Abstract
The long-term effects of interferon treatment on cell lines that maintain human papillomavirus type 31 (HPV-31) episomes have been examined. High doses and prolonged interferon treatment resulted in growth arrest of HPV-positive cells, with a high percentage of cells undergoing apoptosis. These effects were not seen with interferon treatment of either normal human keratinocytes or cells derived from HPV-negative squamous carcinomas, which exhibited only slight decreases in their rates of growth. Within 2 weeks of the initiation of treatment, a population of HPV-31-positive cells that were resistant to interferon appeared consistently and reproducibly. The resistant cells had growth and morphological characteristics similar to those of untreated cells. Long-term interferon treatment of HPV-positive cells also resulted in a reduction in HPV episome levels but did not significantly decrease the number of integrated copies of HPV. Cells that maintained HPV genomes lacking E5 were sensitive to interferon, while cells expressing only the E6/E7 genes were resistant. In contrast, cells that expressed E2 from a tetracycline-inducible promoter were found to be significantly more sensitive to interferon treatment than parental cells. This suggests that at least a portion of the sensitivity to interferon could be mediated through the E2 protein. These studies indicate that cells maintaining HPV episomes are highly sensitive to interferon treatment but that resistant populations arise quickly.
Human papillomaviruses (HPV) are small double-stranded DNA viruses that infect epithelial tissues. More than 85 subtypes have been identified, and each of these types exhibits strict tissue specificity (13). About one-third of HPV types infect the anogenital epithelia and induce the most common form of sexually transmitted disease (71). HPV infect cells in the basal layer of epithelia and establish a latent infection in these cells. Production of HPV virions, however, requires infected cells to migrate away from the basal layer and undergo differentiation (28, 31, 71). The HPV that infect the anogenital region can be divided into high-risk and low-risk HPV types depending on their association with malignancy. The low-risk HPVs, such as HPV type 6 (HPV-6) and HPV-11, cause hyperproliferative lesions of external genitalia and are rarely associated with malignancies. In contrast, the high-risk HPVs, HPV-16, -18, -31, -33, and -45, are the etiological agents of cervical cancer (28, 31, 35, 71). The difference in clinical outcome between low- and high-risk HPV infections has been an area of major research interest.
The genomes of all genital HPV types encode 8 to 10 proteins. In the high-risk HPVs, E6 and E7 function as oncogenes. E6 binds to the cellular ubiquitin ligase, E6AP, which then targets p53 for degradation (29, 53, 54, 67). In addition, E6 activates the expression of htert, the catalytic subunit of telomerase. E7 binds to and inhibits the activity of retinoblastoma protein, pRB, and promotes the constitutive activation of E2F family members (9, 16, 38, 44). The E1 and E2 genes encode the viral replication proteins. E1 has been shown to bind to the viral origin in a complex with E2, resulting in the recruitment of cellular DNA replication proteins and initiation of viral DNA synthesis (18, 25, 30). In addition to replication, E2 is also involved in transcriptional regulation of the early viral promoter (6, 39, 43, 69). The functions of E4 and E5 are largely unknown. E4 has been shown to cause cytoskeleton collapse and may play a role in viral particle egress (15). E5 has been shown to down-regulate the rate of epidermal growth factor receptor turnover and presumably may affect the cellular response to this growth factor (32, 58). Finally, the L1 and L2 genes, which are expressed late in the viral life cycle, encode capsid proteins.
Interferons (IFNs) are cytokines that have important antiviral, antiproliferative, and immunomodulatory effects (2, 49, 57). The antiviral activities of IFNs are executed at two levels: one is a growth-inhibitory effect and the second involves stimulation of the host immune response to clear the infected cells (4, 50, 52, 65). IFN is one of the options, either alone or in combination with other therapies, for the treatment of HPV induced lesions (63). Results from clinical trials indicate that IFN therapy has some efficacy in treating low-risk-HPV-induced lesions; however, it is not clear if similar effects are seen in high-risk-HPV infection, as the results remain controversial (12, 21). It has been reported that long-term treatment (50 to 60 days) of mouse C127 cells transformed with bovine papillomavirus type 1 (BPV-1) with mouse IFN could reverse the transformed phenotype, inducing a flat cell morphology typical of uninfected cells (64). These reverted cells also were found to have lost episomal BPV genomes and were assumed to be “cured” of BPV infection. No similar analyses have been performed on keratinocytes that maintain episomal copies of high-risk HPV types.
In a previous study, we used microarray analysis to investigate the changes in cellular transcription induced by the presence of the complete HPV-31 genome (7). In that study, IFN-α/β-inducible genes were identified as major transcriptional targets of high-risk HPV types. We observed that a large number of IFN-inducible genes, including Stat-1, are suppressed in cells that maintain HPV-31 genomes. When treated with IFN-α, HPV-31-positive cells exhibit a delayed response in the induction of expression of downstream IFN-inducible genes such as MxA and Stat-1. However, initial studies suggested that this impaired response could be overcome if the cells were treated with IFN-α at high doses for an extended period (7). These observations indicate that the IFN signal transduction pathway can still be activated in HPV-31-positive cells. Our initial studies failed to determine if the reduced basal level of expression of IFN-inducible genes alters the long-term response of HPV-31-positive cells to IFN. In the present study, we have investigated the long-term effect of IFN on cells that express high-risk-HPV genes in a tissue culture model system.
MATERIALS AND METHODS
Cell culture.
Normal human keratinocytes (NHKs) were isolated from foreskin circumcisions and maintained in keratinocyte growth medium (KGM) supplemented with bovine pituitary extract (Biowhittaker, Walkersville, Md.). In experiments where NHKs were used for comparison with other keratinocyte cell lines, all cells were grown in E medium with fibroblast feeders (41). The HPV-31-positive cell lines discussed in this study include LKP31 and CIN 612 9E, as well as HPV-31 E5KO cells. LKP31 cells were derived from human foreskin keratinocytes transfected and immortalized with the HPV-31 genome as described previously (19, 20, 40, 42). CIN 612 9E cells were isolated from a biopsy specimen of a low-grade cervical lesion and contain episomal HPV-31b genomes (40). 16-E6/E7 cells were generated by infecting NHKs with retroviral vectors containing the HPV-16 E6 and E7 genes as described previously (27). E5KO cells were generated by transfecting foreskin keratinocytes with HPV-31 genomes containing a premature translation termination mutation in the E5 open reading frame (F. Fehrmann, unpublished data). SCC-13 cells were derived from a squamous cell carcinoma (51, 68). Murine 3T3 J2 fibroblasts were maintained in Dulbecco modified Eagle medium(DMEM) supplemented with 10% calf serum (Gibco BRL, Grand Island, N.Y.). All keratinocytes used in these experiments (LKP31, CIN 612 9E, SCC-13, NHK, E5KO, and 16-E6/E7 cells) were maintained in E medium containing serum in the presence of mitomycin C (Roche, Indianapolis, Ind.)-treated 3T3 J2 fibroblast feeder cells (41). Other cells used in this study included HeLa cells (maintained in DMEM supplemented with 10% fetal bovine serum [FBS; HyClone, Logan, Utah]), A431 cells (maintained in DMEM supplemented with 10% FBS and 300 μg of G418/ml), and doxycycline-inducible E2-expressing cells derived from A431 cells (maintained in the same medium as A431 cells with the addition of 1 μg of puramycin/ml).
Generation and screening of E2-expressing cells.
The Tet-On system (Clontech, Palo Alto, Calif.) was used to generate E2-inducible lines. A431 cells stably transfected with the pTet-On vector were a kind gift from K. Green, Northwestern University (23; A. Huen and K. Green, unpublished data). The HPV-31 E2 coding sequence was cloned into the BamHI site of the pTRE vector. Subconfluent cultures of A431 cells were then transfected by the calcium phosphate transfection method with plasmid pTRE/E2 and a puramycin resistance gene containing plasmid pBSpacδP (11). Briefly, plasmids pTRE/E2 and pBSpacδP precipitated in a 20:1 ratio were resuspended in 112.5 μl of water. To this solution 12.5 μl of 2.5 M CaCl2 and 125 μl of 2× HEPES-buffered saline were added. The DNA solution was incubated for 20 min at room temperature before being added directly to the cell culture medium and maintained in it for 16 h. The cells were washed with phosphate-buffered saline (PBS) plus CaCl2 (Mediatech, Herndon, Va.) and maintained in medium (DMEM with 10% FBS and 300 μl of G418) overnight. Cells were then trypsinized, seeded to a new dish, and subjected to selection with 1 μg of puromycin (Sigma, St. Louis, Mo.)/ml. Colonies were expanded and tested for expression of E2 by reverse transcription-PCR (RT-PCR) and luciferase activity upon induction with doxycycline (Sigma). For RNA analysis, clones of E2 transfectants were treated with 3 μg of doxycycline (Sigma)/ml for 24 h, and total RNA was harvested with the Trizol reagent (Gibco). A 250-ng portion of total RNA from each clone was used in the RT-PCR (GeneAmp EZ rTth RNA PCR system; Roche) with primers E2 2700T (5′ GGG GGG TCT TTC TCA ACG TTT AAA TGT GTG T) and E2 3000B (5′ GGG GGG AAA TAC AGT TCA AGA CTT GTT TGC TG). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was also examined as an internal control by RT-PCR by using primers K136 (5′ CTC AGA CAC CAT GGG GAA GGT GA) and K137 (5′ ATC TTG AGG CTG TTG TCA TG). For reporter analysis, clones of E2 transfectants were treated with 3 μg of doxycycline/ml for 24 h and then transiently transfected with 500 ng of a luciferase reporter construct (pGL3Basic or pGL3Basic/6XE2BS) and 500 ng of pSVβGal plasmids (both from Promega, Madison, Wis.). At 24 h posttransfection, lysates were harvested, and luciferase activity was assayed with the Dual-Light luminescent reporter gene assay system (Perkin-Elmer Applied Biosystems, Foster City, Calif.) and a luminometer (Monolight Luminometer; Analytical Luminescence Laboratory, San Diego, Calif.).
Cell counting and FACS analysis.
Keratinocytes were seeded onto 6-cm culture dishes (at 4 × 105 cells/dish) 1 day prior to IFN treatment. The next day, IFN-β was added at a concentration of 1,000 U/ml. For A431 and doxycycline-inducible E2 cells, 3 μg of doxycycline/ml was added soon after plating and was maintained in the culture medium. At various times after initiation of IFN treatment, fibroblast feeder cells were removed by washing in 0.5 mM EDTA in PBS, and keratinocytes were detached from the dish by trypsin. Cells were incubated with trypan blue, and viable cells were counted using a hemocytometer. The remainder of the cells were spun down, fixed in 70% cold ethanol, stained with propidium iodide (50 μg/ml in PBS; Sigma), and subjected to fluorescence-activated cell sorter (FACS) analysis.
TUNEL and DAPI staining.
Apoptosis was analyzed by monitoring the activity of terminal deoxynucleotidyl transferase by use of the Apoptosis detection kit from Roche. DNA breakage induced by apoptosis was detected by labeling cells with fluorescein-conjugated dUTP. Cells grown on coverslips were treated with IFN-β at 1,000 U/ml for 6 days. Cells were fixed in 4% paraformaldehyde, permeabilized with PBS containing 0.1% Triton X-100 and 0.1% sodium citrate, and incubated with 50 μl of TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) enzyme mixture. After a 1-h incubation at 37°C, coverslips were rinsed in PBS, and cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma) at 2 μg/ml for 5 min. The fluorescein and DAPI signals were detected with a fluorescent microscope, and images were taken with a Spot camera (Diagnostic Instruments Inc., Sterling Heights, Mich.).
Annexin V staining.
The annexin V assay system was purchased from Immunotech (Marseille, France) and used as an assay for apotosis. Cells were either left untreated or treated with IFN-β at 1,000 U/ml for various times. The culture medium that contained floating cells was first collected, and adherent keratinocytes were trypsinized after removal of fibroblast feeder cells. Trypsinized keratinocytes were combined with the floating cells, spun down, and washed once in cold PBS. Pelleted cells were resuspended in 1× binding buffer provided by the manufacturer and were incubated on ice with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide in the dark for 10 min. The stained cells were then diluted with 1× binding buffer and analyzed by flow cytometry.
Western blot analysis.
Subconfluent NHK, LKP31, and CIN 612 9E cells were grown in E medium and were treated with IFN-β at 1,000 U/ml for various periods of time. Keratinocytes were trypsinized after the removal of feeders, spun down, resuspended in lysis buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% deoxycholate, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 1% Triton X-100, 1 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium orthovanadate, 20 mM p-nitrophenyl phosphate), and allowed to stand on ice for 10 min. Insoluble cell debris was removed by brief centrifugation. Equal amounts of protein lysates were separated by SDS-10% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.). The membrane was suspended in blocking solution (5% nonfat dry milk, 0.1% Tween 20 in PBS) for 30 min and incubated with the primary antibody for 1 h. The primary antibodies used in this study were anti-human TRAIL (BD PharMingen, San Diego, Calif.), anti-human caspase 1 (specific for both procaspase p45 and activated caspase p20; Upstate Biotechnology, Lake Placid, N.Y.), anti-human Fas L, and anti-human BclX-s/l (Santa Cruz Biotechnology, Santa Cruz, Calif.). The membrane was washed in wash buffer (0.1% Tween 20 in PBS) for 1 h with four changes of buffer and incubated with the proper secondary antibody. The secondary antibody used in this study was horseradish peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit whole immunoglobulin G (IgG; Amersham Pharmacia, Piscataway, N.J.). After incubation with the secondary antibody, the membrane was washed again and the signal was detected by enhanced chemiluminescence (ECL Western blotting detection reagents; Amersham Pharmacia). After the signals were detected by autoradiography (Hyperfilm; Amersham Pharmacia), the intensity of the signal was quantified by using ChemiImager 5500 from Alpha Innotech Corporation (San Leandro, Calif.).
β-Gal staining.
Cells (LKP31 or CIN 612 9E) were grown on coverslips to 50% confluence. Cells were then either left untreated and stained when they reached 80% confluence or treated with IFN-β (1,000 U/ml) for 6 days. Coverslips were rinsed with PBS and fixed in 3% formaldehyde. Senescence-specific staining for β-galactosidase (β-Gal) activity at pH 6 was performed as described previously (14). Briefly, coverslips were incubated at 37°C with a staining solution containing 1 mg of 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal; Sigma)/ml, 40 mM citric acid-sodium phosphate (pH 6), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2. After overnight incubation with the staining solution, coverslips were washed in PBS and mounted on slides.
Southern blot analysis.
Low-molecular-weight DNA was harvested from keratinocytes at different passages by the following procedure. After removal of fibroblast feeders with 0.5 mM EDTA in PBS, keratinocytes were trypsinized and harvested by centrifugation. Cells were then resuspended in buffer A (400 mM NaCl, 10 mM Tris-HCl [pH 7.5], 10 mM EDTA), followed by RNase and proteinase digestion (50 μg of RNase A/ml, 50 μg of proteinase K/ml, 0.2% SDS) at 37°C overnight. Chromosomal DNA was sheared by being passed through an 18-gauge needle several times, and low-molecular-weight DNA was purified by phenol-chloroform extraction and ethanol precipitation. Purified DNA was digested with XbaI, electrophoretically separated on a 0.8% agarose gel, transferred to a nylon membrane (Gene Screen; NEN Life Science Products Inc., Boston, Mass.) by use of a vacuum transfer apparatus (Boekel Scientific, Feasterville, Pa.), and hybridized with High Prime (Roche) an [α-32P]dCTP-labeled HPV-31-specific probe (Amersham Pharmacia).
RESULTS
Long-term treatment with IFN-β severely retards the growth of HPV-31-positive cells.
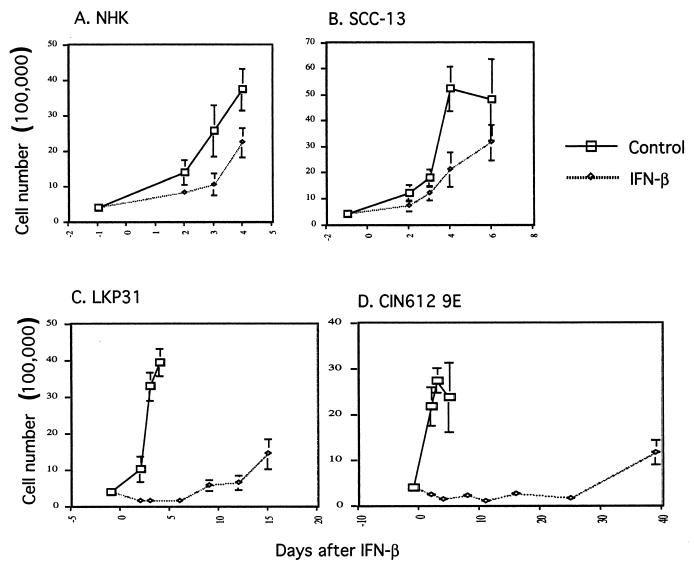
In order to investigate the effects of long-term treatment with IFN on keratinocytes that stably maintain the complete HPV-31 genome, a series of tissue culture experiments were initiated. Both IFN-α and IFN-β have been shown to induce apoptosis in sensitive cells, and this is believed to occur in a cell type-specific manner (5, 8, 70). In addition, IFN-β has been shown to have a higher efficacy in controlling HPV-derived lesions in clinical trials than IFN-α (26, 37). In this analysis, we have focused our efforts on the effects of IFN-β on HPV-31-positive cells. Proliferating monolayer cultures of NHKs, SCC-13 cells (an immortal keratinocyte line that is HPV negative), and HPV-31-positive keratinocytes that stably maintain viral episomes (LKP31 cells) (20) were treated with 1,000 U of IFN-β/ml. After 3 days of treatment with IFN-β, we observed that the growth of LKP31 cells, but not that of NHKs, was significantly altered as determined by counting the number of viable cells. As seen in Fig. 1, the growth rate of LKP31 cells in the presence of IFN was significantly more retarded than that of NHK or SCC-13 cells, which exhibited only slight decreases (Fig. 1A, B, and C). Similar effects were seen in an HPV-31-positive cell line derived from a biopsy specimen of a low-grade cervical lesion (CIN 612 9E) (Fig. 1D), as well as in three other cell lines that maintain HPV-31 episomes (data not shown). In contrast to HPV-31-positive cells, which were severely impaired in their ability to proliferate, NHKs and SCC-13 cells continued to grow in the presence of IFN, though at a slightly reduced rate compared to untreated cells (Fig. 1). This modest reduction in the growth rate seen with HPV-negative cells is consistent with the reported global effects of IFN on cells in tissue culture (23).
FIG. 1.
Growth of HPV-31-positive cells is severely retarded by IFN-β. Multiple dishes of keratinocytes were seeded at 4 × 105 cells per 6-cm dish 1 day prior to IFN treatment. Beginning on the next day (day 0), cells were either left untreated or treated with IFN-β at 1,000 U/ml. One dish from control or IFN-treated cells was harvested on the indicated date, and viable cells were counted. (A) NHKs; (B) SCC-13 carcinoma cell line; (C) LKP31 cell line; (D) CIN 612 9E cell line.
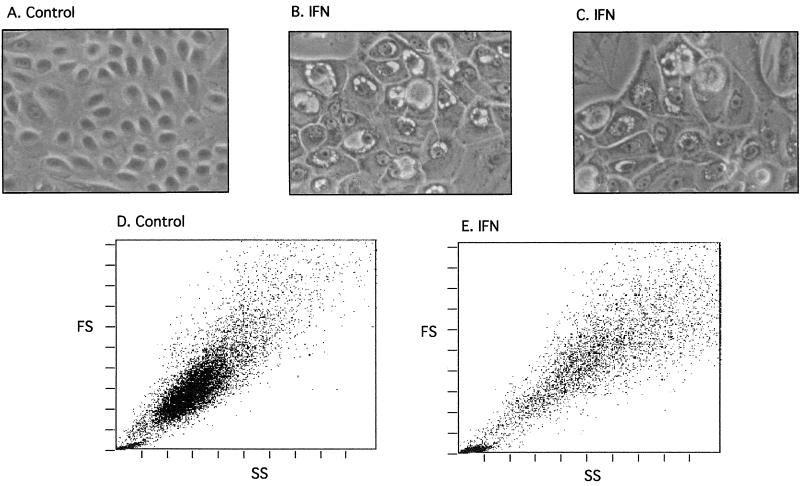
Closer examination of the HPV-31-positive cells revealed them to be growth arrested. After 6 days of IFN treatment the majority of NHKs and SCC-13 cells remained viable, as determined by trypan blue staining, while the number of viable HPV-31-positive cells was reduced by 96% in the case of LKP31 cells and by 94% in the case of CIN 612 9E cells. Furthermore, the cell morphology of IFN-β-treated NHKs was indistinguishable from that of untreated cells (data not shown); however, the appearance of IFN-β-treated LKP31 and CIN 612 9E cells was dramatically changed. HPV-31-positive cells treated with IFN-β became enlarged and often contained vacuoles on the periphery of the nucleus (Fig. 2; compare panels A, B, and C). When the IFN-treated CIN 612 9E cells were examined by flow cytometry, the change in morphology was also reflected by the aberrant forward- and side-scatter patterns, which indicate increased heterogeneity in cell size and complexity (Fig. 2D and E). Similar effects were seen in LKP31 cells and three other HPV-31-positive cell lines (data not shown). We conclude from these experiments that high doses of IFN induce severe growth inhibition and altered morphology of cells that contain HPV-31 genomes. No such effects were seen in three different isolates of NHKs or in the HPV-negative squamous carcinoma cell line SCC-13.
FIG. 2.
The morphology, size, and shape of HPV-31-positive cells are dramatically changed upon IFN treatment. CIN 612 9E cells were either left untreated (A and D) or treated with IFN-β for 6 days (B, C, and E), and the morphology of cells was observed with a phase-contrast microscope (magnification, ×70) (A through C). The same samples were analyzed by flow cytometry, and the forward-scatter (FS) and side-scatter (SS) patterns of control and IFN-treated cells are shown (D and E).
Treatment of HPV-31 cells with IFN-β rapidly induces resistance to IFN.
After 10 to 14 days of continuous treatment of HPV-31-positive cells with 1,000 U of IFN-β/ml, a resistant cell population was detected in multiple experiments (Fig. 1C and D). The growth rate and morphology of these resistant cells in the presence of IFN-β were indistinguishable from those of untreated cells (data not shown). Similar effects were seen in a total of three HPV-31-positive cell lines and upon repeated experiments using the same cell line.
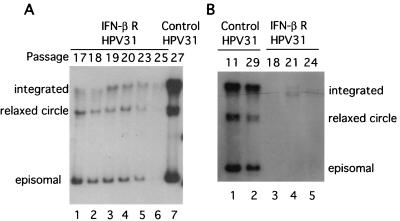
Long-term treatment of BPV-1-transformed mouse cells with IFN has been reported to result in a complete loss of replicating BPV DNA and to reverse the transformed phenotype of these cells (64). We therefore investigated if similar effects were seen following IFN-β treatment of cells containing HPV episomes. Prior to IFN treatment, LKP31 cells contained episomal copies of the HPV-31 genome as well as a small amount of integrated DNA. In the continuous presence of IFN-β at 1,000 U/ml, LKP31 cells first became growth arrested, followed by a recovery period and then the rapid appearance of cells that were resistant to IFN (see Fig. 1C). These resistant cells were propagated for an additional 9 passages (to passage 25) in the presence of IFN, and the state of viral DNA was examined. In the IFN-resistant LKP31 population, the average copy number of HPV-31 episomes was found to decrease as the passage number increased (Fig. 3). No significant decrease in copy number was seen in untreated LKP31 cells upon passaging (Fig. 3B). Interestingly, the integrated copies of the HPV-31 genome were retained throughout the course of IFN treatment, with only a modest reduction in levels. This suggests that cells with episomal copies of HPV-31 are more sensitive to the effects of IFN than those with integrated copies. The analyses shown in Fig. 3A and B were performed on the IFN-resistant populations that arose after 15 and 37 days, respectively. These cells were not undergoing apoptosis, so fragmented DNA was not present during the analysis. In the experiment for which results are shown in Fig. 3A, the entire process consisted of approximately 60 cell doublings that spanned a 2-month period. This experiment was repeated four times with two different HPV-31-positive cell lines. In three of four cases we observed dramatic decreases in episomal viral DNA levels in IFN-β-resistant cells. In the fourth case, we saw a moderate reduction in viral episome levels (data not shown).
FIG. 3.
Levels of episomal HPV-31 DNA gradually decrease upon prolonged IFN treatment. LKP31 cells were treated with IFN-β at passage 15. The IFN-resistant population was expanded, and the low-molecular-weight DNA was harvested at different passages. Ten micrograms of harvested DNA was examined by Southern blot analysis with an HPV-31-specific probe. The positions of episomal, relaxed-circle, and integrated viral DNAs are indicated at the left or right of each panel. Panels A and B represent two independent experiments. Levels of viral DNA from control untreated LKP31 cells at passage 27 (A) or passages 11 and 29 (B) are also shown.
To address whether HPV-31 episomal maintenance could be restored by removal of IFN-β, experiments were performed in which IFN treatment was stopped at passage 27 and the state of viral DNA was analyzed for 6 additional passages. Removal of IFN-β did not reverse the negative effect of IFN on episomal maintenance and failed to restore the levels of episomes to those of parental untreated cells (data not shown). We conclude that high doses of IFN-β impose negative selection on cells that maintain HPV-31 episomes.
IFN-β induces apoptosis in HPV-31 cells.
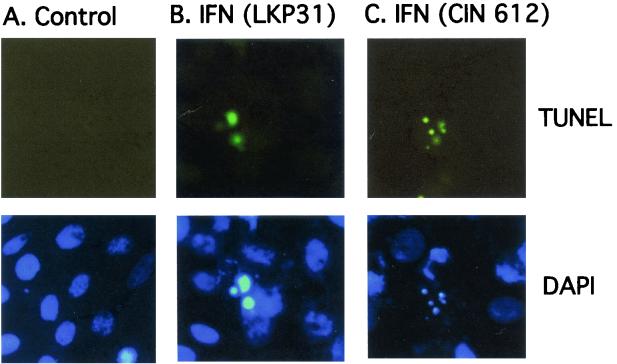
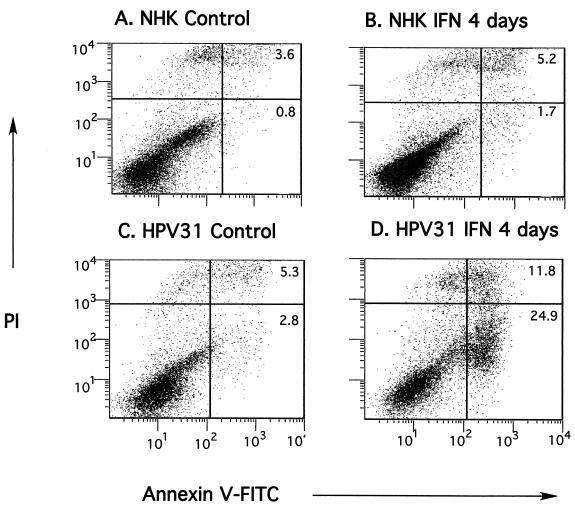
Apoptosis has been implicated as one of the major mechanisms of the antiviral activity of IFN (34, 57). In order to determine whether the inhibition of growth and the morphology changes induced in HPV-31-positive cells were a result of IFN-induced apoptosis, we performed TUNEL staining and annexin V staining to detect late and early apoptotic markers, respectively. Positive TUNEL-staining apoptotic cells were readily detectable among both CIN 612 9E and LKP31 cells treated with IFN-β for 6 days (Fig. 4). By TUNEL staining we observed similar levels of apoptosis in LKP31 and CIN 612 9E cells, both of which are significantly elevated above those in NHKs. To further examine the level of apoptosis, flow cytometry analysis of annexin V-FITC labeling was performed on LKP31 cells treated with IFN-β at 1,000 U/ml. After 4 days of treatment, cells were harvested, incubated with FITC-conjugated annexin V and propidium iodide, and subjected to FACS analysis. As shown in Fig. 5C and D, a ∼10-fold increase in the proportion of annexin V-positive cells (from 2.8 to 24.9%) was observed after 4 days of IFN treatment. Analysis of IFN-β-treated CIN 612 9E cells yielded similar results, although the increase (from 6.8 to 17.4% [data not shown]) was not as dramatic as that in LKP31 cells. In contrast, the number of NHKs positive for annexin V was largely unaffected by IFN (increasing from 0.8 to 1.7%) at 4 days posttreatment (Fig. 5A and B).
FIG. 4.
IFN induces apoptosis in HPV-31-positive cells. LKP31 cells grown on coverslips were either left untreated (A) or treated with IFN-β at 1,000 U/ml for 6 days (B). Coverslips were processed for a TUNEL apoptosis assay (upper panels) and counterstained with DAPI (lower panels). Images of TUNEL and DAPI staining of CIN 612 9E cells treated with IFN-β under the same conditions are also shown (C).
FIG. 5.
IFN induces apoptosis in HPV-31-positive cells. NHK or LKP31 cells were either left untreated (A and C) or treated with IFN-β at 1,000 U/ml for 4 days (B and D). Cells were harvested and processed for annexin V-FITC-propidium iodide (PI) staining. The percentage of cells that were FITC positive or FITC-PI double positive is indicated in each quadrant.
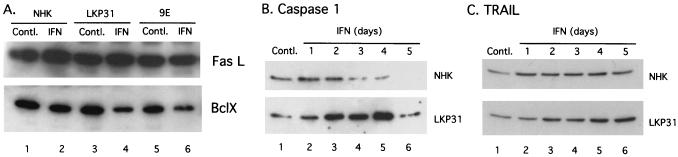
We next investigated which apoptotic pathway was activated by IFN-β in HPV-31-positive cells. For these studies the levels of several apoptosis-related genes were examined by Western blot analysis. Bcl-X is a Bcl-2-related protein that is repressed in several apoptotic pathways (47, 48, 60). As shown in Fig. 6A, levels of Bcl-X were decreased by 55% in LKP31 cells and 54% in CIN 612 9E cells after 4 days of incubation with IFN-β, whereas they were reduced only by 27% in NHKs. This result is consistent with the observation that IFN-β induces a higher frequency of apoptosis in HPV-31-positive cells than in NHKs. When the induction kinetics of proapoptotic genes was examined, we found that IFN-β induced a weak and transient induction of activated caspase 1 (p20) in NHKs (peak at 1 day posttreatment, 1.2-fold increase), whereas the induction of activated caspase 1 in LKP31 cells was more significant and was sustained for a longer period (peak at 4 days posttreatment, 3.4-fold increase) (Fig. 6B) (10).
FIG. 6.
Western blot analysis of the expression of apoptosis-related genes in cells treated with IFN. (A) NHKs, LKP31 cells, or CIN 612 9E cells were either left untreated or treated with IFN-β at 1,000 U/ml for 4 days. Cells were harvested, and protein lysates were separated by SDS-polyacrylamide gel electrophoresis. Proteins were electrophoretically transferred onto a membrane and probed with a FasL-specific antibody. The same blot was stripped and probed with an anti-Bcl-X antibody. (B and C) NHKs or LKP31 cells were either left untreated or treated with IFN-β at 1,000 U/ml for various times. Protein lysates were harvested at the indicated times posttreatment and subjected to Western blot analysis for the expression of caspase 1 (B) and TRAIL (C).
IFN-β has been shown to induce the expression of TRAIL, a member of the tumor necrosis factor family, in IFN-sensitive cells (8, 62). When IFN-β-treated HPV-31-positive cells were examined, an elevated level of TRAIL (2.5-fold) compared to that seen in treated NHKs (1.5-fold) was observed (Fig. 6C). In contrast, the level of Fas ligand (FasL) was largely unchanged upon IFN treatment (Fig. 6A), suggesting that the apoptotic effect of IFN-β is more likely mediated through the TRAIL than the Fas pathway in HPV-31-containing cells. This is consistent with previous observations that IFN-β induces apoptosis via the TRAIL pathway instead of the Fas pathway. We conclude that in agreement with the differential effect on cell growth, IFN-β altered the expression of apoptosis-related genes to a significantly greater degree in HPV-31-positive cells than in NHKs.
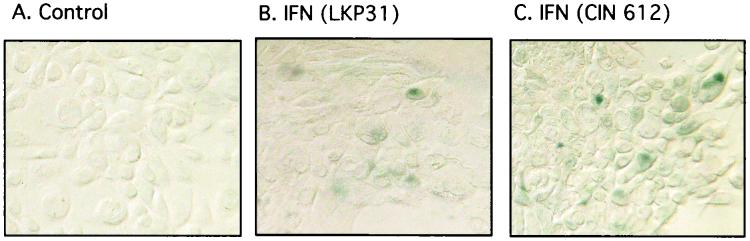
The enlarged and flattened morphology seen in HPV-31-positive cells treated with IFN-β is also reminiscent of cells undergoing senescence, and this prompted us to investigate whether these cells also express senescence-specific markers, such as β-Gal. β-Gal activity at pH 6 was measured as a marker for senescence (14). Six days after IFN treatment, approximately 2 to 5% of HPV-31-positive cells expressed β-Gal activity at pH 6 (Fig. 7; compare panels A, B, and C), whereas no increase in β-Gal activity at pH 6 was detected in NHKs upon IFN treatment (data not shown). We conclude that a small number of HPV-31-positive cells treated with IFN express markers of senescence.
FIG. 7.
IFN induces senescence in HPV-31-positive cells. (A and C) CIN 612 9E cells grown on coverslips were either left untreated (A) or treated with IFN-β at 1,000 U/ml for 6 days (C). The cells were then processed for β-Gal staining. (B) LKP31 cells treated under the same conditions were stained for β-Gal.
HPV E2 protein sensitizes keratinocytes to the negative growth effect of IFN-β.
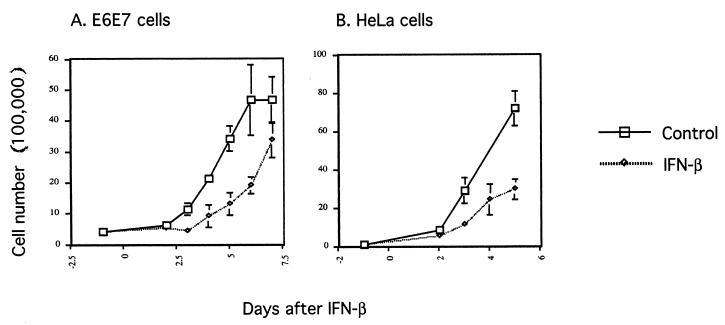
In our experiments primary NHKs and HPV-negative transformed keratinocytes (SCC-13) were found to be resistant to IFN-β-induced growth arrest and apoptosis, whereas cells containing HPV-31 episomes were highly sensitive (Fig. 1). Similar resistance to IFN was seen with the HPV-negative vulvar carcinoma-derived cell line A431 (data not shown). From this we conclude that immortalization or transformation alone does not sensitize cells to IFN-β. It is likely that the presence of one or more viral genes may prime cells to the negative effect of IFN-β. To investigate which viral genes are involved, genetic analysis was performed by using either keratinocyte cell lines containing HPV-31 genomes with mutations in a specific viral gene or cell lines expressing only single or multiple viral genes. These cell lines were tested for sensitivity to IFN-β in a growth assay. For these studies we used retroviruses expressing E6 and E7 proteins rather than genetically mutagenizing the HPV-31 genome, since mutation of these genes often leads to integration of the genome (61). We first examined the effects of IFN-β on cells expressing the E6 and E7 oncoproteins of HPV-16 and found that the effects were similar to NHKs (Fig. 8A). Furthermore, HeLa cells, which express the E6 and E7 genes of HPV-18 (24), were also more resistant to IFN-β-induced apoptosis than cells that maintained the complete HPV episome (Fig. 8B).
FIG. 8.
Growth rate assays with E6E7 (A) and HeLa (B) cells. Cells were seeded at 4 × 105 (E6E7) or 1 × 105 (HeLa) per 6-cm dish 1 day prior to IFN treatment. The next day (day 0), cells were either left untreated or treated with IFN-β at 1,000 U/ml. One dish each from control or IFN-treated cells was harvested, and the number of viable cells was counted at the indicated times.
We next examined whether the E5 gene could confer sensitivity to IFN-β. For these studies, we used cell lines which contain HPV-31 genomes with translational termination codons inserted at amino acid number 1 of the E5 open reading frame (E5KO). Cell lines containing E5 knockout mutations showed identical sensitivity to IFN-β as cell lines containing the wild-type HPV-31 genome (data not shown). Therefore, the factor in the HPV-31 genome that confers sensitivity to IFN-β is not E5. We conclude that neither E5, E6, nor E7 is the factor that confers sensitivity to IFN-β but rather that either E1, E2, E4, or a combination of factors is responsible for sensitivity to IFN-β.
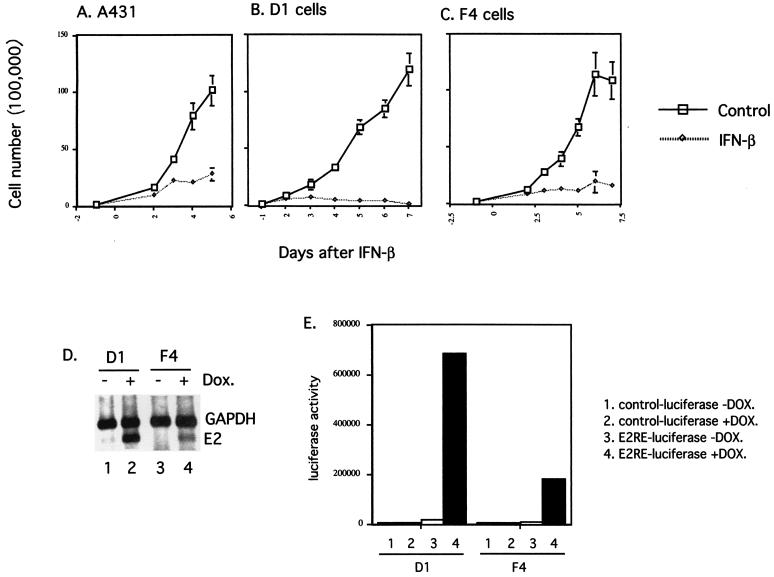
Knockout mutations of E1 or E2 in the context of the complete genome have previously been shown to lead to integration of viral DNA into host chromosomes, which results in variable expression of viral genes. We therefore sought to examine the IFN sensitivity of cells that express E2 from a heterologous promoter. However, expression of HPV E2 from a heterologous promoter in cells that have wild-type p53 often results in growth arrest (66). To overcome this problem, we utilized cell lines derived from A431 cells in which the expression of E2 is directed from the tetracycline-inducible promoter. A431 cells were originally isolated from a vulvar carcinoma and are p53 negative. A series of E2-inducible A431 cell lines were isolated and examined for the level of induction of E2 expression by RT-PCR following doxycycline treatment (Fig. 9D). In addition, we examined the ability of these cell lines to activate a transiently transfected luciferase reporter plasmid containing multimerized E2 binding sites (Fig. 9E) (18, 59). From this analysis, we were able to identify two clonal cell lines derived from the parental A431 cells that express E2 at high (clone D1) or low (clone F4) levels upon doxycycline induction (Fig. 9D and E). Most importantly, in these cells E2 expression did not induce growth arrest (data not shown).
FIG. 9.
(A through C) Growth rate assays with A431, D1, and F4 cells. Cells were seeded at 105 per 6-cm dish 1 day prior to IFN treatment. From the next day (day 0) on, cells were either left untreated or treated with IFN-β at 1,000 U/ml. One dish from either control or IFN-treated cells was harvested, and the number of viable cells was counted, at the indicated times. (D) RT-PCR analysis of E2 expression in two independent E2 clones upon induction with doxycycline (Dox). (E) Luciferase assay of two E2-expressing clones transiently transfected with either the control reporter construct (pGL3Basic) or the luciferase reporter construct with multiple E2 binding sites (pGL3Basic/6XE2BS) upon induction with doxycycline. Control-luciferase, pGL3Basic; E2RE-luciferase, pGL3Basic/6XE2BS.
We next examined the parental A431 cells, the high-E2-expressing clone (D1), and the low-E2-expressing clone (F4) for sensitivity to IFN. In the presence of doxycycline and IFN-β (1,000 U/ml), parental A431 cells continued to divide, although at a reduced rate (at 5 days posttreatment, the number of viable cells was 27% of the number of viable untreated A431 cells [Fig. 9A]). The morphology of parental A431 cells treated with IFN was indistinguishable from that of untreated cells (data not shown). In contrast, the growth rate of D1 cells in the presence of doxycycline and IFN-β was severely retarded (0.75% of the number of viable untreated D1 cells at 7 days posttreatment [Fig. 9B]). Eventually, all E2-expressing cells died and detached from the dish, in contrast to parental cells, which continued to proliferate. The low-level-E2-expressing clone F4 showed an intermediate response to IFN-β (14.3% of the number of viable untreated F4 cells at 7 days posttreatment [Fig. 9C]). These studies suggest that E2 may act to sensitize HPV-31-positive cells to IFN.
DISCUSSION
Previously we examined gene expression profiles in proliferating monolayer cultures of cells maintaining HPV-31 episomes in the absence of exogenous IFN, and we found that IFN-α/β-inducible genes, but not IFN-γ-inducible genes, are a major transcriptional target of HPV (7), in agreement with the results of others (45). This led us to investigate whether this repression of basal expression of IFN genes by HPV gene products modified the long-term response of cells to IFN. Despite the repression of basal expression, we found that upon prolonged treatment with IFN, IFN-responsive genes were activated to levels comparable to those seen in normal keratinocytes treated with IFN (7). Surprisingly, we also observed that cells harboring episomal copies of HPV-31 are more sensitive than NHKs to long-term treatment with IFN.
In this study, we compared NHKs with NHKs transfected with the complete HPV-31 genome, or NHKs immortalized by viral E6/E7 genes. We observed consistent and reproducible results indicating that cells containing the complete HPV-31 genome underwent apoptosis upon exposure to IFN, while normal or E6/E7-expressing cells did not. Since all these cells were derived from similar parental cells, we believe it is valid to make comparisons between different cell lines. We also investigated whether the effects of IFN were specific to transfected NHKs and observed similar effects in an HPV-31-positive cell line derived from a cervical biopsy specimen, CIN 612 9E. Furthermore, we determined that immortalization or transformation alone does not render cells sensitive to IFN, since NHKs and carcinoma-derived SCC-13 cells were found to be equally resistant to the effects of IFN-β.
Continuous IFN treatment resulted in apoptosis of a large portion of HPV-31-positive cells, but a resistant population quickly and consistently arose within a short time. These effects were seen in cells generated by transfection of HPV DNA in tissue culture as well as in cells derived from HPV-positive biopsy specimens of low-grade lesions. Our studies indicate that while IFN treatment can arrest the growth of HPV-positive cells, resistant populations quickly appear.
We observed that apoptosis occurs at a high rate in HPV-positive cells and that this is at least one mechanism by which IFN induces retardation of growth. The apoptosis induced in HPV-31-positive cells correlated with a reduction in the levels of the antiapoptosis gene Bcl-X and with induction of the proapoptosis genes TRAIL and caspase 1. Apoptosis induced by IFN-β is a rather slow process, and it is usually mediated through the TRAIL pathway (8). Our results are consistent with these findings. Interestingly, we also detected an increased level of cells expressing markers of senescence, though the numbers were small (2 to 5% of total cells). The appearance of these two populations of cells following IFN treatment could reflect an intrinsic difference among HPV-31-positive cells. It is possible that a subpopulation of HPV-positive cells responded to IFN-β by activating markers of senescence instead of undergoing apoptosis and that this activation allowed cells to escape cell death. The cells starting to undergo senescence could serve as a reservoir for resistant cells. Alternatively, a subpopulation of cells could quickly acquire mutations or undergo other epigenetic changes such as alterations in the state of promoter methylation that may result in resistance to IFN.
If the resistant population arose from cells that never become growth arrested, then from the kinetics of appearance of resistant cells, we calculate that about 1% of cells are intrinsically resistant to IFN. If the resistant population of cells is arrested for a time and then begins to grow, this number would, of course, be higher. Whatever the nature of the initial resistant population, it seems unlikely that these cells arose as a result of a simple mutation, since this would have to occur at a very high frequency. In addition, since the 9E isolate of the CIN 612 cell line used in this study is clonal in origin, it is unlikely that the resistant population grew out of a subset of cells that were present at the time of isolation. We favor the idea that some epigenetic changes leading to altered responses to IFN were responsible for resistance. Interestingly, we detected no differences between the levels of Stat-1 protein in resistant and sensitive populations of HPV-31-positive cells upon treatment with IFN-β (data not shown). The mechanism leading to IFN resistance is likely to be complex, and it is currently under investigation.
Our studies further suggest that the viral E2 protein alone can confer sensitivity to IFN-β when expressed at high levels. Whether this level of E2 expression is comparable to the E2 activity seen in cells that maintain HPV episomes is, however, not clear. E2 plays important roles in viral genome replication and transcriptional regulation (6, 17, 43, 56). It is possible that E2 may also regulate the expression of cellular genes that augment the response to IFN-β (46). Interestingly, HeLa cells as well as other cell lines isolated from high-grade HPV lesions often contain integrated high-risk-HPV genomes (1, 55). The consequences of such integration are not only up-regulation of E6 and E7 expression but also abolishment of E2 expression (55). Based on our results, the importance of controlling E2 expression during the malignant progression of high-risk-HPV-containing cells could be twofold: removing the negative effect of E2 on viral oncogene expression and eliminating E2-induced sensitivity to IFN. It is not clear whether E2 is the only viral factor that contributes to the phenomenon; it is possible that other factors may also play a role. For instance, it is possible that the up-regulation of E6/E7 expression in HeLa and E6/E7 cells could also contribute to resistance to the effects of IFN.
Previously we have shown that the basal levels of expression of IFN-inducible genes are suppressed in HPV-31-positive cells in the absence of exogenous IFN. The mechanism that contributes to this repression is not clear. Several studies have shown that the E6 and E7 proteins can act to interfere with the IFN signal transduction pathways by altering the activity of Tyk-2 kinase and blocking nuclear translocation of p48 (3, 33). However, we observed that, upon high-dose and long-term IFN treatment, HPV-31-positive cells are able to mount a response comparable to that of control cells (7). In fact, after the initial delay, the rate of response to IFN in HPV-31-positive cells is actually accelerated, which may explain the higher sensitivity to IFN observed in these HPV-31-positive cells (7). The basal-level repression of IFN-inducible genes by HPV proteins may act to delay a response in vivo, where the levels of IFN may not be as high as the amounts used in our tissue culture studies. In addition, repression of several IFN-inducible genes such as Stat-1 may play other important roles in the suppression of genes involved in immune recognition, such as genes that encode major histocompatibility complex proteins (4, 36).
IFN has been used in clinical trials to treat HPV-derived lesions. Some reports indicate success in using IFN to treat low-risk-HPV-derived lesions; however, the results with high-risk HPV remain controversial (12, 21, 26). IFN treatment is able to cause regression of high-risk-HPV-derived lesions, but the rate of recurrence is high. Our results are consistent with these clinical observations and may provide a model system for study of this clinically observed IFN resistance. It will be interesting to examine the long-term effect of IFN on cells that maintain low-risk-HPV genomes in this tissue culture model.
Long-term treatment of BPV-1-transformed C127 cells with IFN has been shown to greatly decrease the number of transformed cells and cause the cellular phenotype to revert to a flat, untransformed morphology (64). BPV DNA was reduced to nondetectable levels in these clonal isolates of revertant cells. Similarly, in our cultures of IFN-resistant HPV-31-positive cells, episomal HPV DNA was reduced to undetectable levels within a comparable time frame. However, small amounts of integrated viral DNA were always detected. In our study, IFN treatment had little effect on cells containing only integrated viral DNA. A similar situation has been described for established simian virus 40 (SV40)-transformed cells. Prolonged treatment with IFN decreases the number of SV40 transformants in acutely infected cultures, whereas it has no effect on the maintenance of the integrated SV40 genome in established SV40-transformed cells (22, 48). We observed a loss of episomal HPV DNA, while integrated copies remained detectable up to 4 months after resistant cells appeared (data not shown). We conclude that IFN has a negative effect on HPV episomes but that the elimination of episomal viral DNA is not a prerequisite for the establishment of IFN resistance in HPV-positive cells. This suggests the possibility that IFN treatment in vivo could select for cells that maintain integrated copies of HPV DNA. While IFN has been proposed as a therapy for HPV-derived lesions, the ability of HPV-positive cells to readily acquire resistance to IFN upon prolonged treatment suggests a cautionary approach.
Acknowledgments
We thank K. Green for the A431 cells and valuable comments on the manuscript and M. Paniagua for assistance in flow cytometry. We also thank F. Fehrmann for E5KO HPV-31-positive cells.
This study was supported by grants from the National Cancer Institute to L.A.L. (CA59655) and G.C.S. (CA68782) and by a grant from the National Institutes of Health to Y.E.C. (AI10559-01).
REFERENCES
- 1.Baker, C. C., W. C. Phelps, V. Lindgren, M. J. Braun, M. A. Gonda, and P. M. Howley. 1987. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 61:962-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, G. N. 2001. Host defense, viruses and apoptosis. Cell Death Differ. 8:113-126. [DOI] [PubMed] [Google Scholar]
- 3.Barnard, P., and N. A. J. McMillan. 1999. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-α. Virology 259:305-313. [DOI] [PubMed] [Google Scholar]
- 4.Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749-795. [DOI] [PubMed] [Google Scholar]
- 5.Borden, E. C., T. F. Hogan, and J. G. Voelkel. 1982. Comparative antiproliferative activity in vitro of natural interferons alpha and beta for diploid and transformed human cells. Cancer Res. 42:4948-4953. [PubMed] [Google Scholar]
- 6.Bouvard, V., A. Storey, D. Pim, and L. Banks. 1994. Characterization of the human papillomavirus E2 protein: evidence of trans-activation and trans-repression in cervical keratinocytes. EMBO J. 13:5451-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. E., and L. A. Laimins. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 74:4147-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chawla-Sarkar, M., D. W. Leaman, and E. C. Borden. 2001. Preferential induction of apoptosis by interferon (IFN)-β compared with IFN-α2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin. Cancer Res. 7:1821-1831. [PubMed] [Google Scholar]
- 9.Cheng, S., G. D. Schmidt, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 10.Chin, Y. E., M. Kitagawa, K. Kuida, R. A. Flavell, and X. Y. Fu. 1997. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol. Cell. Biol. 17:5328-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Luna, S., and J. Ortin. 1992. pac gene as efficient dominant marker and reporter gene in mammalian cells. Methods Enzymol. 216:376-385. [DOI] [PubMed] [Google Scholar]
- 12.De Palo, G., B. Stefanon, F. Rilke, S. Pilotti, and M. Ghione. 1985. Human fibroblast interferon in cervical and vulvar intraepithelial neoplasia associated with viral cytopathic effects. A pilot study. J. Reprod. Med. 30:404-408. [PubMed] [Google Scholar]
- 13.de Villiers, E. M. 1994. Human pathogenic papillomavirus types: an update. Curr. Top. Microbiol. Immunol. 186:1-12. [DOI] [PubMed] [Google Scholar]
- 14.Djmri, G., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, M. Peacocke, and J. Campisi. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 16.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 17.Frattini, M. G., and L. A. Laimins. 1994. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. USA 91:12398-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frattini, M. G., and L. A. Laimins. 1994. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology 204:799-804. [DOI] [PubMed] [Google Scholar]
- 19.Frattini, M. G., H. B. Lim, J. Doorbar, and L. A. Laimins. 1997. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 71:7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frost, L., K. Skajaa, L. E. Hvidman, S. J. Fay, and P. M. Larsen. 1990. No effect of intralesional injection of interferon on moderate cervical intraepithelial neoplasia. Br. J. Obstet. Gynaecol. 97:626-630. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Blanco, M. A., P. K. Ghosh, B. M. Jayaram, S. Ivory, P. Lebowitz, and P. Lengyel. 1985. Selectivity of interferon action in simian virus 40-transformed cells superinfected with simian virus 40. J. Virol. 53:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaudry, C. A., H. L. Palka, R. L. Dusek, A. C. Huen, M. J. Khandekar, L. G. Hudson, and K. J. Green. 2001. Tyrosine-phosphorylated plakoglobin is associated with desmogleins but not desmoplakin after epidermal growth factor receptor activation. J. Biol. Chem. 276:24871-24880. [DOI] [PubMed] [Google Scholar]
- 23a.Goodburn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 24.Goodwin, E. C., L. K. Naeger, D. E. Breiding, E. J. Androphy, and D. DiMaio. 1998. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J. Virol. 72:3925-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopalakrishnan, V., and S. A. Khan. 1994. E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication. Proc. Natl. Acad. Sci. USA 91:9597-9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross, G. 1995. Treatment of human papillomavirus infection. St Edmundsbury Press, London, United Kingdom.
- 27.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howley, P. M. (ed.). 1996. Papillomavirinae: the viruses and their replication, p. 947-978. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 29.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, S. R., J. S. Liu, T. R. Broker, and L. T. Chow. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058-24065. [PubMed] [Google Scholar]
- 31.Laimins, L. A. 1993. The biology of human papillomaviruses: from warts to cancer. Infect. Agents Dis. 2:74-86. [PubMed] [Google Scholar]
- 32.Leechanachai, P., L. Banks, F. Moreau, and G. Matlashewski. 1992. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene 7:19-25. [PubMed] [Google Scholar]
- 33.Li, S., S. Labrecque, M. C. Gauzzi, A. R. Cuddihy, A. H. Wong, S. Pellegrini, G. J. Matlashewski, and A. E. Koromilas. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727-5737. [DOI] [PubMed] [Google Scholar]
- 34.Lokshin, A., J. E. Mayotte, and M. L. Levitt. 1995. Mechanism of interferon beta-induced squamous differentiation and programmed cell death in human non-small-cell lung cancer cell lines. J. Natl. Cancer Inst. 87:206-212. [DOI] [PubMed] [Google Scholar]
- 35.Lowy, D. R., R. Kirnbauer, and J. T. Schiller. 1994. Genital human papillomavirus infection. Proc. Natl. Acad. Sci. USA 91:2436-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mach, B., V. Steimle, E. Martinez-Soria, and W. Reith. 1996. Regulation of MHC class II genes: lessons from a disease. Annu. Rev. Immunol. 14:301-331. [DOI] [PubMed] [Google Scholar]
- 37.Marcante, M. L., and A. Venuti. 1991. Human papillomavirus DNA as a possible index of invasiveness in female genital tract carcinomas. Eur. J. Cancer 27:187-190. [DOI] [PubMed] [Google Scholar]
- 38.Martin, L. G., G. W. Demers, and D. A. Galloway. 1998. Disruption of the G1/S transition in human papillomavirus type 16 E7-expressing human cells is associated with altered regulation of cyclin E. J. Virol. 72:975-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride, A. A., H. Romanczuk, and P. M. Howley. 1991. The papillomavirus E2 regulatory proteins. J. Biol. Chem. 266:18411-18414. [PubMed] [Google Scholar]
- 40.Meyers, C., M. G. Frattini, J. B. Hudson, and L. A. Laimins. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971-973. [DOI] [PubMed] [Google Scholar]
- 41.Meyers, C., and L. A. Laimins. 1994. In vitro systems for the study and propagation of human papillomaviruses. Curr. Top. Microbiol. Immunol. 186:199-215. [DOI] [PubMed] [Google Scholar]
- 42.Meyers, C., T. J. Mayer, and M. A. Ozbun. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J. Virol. 71:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 44.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nees, M., J. M. Geoghegan, T. Hyman, S. Frank, L. Miller, and C. D. Woodworth. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-κB-responsive genes in cervical keratinocytes. J. Virol. 75:4283-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newhouse, C. D., and S. J. Silverstein. 2001. Orientation of a novel DNA binding site affects human papillomavirus-mediated transcription and replication. J. Virol. 75:1722-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ortiz, A., F. N. Ziyadeh, and E. G. Neilson. 1997. Expression of apoptosis-regulatory genes in renal proximal tubular epithelial cells exposed to high ambient glucose and in diabetic kidneys. J. Investig. Med. 45:50-56. [PubMed] [Google Scholar]
- 48.Oxman, M. N., S. Baron, P. H. Black, K. K. Takemoto, K. Habel, and W. P. Rowe. 1967. The effect of interferon on SV-40 T antigen production in SV-40-transformed cells. Virology 32:122-127. [DOI] [PubMed] [Google Scholar]
- 49.Pfeffer, L. M., C. A. Dinarello, R. B. Herberman, B. R. Williams, E. C. Borden, R. Bordens, M. R. Walter, T. L. Nagabhushan, P. P. Trotta, and S. Pestka. 1998. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Res. 58:2489-2499. [PubMed] [Google Scholar]
- 50.Qin, X. Q., L. Runkel, C. Deck, C. DeDios, and J. Barsoum. 1997. Interferon-beta induces S phase accumulation selectively in human transformed cells. J. Interferon Cytokine Res. 17:355-367. [DOI] [PubMed] [Google Scholar]
- 51.Rheinwald, J. G., and H. Green. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6:331-343. [DOI] [PubMed] [Google Scholar]
- 52.Sangfelt, O., S. Erickson, J. Castro, T. Heiden, A. Gustafsson, S. Einhorn, and D. Grander. 1999. Molecular mechanisms underlying interferon-α-induced G0/G1 arrest: CKI-mediated regulation of G1 Cdk-complexes and activation of pocket proteins. Oncogene 18:2798-2810. [DOI] [PubMed] [Google Scholar]
- 53.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 54.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 55.Shirasawa, H., Y. Tomita, S. Sekiya, H. Takamizawa, and B. Simizu. 1987. Integration and transcription of human papillomavirus type 16 and 18 sequences in cell lines derived from cervical carcinomas. J. Gen. Virol. 68:583-591. [DOI] [PubMed] [Google Scholar]
- 56.Spalholz, B. A., Y. C. Yang, and P. M. Howley. 1985. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42:183-191. [DOI] [PubMed] [Google Scholar]
- 57.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 58.Straight, S. W., B. Herman, and D. J. McCance. 1995. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. J. Virol. 69:3185-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stubenrauch, F., A. M. Colbert, and L. A. Laimins. 1998. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 72:8115-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang, D. G., L. Li, D. P. Chopra, and A. T. Porter. 1998. Extended survivability of prostate cancer cells in the absence of trophic factors: increased proliferation, evasion of apoptosis, and the role of apoptosis proteins. Cancer Res. 58:3466-3479. [PubMed] [Google Scholar]
- 61.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 96:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tiefenbrun, N., D. Melamed, N. Levy, D. Resnitzky, I. Hoffman, S. I. Reed, and A. Kimchi. 1996. Alpha interferon suppresses the cyclin D3 and cdc25A genes, leading to a reversible G0-like arrest. Mol. Cell. Biol. 16:3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tindle, R. 1999. Immunomodulation of HPV infection and disease: an overview. Landes Company, Georgetown, Tex.
- 64.Turek, L. P., J. C. Byrne, D. R. Lowy, I. Dvoretzky, R. M. Friedman, and P. M. Howley. 1982. Interferon induces morphologic reversion with elimination of extrachromosomal viral genomes in bovine papillomavirus-transformed mouse cells. Proc. Natl. Acad. Sci. USA 79:7914-7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vilcek, J., and G. Sen. 1996. Interferons and other cytokines, p. 341-365. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 66.Webster, K., J. Parish, M. Pandya, P. L. Stern, A. R. Clarke, and K. Gaston. 2000. The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. J. Biol. Chem. 275:87-94. [DOI] [PubMed] [Google Scholar]
- 67.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 68.Wu, Y. J., L. M. Parker, N. E. Binder, M. A. Beckett, J. H. Sinard, C. T. Griffiths, and J. G. Rheinwald. 1982. The mesothelial keratins: a new family of cytoskeletal proteins identified in cultured mesothelial cells and nonkeratinizing epithelia. Cell 31:693-703. [DOI] [PubMed] [Google Scholar]
- 69.Yang, L., R. Li, I. J. Mohr, R. Clark, and M. R. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, H., P. P. Koty, J. Mayotte, and M. L. Levitt. 1999. Induction of multiple programmed cell death pathways by IFN-β in human non-small-cell lung cancer cell lines. Exp. Cell Res. 247:133-141. [DOI] [PubMed] [Google Scholar]
- 71.zur Hausen, H., and E. M. de Villiers. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]