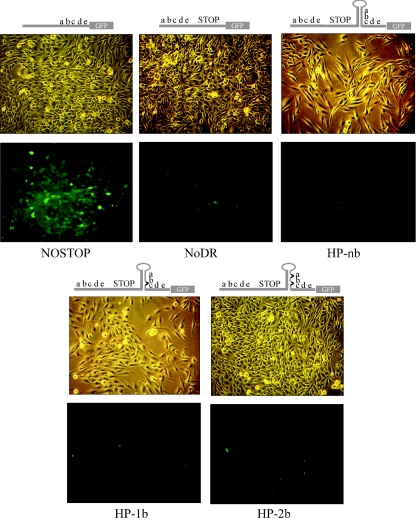
To confirm that the proviral constructs produced the predicted GFP phenotype, D17 cells (which serve as the target cells for infection) (see below) were transfected with each plasmid and selected for G418 resistance (see Materials and Methods). A mock transfection resulted in no G418-resistant cells (data not shown). Figure 3 shows representative data comparing light and fluorescence images for G418-resistant cells transfected by NOSTOP, NoDR, HPnb, HP-1b, and HP-2b. The NOSTOP construct induced cell-associated GFP fluorescence as expected, whereas all other constructs showed virtually no fluorescence. Recombination resulting in GFP expression at the plasmid DNA level was not detected, presumably because the direct repeat is only 61 nucleotides in length and the repeats are separated by only 80 nucleotides. These data indicate that the inserted STOP sequence and resulting frameshifts were sufficient to prevent GFP expression. They also demonstrate that the background due to the internal initiation of GFP translation was negligible.
FIG. 3.
Comparison of GFP expression in D17 cells transfected with the proviral vectors shown in Fig. 1 and selected for G418 resistance for 10 days. Light microscopy (above) and fluorescence microscopy (below) show that only the NOSTOP positive control construct produces cell associated green fluorescence. Above each set of images is a schematic diagram of the construct used for transfection (Fig. 1).
Unpaired nucleotides decrease pausing and strand transfer in vitro.
Although previous work with similar stem-loop structures demonstrated that increased stem base pair contiguity increased HIV-1 RT pausing (33), these structures have not been characterized with M-MLV RT. To test this for MLV RT pausing, in vitro RNA synthesis was performed by cloning the recombination region of each of the constructs in Fig. 1 into pGEM9Zf(−). RNA synthesized in vitro was used as template in a primer extension assay with M-MLV RT. Predicted RNA structures were confirmed using RNase T1 and RNase V1 sensitivity profiles (data not shown). Figure 4A shows the primer extension assays for the various RNAs. The NOSTOP construct induced little pausing with rapid accumulation of the full-length product (Fig. 4A, lanes 1 to 6). Synthesis through HPnb, however, resulted in significant pausing at +5 and +14/+15 within the duplex stem (Fig. 4A, lanes 7 to 12; pause site numbering is in relation to the first base pair of the duplex stem, which is designated as +1). Some weaker pausing at +6, +7, and + 8 was also observed. The M-MLV RT pause pattern was different from that of HIV-1 RT (33) in that the +5 pause was stronger than the +14/+15 pausing for HIV-1 RT. The reason for this difference is unknown. We suspected that the strong band that migrates near the position of the full-length NOSTOP extension product resulted from strand transfer to the distal direct repeat. That this band was indeed the result of a strand transfer event was verified by gel purifying the putative strand transfer products from the HPnb and HP-1b extension reactions as well as the full-length extension product from the NOSTOP reaction and sequencing a PCR amplification product (see Materials and Methods). When a bulge was engineered in the region of the +5 pause by deleting the nucleotide on the distal strand complementary to +5 (HP-1b), a decrease in +5 pausing was observed, but the +14/+15 pauses and strand transfer product still accumulated (Fig. 4A, lanes 13 to 18). To promote synthesis through the +14/+15 pauses, therefore, the nucleotide that base pairs with the +17 residue of the proximal direct repeat was deleted. This nucleotide was chosen based on the normal structure of the HIV-1 TAR element, which has a bulge at this position. The unpaired nucleotide was predicted to not only weaken pausing at +14 and +15 but also decrease the formation of strand transfer product (33). As shown in Fig. 4A, lanes 19 to 24, introducing the bulge produced the predicted results with a concomitant increase in extension to the end of the template. The proportion of strand transfer product generated in each reaction was then determined and normalized to the NOSTOP full-length product. Fig. 4B graphically represents the results of this comparison. Both the HPnb and HP-1b templates generated approximately ∼50% strand transfer product in relation to NOSTOP full-length, whereas HP-2b generated only 10% strand transfer product. Together, these data set a baseline from which we can predict the outcome of synthesis through these structures during viral infection. According to our hypothesis, since reverse transcription should be difficult through HPnb and HP-1b hairpins compared to synthesis through the HP-2b hairpin, there should be a higher rate of recombination and therefore a larger proportion of GFP-positive colonies after infection with HPnb and HP-1b virus compared to the proportion observed after infection with HP-2b virus.
Pausing in vitro directly correlates to an increase in recombination during M-MLV reverse transcription in vivo.
The NIH 3T3-derived clone PG13 produces MLV pseudotyped with the GALV envelope (39). Since the GALV envelope cannot mediate infection of NIH 3T3 cells and its derivatives, the PG13 line is an ideal packaging cell for preventing reinfection and the potential confounding effects of an integrated provirus that has undergone reverse transcription-mediated recombination. Stably transfected PG13 clones producing vNOSTOP, vNoDR, vHPnb, vHP-1b, and vHP-2b (the “v” denotes “virus”) were characterized for their pattern of GFP expression, virus production, and the nature of the integrated proviral sequences. Flow cytometric analysis of the virus-producing cell lines showed that the NOSTOP clones expressed GFP as expected, while the NoDR, HPnb, HP-1b, and HP-2b clones were negative and similar to the untransfected PG13 control cell line (data not shown). Genomic DNA was harvested from the various clones, and the region encompassing the inserted hairpin sequences was amplified by PCR using flanking primers. Only cell clones that had the correct GFP expression pattern, an unmodified PCR product sequence, and a sufficient titer were used for infection of D17 cells for flow cytometry analysis.
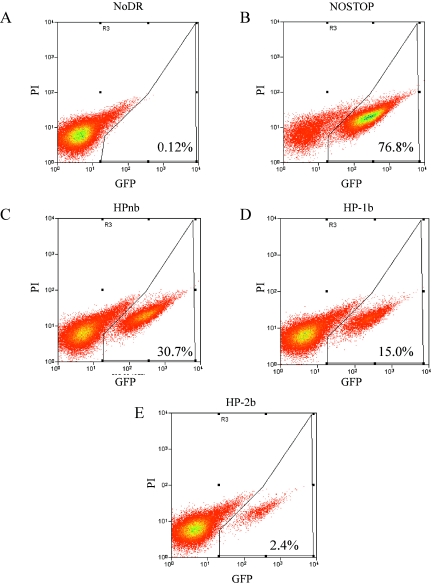
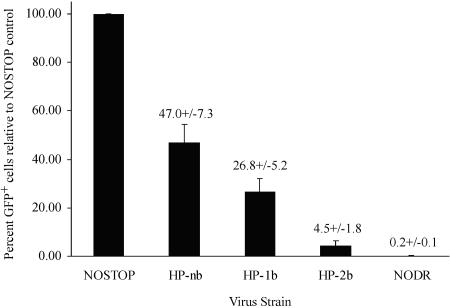
As outlined in Fig. 5, virus derived from these packaging clones was used to singly infect D17 cells, a dog osteosarcoma-derived line susceptible to GALV envelope-mediated MLV infection. Figure 6 shows a representative analysis of G418-resistant D17 cells infected with virus derived from each construct. The data in Fig. 6A and 6B define the lower (NoDR, 0.12% GFP+) and upper (NOSTOP, 76.8% GFP+) limits of detection of GFP+ D17 cells. When virus containing a hairpin structure was used to infect D17 cells, there was a mix of GFP+ and GFP− cells (Fig. 6C, 6D, and 6E). In this particular experiment, approximately 31% of D17 cells infected with HPnb were GFP+, whereas only 2.4% of cells infected with HP-2b were GFP+. For each construct, in a total of four separate experiments, virus from at least three separate packaging lines was used in at least nine independent infections. Normalizing the number of GFP+ cells in each experiment to the corresponding NOSTOP positive control revealed that recombination during reverse transcription of the HPnb genome occurred nearly 50% of the time, whereas for HP-1b, it was 27% and for HP-2b, it was only 4.5% (Fig. 7). Thus, the flow cytometry data show a direct relationship between the amount of pausing within the stem-loop structure observed with in vitro primer extension assays and the number of GFP+ cells detected by flow cytometry.
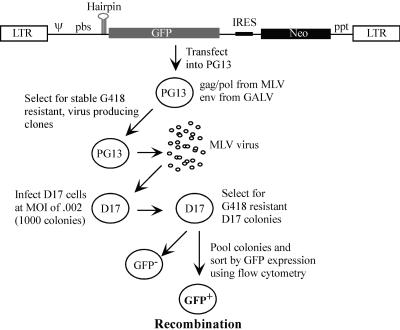
FIG. 5.
Overview of infection assay to determine the proportion of GFP+ D17 cells by flow cytometry. Above is the general proviral structure of plasmids used to create stably transfected PG13 packaging lines. LTR, long terminal repeat.
FIG. 6.
Representative flow cytometry analysis of D17 cells infected with virus derived from virus packaging lines producing NoDR (A) , NOSTOP (B), HPnb (C), HP-1b (D), or HP-2b (E). Horizontal axis shows GFP fluorescence (GFP); vertical axis shows propidium iodide (PI) signal. Percentages of live cells expressing GFP (GFP+ in the text) are indicated in the lower right corner of each panel.
FIG. 7.
Graph showing the mean percentage of GFP+ cells relative to the NOSTOP positive control ± 1 standard deviation. In four separate experiments, virus from at least three independent PG13 packaging lines was used for each construct. Total numbers of live cells analyzed in all four experiments were 389,738 for NOSTOP, 425,736 for NoDR, 429,402 for HPnb, 369,837 for HP-1b, and 511,857 for HP-2b.
GFP expression in infected D17 cells results from recombination-mediated deletion of the stem-loop and STOP sequences.
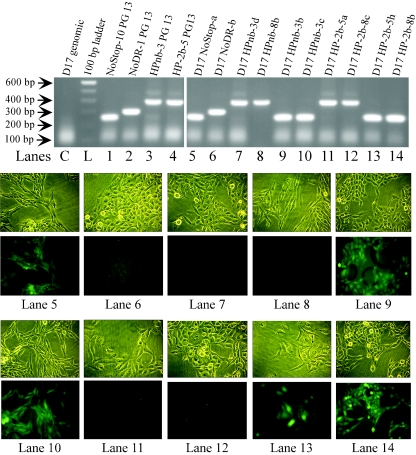
To ensure that our interpretation of the flow cytometry data was correct, single-cell-derived clones produced by infection with vHPnb and vHP-2b were obtained and compared to controls. Each clone was analyzed by fluorescence microscopy to determine the GFP phenotype, and genomic DNA was harvested for PCR amplification of the recombination region. In the absence of recombination, the clone should not express GFP and the PCR product should be approximately 400 bp in size. If recombination occurred by the mechanism predicted in Fig. 2B, the cell line should be GFP+ and the PCR product should be approximately 250 base pairs since the hairpin and STOP sequence would be deleted. Figure 8 shows representative results of this analysis. Lanes 1 to 4 of the agarose gel at the top of the figure are control PCR analyses of stably transfected PG13 virus-packaging cell lines which gave the expected sized PCR products.
FIG. 8.
Comparison of genomic PCR products and GFP phenotypes of cloned, infected D17 cell lines. At the top is an ethidium bromide-stained 1% agarose gel comparing PCR products amplified from genomic DNA from the indicated PG13 and D17 clones. Clone names are indicated above each lane. Lane C shows noninfected D17 genomic DNA, and lane L shows the 100-bp ladder (Invitrogen). Lanes 1 to 4 are PG13 packaging lines for NOSTOP, NoDR, HPnb, and HP-2b, respectively. The right hand portion of gel shows PCR products from D17 clones infected by NOSTOP virus (lane 5), NoDR virus (lane 6), HPnb virus (lanes 7 to 10), and HP-2b virus (lanes 11 to 14). Below the gels are light (top rows) and fluorescent (bottom rows) microscopy comparisons for each of the D17 clones from which genomic DNA was derived for PCR in lanes 5 to 14 of the agarose gel. Below each image set is the corresponding lane on the agarose gel.
Reactions using genomic DNA from infected D17 cloned cell lines demonstrated a direct correspondence between the GFP expression phenotype and the size of the PCR product. Of the 18 D17 clones derived from HPnb infection, 9 were GFP+ and 9 were GFP−, and among 19 HP-2b-derived clones, 2 were GFP+ and 17 were GFP−. PCR amplification from genomic DNA produced a 250-bp fragment only if the cell line was GFP+ (Fig. 8, lanes 9, 10, 13, and 14, and data not shown). All GFP− lines, however, produced only the 400-bp fragment (Fig. 8, lanes 7, 8, 11, and 12, and data not shown). Ten NOSTOP- and 10 NoDR-derived D17 clones all produced a 250- or 300-bp fragment, respectively (Fig. 8, lane 5 and 6, respectively, and data not shown). In total, these data indicated that the D17 GFP+ phenotype correlates completely with deletion of the hairpin and STOP sequences. As further verification, the PCR products from the clones analyzed in Fig. 8 were sequenced. As predicted, the 250-bp band in the recombinants yielded a sequence identical to the sequence observed for the 250-bp NOSTOP control band, whereas the 400-bp band observed for the nonrecombinants maintained the direct repeats, with the only difference between HPnb and HP-2b clones being the deleted nucleotides in the distal hairpin strand for HP-2b (data not shown).
DISCUSSION
This study is the first direct examination of the effects of pausing on retroviral recombination. A previous report correlated the presence of pause sites with crossover points in HIV recombination (21); however, quantitative effects were not determined. Work by Zhang et al. (66) suggested that the presence of a stable hairpin between two direct repeats increases the rate of homologous recombination in MLV. Their data led to the conclusion that stable secondary structures decrease processive RT synthesis and, further, that a functional NC can increase RT processivity. However, in this case, there were no corresponding in vitro data (such as primer extension assays) to correlate with the in vivo observations. Here, we have demonstrated for the first time a direct relationship between pausing in vitro and viral recombination in vivo.
The dynamic copy choice model describing retroviral recombination proposes that an equilibrium between DNA polymerization and RNase H degradation determines the rate of crossover during reverse transcription (57, 67). Factors such as pausing (52, 53), nucleotide misincorporation (19, 45), nontemplated base addition (24), the presence of a functional NC (44, 52, 66), and nucleotide availability (17, 48, 57) all determine the rate of polymerization and whether branch migration will “catch up” to the DNA 3′ terminus. Our in vitro primer extension results demonstrate that pausing increases the proportion of strand transfer product. Extension on the HPnb, HP-1b, and HP-2b RNA templates showed a relative transfer frequency of 46%, 46%, and 10%, respectively. In vivo, the recombination rates during viral reverse transcription generated from these same constructs were 47%, 27%, and 4.5%, respectively. Based on the in vivo results, disrupting RNA base pair contiguity with two unpaired nucleotides near pause sites (Fig. 1B) resulted in a 10-fold decrease in recombination (47% versus 4.5%) while the construct with one unpaired nucleotide (HP-1b) gave an intermediate result. HP-1b generates a greater proportion of strand transfer in vitro than in vivo (46% versus 27%), suggesting that the in vivo assay may be more sensitive at detecting differences in pausing and recombination within a direct repeat. Overall, our data clearly demonstrate a trend between decreased pausing within the stem of a stem-loop structure in vitro and decreased direct repeat recombination in vivo. These results provide additional support for the dynamic copy choice model for retroviral recombination.
It should be noted that strand transfer could have been initiated regardless of pausing, with complete crossover occurring beyond the distal direct repeat. Since this would not restore GFP translation, this type of event would not be detected with our system and, therefore, the overall recombination frequency is likely underestimated. The observation that increased pausing increases the proportion of GFP+-infected D17 cells confirms that pausing can direct 3′-terminal transfer to a location on the acceptor template in close proximity to the pause site in vivo (15, 33).
The results from this work provide further support for the hypothesis that the evolution of secondary structures in the retroviral genome was strongly influenced by the presence of unpaired nucleotides that disrupt base pair contiguity in the stems of stem-loop structures (33, 34). The completely duplex stem used in this study was derived from the HIV-1 TAR element, and both HIV-1 RT and MLV RT-mediated RNA displacement syntheses are impeded at identical positions within the stem that normally have single-nucleotide bulges in close proximity (Fig. 4). We also found that the frequency of homologous recombination is substantially increased when unpaired nucleotides are absent due to increased pausing within the RNA duplex stem (Fig. 6 and 7). Together, these data suggest that the spectrum of possible secondary structures that can form within a retroviral genome is limited by how well the RT can replicate through the structure. If there were strong pausing within the duplex stem, then there would be a greater probability that the secondary structure would be lost due to either abortive synthesis or, more likely, recombination. Stem-loop structures possessing unpaired bulges in the duplex portion, however, would be readily replicated, and the secondary structure would be maintained in the genomes of subsequent generations of virus. In turn, the “replicability” of secondary structures would delineate the pool of RNA conformations available for the development of RNA-protein interactions, as is seen with the HIV-1 TAR sequence and poly(A) signal (6, 32).
The data from this and previous work suggest that retroviral genomes are optimized for replication during reverse transcription (33, 34). Genomes possessing stem-loop structures with strong pause sites would likely exhibit a tendency to modify the stem-loop either by deletion due to recombination or perhaps by the formation of bulges due to RT-mediated mutagenesis. Both processes would facilitate replication of the affected structures in the genome by decreasing the pausing associated with the structure. Since retrovirus-based vectors are used for gene therapy, it may therefore be necessary to evaluate whether a nonretroviral sequence, such as a human gene, should be optimized for reverse transcription. Assuming that human RNA structures do not undergo the same selective processes as retroviral secondary structures, there may be stem-loop or other RNA secondary structures within human sequences that induce significant RT pausing in the context of reverse transcription. For retroviruses used in gene therapy, this may cause abortive reverse transcription or unwanted modification of the exogenous human sequence through recombination, which would potentially decrease transduction efficiency. Our work suggests that by destabilizing the duplex portions of RNA secondary structures, the negative effects of pausing can be avoided. Importantly, this could be accomplished without modifying the coding sequence because a nucleotide involved in base pairing can be changed without altering the encoded amino acid. By generating a mismatch that disrupts base pair contiguity, pausing can be reduced and replication of the exogenous human sequence by RT enhanced.
Acknowledgments
This work was supported by National Institutes of Health grant CA51605.
We thank V. K. Pathak, M. Lagunoff, M. Linial, T. Pham, P. Carroll, R. Life, and J. Karlinsey for assistance, reagents, and equipment use. Thanks to M. Zhang for excellent RNA structural probing and to David Coil for extremely helpful discussions. Also, we thank M. Linial, J. B. Leppard, J. Carey, B. T. Paulson, S. Schultz, and H. Interthal for discussion of data and for review of the manuscript prior to submission.
REFERENCES
- 1.Andersen, E. S., R. E. Jeeninga, C. K. Damgaard, B. Berkhout, and J. Kjems. 2003. Dimerization and template switching in the 5′ untranslated region between various subtypes of human immunodeficiency virus type 1. J. Virol. 77:3020-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. A., V. K. Pathak, and W. S. Hu. 2000. Effect of the murine leukemia virus extended packaging signal on the rates and locations of retroviral recombination. J. Virol. 74:6953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. A., R. J. Teufel II, P. D. Yin, and W. S. Hu. 1998. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J. Virol. 72:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakrishnan, M., P. J. Fay, and R. A. Bambara. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482-36492. [DOI] [PubMed] [Google Scholar]
- 5.Balakrishnan, M., B. P. Roques, P. J. Fay, and R. A. Bambara. 2003. Template dimerization promotes an acceptor invasion-induced transfer mechanism during human immunodeficiency virus type 1 minus-strand synthesis. J. Virol. 77:4710-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkhout, B., B. Klaver, and A. T. Das. 1997. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 25:940-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair, D. G., W. S. Mason, E. Hunter, and P. K. Vogt. 1976. Temperature-sensitive mutants of avian sarcoma viruses: genetic recombination between multiple or coordinate mutants and avian leukosis viruses. Virology 75:48-59. [DOI] [PubMed] [Google Scholar]
- 8.Brincat, J. L., J. K. Pfeiffer, and A. Telesnitsky. 2002. RNase H activity is required for high-frequency repeat deletion during Moloney murine leukemia virus replication. J. Virol. 76:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y., M. Balakrishnan, B. P. Roques, and R. A. Bambara. 2003. Steps of the acceptor invasion mechanism for HIV-1 minus strand strong stop transfer. J. Biol. Chem. 278:38368-38375. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y., M. Balakrishnan, B. P. Roques, P. J. Fay, and R. A. Bambara. 2003. Mechanism of minus strand strong stop transfer in HIV-1 reverse transcription. J. Biol. Chem. 278:8006-8017. [DOI] [PubMed] [Google Scholar]
- 11.Cheslock, S. R., J. A. Anderson, C. K. Hwang, V. K. Pathak, and W. S. Hu. 2000. Utilization of nonviral sequences for minus-strand DNA transfer and gene reconstitution during retroviral replication. J. Virol. 74:9571-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin, M. P., T. D. Rhodes, J. Chen, W. Fu, and W. S. Hu. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. USA 102:9002-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffin, J. M. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42:1-26. [DOI] [PubMed] [Google Scholar]
- 14.Coffin, J. M., S. H. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 15.Derebail, S. S., and J. J. DeStefano. 2004. Mechanistic analysis of pause site-dependent and -independent recombinogenic strand transfer from structurally diverse regions of the HIV genome. J. Biol. Chem. 279:47446-47454. [DOI] [PubMed] [Google Scholar]
- 16.Derebail, S. S., M. J. Heath, and J. J. DeStefano. 2003. Evidence for the differential effects of nucleocapsid protein on strand transfer in various regions of the HIV genome. J. Biol. Chem. 278:15702-15712. [DOI] [PubMed] [Google Scholar]
- 17.DeStefano, J. J., L. M. Mallaber, L. Rodriguez-Rodriguez, P. J. Fay, and R. A. Bambara. 1992. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J. Virol. 66:6370-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeStefano, J. J., W. Wu, J. Seehra, J. McCoy, D. Laston, E. Albone, P. J. Fay, and R. A. Bambara. 1994. Characterization of an RNase H deficient mutant of human immunodeficiency virus-1 reverse transcriptase having an aspartate to asparagine change at position 498. Biochim. Biophys. Acta 1219:380-388. [DOI] [PubMed] [Google Scholar]
- 19.Diaz, L., and J. J. DeStefano. 1996. Strand transfer is enhanced by mismatched nucleotides at the 3′ primer terminus: a possible link between HIV reverse transcriptase fidelity and recombination. Nucleic Acids Res. 24:3086-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duch, M., M. L. Carrasco, T. Jespersen, L. Aagaard, and F. S. Pedersen. 2004. An RNA secondary structure bias for non-homologous reverse transcriptase-mediated deletions in vivo. Nucleic Acids Res. 32:2039-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dykes, C., M. Balakrishnan, V. Planelles, Y. Zhu, R. A. Bambara, and L. M. Demeter. 2004. Identification of a preferred region for recombination and mutation in HIV-1 gag. Virology 326:262-279. [DOI] [PubMed] [Google Scholar]
- 22.Galetto, R., A. Moumen, V. Giacomoni, M. Veron, P. Charneau, and M. Negroni. 2004. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J. Biol. Chem. 279:36625-36632. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh, M., K. J. Howard, C. E. Cameron, S. J. Benkovic, S. H. Hughes, and S. F. Le Grice. 1995. Truncating alpha-helix E' of p66 human immunodeficiency virus reverse transcriptase modulates RNase H function and impairs DNA strand transfer. J. Biol. Chem. 270:7068-7076. [DOI] [PubMed] [Google Scholar]
- 24.Golinelli, M. P., and S. H. Hughes. 2002. Nontemplated base addition by HIV-1 RT can induce nonspecific strand transfer in vitro. Virology 294:122-134. [DOI] [PubMed] [Google Scholar]
- 25.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 26.Hu, W. S., T. Rhodes, Q. Dang, and V. Pathak. 2003. Retroviral recombination: review of genetic analyses. Front Biosci. 8:d143-d155. [DOI] [PubMed] [Google Scholar]
- 27.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 28.Hwang, C. K., E. S. Svarovskaia, and V. K. Pathak. 2001. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc. Natl. Acad. Sci. USA 98:12209-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, J. K., C. Palaniappan, W. Wu, P. J. Fay, and R. A. Bambara. 1997. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 272:16769-16777. [DOI] [PubMed] [Google Scholar]
- 31.Klarmann, G. J., C. A. Schauber, and B. D. Preston. 1993. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J. Biol. Chem. 268:9793-9802. [PubMed] [Google Scholar]
- 32.Klaver, B., and B. Berkhout. 1994. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 13:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanciault, C., and J. J. Champoux. 2005. Effects of unpaired nucleotides within HIV-1 genomic secondary structures on pausing and strand transfer. J. Biol. Chem. 280:2413-2423. [DOI] [PubMed] [Google Scholar]
- 34.Lanciault, C., and J. J. Champoux. 2004. Single unpaired nucleotides facilitate HIV-1 reverse transcriptase displacement synthesis through duplex RNA. J. Biol. Chem. 279:32252-32261. [DOI] [PubMed] [Google Scholar]
- 35.Li, T., and J. Zhang. 2000. Determination of the frequency of retroviral recombination between two identical sequences within a provirus. J. Virol. 74:7646-7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linial, M., and S. Brown. 1979. High-frequency recombination within the gag gene of Rous sarcoma virus. J. Virol. 31:257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCutchan, F. E., J. K. Carr, M. Bajani, E. Sanders-Buell, T. O. Harry, T. C. Stoeckli, K. E. Robbins, W. Gashau, A. Nasidi, W. Janssens, and M. L. Kalish. 1999. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology 254:226-234. [DOI] [PubMed] [Google Scholar]
- 38.Mikkelsen, J. G., S. V. Rasmussen, and F. S. Pedersen. 2004. Complementarity-directed RNA dimer-linkage promotes retroviral recombination in vivo. Nucleic Acids Res. 32:102-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moumen, A., L. Polomack, B. Roques, H. Buc, and M. Negroni. 2001. The HIV-1 repeated sequence R as a robust hot-spot for copy-choice recombination. Nucleic Acids Res. 29:3814-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moumen, A., L. Polomack, T. Unge, M. Veron, H. Buc, and M. Negroni. 2003. Evidence for a mechanism of recombination during reverse transcription dependent on the structure of the acceptor RNA. J. Biol. Chem. 278:15973-15982. [DOI] [PubMed] [Google Scholar]
- 42.Negroni, M., and H. Buc. 2000. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. USA 97:6385-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negroni, M., and H. Buc. 2001. Mechanisms of retroviral recombination. Annu. Rev. Genet. 35:275-302. [DOI] [PubMed] [Google Scholar]
- 44.Negroni, M., and H. Buc. 1999. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J. Mol. Biol. 286:15-31. [DOI] [PubMed] [Google Scholar]
- 45.Palaniappan, C., M. Wisniewski, W. Wu, P. J. Fay, and R. A. Bambara. 1996. Misincorporation by HIV-1 reverse transcriptase promotes recombination via strand transfer synthesis. J. Biol. Chem. 271:22331-22338. [DOI] [PubMed] [Google Scholar]
- 46.Parthasarathi, S., A. Varela-Echavarria, Y. Ron, B. D. Preston, and J. P. Dougherty. 1995. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J. Virol. 69:7991-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pathak, V. K., and H. M. Temin. 1992. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J. Virol. 66:3093-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfeiffer, J. K., M. M. Georgiadis, and A. Telesnitsky. 2000. Structure-based moloney murine leukemia virus reverse transcriptase mutants with altered intracellular direct-repeat deletion frequencies. J. Virol. 74:9629-9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfeiffer, J. K., R. S. Topping, N. H. Shin, and A. Telesnitsky. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 73:8441-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodes, T., H. Wargo, and W. S. Hu. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhodes, T. D., O. Nikolaitchik, J. Chen, D. Powell, and W. S. Hu. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roda, R. H., M. Balakrishnan, M. N. Hanson, B. M. Wohrl, S. F. Le Grice, B. P. Roques, R. J. Gorelick, and R. A. Bambara. 2003. Role of the reverse transcriptase, nucleocapsid protein, and template structure in the two-step transfer mechanism in retroviral recombination. J. Biol. Chem. 278:31536-31546. [DOI] [PubMed] [Google Scholar]
- 53.Roda, R. H., M. Balakrishnan, J. K. Kim, B. P. Roques, P. J. Fay, and R. A. Bambara. 2002. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J. Biol. Chem. 277:46900-46911. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Rodriguez, L., Z. Tsuchihashi, G. M. Fuentes, R. A. Bambara, and P. J. Fay. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 270:15005-15011. [DOI] [PubMed] [Google Scholar]
- 55.Stuhlmann, H., and P. Berg. 1992. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J. Virol. 66:2378-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suo, Z., and K. A. Johnson. 1997. Effect of RNA secondary structure on the kinetics of DNA synthesis catalyzed by HIV-1 reverse transcriptase. Biochemistry 36:12459-12467. [DOI] [PubMed] [Google Scholar]
- 57.Svarovskaia, E. S., K. A. Delviks, C. K. Hwang, and V. K. Pathak. 2000. Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J. Virol. 74:7171-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takebe, Y., K. Motomura, M. Tatsumi, H. H. Lwin, M. Zaw, and S. Kusagawa. 2003. High prevalence of diverse forms of HIV-1 intersubtype recombinants in Central Myanmar: geographical hot spot of extensive recombination. AIDS 17:2077-2087. [DOI] [PubMed] [Google Scholar]
- 59.Takehisa, J., L. Zekeng, E. Ido, Y. Yamaguchi-Kabata, I. Mboudjeka, Y. Harada, T. Miura, L. Kaptu, and M. Hayami. 1999. Human immunodeficiency virus type 1 intergroup (M/O) recombination in Cameroon. J. Virol. 73:6810-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, W., B. M. Blumberg, P. J. Fay, and R. A. Bambara. 1995. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J. Biol. Chem. 270:325-332. [DOI] [PubMed] [Google Scholar]
- 61.Xu, H., and J. D. Boeke. 1987. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84:8553-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin, P. D., V. K. Pathak, A. E. Rowan, R. J. Teufel II, and W. S. Hu. 1997. Utilization of nonhomologous minus-strand DNA transfer to generate recombinant retroviruses. J. Virol. 71:2487-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, J., L. Y. Tang, T. Li, Y. Ma, and C. M. Sapp. 2000. Most retroviral recombinations occur during minus-strand DNA synthesis. J. Virol. 74:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, J., and H. M. Temin. 1993. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science 259:234-238. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, J., and H. M. Temin. 1994. Retrovirus recombination depends on the length of sequence identity and is not error prone. J. Virol. 68:2409-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, W. H., C. K. Hwang, W. S. Hu, R. J. Gorelick, and V. K. Pathak. 2002. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J. Virol. 76:7473-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhuang, J., A. E. Jetzt, G. Sun, H. Yu, G. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76:11273-11282. [DOI] [PMC free article] [PubMed] [Google Scholar]