Abstract
Polyphosphate (polyP), a linear polymer of hundreds of orthophosphate residues, exists in all tested cells in nature, from pathogenic bacteria to mammals. In bacteria, polyP has a crucial role in stress responses and stationary-phase survival. Polyphosphate kinase (PPK) is the principal enzyme that catalyses the synthesis of polyP in bacteria. It has been shown that PPK is required for bacterial motility, biofilm formation and the production of virulence factors. PPK inhibitors may thus provide a unique therapeutic opportunity against antibiotic-resistant pathogens. Here, we report crystal structures of full-length Escherichia coli PPK and its complex with AMPPNP (β-γ-imidoadenosine 5-phosphate). PPK forms an interlocked dimer, with each 80 kDa monomer containing four structural domains. The PPK active site is located in a tunnel, which contains a unique ATP-binding pocket and may accommodate the translocation of synthesized polyP. The PPK structure has laid the foundation for understanding the initiation of polyP synthesis by PPK.
Keywords: polyphosphate, polyphosphate kinase, antibiotic, crystal structure, polymerase
Introduction
Inorganic polyphosphate (polyP), a linear polymer of Pi linked by phosphoanhydride bonds, is found in every cell in nature and possibly is conserved from prebiotic times. The importance of polyP in bacterial pathogenesis has been well documented (Kornberg et al, 1999; Brown & Kornberg, 2004). Under nutritional stringencies and environmental stresses, many Gram-negative bacteria rapidly produce polyP. PolyP regulates rpoS and recA expression at the transcriptional level and affects the expression of many stress-inducible and stationary-phase-inducible genes (Loewen et al, 1998; Shiba et al, 2000; Tsutsumi et al, 2000). PolyP also controls the degradation of ribosomal proteins by means of the Lon protease in response to nutritional downshift (Kuroda et al, 2001). It remains unclear how these deficiencies and stress signals lead to polyP accumulation. Polyphosphate kinase (PPK, also known as PPK1) is the key enzyme responsible for catalysing the reversible conversion of the γ-phosphate of ATP to polyP (Fig 1A). The importance of polyP and PPK in bacterial pathogenesis has been tested through the depletion of the ppk gene in principal human pathogens such as Pseudomonas aeruginosa. It has been shown that PPK is essential for motility, quorum sensing and biofilm formation, which make important contributions to antibiotic resistance of clinically important pathogens (Rashid et al, 2000; Chen et al, 2002). Importantly, PPK is also required for the release of virulence factors. Studies with both the ppk-knockout bacteria and PPKspecific inhibitors indicate that PPK is an attractive antimicrobial drug target, and the mechanism of action of PPK inhibitors may be distinct from that of existing antibiotics (S. Lee, unpublished data). Intriguingly, as polyP is required for stationary-phase survival, PPK inhibitors may be useful to eradicate non-multiplying bacteria.
Figure 1.
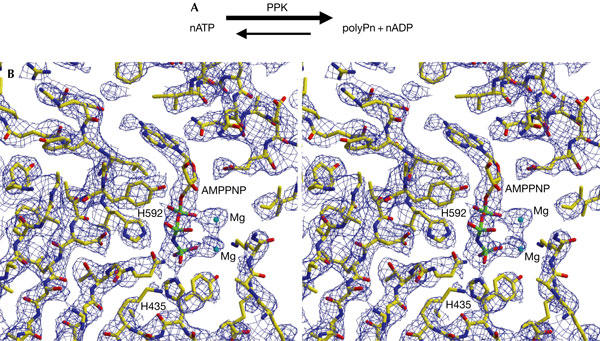
Chemical reaction and experimental electron density map. (A) The chemical reaction catalysed by polyphosphate kinase (PPK). (B) The stereo view of experimental map, showing the AMPPNP (β-γ-imidoadenosine 5-phosphate)-binding region of PPK, calculated from the Se-Met multiwavelength anomalous diffraction data set at 3.0 Å resolution and contoured at 1.0σ. Residues H435 and H592 form part of the active site, with an ordered γ-phosphate group of AMPPNP sitting in between.
Although highly conserved among both Gram-negative and Gram-positive bacteria, including Bacillus anthracis (Tzeng & Kornberg, 1998), PPK does not contain obvious sequence homology with other protein families including ATP- or GTP-binding proteins. With the exception of Dictyostelium discoideum, the enzyme that catalyses polyP synthesis in eukaryotes may be distinct from bacterial PPK and remains to be discovered (Gomez-Garcia & Kornberg, 2004). Among Gram-negative bacterial PPKs, the Escherichia coli PPK has been the fully characterized enzyme. The dimerization of PPK is crucial for the synthesis of polyP (forward reaction) and ATP (reverse reaction; Tzeng & Kornberg, 2000). Autophosphorylation of PPK is the initial step of polyP synthesis (Ahn & Kornberg, 1990). Interestingly, purified recombinant PPK catalyses the elongation of polyP in a highly processive way and terminates the polyP synthesis when the polyP chain reaches about 750 phosphate groups (Kumble et al, 1996).
Here, we report the crystal structures of full-length E. coli PPK and its complex with AMPPNP (β-γ-imidoadenosine 5-phosphate). These structures have shown, for the first time, the architecture of the PPK family proteins and how a PPK can catalyse its autophosphorylation. The crystal structure shows that the PPK catalytic reactions are carried out in a highly conserved structural tunnel. Catalysis inside a tunnel is a common feature observed in other polymerases such as the ribosome and RNA polymerases. Thus the PPK structure may shed light on polymerase evolution, as PPK can be characterized as a polymerase without a template. Furthermore, the PPK structure also expanded our view on how ATP is used in the cells by showing yet another unique ATP-binding site.
Results and Discussion
Overall structure of Escherichia coli PPK
The crystal structures of both unliganded and AMPPNP-bound forms of full-length E. coli PPK were determined in the same crystal lattice at 2.5 Å resolution (Fig 1; supplementary Fig 1S and supplementary Table 1 online). In the crystal lattice, each asymmetric unit contains two PPK monomers, which are related by a pseudo two-fold symmetry and form an interlocked dimer structure (Fig 2A). The two PPK subunits in the asymmetric unit are essentially identical in structure with root mean square deviation (r.m.s.d.) for all PPK Cα atoms of 0.23 Å. The two PPK monomers in the asymmetric unit share a buried interface of 4,412.4 Å, which accounts for 9% of the total surface area of the dimer. There are only minor changes in the conformation of the monomer or the organization of the dimer when AMPPNP is bound to PPK. The r.m.s.d. of all 687 Cα atoms of the unliganded PPK and its AMPPNP complex is 1.5 Å. Each PPK monomer shows an Lshaped structure with four structural domains (Fig 2B). These domains include the amino-terminal domain (N domain), the ‘head' domain (H domain) and two closely related carboxy-terminal domains (C1 and C2 domains). The N domain, residues 2–106, forms a bundle of three long antiparallel α-helices, which lie on the upper surface of the C-terminal domains. The N domain is a highly conserved region of PPKs (Fig 3) and provides the upper binding interface for the adenine ring of the ATP (Fig 2B). The H-domain (residues 107–321), which shows the lowest degree of homology among these domains, has a core α/β/α fold in the middle and forms the outward-facing ‘head' of the PPK monomer. The H-domain interacts with the C1-domain of the other PPK monomer in the asymmetric unit and is involved in dimerization (Fig 2A). Both the C1 and C2 domains (residues 322–502 and 503–687, respectively) consist of a sevenstranded mixed β-sheet flanked by five α-helices. However, the structural topology and relative orientations of the helices to the β-sheet in these two domains are different (Fig 2B). The C1 and C2 domains are highly conserved in the PPK family (Fig 3). Some of the residues previously shown to be crucial for the enzyme catalytic activity are located in these two domains (Tzeng & Kornberg, 2000).
Figure 2.
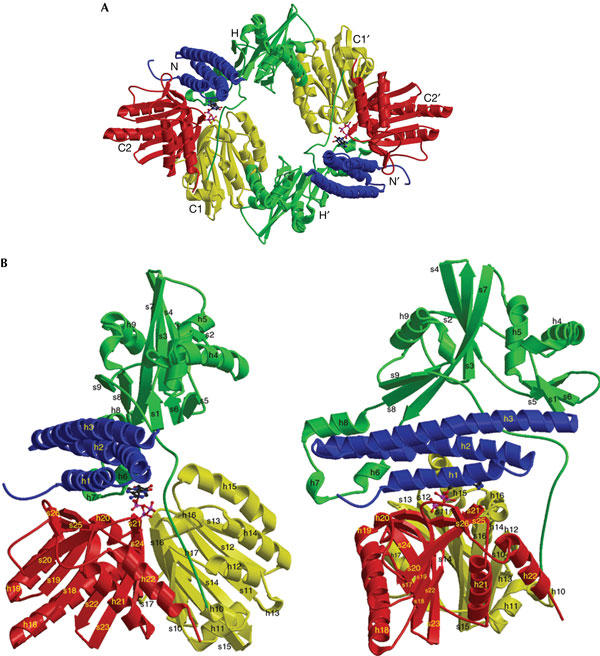
Overall structure of Escherichia coli polyphosphate kinase (PPK). (A) Structure of the PPK dimer in the asymmetric unit. The N-terminal domain of both subunits (residues 2–106) is coloured in blue, the head domain (residues 107–321) in green, the C-terminal domain C1 (residues 322–502) in yellow and domain C2 (residues 503–687) in red. The head domain of one PPK molecule (H) interacts with the C1-domain of the other PPK molecule (C1′). Each AMPPNP (β-γ-imidoadenosine 5-phosphate) molecule is depicted in a ball-andstick representation, coloured in blue and magenta. (B) Structure of PPK monomer (80 kDa, 687 amino acids), viewed from the side (left) and after a 90° rotation from the side (right). Helices (h) and strands (s) are labelled consecutively from the N to C termini. All domains and AMPPNP are coloured in the same code as (A).
Figure 3.
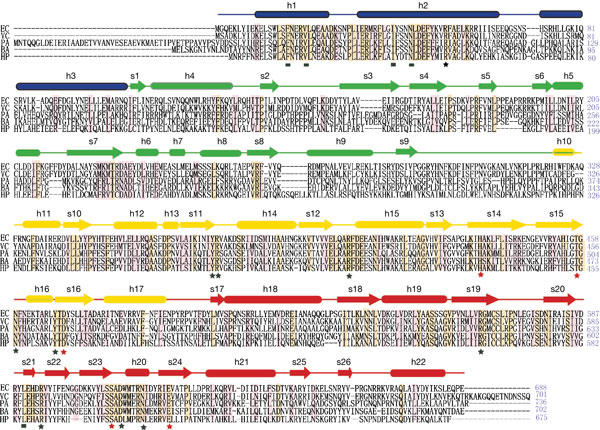
Sequence alignment of polyphosphate kinase (PPK). Identical residues are shaded in yellow, and partially conserved residues are shaded in pink. Secondary structure elements derived from the Escherichia coli PPK structure are indicated above the sequence; α-helices are presented as tubes and βstrands as arrows. The segments of the four domains are shown in the same colour code as in Fig 2A,B. Residues involved in Mg-AMPPNP (β-γ-imidoadenosine 5-phosphate) binding are indicated by the squares and stars under the sequence. Black squares identify the residues involved in side-chain contacts with the adenine ring of the AMPPNP, and stars identify the residues involved in side-chain contacts with the phosphate groups of AMPPNP. The phosphorylated site H435 and the residues involved in the autophosphorylation (D470 and E623) are indicated by red stars. EC, Escherichia coli; VC, Vibrio cholerae; PA, Pseudomonas aeruginosa; BA, Bacillus anthracis; HP, Helicobacter pylori.
Structural tunnel in each PPK subunit
A crucial structural feature of PPK is the tunnel that penetrates the centre of each PPK monomer. This tunnel is mostly formed by the intersection of the N, C1 and C2 domains. Helices h6 and h7 of the H domain also form part of the tunnel wall. The tunnel runs under the three-helix bundle (N domain) and is roughly perpendicular to the PPK dimer plane (Fig 4; supplementary Fig 2S online). Strikingly, 56 of the 108 conserved PPK residues are located within the tunnel, suggesting its functional importance. One side of the tunnel can accommodate only one ATP molecule, whereas the other side contains highly conserved, positively charged residues along the tunnel. These positively charged residues may interact with the polyP chain during polyP elongation. When ATP binds to the active site, the dimension of the rest of the tunnel can accommodate only one linear polyP chain of about 6–10 residues, indicating that the polyP chain is unlikely to elongate on the autophosphorylated H435. It is plausible that ATP enters from one side of the tunnel and the synthesized polyP chain exits from the other side, with the newly synthesized end of the polyP chain remaining at the active site to accept the γ-phosphate group from the next ATP bound to the tunnel. The tunnel may constantly hold the polyP chain during its elongation and allow for its growth on one end, and thus allow PPK to synthesize polyP in a processive way. The mechanism of polyP elongation and termination will require further structural and biochemical studies.
Figure 4.

Cutaway illustration of the solvent-accessible surface of polyphosphate kinase (PPK) structure. The front portion of the protein has been cut away to show the tunnel, which contains the AMPPNP (β-γ-imidoadenosine 5-phosphate) molecule. The PPK structure is oriented with the three long helices in the N-terminal domain roughly in parallel with the paper surface. The cutaway surface is roughly parallel with the C1- and C2-domain interface, with the C2 domain in the back. The N and C termini of PPK are indicated by arrows. The autophosphorylation site H435 is labelled in its position. The surface of PPK is coloured by element with carbon, oxygen, nitrogen and sulphur atoms in white, red, blue and orange, respectively. The dimension of the tunnel is indicated in the box with dashed lines.
The N and H domains, as well as the C1 and C2 domains, form extensive interfaces. In comparison, the interface between these two blocks (N and H domain, C1 and C2 domain, respectively) is small because of the intervening tunnel (Figs 2B and 4). The PPK monomer is probably composed of two relatively rigid structural blocks. It is plausible that the dimerization of PPK contributes to the stabilization of the relative orientation between these two structural blocks and thus stabilizes the tunnel structure in the observed active conformation.
Unique ATP-binding pocket of PPK
In the PPK–AMPPNP complex, the AMPPNP molecule, a non-hydrolysable ATP analogue, occupies one side of the tunnel, with phosphate groups extending deeply into the tunnel. The adenine ring of AMPPNP binds to a highly hydrophobic pocket inside the PPK tunnel under the three-helix bundle of the N domain, which provides the ‘roof' of the adenine-binding site (Figs 1 and 2; supplementary Fig 3S online). Residues I41, F17 and N45 in the PPK N domain form part of this hydrophobic pocket (Fig 5A). The ‘bottom' of the adenine ring is supported by L589 from the C2 domain. It was shown that PPK is specific for ATP as the substrate in the formation of polyP (Tzeng & Kornberg, 2000). There is only one hydrogen bond between the adenine ring and PPK, formed between the side chain of PPK N45 and N7 of AMPPNP, which cannot distinguish ATP from GTP (supplementary Fig 3S online). The distance between adenine N6 and the carbonyl oxygen of PPK N587 is too long for a hydrogen bond (4.23 Å), but the replacement of adenine N6 by guanine O6 may cause unfavourable polar–polar interactions. Thus, this interaction may contribute to the PPK substrate specificity. The phosphate groups of AMPPNP bind to the highly positively charged ‘bottom' of the pocket formed by the C1 and C2 domains. The key residues in this pocket are held in place by an extensive network of hydrogen bonds (Fig 5A). Two magnesium ions coordinate with all three phosphate groups; this is consistent with the requirement of the magnesium ion in PPK catalysis (Ahn & Kornberg, 1990). All the residues interacting with ATP phosphate groups are highly conserved in all PPKs identified thus far (Fig 3). Previous studies have shown that PPK mutations R375A and R564A completely abolish PPK enzymatic activity (Tzeng & Kornberg, 2000). In our structure, both R375 and R564 interact directly with ATP phosphate groups (Fig 5A).
Figure 5.
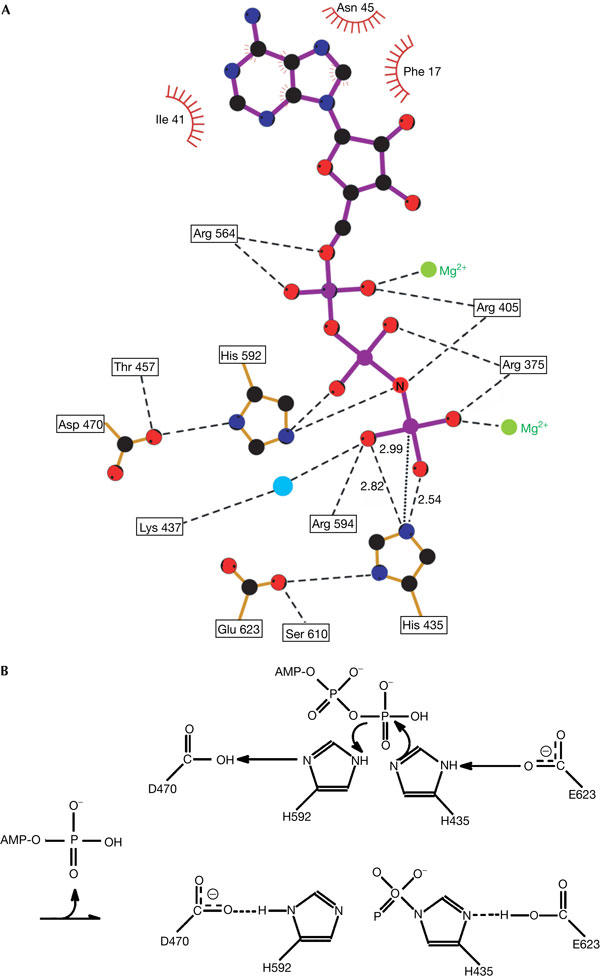
ATP-binding site of polyphosphate kinase (PPK) and the chemical mechanism for PPK autophosphorylation. (A) The hydrogen-bonding network present in the PPK–AMPPNP (β-γ-imidoadenosine 5-phosphate) complex. The dashed lines indicate hydrogen bonds formed among phosphate groups of AMPPNP, Mg2+ and PPK activesite residues, with distances shown in angstroms. Hydrophobic contacts are indicated as radial lines around residues. The dotted line shows the distance between the γ-phosphorus atom of AMPPNP and the Nɛ2 atom of PPK His 435. (B) The proposed chemical mechanism for PPK autophosphorylation.
Structure similarity between PPK and the PLD family
The first step of PPK-catalysed polyP synthesis involves the autophosphorylation of its histidine residues, and PPK is therefore a histidine kinase. We found no structural similarity between PPK and other histidine kinases. However, in spite of the lack of sequence homology, the C1 and C2 domains of PPK, a protein histidine kinase, are structurally similar to the catalytic domain of phospholipase D (PLD), a lipid phosphatase (Leiros et al, 2000). PLD also contains two repetitive catalytic domains, and each PLD domain consists of an eightstranded mixed β-sheet flanked by five α-helices, with a topological arrangement different from that of PPK (supplementary Fig 4S online). When we superimposed the PPK and PLD structures using the program TOPP (CCP4), the r.m.s.d. of 172 Cα atoms position (out of 367 residues in PPK C1 and C2 domains) was 1.77 Å and 35 out of these 367 residues (9.5%) were identical in sequence. Most of the residues that can be superimposed belong to the central β-sheet. PPK structure is less similar to two other PLD family members Nuc1 (Stuckey & Dixon, 1999) and Tdp1 (Davies et al, 2002). Some residues in the activesite motif HxK(x)4D(x)6GSxN of the PLD superfamily are spatially conserved in the active site of PPK (supplementary Fig 5S online). It is possible that PPK and the PLD family have a common ancestor and diverged early in evolution. It should be noted that, whereas the first step of the PPK reaction is autophosphorylation of a histidine residue, the first step of the PLD reaction is the transfer of a phosphate moiety to a PLD histidine residue.
Chemical mechanism of PPK autophosphorylation
It has been shown that the first step of polyP synthesis is the autophosphorylation of PPK. Previous mutagenesis studies showed H435 and H454 as possible autophosphorylation site(s) of PPK (Kumble et al, 1996). In the PPK–AMPPNP crystal structure, H435 directly interacts with the AMPPNP γ-phosphate group, from a ‘back-on' attacking position, whereas H454 is totally buried in the hydrophobic core of the C1-domain (Fig 1B). This observation strongly suggests that H435 is the only autophosphorylation site of PPK.
We propose that H435 of PPK functions as a nucleophile attacking the phosphodiester bond of the γ-phosphate group of ATP, whereas H592 functions as a general acid protonating the oxygen atom between the β- and γ-phosphate (Fig 5B). There are four highly conserved amino acids of the C1 and C2 domains that form crucial hydrogen bonds: E623 with H435, and D470 with H592. It is plausible that E623 has a role in selecting the correct rotamer of H435 and lowering the pKa for attacking ATP. The probable role of D470 is to bind and correctly orientate the general acid H592. This model is consistent with previous biochemical studies, which showed that mutants H435Q and H592Q failed to autophosphorylate (Kumble et al, 1996).
Implications for drug design
The unique ATP-binding pocket of PPK is an obvious target for developing PPK inhibitors, which may be a new type of antibiotic. The hydrophobic pocket for the adenine ring under the three-helix bundle can be particularly important for designing PPK inhibitors. The rest of the tunnel could also provide an extended drug pocket for the ATP competitive inhibitors. In addition, compounds that can disrupt the PPK dimer interface may also be potent PPK inhibitors, as the formation of the PPK dimer is crucial for intact PPK activity.
Methods
Crystallization and data collection. Full-length His-tagged E. coli PPK protein was overexpressed and purified as described (Zhu et al, 2003). PPK crystals were produced by hanging-drop vapour diffusion from a solution containing 12–16% 1,6-hexanediol, 0.1 M Tris–HCl, pH 7.5, 14–16% (v/v) glycerol and 10 mM dithiothreitol at 23°C. Specific procedures, including extensive dehydration of PPK crystals, were developed to improve PPK crystal diffraction quality to beyond 2.5 Å resolution (Zhu et al, 2003). The crystals in complex with AMPPNP were obtained by co-crystallizing in similar conditions with the addition of 2 mM AMPPNP and 5 mM MgCl2. A 2.8 Å resolution selenium multiwavelength anomalous diffraction (MAD) data set was collected at APS beamline 19ID and processed with HKL2000 (Otwinowski & Minor, 1997). All crystals were in space group P42212 with cell dimensions of a=b=152.0 Å and c=150.0 Å.
Structural determination and structural analysis. The structure of the PPK–AMPPNP complex was determined by MAD using a selenomethionine derivative (supplementary Table 1 online). There are two molecules in the asymmetric unit. A total of 21 out of 30 possible selenium sites were found and initial phases were calculated with SOLVE and RESOLVE (Terwilliger & Berendzen, 1999); the initial density map was of high quality and allowed for unambiguous model building with the program XtalView (McRee, 1999) for about 90% of the main chain and more than 60% of the side chains of all PPK residues. The structure refinement was carried out with CNS (Brunger et al, 1998) against a 2.5 Å PPK–AMPPNP data set. With anisotropic B-factor and bulk solvent corrections, the final R-factors are Rwork=24.8% and Rfree=27.4%. The model quality was analysed with PROCHECK, and 97.9% of the residues were in the favoured and additionally allowed regions of the Ramachandran plot. Every PPK residue and the bound AMPPNP, with the exception of the first methionine and the C-terminal His6 tag, were visible in the final electron density map.
The structure of unliganded PPK was determined by molecular replacement using the program AmoRe (Navaza, 1994). The statistics on data processing, structure determination and refinement are summarized in supplementary Table 1 online. To analyse the PPK quaternary structure, the area of interface burial between PPK subunits was calculated with GRASP (Nicholls et al, 1991).
Protein Data Bank accession numbers. The atomic coordinates and structure factors of E. coli PPK and its complex with AMPPNP have been deposited in the Protein Data Bank under the accession codes 1XDO and 1XDP.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr R. Zhang at beamlines 19ID and 19BM (APS) and the staff at 5.0.1, 5.0.2 and 8.0.2 (ALS) for facilitating X-ray data collection and Dr R. Stenkamp, Dr R. Klevit, Dr E. Merritt and Dr N. Zheng for critical comments on the manuscript. This work was supported by NIH Grant AI056253 to W.X., and in part by the W.M. Keck Foundation Center for Microbial Pathogens at the University of Washington.
References
- Ahn K, Kornberg A (1990) Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem 265: 11734–11739 [PubMed] [Google Scholar]
- Brown MR, Kornberg A (2004) Inorganic polyphosphate in the origin and survival of species. Proc Natl Acad Sci USA 101: 16085–16087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chen W, Palmer RJ, Kuramitsu HK (2002) Role of polyphosphate kinase in biofilm formation by Porphyromonas gingivalis. Infect Immun 70: 4708–4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DR, Interthal H, Champoux JJ, Hol WG (2002) The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure (Camb) 10: 237–248 [DOI] [PubMed] [Google Scholar]
- Gomez-Garcia MR, Kornberg A (2004) Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc Natl Acad Sci USA 101: 15876–15880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D (1999) Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem 68: 89–125 [DOI] [PubMed] [Google Scholar]
- Kumble KD, Ahn K, Kornberg A (1996) Phosphohistidyl active sites in polyphosphate kinase of Escherichia coli. Proc Natl Acad Sci USA 93: 14391–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A (2001) Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293: 705–708 [DOI] [PubMed] [Google Scholar]
- Leiros I, Secundo F, Zambonelli C, Servi S, Hough E (2000) The first crystal structure of a phospholipase D. Struct Fold Des 8: 655–667 [DOI] [PubMed] [Google Scholar]
- Loewen PC, Hu B, Strutinsky J, Sparling R (1998) Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol 44: 707–717 [DOI] [PubMed] [Google Scholar]
- McRee DE (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York, NY, USA: Academic Press [DOI] [PubMed] [Google Scholar]
- Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A (2000) Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97: 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T, Tsutsumi K, Ishige K, Noguchi T (2000) Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications. Biochemistry (Mosc) 65: 315–323 [PubMed] [Google Scholar]
- Stuckey JA, Dixon JE (1999) Crystal structure of a phospholipase D family member. Nat Struct Biol 6: 278–284 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi K, Munekata M, Shiba T (2000) Involvement of inorganic polyphosphate in expression of SOS genes. Biochim Biophys Acta 1493: 73–81 [DOI] [PubMed] [Google Scholar]
- Tzeng CM, Kornberg A (1998) Polyphosphate kinase is highly conserved in many bacterial pathogens. Mol Microbiol 29: 381–382 [DOI] [PubMed] [Google Scholar]
- Tzeng CM, Kornberg A (2000) The multiple activities of polyphosphate kinase of Escherichia coli and their subunit structure determined by radiation target analysis. J Biol Chem 275: 3977–3983 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Lee SS, Xu W (2003) Crystallization and characterization of polyphosphate kinase from Escherichia coli. Biochem Biophys Res Commun 305: 997–1001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


