Abstract
Adenosine-to-inosine (A-to-I) RNA editing increases the complexity of the human transcriptome and is essential for maintenance of normal life in mammals. Most A-to-I substitutions occur within repetitive elements in the genome, mainly in Alu repeats. The phenomenon of A-to-I editing is far less abundant in mice, rats, chickens and flies than in humans, which correlates with the relative under-representation of Alu repeats in these non-primate genomes. Here, we review the recent results of bioinformatic and laboratory approaches that have estimated the extent of the editing phenomenon. We discuss the possible biological relevance of the editing pathway, its possible interaction with other cellular pathways that respond to double-stranded RNA and its possible contribution to the accelerated evolution of primates.
Keywords: RNA editing, Alu, human genome, primate evolution
Introduction
RNA editing markedly increases the complexity of the human transcriptome. The essence of this widespread phenomenon is the enzymatic alteration of single or multiple nucleotides, either in coding or non-coding sequences of RNA, which can occur concomitantly with transcription and splicing processes.
Adenosine-to-inosine (A-to-I) nucleoside modification is performed by the adenosine deaminases that act on RNA (ADAR) family of proteins and, until recently, it was believed to occur in only a small number of genes. The outcome of this editing is usually amino-acid substitution in the resulting protein or the formation of a new transcript through the alteration of splice signals. In some cases, editing is functionally important and, accordingly, the disruption of ADAR genes in animal models is incompatible with normal life (Higuchi et al, 2000; Wang et al, 2000; Hartner et al, 2004). Several groups have recently used various computational approaches that have predicted A-to-I editing to be far more abundant than previously thought, and that it in fact affects thousands of human genes. It mostly occurs in non-coding regions—that is, introns and untranslated regions (UTRs)—and is preferentially clustered in Alu repetitive elements (Athanasiadis et al, 2004; Blow et al, 2004; Kim et al, 2004; Levanon et al, 2004). A-to-I substitutions are significantly less abundant in mice, rats, flies and chickens than in humans, mainly owing to the low representation of Alu repeats in these genomes (Kim et al, 2004; Eisenberg et al, 2005). This review summarizes our current knowledge about A-to-I editing within Alu repeats, possible clues to the biological function of the human 'editosome', and the directions of future research efforts in this area.
The human editosome
There are two known families of RNA-editing enzymes in humans: the ADAR family, as mentioned above, and the apoB mRNA-editing catalytic peptide (APOBEC) family, which includes the related activation-induced deaminases (AID) family and which induce the cytosine (C) to uracil (U) transformation in RNA and DNA. Both enzyme families have a highly homologous zinc-coordinating catalytic domain, which emerged from a common cytidine deaminase during evolution. Interestingly, the APOBEC family underwent marked expansion in the primate genome owing to duplication events: 11 APOBEC enzymes were identified in humans compared with only four in mice (Jarmuz et al, 2002; Wedekind et al, 2003). C-to-U editing is involved in several important biological functions: the creation of two variants from the same gene, such as in the human apoB and NF1 (Skuse et al, 1996) mRNA; protection against retrotransposition (Esnault et al, 2005); antiviral activity, including protection against human immunodeficiency virus (HIV; Sheehy et al, 2002) and hepatitis B virus (HBV; Turelli et al, 2004); and the diversification of the vertebrate antibody repertoire (Nussenzweig & Alt, 2004).
The ADAR family acts on double-stranded RNA (dsRNA), which is formed by the coupling of complementary regions within a single transcript. Adar1-knockout mice show an embryonic lethal phenotype (Wang et al, 2000; Hartner et al, 2004), whereas Adar2-knockout mice suffer from convulsions and die prematurely (Higuchi et al, 2000). A third enzyme, ADAR3, contains both single- and doublestranded RNA-binding domains, although its deaminating activity has not yet been proven (Chen et al, 2000).
The nucleoside inosine (I) is interpreted as guanosine (G) by the translation and splicing machinery, therefore there are several biological consequences of A-to-I substitutions: a change of codon, which leads to amino-acid substitution (as exemplified by the glutamate receptor GluR-B; reviewed in Seeburg et al, 1998); the insertion or elimination of a splice site (Rueter et al, 1999); or, potentially, the disruption of a stop codon. In addition to these recoding events, A-to-I editing might have other regulatory effects. For example, interferon-induced ADAR1 is active on viral RNA (Patterson & Samuel, 1995). Although it has been proposed that viruses benefit from A-to-I substitutions (Polson et al, 1996), massive hyper-editing of viral transcripts could be a cellular protection mechanism (Scadden & Smith, 1997). Recent works have also emphasized the multitude of A-to-I editing events in repetitive elements in the genome, the meaning of which has not yet been elucidated.
Repetitive elements in the human genome
Almost half of the human genome comprises mobile repetitive elements that were introduced by retrotransposition through RNA intermediates (Lander et al, 2001). These repetitive elements encompass autonomous long terminal repeats (LTRs), long interspersed elements (LINEs), L1 elements, processed pseudogenes and, most abundantly, non-autonomous short interspersed elements (SINEs). SINEs depend on the open reading frame of LINEs for amplification, as they have no protein coding ability (Dewannieux et al, 2003). By far the most common SINE in humans is the primatespecific Alu sequence, which, at more than one million copies, accounts for greater than 10% of the human genome. The Alu element is about 280 bp long and is composed of two similar monomers connected by an A-rich region.
During the early steps of primate evolution, the rate of Alu amplification was about one for every primate birth (Shen et al, 1991) but this rate has now decreased by two orders of magnitude (Deininger & Batzer, 1999). Indeed, several thousand Alu elements have integrated into the human genome since the divergence of humans from the African apes (Roy et al, 1999), although few are potentially still active (Deininger et al, 1992).
Being clustered in gene-rich regions, Alu repeats are able to modify genes through the insertion of mutations, gene conversion and recombination. They can also disrupt promoter regions, change methylation status, insert new regulatory features, interfere with alternative splicing and regulate the initiation of translation (Deininger & Batzer, 1999). Alu elements can also be incorporated into exons (Lev-Maor et al, 2003) and therefore directly influence the open reading frame in a mature mRNA. Intriguingly, A-to-I editing of Alu repeats in the primate genome is a unique phenomenon, which implies that it is connected to the evolution of this mammalian branch.
Large-scale survey of editing in human transcripts
Until recently, despite the detection of high levels of inosine in mRNAs (Paul & Bass, 1998), only a few edited transcripts had been characterized. Using an Ispecific cleavage reaction, Morse et al (2002) were the first to conduct a targeted search for additional A-to-I substitutions and revealed clusters of editing sites in 19 mRNAs derived from human brain. Of the clusters, 15 out of 19 occurred in repetitive elements, mainly in Alu sequences, within introns and UTRs. In the same year, clusters of predominantly A-to-G substitutions within full-length mRNAs were also reported by the human unidentified gene-encoded (HUGE) protein database (Kikuno et al, 2002).
Three independent groups recently launched systematic searches using computational algorithms, and these corroborated the existence of abundant A-to-I substitutions, mainly in Alu repetitive elements (Fig 1). All of these methods are based on the alignment of RNA transcripts with the genome, the identification of mismatches and the elimination of sequencing errors, single-nucleotide polymorphisms (SNPs), mutations and other 'contaminating mismatches'. The main difference between the three approaches is their use of either the database of more than five million expressed sequence tags (ESTs), which has up to 3% sequencing errors (Hillier et al, 1996), or the more accurate, but smaller, set of longer mRNAs (about 100,000 transcripts).
Figure 1.
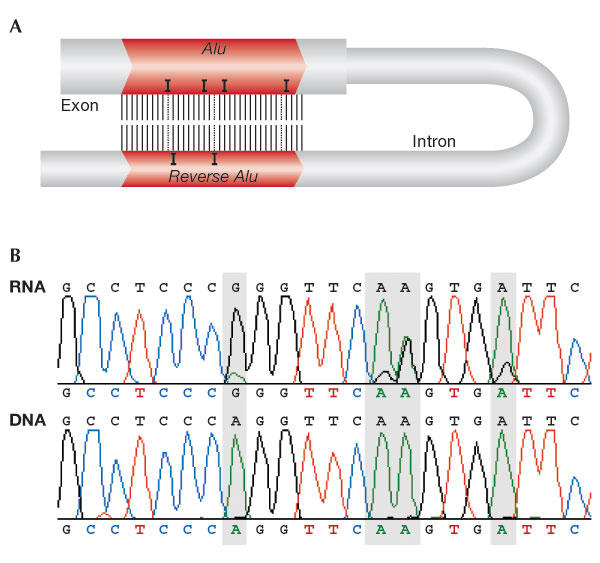
Adenosine-to-inosine editing in Alu repeats. (A) Extended dsRNA are ubiquitously formed between two inverted Alu repeats originating from exons or introns in the pre-mRNA. Adenosine deaminases that act on RNA (ADAR) enzymes perform the A-to-I substitutions in such dsRNA. (B) Editing signature. Matching DNA and RNA sequences from the same source. Editing is characterized by a trace of guanosine in the RNA sequence and a corresponding adenosine in the DNA sequence (highlighted by grey shading).
One group extracted 429,000 candidate dsRNA regions from 14,512 different genes (Levanon et al, 2004). Laborious cleaning procedures yielded 12,723 A-to-I substitutions in 1,637 human genes. A second method (Kim et al, 2004) revealed 30,085 A-to-I substitutions in 2,674 human transcripts. The third study (Athanasiadis et al, 2004) screened 103,723 human mRNAs for clusters of A-to-I substitutions, and searched for Alu repeats in the edited transcripts. They identified 1,445 human mRNAs that are subject to RNA editing at more than 14,500 sites. These discrepancies might have arisen because of differences in fidelity between the methods used in each search. In another study, Blow et al (2004) conducted a survey of RNA editing by comparing sequences from a library of human brain cDNA clones that cover a 3 Mb genomic locus with the reference human genome sequence and genomic DNA of the same individual.
In all four studies, A-to-I substitutions formed more than 80% of the 12 possible types of mismatch in the selected set of transcripts. Virtually all editing events occurred within Alu repetitive elements, which formed only 20% of the total length of the tested transcripts. More than 50% of the substitutions occurred in 3'-UTR regions, 12% in 5'-UTRs and 33% in introns. Many of the edited exons showed non-conformist characteristics, such as intron retention, extension beyond the known UTR and a marked deviation from those listed in RefSeq, all of which are related to defective splicing (Kim et al, 2004).
The characterization of a typical editing site is not straightforward as there is no consensus sequence to which the editing complex binds. Examining nucleotide triplets centred around the edited A, it is possible to identify more favourable sites: C and T are significantly overrepresented one nucleotide 5' to the edited A, whereas G is underrepresented; at the 3' nucleotide, G is significantly overrepresented. This finding is consistent with the previously described pattern of action of both ADAR1 and ADAR2 (Lehmann & Bass, 2000). Focusing on the Alu sequence, there are several hotspots that are prone to editing. In particular, the A nucleotides at positions 27, 28, 136 and 162 of the consensus Alu-J sequence account for 25% of the editing events within Alu repeats (Levanon et al, 2004). The internal poly-A region is also prone to editing.
The level of editing depends strongly on tissue type. Higher levels of editing have been reported in different brain tissues and in the thymus, whereas lower editing levels were generally found in transformed cell lines (Athanasiadis et al, 2004; Kim et al, 2004; Levanon et al, 2004; Eisenberg et al, 2005).
An absolute prerequisite for the editing process is the temporary existence of a dsRNA. Therefore, two groups have investigated the correlation between the proximity of an inverted Alu repeat with the rate of A-to-I substitution (Athanasiadis et al, 2004; Blow et al, 2004). They found that the distance of an Alu sequence to an adjacent inverted Alu is indeed shorter for edited Alu repeats than for unedited ones. The shorter the distance (up to 2 kb), the more probable is the editing of the Alu element. Moreover, the editing frequency correlates with the number of Alu sequences in opposite orientation and close proximity.
Nearly all A-to-I substitutions result in changing the stability of the dsRNAs formed by Alu sequences. About 80% resulted in the destabilization of an A-U pair (changed to I-U) and 20% in the stabilization of an A-C pair (changed to I-C; Blow et al, 2004; Levanon et al, 2004). Nevertheless, the proportion of editing events that 'correct a mismatch' within dsRNA structures significantly outnumbers the prevalence of such mismatches, a fact that substantiates previous claims about the stabilizing effect of editing (Wong et al, 2001).
Despite the use of different computational algorithms to estimate their prevalence, all studies conclude that A-to-I substitutions within Alu repeats are widespread. Validation of the results by concomitant sequencing of both DNA and RNA of candidate genes predicted to have a cluster of editing sites, also adds to the degree of confidence in the computational results. However, the prevalence of the editing phenomenon is still probably underestimated owing to the use of strict 'cleaning' procedures of the sequences, as demonstrated by the only partial overlap between various editing data sets; a failure to predict already known editing sites (within coding regions); and the existence of more editing sites in highly edited sequences than predicted. Direct sequencing of 3 Mb of human brain cDNA has shown that the average editing rate within intronic and intergenic regions is approximately 1:1,000 bp (Blow et al, 2004). Furthermore, the prevalence of Alu elements in the human genes suggests that most human gene transcripts are subject to A-to-I RNA editing (Athanasiadis et al, 2004).
It is worth noting that in addition to the A-to-I substitutions that occur in poorly evolutionarily conserved Alu repeats, five novel editing targets were recently identified by computational methods that targeted the most evolutionarily conserved regions of the genome across various species (Hoopengardner et al, 2003; Clutterbuck et al, 2005; Levanon et al, 2005). Thus, editing is found in both extremes of the evolutionary conservation curve.
Primate specificity of editing
All mammalian genomes contain a high proportion of retro-elements, although their character and number vary significantly. The dominance of Alu among the repetitive elements in the human genome is unique. All mammalian genomes sequenced so far have some LTRs or LINEs to support propagation of non-autonomous SINEs; however, it is possible that some species have not created suitably efficient retroelements or acquired potent means for negative selection against highly active elements. The mouse genome, for instance, contains four types of SINE besides Alu repeats, and their total number is similar to that of Alu elements alone in humans (Waterston et al, 2002). Hence, the formation of dsRNA by two adjacent, identical and inverted SINEs within a single transcript is far more likely to occur in the human genome.
A natural extension of this is to examine whether the editing phenomenon is primate specific. The computational algorithms that were used to explore A-to-I editing in humans were applied to the mouse, rat, chicken and fly transcriptome databases to estimate the abundance of editing in Alu-deficient organisms. Indeed, there are about 40-fold less A-to-I substitutions in the mouse 4 million mRNA and ESTs data set (Eisenberg et al, 2005). By comparing human and mouse mRNA sequences with their genomes, a 35-fold increase in editing events in humans was found (Fig 2). Furthermore, the number of multi-edited transcripts in humans is estimated to be about 30-fold higher than in mice (Kim et al, 2004; Eisenberg et al, 2005). Therefore, the available data imply that abundant editing might be primate specific and strongly interlinked with Alu sequences (Fig 3).
Figure 2.
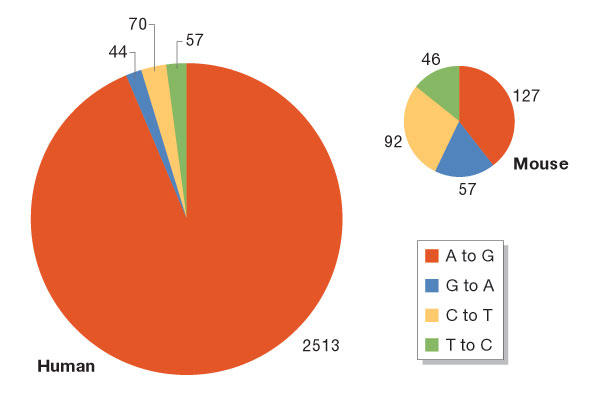
Primate specificity of adenosine-to-inosine editing. The four most prevalent types of mismatch between genomic and mRNA sequences in either human or mouse are presented. In humans, A-to-G sets of mismatches are significantly overrepresented, whereas no such tendency is seen in mice. Only clusters of consecutive mismatches of the same type in a single transcript were included in this analysis. A, adenosine; C, cytosine; G, guanosine; T, thymine.
Figure 3.
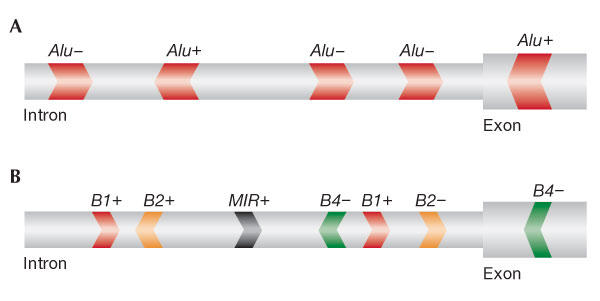
Schematic of the distribution of short interspersed elements in human and mouse genomes. (A) The Alu repetitive element is highly abundant in the human genome, and inverse copies tend to form transient double-stranded RNA (dsRNA) structures. (B) The mouse genome contains four main, divergent, shorter types of short interspersed element, named B1, B2, B4 and MIR. This means that the intramolecular coupling of adjacent, inverted repeats from the same family is significantly less likely compared with humans. Such intramolecular dsRNA structures are essential for adenosine deaminases that act on RNA (ADAR) activity.
The role of editing
The alteration of A-to-I editing has been ascribed to several pathological conditions, mainly to central nervous system-related abnormalities such as amyotrophic lateral sclerosis (ALS; Kawahara et al, 2004), epilepsy (Brusa et al, 1995), major depression disorder (MDD; Gurevich et al, 2002) and glioblastoma multiforme (GBM; Maas et al, 2001). Editing therefore could have physiological significance, but its precise role is still speculative.
Editing in coding regions can result in amino-acid substitutions, but what might be the effects of editing in noncoding parts of the transcript? First, heavily edited transcripts are often retained in the nucleus. This might be interpreted as a means of protection against abnormal transcripts (Zhang & Carmichael, 2001). The fact that clusters of editing sites are abundant in splicing-defective transcripts also support this assumption (Kim et al, 2004). Furthermore, Ispecific cleavage of RNAs can lead to the selective destruction of edited RNAs (Scadden & Smith, 2001). Second, non-viral dsRNA can enter the RNA interference (RNAi) pathways for either cleavage and degradation or RNA-mediated gene silencing at the heterochromatin level. As RNA editing alters the stability of dsRNA complexes, it can augment or counteract these RNAi mechanisms (Knight & Bass, 2002). Third, a direct role for editing in gene silencing was recently suggested from the observation of heterochromatin localization of complexes that contain the Vigilin protein, I-rich transcripts and ADAR1 enzymes (Wang et al, 2005). Fourth, editing can also be viewed as an anti-retroelement mechanism. In fact, the abundance of editing in repetitive elements such as Alu and the evidence for the involvement of APOBEC3G (Esnault et al, 2005) in anti-retrotransposition activity raises the possibility that A-to-I editing defeated the enormous deleterious spread of Alu in the human genome. Finally, editing in 3' UTR regions and introns might alter the stability, transport and handling of RNA transcripts by the translation machinery.
With deeper insight into other aspects of the dsRNA phenomena, an additional potential source of dsRNA are putative RNA complexes resulting from the abundant sense-antisense transcript pairs (Yelin et al, 2003). However, almost all known A-to-I substitutions have occurred in dsRNA that has arisen from intra-molecular pairing within the transcript, leaving this option as anecdotal at most (Athanasiadis et al, 2004). Yet another source of dsRNA are primary and precursor microRNA molecules, which are underrepresented in RNA databases. Indeed, there is emerging evidence for post-transcriptional editing of these regulatory RNA transcripts (Luciano et al, 2004).
As editing controls the structure and stability of the dsRNA, it could cause a transition between distinct secondary structure conformations of a transcript. The increased frequency of editing in Alu elements with more than one inverted neighbouring Alu supports this idea.
Current studies are aimed at revealing the roles and mechanisms of RNA editing. Which ADAR enzyme is responsible for editing the Alu repeats? Is there any cross talk between different pathways that involve dsRNA? Can RNA editing change the stability of dsRNA in a manner that affects its further processing? How relevant is the mechanism of editing to the understanding of disease? What is the exact role of editing as an anti-retroviral and anti-retroelement tool? Does the remarkable diversity of the transcriptome as a result of editing have any meaning at all? And— maybe most intriguingly—did editing of primate repetitive elements contribute to the evolution of mankind?




Acknowledgments
We thank R. Sorek for helpful discussions, and M.F. Jantsch for critical reading of the manuscript. E.E. is supported by an Alon fellowship at Tel Aviv University.
References
- Athanasiadis A, Rich A, Maas S (2004) Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR (2004) A survey of RNA editing in human brain. Genome Res 14: 2379–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R (1995) Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270: 1677–1680 [DOI] [PubMed] [Google Scholar]
- Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K (2000) A third member of the RNAspecific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck DR, Leroy A, O'Connell MA, Semple CA (2005) A bioinformatic screen for novel A-I RNA editing sites reveals re-coding editing in BC10. Bioinformatics 21: 2590–2595 [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA (1999) Alu repeats and human disease. Mol Genet Metab 67: 183–193 [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, Hutchison CA III, Edgell MH (1992) Master genes in mammalian repetitive DNA amplification. Trends Genet 8: 307–311 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T (2003) LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48 [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Nemzer S, Kinar Y, Sorek S, Rechavi G, Levanon EY (2005) Is abundant A-to-I RNA editing primatespecific. Trends Genet 21: 77–81 [DOI] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O (2005) APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433: 430–433 [DOI] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C (2002) Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34: 349–356 [DOI] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH (2004) Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279: 4894–4902 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81 [DOI] [PubMed] [Google Scholar]
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W (1996) Generation and analysis of 280,000 human expressed sequence tags. Genome Res 6: 807–828 [DOI] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R (2003) Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832–836 [DOI] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N (2002) An anthropoidspecific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79: 285–296 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S (2004) Glutamate receptors: RNA editing and death of motor neurons. Nature 427: 801. [DOI] [PubMed] [Google Scholar]
- Kikuno R, Nagase T, Waki M, Ohara O (2002) HUGE: a database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res 30: 166–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A (2004) Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 14: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL (2002) The role of RNA editing by ADARs in RNAi. Mol Cell 10: 809–817 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lehmann KA, Bass BL (2000) Doublestranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 39: 12875–12884 [DOI] [PubMed] [Google Scholar]
- Levanon EY et al. (2004) Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Hallegger M, Kinar Y, Shemesh R, Djinovic-Carugo K, Rechavi G, Jantsch MF, Eisenberg E (2005) Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res 33: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Maor G, Sorek R, Shomron N, Ast G (2003) The birth of an alternatively spliced exon: 3' splicesite selection in Alu exons. Science 300: 1288–1291 [DOI] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S (2004) RNA editing of a miRNA precursor. RNA 10: 1174–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Patt S, Schrey M, Rich A (2001) Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci USA 98: 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL (2002) RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci USA 99: 7906–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig MC, Alt FW (2004) Antibody diversity: one enzyme to rule them all. Nat Med 10: 1304–1305 [DOI] [PubMed] [Google Scholar]
- Patterson JB, Samuel CE (1995) Expression and regulation by interferon of a doublestranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol 15: 5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MS, Bass BL (1998) Inosine exists in mRNA at tissuespecific levels and is most abundant in brain mRNA. EMBO J 17: 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson AG, Bass BL, Casey JL (1996) RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380: 454–456 [DOI] [PubMed] [Google Scholar]
- Roy AM, Carroll ML, Kass DH, Nguyen SV, Salem AH, Batzer MA, Deininger PL (1999) Recently integrated human Alu repeats: finding needles in the haystack. Genetica 107: 149–161 [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, Emeson RB (1999) Regulation of alternative splicing by RNA editing. Nature 399: 75–80 [DOI] [PubMed] [Google Scholar]
- Scadden AD, Smith CW (1997) A ribonuclease specific for inosine-containing RNA: a potential role in antiviral defence? EMBO J 16: 2140–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD, Smith CW (2001) Specific cleavage of hyper-edited dsRNAs. EMBO J 20: 4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg PH, Higuchi M, Sprengel R (1998) RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res Rev 26: 217–229 [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418: 646–650 [DOI] [PubMed] [Google Scholar]
- Shen MR, Batzer MA, Deininger PL (1991) Evolution of the master Alu gene(s). J Mol Evol 33: 311–320 [DOI] [PubMed] [Google Scholar]
- Skuse GR, Cappione AJ, Sowden M, Metheny LJ, Smith HC (1996) The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res 24: 478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D (2004) Inhibition of hepatitis B virus replication by APOBEC3G. Science 303: 1829. [DOI] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K (2000) Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290: 1765–1768 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Z, Blackwell K, Carmichael GG (2005) Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol 15: 384–391 [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R et al. , (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Wedekind JE, Dance GS, Sowden MP, Smith HC (2003) Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet 19: 207–216 [DOI] [PubMed] [Google Scholar]
- Wong SK, Sato S, Lazinski DW (2001) Substrate recognition by ADAR1 and ADAR2. RNA 7: 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R et al. (2003) Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol 21: 379–386 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG (2001) The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475 [DOI] [PubMed] [Google Scholar]


