Abstract
The genomic RNA of tobacco mosaic virus (TMV), like that of other positive-strand RNA viruses, acts as a template for both translation and replication. The highly structured 3′ untranslated region (UTR) of TMV RNAs plays an important role in both processes; it is not polyadenylated but ends with a tRNA-like structure (TLS) preceded by a conserved upstream pseudoknot domain (UPD). The TLS of tobamoviral RNAs can be specifically aminoacylated and, in this state, can interact with eukaryotic elongation factor 1A (eEF1A)/GTP with high affinity. Using a UV cross-linking assay, we detected another specific binding site for eEF1A/GTP, within the UPDs of TMV and crucifer-infecting tobamovirus (crTMV), that does not require aminoacylation. A mutational analysis revealed that UPD pseudoknot conformation and some conserved primary sequence elements are required for this interaction. Its possible role in the regulation of tobamovirus gene expression and replication is discussed.
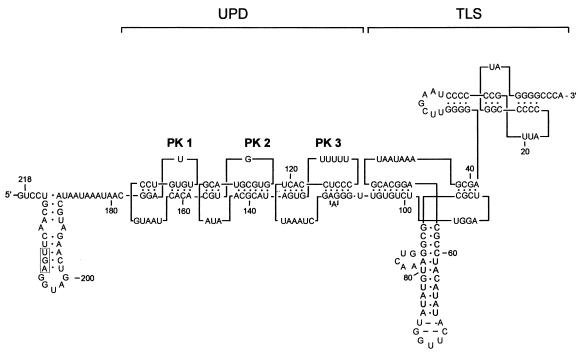
Tobacco mosaic virus (TMV), a positive-strand plant RNA virus, is the type member of the tobamovirus family in the alphavirus-like superfamily. The genomic RNA of TMV strain vulgare (U1) is 6,395 nucleotides (nt) long and encodes at least four proteins. The full-length RNA is used to produce 126- and 183-kDa RNA-dependent replicase proteins, while the 30-kDa movement protein (MP) and the 17.5-kDa coat protein are translated from 3′-coterminal subgenomic mRNAs. The coding region is flanked by the 5′ untranslated region (5′-UTR, or Ω) and the 3′-UTR, both of which are required for viral replication. TMV RNA is capped, but it lacks a 3′ poly(A) tail. Instead, the 3′-UTR contains a highly structured and conserved sequence composed of several pseudoknots (PKs) of the hairpin loop type (Fig. 1) (50, 54, 61). The TMV U1 3′-UTR is comprised of two structural domains: a 3′-terminal domain containing two PKs important for formation of a tRNA-like structure (TLS) (21, 53), linked to an upstream PK domain (UPD)—a quasicontinuous double-helical stalk comprising three consecutive PKs (61). A similar tandem arrangement of structural units (UPD-TLS) is found in all tobamoviruses and satellites of TMV, although the sizes and numbers of PKs of the UPD are variable (26). The TLS of TMV can be aminoacylated and binds to several tRNA-specific enzymes (reviewed in reference 42). Interestingly, two of the PKs in the U1 UPD are phylogenetically conserved among all tobamoviruses, TMV satellites, and hordeiviruses in location, in structure, and even in several positions of primary sequence, strongly suggesting their functional importance (38, 61).
FIG. 1.
Primary sequence and secondary structure of the TMV U1 3′-UTR. Major structural domains are shown: the L-shaped TLS (nt 1 to 105 counted from the 3′ end) and the UPD, consisting of three consecutive PKs. The coat protein translational termination codon is boxed. Numbering of the PKs in the UPD is as indicated. This structure is derived from chemical and enzymatic probing data (21, 54, 61).
Whether ending with a poly(A) tail or a TLS, the 3′ terminus of the genomic RNA of positive-strand RNA viruses is generally thought to contain elements of the promoter for initiation of minus-strand viral RNA synthesis. The minimal 3′ cis-acting element required for the initiation of negative-strand RNA synthesis on TMV genomic RNA includes the 3′-terminal TLS and the 3′-most-proximal PK structure (PK3) of the UPD (12, 49, 59). Destabilization of PKs in the TLS of turnip yellow mosaic virus (TYMV), brome mosaic virus (BMV), and TMV results in a reduction in infectivity of these viruses (12, 15, 19). Host cell proteins that bind to cis-acting sequences known to be required for viral replication of both positive- or negative-strand viral RNA have been identified (4, 6, 16, 27, 36, 58), and several host proteins have been found associated with template-specific replicase purified from TMV-infected plants (48), including a 56-kDa protein identified as being immunologically related to GCD10, a subunit of yeast initiation factor eIF3 (48). However, the functional role of such proteins in viral replication remains unclear.
Specific interactions of the TMV 3′-UTR with CTP, ATP, nucleotidyltransferase, an aminoacyl-tRNA synthetase, and translational eukaryotic elongation factor 1A (eEF1A) have been demonstrated (40, 41). It is not known whether these interactions are involved in virus replication or in translation, although they apparently play a role in increasing mRNA stability and maintenance of intact 3′ termini (41).
Functional interaction between 5′- and 3′-UTRs involved in promoting efficient mRNA translation has been demonstrated for a number of plant and animal viruses as well as for some cellular mRNAs (24). In the latter case, it is believed that the 5′-UTR-3′-UTR interaction is mediated by eukaryotic translation initiation factor 4G (eIF-4G) and poly(A)-binding protein (PABP). Moreover, different RNA structural elements in the 3′-UTR of several viral and cellular mRNAs that functionally substitute for a poly(A) tail in both cap-dependent and cap-independent translation have been identified; in some cases, specific complexes of these 3′-UTRs with certain protein factors can functionally replace the PABP/poly(A)-tail complex (29, 30, 62). However, for nonpolyadenylated plant RNA viruses, such functional analogues of PABP have not yet been identified.
In TMV, the capped 5′ leader sequence (Ω) and the 3′-UTR functionally interact to effect high-level translation of viral polymerase proteins, whereby the UPD appears to functionally substitute for a poly(A) tail (22, 66). Both the conserved primary sequence within PK2 and PK3 of the UPD and its higher-order structure were found to be necessary for translation (38), although the 3′-terminal TLS also played a role (23, 38).
Thus, replication of RNA viruses involves both viral and host cell protein components. Many of the latter have been identified as having been subverted from host translational and RNA processing machineries (reviewed in reference 36), highlighting the intimate coupling of viral translation and RNA replication. Since the cis-acting signals controlling translation and initiation of negative-strand RNA synthesis overlap in the TMV 3′-UTR, we have sought to identify host protein factors that interact with this region and are required for these two viral functions. Using a UV cross-linking assay, we have identified a novel site within the UPDs of the TMV and crucifer-infecting tobamovirus (crTMV) RNA 3′-UTRs that interacts specifically in the nonaminoacylated state with host cell protein eEF1A in the presence of GTP. Extensive mutagenesis analysis showed that both the structure and the conserved primary sequence of two coterminal PKs of the UPD, but not the TLS, were required for this interaction.
MATERIALS AND METHODS
Plasmid construction and site-directed mutagenesis.
T7-based transcription vectors containing cDNA fragments representing 3′-terminal parts of TMV U1 or crTMV genomic RNAs were constructed by standard cloning techniques. The numbering of the nucleotides in cDNAs of TMV U1 and crTMV in this study starts from the 3′ end. cDNA fragments representing the 3′-terminal part of TMV U1 (nt 1 to 218) or the corresponding region of crTMV (nt 1 to 261) were amplified by PCR from appropriate cDNA-containing plasmids (kindly provided by Y. Dorokhov, Moscow State University, Moscow, Russia). Sense and antisense primers were 5′-CAGGAATTCGTCCTGCAACTTGAGGTAGT-3′ and 5′-CGTGGATCCACCTGGGCCCCTACCGGGGGTAAC-3′, respectively, for the TMV U1 3′-UTR and 5′-CAGGAATTCCTTTGGTGCCATGGACTAC-3′ and 5′-CGTGGATCCACCTGGGCCCCTACCGGGGGTAAC-3′, respectively, for the crTMV 3′-UTR (EcoRI and BamHI sites are underlined). The PCR products were digested with EcoRI and BamHI, purified after gel electrophoresis with a gel extraction kit (Qiagen), and ligated into vector pT7T3LacZ (Roche) prepared by digestion with the same enzymes. The resulting recombinant plasmids were named pTZ1 and pCTZ. Additional plasmid constructs pTZ2, pTZ3, pTZ4, corresponding to cDNA fragments representing 5′-truncated versions of the TMV U1 3′-UTR were obtained in the same way, using one of the following sense primers: 5′-CAGGAATTCCACGTGGTGCGTACGATAAC-3′, 5′-CAGGAATTCCATAGTGTTTTTCCCTCCAC-3′, and 5′-CAGGAATTCGGGTTGGTC TTGGATGG-3′ annealing to nt 62 to 179, nt 137 to 160, and nt 106 to 62, respectively, and the same antisense primer as for pTZ1. Plasmid constructs containing internal deletions within the TMV U1 3′-UTR (nt 162 to 138 or nt 106 to 138) were constructed by PCR-based site-directed mutagenesis using the sense primers 5′-AAGATGCATAATAAATAACGGATTGTGTCCGTAATCACAAGTGTTTTTCCCTCCACTTA-3′ or 5′-GGTGCGTACGATAACGCATTTGTGTCTTGGATCGCGCGG-3′, respectively, and the same antisense primer as for pTZ1. The resulting fragments were digested with NsiI and BamHI or BsiWI and BamHI and ligated into the correspondingly digested plasmid pTZ1 to produce plasmids pTZ5 and pTZ6, respectively.
To prepare the plasmid series containing cDNA fragments including PK2 and PK3 and the TLS (nt 162 to 1), the TMV U1 3′-UTR sequence in pTZ2 was amplified with sense oligonucleotide primers incorporating the required mutation and the same antisense primer as for pTZ1. The cDNA fragments with mutations were inserted into the vector pT7T3LacZ prepared by digestion with EcoRI and BamHI. The resulting plasmids were used to produce the RNA transcripts shown in Fig. 7 and 8. The nucleotide sequences of all constructs were verified by DNA sequencing.
FIG. 7.
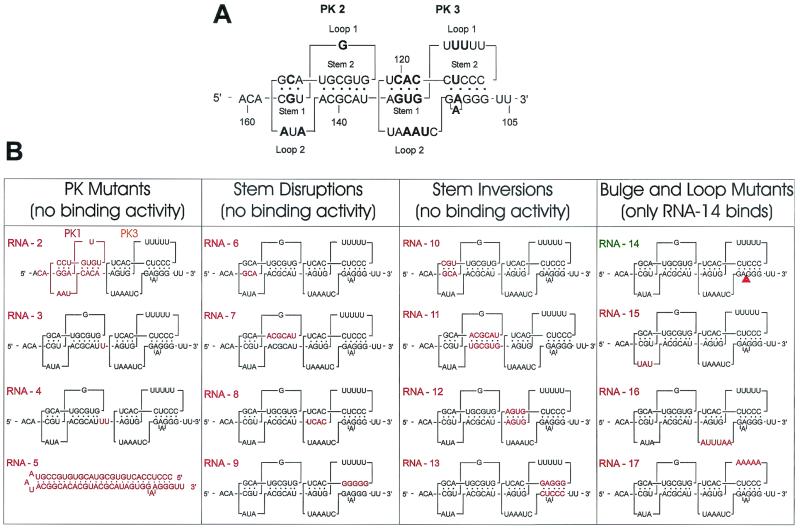
Fine analysis of eEF1A binding to PK2-PK3. (A) Primary and secondary structures of the wild-type sequence (RNA-1) of the PK2-PK3 region showing the stems and loops involved in PK formation. Conserved nucleotides are in bold. (B) Maps of the mutants analyzed. The modified sequences or structures are shown in red. Construct names (RNA-2 to RNA-17) are in red or green to indicate no binding activity or normal binding activity, respectively.
FIG. 8.
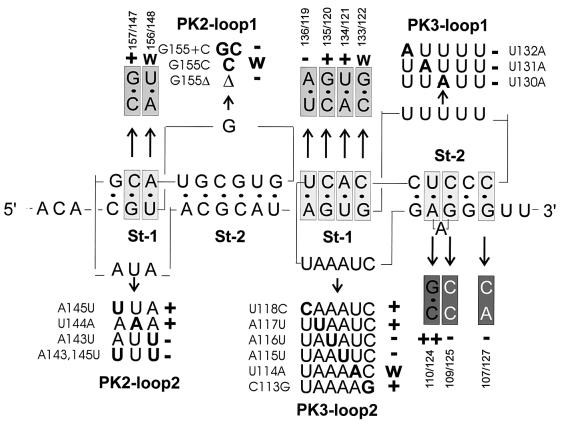
Analysis of single mutants in the loops and base pair mutants in the stems of PK2 and PK3 affecting eEF1A binding. Shown are primary and secondary structures of the region, with mutants analyzed indicated. Mutated nucleotides in the loops are shown in bold, and their effects in the UV cross-linking assay are indicated by + (wild-type-like), w (weakened binding), and − (loss of binding activity). Analyzed base pairs (boxed) were exchanged for flipped base pairs (light grey), strengthened base pairs (dark grey), or unpaired sequences (white on grey). Results are indicated as + (active), ++ (strongly active), w (weakly active), and − (inactive).
Preparation of RNA transcripts.
To prepare templates for runoff transcription with T7 phage RNA polymerase, plasmid DNA was linearized as follows: plasmids pTZ1 and pCTZ containing the wild-type sequences of TMV U1 and crTMV, respectively, plasmids pTZ2, pTZ3, pTZ4, pTZ5, pTZ6, containing 5′ truncation or internal deletion of the TMV U1 3′-UTR, and all plasmids of series pTZ2.N were linearized at the BamHI site. To generate RNA transcripts corresponding to nt 218 to 97, nt 162 to 97, or nt 137 to 97 of the TMV U1 3′-UTR, plasmids pTZ1, pTZ2, or pTZ3 were linearized by digestion with Sau3AI. To generate RNA transcripts corresponding to nt 218 to 113 and nt 162 to 113 of the TMV U1 3′-UTR, plasmids pTZ1 and pTZ2 were linearized by digestion with TaqI. Other plasmids used in this study were linearized with restriction endonucleases as follows. pTZ1 was linearized with RsaI and PmlI to generate 5′-truncated fragments of the TMV U1 3′-UTR. An antisense RNA to the TMV U1 3′-UTR was transcribed from pTZ1 with T3 RNA polymerase after linearization at the EcoRI site. pT7T3lacZ was digested with PvuII to produce an RNA probe of approximately the same length as that from the TMV U1 3′-UTR for use as a control. 32P-labeled RNAs were prepared by in vitro transcription using 20 μCi of [32P]UTP (800 Ci/mmol; NEN) in a 20-μl reaction mixture containing 80 mM HEPES (pH 7.8), 20 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 0.025 mM UTP, 0.5 mM concentrations each of ATP, CTP, and GTP, 200 U of RNasin (Promega) per ml, 1,500 U of T7 or T3 RNA polymerase (Biofinex) per ml, and 30 μg of linearized plasmid DNA per ml. After 2 h of incubation at 37°C, the reactions were stopped and precipitated with ethanol, and RNA transcripts were purified from nonincorporated nucleotides and DNA fragments by electrophoresis with 6% polyacrylamide gels containing 7 M urea in 1× Tris-borate EDTA (89 mM Tris-boric acid [pH 8.2], 2 mM EDTA). RNA was eluted from the gel slices overnight at 24°C in 0.5 ml of elution buffer (0.5 M ammonium acetate, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA) and precipitated with ethanol. The radioactivity in the RNA transcripts was quantified by Cherenkov counting in a liquid scintillation analyzer TRI-CARB 2100 TR (Packard). The labeled RNAs were resuspended in water to 100,000 cpm/μl (about 20 to 30 fmol per μl). Unlabeled RNA transcripts, used in competition experiments, were synthesized in a volume of 0.2 ml under similar reaction conditions, except that the concentrations of ATP, CTP, GTP, and UTP were 3 mM. These RNAs were also purified as described above. The RNA concentration was determined spectrophotometrically, and transcripts were resuspended in water to 500 ng of RNA per μl. Labeled and unlabeled RNAs were renatured by heating and cooling down in the presence of 1 mM MgCl2 and stored at −20°C.
Preparation of plant cell extracts.
Cytoplasmic S-30 and S-100 wheat germ extract was prepared essentially as described by Lax et al. (37). To prepare cytoplasmic extracts from Nicotiana benthamiana (tobacco-related plants), 50 g of young leaves was homogenized by using a mortar and pestle with 50 ml of an ice-cold solution containing 20 mM HEPES (pH 7.8), 5 mM MgCl2, 100 mM KCl, 2 mM DTT, 0.1 mM EDTA, 10% (vol/vol) glycerol, and EDTA-free protease inhibitor cocktail (Roche). The extract was centrifuged at 30,000 × g for 15 min in an SS-34 Sorval rotor, and the supernatant was recentrifuged at 100,000 × g for 3 h in a Beckman Ti 50 rotor to obtain S-100 extract. The total protein concentration in the extracts was determined by Bradford analysis using a Bio-Rad protein assay kit. Aliquots of extracts were stored at −80°C.
Gel electrophoresis.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli (35). The stacking gel contained 4% acrylamide-bisacrylamide, and the separating gel contained 12% acrylamide-bisacrylamide in a ratio of 40 to 0.6. The gels were fixed and stained with 0.25% (wt/wt) Coomassie brilliant blue G250 in 40% methanol and 9% acetic acid and destained with the same solution without Coomassie brilliant blue. Two-dimensional nonequilibrium pH gradient (NEPHGE)-SDS-PAGE was carried out according to the method of O'Farrell et al. (47) by using ampholine pH 3.5 to 10 (Bio-Rad) and pH 8 to 10 (Pharmacia) at a 1:1 ratio. The second dimension was performed by SDS-PAGE on a 12% gel.
UV-induced cross-linking of RNA to proteins.
The amount of protein of different extracts or purified proteins indicated was preincubated with 0.1 mM GTP and 2 mM MgCl2 (except where indicated otherwise) in buffer B (20 mM HEPES [pH 7.6], 50 mM KCl, 1 mM DTT, 0.1 mM EDTA, 20% [vol/vol] glycerol) with the addition of 20 U of RNase inhibitor (Promega) on ice for 10 min. Uniformly labeled RNA transcripts (30 fmol; ∼100,000 cpm) and 2 μg of nonspecific competitor (Escherichia coli rRNAs) were then added, and the reaction mixture was incubated for 15 min at 24°C. Heparin was added to a final concentration of 0.3 mg/ml, and the mixture was further incubated for 10 min in a final volume of 10 μl. In competition experiments, in addition to rRNAs, an excess of unlabeled RNA was added to the reaction mixture simultaneously with the labeled probes. Competition experiments with single-stranded RNA p(U), p(A), p(C), p(G), or p(UC), p(AC), p(UA), p(CG), p(GA), or double-stranded RNA p(A)-p(U) were performed in the same way, with a final competitor concentration of 0.1 mg/ml. The binding reactions were irradiated with UV light (254 nm) from a Stratalinker 1800 (Stratagene) at a distance of 8 cm for 15 min at 120 mJ/cm2 on ice. After irradiation, samples were treated with a mixture of 10 μg of RNase A, 25 U of RNase T1, and 0.2 U of cobra venom RNase V1 (Pharmacia Biotech) for 20 min at 37°C to digest unprotected and unbound RNA and analyzed by electrophoresis on SDS-12% PAGE. Gels were fixed in acetic acid-methanol and then dried. UV cross-linked proteins were visualized by autoradiography on Kodak BIOMAX MS X-ray film or by using a PhosphorImager.
Purification of the p52 protein.
Wheat germ S100 extract prepared as described above was used as starting material for the purification of p52 with buffer A (20 mM HEPES-KOH [pH 7.6], 50 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, and 20% [vol/vol] glycerol). At each step of the purification procedure, an aliquot of each fraction was dialyzed against buffer A (if required) and assayed for specific binding activity to the 32P-labeled TMV U1 RNA 3′-UTR in the UV cross-linking assay. Fractions were also analyzed by SDS-12% PAGE, and protein concentrations were determined by Bradford assay. All purification procedures were performed at 4°C. Ammonium sulfate was added to the wheat germ extract S100 fraction to 40% saturation (22.9 g of ammonium sulfate to 100 ml of extract, with a total of 5 g of protein). Ammonium sulfate was added in small portions under constant stirring, the mixture was stirred for another 60 min, and precipitated proteins were removed by centrifugation at 12,000 × g for 30 min in an SA-600 Sorval rotor. p52 was detected in the supernatant and was subsequently precipitated by the further addition of 19 g of ammonium sulfate (70% saturation). The precipitate was collected by centrifugation as described above. The pellet (2.7 g) was dissolved in 100 ml of buffer A and dialyzed against the same buffer overnight before loading onto a Q-Sepharose Fast-Flow (Pharmacia Biotech) column (2.6 by 20 cm) equilibrated with buffer A. Unbound material was removed by being washed with the same buffer. A 600-ml linear gradient of 50 to 500 mM KCl in buffer A was applied at a flow rate of 3.5 ml/min. Specific binding activity was detected in the flowthrough and wash fractions from this column. The pooled and concentrated flowthrough and wash fractions from the Q-Sepharose step (600 mg) were applied to an SP-Sepharose High-Performance (Pharmacia Biotech) column (1.6 by 20 cm) preequilibrated with buffer A containing 30% (vol/vol) glycerol. After the column was washed with the same buffer, a 400-ml linear gradient of 50 to 800 mM KCl was applied at a flow rate of 1 ml/min. A peak of specific binding activity was eluted between 160 and 350 mM KCl. Fractions active in the UV cross-linking assay were dialyzed against buffer A containing 30% (vol/vol) glycerol, and the proteins were concentrated with a Centricon-30 microconcentrator (Amicon). The concentrated fraction eluted from SP-Sepharose at 200 mM KCl (SP-200) with high specific UV cross-linking activity was used in all subsequent experiments and consisted of a single 52-kDa protein, which was approximately 90% pure, as judged by SDS-PAGE stained with Coomassie blue.
Peptide sequencing.
Purified p52 protein (2 μg of protein from fraction SP-200) was separated by SDS-PAGE, excised from the gel, reduced with DTT, alkylated with iodoacetamide, and cleaved with trypsin (sequencing grade; Promega). The resulting peptides were fractionated and analyzed by liquid chromatography interfaced with electrospray mass spectrometry (LC-MS) using a Rheos 4000 chromatograph equipped with a 1 by 250 mm Vydac (Hesperia, Calif.) C8 column and interfaced with a Sciex API 300 mass spectrometer (PE Sciex, Toronto, Ontario, Canada) operated in the single quadrupole operating mode as described by Krieg et al. (34). The mass range from 300 to 2,400 Da was scanned with a step size of 0.5 Da and a dwell time of 3.15 s per scan. The column was equilibrated in 95% solvent A (2% CH3CN, 0.05% trifluoroacetic acid in H2O) and 5% solvent B (80% CH3CN, 0.045% trifluoroacetic acid in H2O), and a linear gradient was developed from 5 to 50% solvent B in 60 min at a flow rate of 0.05 ml/min. N-terminal sequence analysis was carried out on a model 477A protein sequencer (Applied Biosystems, Foster City, Calif.) according to the recommendations of the manufacturer. The amino acid sequences of peptides obtained were used to identify the protein using the Swiss-Prot protein sequence database with the BLAST program.
Western blotting analysis for detection of eEF1A.
Protein samples were separated on SDS-12% PAGE and then transferred onto a nitrocellulose membrane (Schleicher & Schuell) using semidry electroblotting. The membrane was blocked with 5% nonfat dried milk (Bio-Rad) overnight at 4°C and probed with polyclonal rabbit anti-wheat eEF1A antibodies at a dilution of 1:5,000 (antibody preparation has been described previously [10]). After 1 h of incubation, the membrane was washed and probed with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (Bio-Rad) and visualized with an ECL kit (Amersham).
Partial proteolytic digestion analyses.
Partial digestion of the proteins of crude wheat germ S100 extract after UV cross-linking to 32P-labeled TMV U1 3′-UTR was performed with trypsin in the buffer for UV cross-linking with 20% glycerol added. UV cross-linking reactions were performed as described above.
Following treatment with RNases, the reaction mixtures were supplemented with 0.5, 1, or 3 μg of trypsin and incubated at 37°C for 30 min before addition of 1 mM phenylmethylsulfonyl fluoride. Partially digested proteins were separated by SDS-16% PAGE and transferred onto nitrocellulose membrane. 32P-labeled protein fragments of p52 were visualized by autoradiography. The membrane was then incubated with rabbit polyclonal anti-wheat eEF1A antibody, and the bound antibody was detected with an ECL kit (Amersham).
Cloning, expression, and purification of glutathione S-transferase-tagged eEF1A protein.
A DNA fragment encoding eEF1A was PCR amplified from a plasmid containing the wheat eEF1A cDNA (44) by using two primers corresponding to the 5′ CGCGGATCCCATATGGGTAAGGAGAAGACTC and 3′ CGGGAATTCCTCATTTCTTCTTGATGGCAGCCTTG ends of the eEF1A coding sequence, with attached BamHI or EcoRI sites (underlined), respectively. The PCR product was digested with BamHI and EcoRI and ligated into the pGEX-2TK (Pharmacia Biotech) expression vector digested with the same enzymes. The resulting construct (pGEX-wheat eEF1A) was confirmed by DNA sequencing. The glutathione S-transferase-eEF1A fusion protein was expressed in E. coli strain BL21(DE3) by induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) after growth to an A600 of 1.0 and harvested after 3 h. Lysis, purification, and cleavage of the fusion protein with thrombine protease on glutathione-agarose beads were performed in accordance with the manufacturer's guidelines (Pharmacia Biotech). Recombinant wheat eEF1A protein was stored at −80°C in elution buffer containing 30% glycerol.
RESULTS
A 52-kDa plant cell protein interacts specifically with the 3′-UTR of TMV U1 and crTMV RNA.
To assess the interaction of cytoplasmic proteins with the 3′-UTRs of TMV U1 and crTMV RNA, in vitro transcripts covering the 218 most-3′-terminal nucleotides of TMV U1 RNA and the corresponding 263 nt of crTMV RNA were prepared. To block the aminoacylation capacity of these molecules, and consequently the aminoacylation-dependent interaction with eEF1A, a stretch of 12 nt was added to the original 3′ CCA end. As controls, an antisense version of the terminal 218 nt of TMV U1 RNA and a 240-nt RNA fragment derived from the plasmid vector pT3T7LacZ were used. The interaction of these RNAs with proteins from wheat germ and N. benthamiana leaf extracts was determined by UV cross-linking.
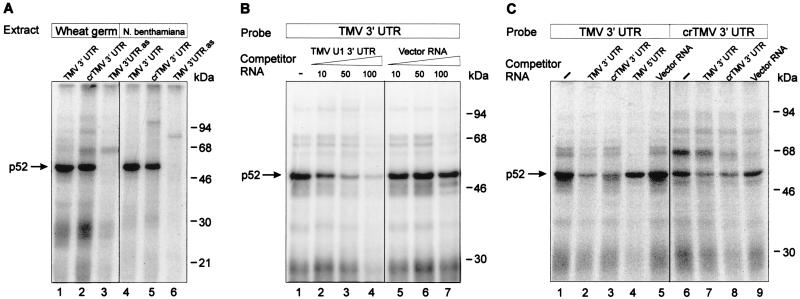
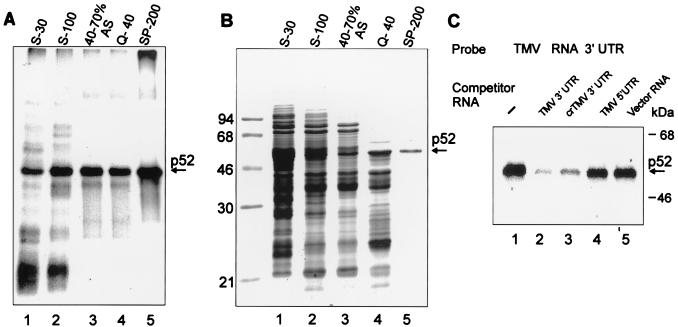
UV irradiation resulted in covalent cross-linking of the 32P-labeled RNA to several proteins (Fig. 2A). The most prominent of these had a mobility of 52 kDa, was found in extracts of both wheat germ (Fig. 2A, lanes 1 and 2) and N. benthamiana leaves (lanes 4 and 5) and cross-linked with both the TMV U1 (lanes 1 and 4) and the crTMV (lanes 2 and 5) 3′-UTRs. Addition of proteinase K (2 mg/ml) eliminated complex formation (not shown), showing that p52 is a protein. Furthermore, no binding of p52 to the control RNAs occurred (Fig. 2A, lanes 3 and 6), demonstrating that the interaction was specific to the tobamovirus 3′-UTRs.
FIG. 2.
Specific binding of p52 to the 3′-UTRs of TMV U1 and crTMV RNAs. (A) Binding of proteins to the 3′-UTRs of TMV U1 and crTMV RNAs. UV light induced cross-linking of wheat germ (lanes 1 to 3) or N. benthamiana (lanes 4 to 6) S-100 extract proteins to 32P-labeled RNA probes as indicated above the lanes. Binding reactions were performed as described in Materials and Methods. The complexes formed were irradiated with UV light and then treated with RNases and analyzed by SDS-12% PAGE followed by autoradiography. The major cross-linked protein (p52) is indicated by an arrow. The positions of size markers are shown on the right. An RNA complementary to the TMV U1 3′-UTR (TMV 3′ UTR.as) was used as the control RNA in lanes 3 and 6. (B) Specificity of p52 binding to the TMV U1 3′-UTR. Binding reactions were carried out with wheat germ extract with 30 fmol (100,000 cpm) of 32P-labeled TMV U1 3′-UTR in the absence (lane 1) or in the presence of increasing amounts of unlabeled specific (lanes 2 to 4) and nonspecific (lanes 5 to 7) RNA competitors. The fold molar excess of competitor is indicated above each lane . (C) Cross-competition of the TMV U1 3′-UTRs, crTMV 3′-UTR, the TMV 5′-UTR and nonviral RNA. Conditions of binding were as described for panel A, except that a 100-fold molar excess of competitor was added in each case.
The specificity of the interaction was further investigated by using competitor RNAs. Binding of p52 to the labeled TMV U1 3′-UTR transcript was significantly reduced with a 50-fold excess of unlabeled transcript (Fig. 2B, lanes 1 to 4), whereas a 100-fold molar excess of nonspecific competitor (vector RNA, lanes 5 to 7) had no effect. Furthermore, no competition was observed for a 100-fold molar excess of other RNA species, such as the 68-nt TMV 5′-UTR sequence (Fig. 2C, lane 4), or a 1,000-fold excess of either single-stranded p(A), p(U), p(C), p(G), or p(CU), p(AC), p(AU) or double-stranded pA-pU (not shown). However, the TMV U1 3′-UTR did compete with the crTMV 3′-UTR and vice versa (Fig. 2C, lanes 2, 3, 7, and 8). These results suggest that p52-binding may be a common property of the 3′-UTRs of the tobamoviruses.
Purification of p52 from wheat germ extract and its identification as eEF1A.
To identify p52, we purified it from a wheat germ extract postribosomal supernatant (S-100) using ammonium sulfate precipitation and two steps of ion-exchange chromatography. At each step of purification, the specific RNA binding activity was determined by UV cross-linking (Fig. 3A), and the protein composition of fractions was analyzed by SDS-PAGE (Fig. 3B). p52 precipitated predominantly in the 40 to 70% saturated ammonium sulfate fraction and was eluted from Q-Sepharose as flowthrough and from SP-Sepharose between 180 and 360 mM KCl. In UV cross-linking competition assays, binding of p52 that was eluted from SP-Sepharose at 200 mM KCl (SP-200; judged as 90% pure protein) to the TMV U1 3′-UTR was effectively blocked by addition of a 30-fold molar excess of unlabeled 3′-UTR RNA—either TMV U1 or crTMV—but not by nonspecific competitor RNAs (Fig. 3C); thus, there is no difference in specificity of the binding whether p52 from crude extract or purified protein is used or not (Fig. 2C and 3C), and p52 apparently interacts directly with the TMV U1 3′-UTR and does not require additional proteins (Fig. 3C).
FIG. 3.
Purification of p52. (A) Specific UV cross-linking activity of protein fractions at different stages of p52 purification. Shown are proteins of crude wheat germ extract (S-30; 50 μg, lane 1) and selected fractions: S-100 postribosomal supernatant (S-100; 40 μg, lane 2), the 40 to 70% ammonium sulfate (AS) cut of S-100 (40 to 70% AS; 15 μg, lane 3), proteins from the 40 to 70% AS cut in the flowthrough from a Q-Sepharose column at 40 mM KCl (Q-40; 6 μg, lane 4), and protein from the Q-40 fraction eluted from a SP-Sepharose column with 200 mM KCl (SP-200; 1 μg, lane 5) were dialyzed to a salt concentration of 50 mM KCl before the assay (if required). Binding reactions, UV cross-linking to the 32P-labeled TMV U1 RNA 3′-UTR, RNase treatment, and SDS-PAGE were performed as described in Materials and Methods. (B) Protein composition of the selected fractions. Coomassie blue-stained SDS-12% PAGE of the same amounts of proteins from the fractions described for panel A. The SP-200 fraction contains one major protein band with a mobility of approximately 52 kDa as indicated. (C) UV cross-linking experiment with TMV U1 3′-UTR. The experiment was conducted as described for Fig. 2C, except that purified p52 was used and competitor RNAs were in 30-fold excess.
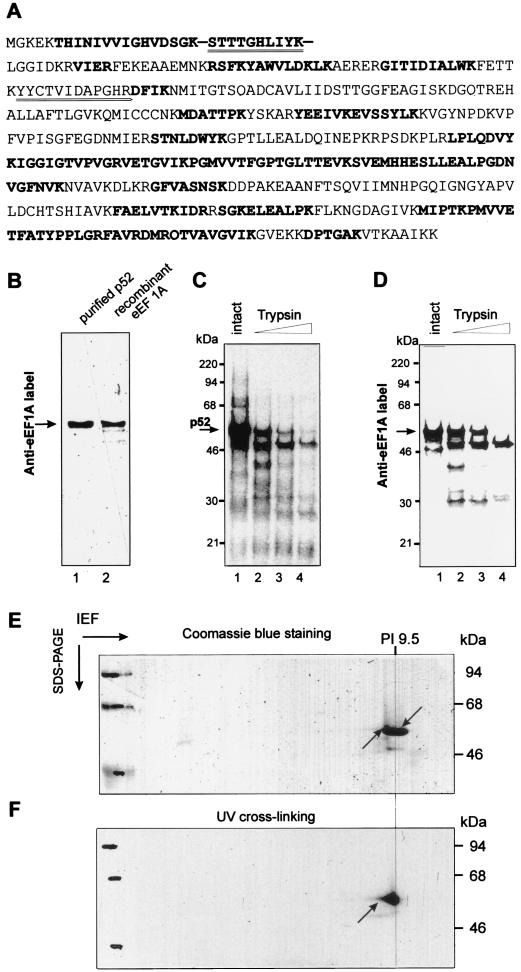
The p52 protein eluted from SP-Sepharose was judged pure enough to sequence. Following SDS-PAGE, p52 was excised from the gel and treated with trypsin. Two tryptic fragments were isolated and sequenced by the Edman degradation method. The two sequences obtained (Fig. 4A, double-underlined peptides) were identical to the corresponding sequences of peptides derived from wheat eEF1A (Swissprot accession number Q03033) (44). Additionally, p52 fragments were analyzed by mass spectroscopical analysis. Both methods revealed p52 peptides to be identical to the predicted peptides of wheat eEF1A (Fig. 4A, bold underlined peptides).
FIG. 4.
Identification of p52 as eEF1A. (A) Comparison of sequences of wheat germ eEF1A (Swissprot Q03033) and of peptides derived from p52. p52 from the SP-200 fraction was excised from the gel, digested with trypsin, and subjected to LC-MS as described in Materials and Methods. Peptides found by LC-MS are shown in bold. Additionally, two of the peptides were isolated and sequenced by Edman degradation (double underlined). (B) Identity of p52 and eEF1A as determined by Western blotting. Equal amounts (0.4 μg) of purified p52 and recombinant wheat eEF1A protein (see Materials and Methods) were fractionated on SDS-12% PAGE, transferred to nitrocellulose membrane, and incubated with rabbit polyclonal anti-wheat eEF1A antibody. Bound antibody was revealed by using an ECL kit (Amersham). (C and D) Comparative protease mapping of p52. Shown are proteins UV cross-linked to the 32P-labeled TMV U1 3′-UTR (C) and Anti-eEF1A activity in wheat germ extracts (S100) (C). (E and F) Two-dimensional isoelectric focusing/SDS-PAGE gel of purified p52. The purified p52 protein was UV cross-linked to 32P-labeled TMV U1 3′-UTR, treated with RNases, and resolved in the first dimension by NEPHGE and in the second dimension by SDS-PAGE. The gel was stained with Coomassie blue and analyzed by autoradiography as described in Materials and Methods. (E) Coomassie blue-stained gel. The two arrows indicate the positions of the Coomassie blue-stained and radiolabeled p52 spot. (F) Autoradiogram of the gel shown in panel E. The arrow indicates the position of the radiolabeled spot corresponding to p52.
The identity of p52 as eEF1A was further confirmed by immunoblotting. Polyclonal rabbit antibodies raised against wheat eEF1A interacted with purified wheat germ p52 as strongly as with purified recombinant wheat eEF1A produced in E. coli (Fig. 4B).
Comparison of specific protein fragments obtained after partial digestion with trypsin also revealed that those originating from wheat germ p52 that were labeled in the UV cross-linking assay and those of wheat eEF1A identified by Western blotting comigrated identically on SDS-PAGE gel at positions corresponding to 46, 40, and 30 kDa (Fig. 4C and D).
The isoelectric point of p52, 32P-labeled by UV cross-linking followed by RNase digestion, was determined by NEPHGE/SDS-PAGE (see Materials and Methods) to be approximately pI 9.5, a value very close to that calculated for wheat eEF1A (pI 10), and the Coomassie blue-stained protein spot closely matched the 32P-labeled protein spot of approximately 52 kDa (Fig. 4E and F).
The identity of digestion patterns of p52 and eEF1A detected by 32P labeling and Western blotting, as well as the similarity of their isoelectric points, excludes the possibility that p52 merely copurifies with eEF1A and another protein of similar mobility, as eEF1A is involved in the interaction with the TMV U1 3′-UTR. Taken together, these data confirm the identity of the p52 protein that specifically interacts with the UPD of the TMV U1 3′-UTR as eEF1A.
Conditions required for the interaction of eEF1A with the TMV 3′-UTR.
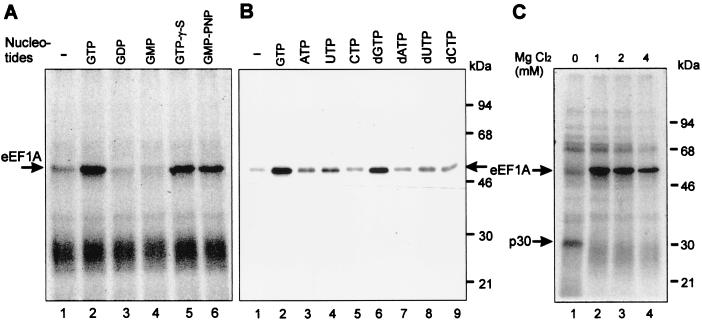
Since eEF1A is a G protein whose conformation state is regulated by GTP and GDP (7), we examined the effects of different nucleotides and nucleotide analogues on TMV 3′-UTR/eEF1A complex formation. The UV cross-linking experiments described above had been performed in the presence of 0.2 mM ATP, 0.2 mM GTP, and 2 mM MgCl2. Omission of nucleotides from the incubation mixture almost abolished eEF1A binding (Fig. 5A and B, lanes 1), with the interaction being restored upon addition of GTP (Fig. 5A and B, lanes 2). Binding was also observed in the presence of dGTP, but no other nucleoside triphosphate (NTP) or deoxynucleoside triphosphate (dNTP) could support the interaction with the purified eEF1A protein as efficiently (Fig. 5B). Moreover, the TMV 3′-UTR can discriminate between the GTP, GDP, or GMP complexes of eEF1A and interacts only with the GTP-activated protein (Fig. 5A). GTP could be replaced by the slowly hydrolyzable GTP-γ-S or GMP-PNP without affecting complex formation, demonstrating that hydrolysis of the high-energy γ-phosphate bond of GTP is not required for the interaction (Fig. 5A).
FIG. 5.
Requirements for p52/eEF1A binding to the TMV 3′-UTR. (A) Effect of various NTPs on UV cross-linking of wheat germ extract proteins (S-100). The nucleotides indicated were added to a final concentration of 0.2 mM in binding reactions containing 2 mM MgCl2. (B) Effects of various NTPs on UV cross-linking of purified wheat eEF1A. Final concentrations of 0.05 mM for the different NTPs in the presence of 2 mM MgCl2 were used. (C) MgCl2 concentration dependence of UV cross-linking of wheat germ proteins to the TMV 3′-UTR. MgCl2 was included in the standard binding reaction at the concentrations shown above each lane in reactions including 0.5 mM EDTA and 0.1 mM GTP. The major cross-linked proteins (p52/eEF1A and p30) are indicated by arrows.
The eEF1A/TMV 3′-UTR interaction specifically requires the presence of Mg2+ ions with an optimum concentration of 1 to 2 mM, whereas the absence of Mg2+ seems to favor binding of an unidentified protein of around 30 kDa rather than eEF1A (Fig. 5C); Mn2+, Zn2+, or Ca2+ did not support binding (not shown). Optimal pH and K+ concentrations were also determined. Complex formation was most efficient at the lowest KCl concentration tested (30 mM), with efficiency of binding reduced to 20% at 300 mM. Binding was observed over a wide pH range, with a rather broad optimum range of pH 7.2 to 7.8 (not shown). Based on these data, we performed all further experiments under standard conditions, in the presence of 0.1 mM GTP and 2 mM MgCl2 and at pH 7.6.
Structural determinants of the TMV U1 3′-UTR required for eEF1A binding.
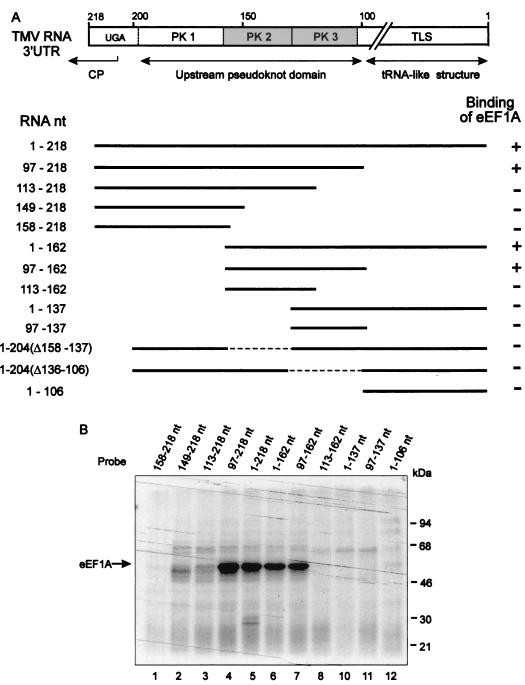
In the experiments described above, RNA consisting of TMV U1 nt 1 to 218 (counted from the 3′ end) with a stretch of 12 nonviral nucleotides masking the aminoacylation site at the 3′ end was used as a probe to detect specific protein binding. This region can be divided into distinct domains (21, 54, 61): a tRNA-like structure, formed by nt 1 to 105, and the UPD, nt 106 to 179, which consists of three consecutive PK structures. The UPD is preceded by a short AU-rich sequence (nt 180 to 190) and a stem-loop (Fig. 1).
To localize the site(s) essential for eEF1A binding, we tested the effect of deleting the predicted structural domains in the UV cross -linking assay (Fig. 6). This analysis showed that the TLS is not required for eEF1A binding, while the UPD is involved in the interaction. Furthermore, deletion of PK1 from the UPD only slightly decreased protein binding (Fig. 6B, lane 6). In contrast, deletion of PK2 (lane 10) as well as deletion or disruption of PK3 (lanes 2, 3, and 8) completely abolished binding. These results suggest that the region between nt 97 and 162 that contains both PK2 and PK3 is the minimal eEF1A-binding site.
FIG. 6.
Mapping of the eEF1A binding sites on the TMV 3′-UTR. (A) Linear map of TMV U1 3′-UTR. The diagram shows the positions of the TLS and PK structures of the UPD. The minimal binding sequence (nt 97 to 162) determined by these experiments is shown in gray. Deletion derivatives used for UV cross-linking assays are shown below. Results of UV cross-linking experiments, some of which are shown in panel B, are summarized on the right.
To determine the role of the PK structures in eEF1A binding, mutations in PK2 and PK3 were tested in the UV cross-linking assay. An RNA fragment consisting of the minimal binding site defined above (nt 97 to 162) with the addition of seven nonviral nucleotides (derived from the plasmid polylinker) upstream of the PKs was used as the reference construct (RNA-1) (Fig. 7A) in this and all subsequent experiments. Features of PK2 and PK3 are shown in detail in Fig. 7A. The lengths of stacked stems in PK2 and PK3 are conserved at 9 bp each. Moreover, the sizes of PK2 and PK3 are conserved at 22 and 30 bases, respectively. PK2 possesses two stacked stems (1 and 2) of 3 and 6 bp and connecting single-stranded loops (1 and 2) of 1 and 3 nt. In PK3, the two PK stems are 4 and 5 bp and the connecting loops are 5 and 6 nt long. The two PKs stacked coaxially generate a quasicontinuous helix of 18 bp. Conserved sequences are found in both the single-stranded and base-paired regions of tobamovirus UPDs (38). Absolutely conserved nucleotides (as determined in reference 38) are indicated in bold in Fig. 7A.
A combination of PK1 and PK3 (Fig. 7B, RNA-2) could not support protein binding, indicating that the specific structure and/or sequence of PK2 is required. The correct coaxial stacking of the PK structures is also critical; insertion of 1 or 2 nt between PK2 and PK3 totally abolished the interaction (Fig. 7B, RNA-3 and RNA-4). A simple stem-loop structure (RNA-5), consisting of an RNA with a base-paired stem of the same length and nucleotide composition as the coaxially stacked stems of the UPD, could not functionally replace the UPD in the interaction, suggesting that specific conformational features of the PK structures determine its ability to bind eEF1A. Destabilization of stem 1 or 2 of either PK by replacing the nucleotides on one strand with their complements (RNA-6 to RNA-9) abolished binding. Since these mutations would disrupt PK formation, this result does not distinguish between sequence and structure requirements of the stems. We therefore restored the stem structures, but not the original sequence, by introducing complementary nucleotides on the opposite strand (RNA-10 to RNA-13). Although PK structures apparently can form in these double mutants (38), we detected no eEF1A-binding. This result indicates that not only the structure but also the specific sequence is required for eEF1A recognition.
Deletion of the conserved A residue that creates a bulge in stem 2 of PK3 (RNA-14) had no effect on binding of eEF1A.
We next turned our attention to the role of the PK loop sequences in the eEF1A interaction. According to the model for the TMV UPD (61), loop 1 of PK2 consists of only one base, G155, which is conserved in all TMV strains. Replacement of this base by C significantly reduced UV cross-linking (Fig. 8, G155C). Moreover, deletion of G155 completely abolished binding, probably due to changes in structure (G155). Increasing the loop size to 2 nt also caused loss of binding of eEF1A (G155+C). To investigate the primary sequence requirement of the other PK loops for binding eEF1A, each was replaced with its complementary sequence (Fig. 7, RNA-15 to RNA-17). None of these RNAs could bind eEF1A. Point mutants in the loop sequences were then made in order to determine precisely which nucleotides play a role in eEF1A binding. Loop 2 of PK2 consists of three bases, AUA, of which the two A residues (A145 and A143) are conserved. Simultaneous mutation of both these residues to U dramatically inhibited binding (Fig. 8, A143,145U), but individual mutations revealed that only A143 is critical (A143U); a U residue was an acceptable replacement for A145 (A145U). Changing the nonconserved U144 to A (U144A) had no effect on binding. Point mutation analyses of loops 1 and 2 of PK3 revealed the importance of some of the conserved nucleotides in binding of eEF1A. In the 5-nt loop 1 of PK3, U130 and U131 are highly conserved and U132 is moderately so. However, U cannot be replaced by A in any of these positions without affecting binding (U132A, U131A, U130A). Mutations were introduced at each of the six positions of PK3 loop 2 (Fig. 8, U118C, etc.). Again, the three nucleotides involved in binding (U114, A115, and A116) are absolutely conserved in this position among tobamoviruses.
Three of the four base pairs in stem 1 of PK3 are conserved among tobamoviruses. As shown above, flipping of this stem abolished binding (Fig. 7, RNA-12). Flipping single base pairs individually had different effects on UV cross-linking of eEF1A. Mutations of the central two base pairs had very little effect on binding (Fig. 8, 135/120; 134/121). However, flipping the distal (136/119) or proximal (133/122) base pairs of the stem eliminated or substantially decreased binding of eEF1A, respectively, indicating that junctions both between the two stems of PK3 and between PK2 and PK3 are important for binding and might represent specific structural determinants directly involved in interaction. Only one of the five base pairs of PK3 stem 2 (U124-A110) is conserved. UV cross-linking of eEF1A was increased in RNA 110/124 when this base pair was replaced with a GC base pair, thus strengthening the stem. Introducing single base mismatches in the middle, or at the end, of stem 2 of PK3 eliminated binding (109/125; 107/127). Thus, stability of PK3 stem 2 is important for binding of eEF1A. In contrast, flipping of the single conserved G157-C147 base pair in the 3-bp stem 1 of PK2 that maintained PK structure had no effect on UV cross-linking of eEF1A. However, cross-linking was reduced by 80% when the U156-A148 base pair of this stem was flipped.
In summary, by studying the effect on UV cross-linking of deletions and site-directed mutations introduced in the TMV U1 3′-UTR, it was shown that PK2 and PK3 of the UPD located immediately upstream of the TLS are involved in interaction with eEF1A. The structures of PK2 and PK3, particularly the conserved primary sequences of PK3 loops 1 and 2 and stem 1 probably serve as the recognition site for eEF1A-binding.
DISCUSSION
Host-derived components are known to be involved in replication of RNA viruses (reviewed in references 11 and 58). This was first shown for the positive-strand bacteriophage Qβ. In addition to phage-encoded proteins, the Qβ replicase complex contains three cellular proteins—ribosomal protein S1 and elongation factors EFTu and EFTs—involved in recognition and initiation of RNA replication of Qβ RNA (5, 9). Subsequently, a number of host cell proteins that play diverse roles in the replication of eukaryotic RNA viruses have been identified. Interestingly, the primary function of the majority of these host proteins is in translation and/or RNA processing, highlighting the intimate coupling of viral translation and RNA replication (reviewed in reference 36). Similar to its prokaryotic counterpart EF-Tu, eEF1A plays an important role in viral RNA replication, interacting with viral RNA and/or proteins. In addition to the well-characterized interaction of plant eEF1A with the 3′-terminal aminoacylated TLS (aa-TLS) of several plant viruses (20, 41), eEF1A was found to specifically interact with the cis-acting replication signal within the nonpolyadenylated 3′-UTR of West Nile virus (4). Strong evidence has also been presented for the binding of eEF1A to RNA polymerase L of vesicular stomatitis virus and the functional involvement of this binding in viral replication (14). Our finding that eEF1A interacts with the TMV 3′-UTR UPD, which is functionally involved in viral RNA replication, additionally suggests that utilization of host translation factors, particularly eEF1A, by RNA viruses may be widespread.
The interaction of eEF1A with the TMV 3′-UTR.
Evidence that translation and replication of positive-sense RNA viruses are coupled is accumulating (1, 36, 65). To coordinate both processes, these viruses probably use common RNA structures that are functionally involved in replication as well as in translation processes (5, 6). These various functions depend on specific interactions between the viral RNA genome and viral and host proteins. In this study, we have shown that the host protein eEF1A specifically interacts with two pseudoknots, PK2 and PK3, within the UPD of the 3′-UTR of TMV U1 RNA.
The important features of the major binding sites for eEF1A in PK2 and PK3 of the UPD are shown in Fig. 9. These PKs are conserved among tobamoviruses in structure and in several positions at the primary sequence level (38, 61). The major primary sequence determinants for binding are located in stem 1 and both loops of PK3. The strong requirement for the conserved nucleotides of loops 1 and 2 of PK3 for eEF1A binding suggests that they might directly contact the protein. Specific primary sequences in the PK stems seem also to be involved. The specific sequence requirements in the loops of PK2 and PK3 might suggest special tertiary interactions in the PKs that are important for protein binding. That PK3 from the U1 UPD might have unusual structural features was already known from structural mapping experiments in the absence of Mg2+; the conformation of the loop regions including nucleotides AAU (115 to 117) and AUA (143 to 145) changed significantly, although those nucleotides are within single-stranded regions in both PK and stem-loop conformations of the TMV UPD (61).
FIG. 9.
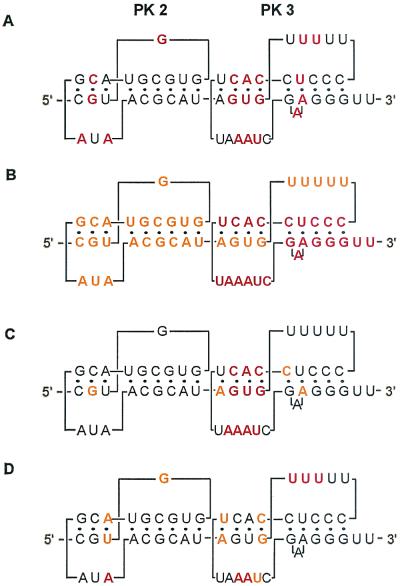
Summary of the important primary sequences in the UPD of TMV U1 3′-UTR. (A) Sequence conserved within tobamovirus family (38). Absolutely conserved sequences are in red. (B) Sequences strongly required for replication of TMV RNA in tobacco plants and protoplasts (12, 49, 59) are in red, and those still significantly affecting replication are in orange. (C) Sequence affecting translation (38). Critical and moderately important sequences are shown in red and orange, respectively. (D) Sequence important for eEF1A binding (this study). Critical and moderately important sequences are shown in red and orange, respectively.
Here, we have demonstrated that eEF1A can interact with the UPD of the TMV 3′-UTR in a nonaminoacylated state. eEF1A/GTP is already known to interact with the aa-TLS located just downstream of the UPD in the 3′-UTR of TYMV RNA (20, 32) and the 3′-aminoacylated TMV RNA (40). A fragment of TYMV TLS as short as 47 nt, possessing only the T-stem-loop and acceptor stem, was sufficient for interaction with eEF1A/GTP (32). However, in the nonaminoacylated state, viral TLSs or tRNAs bind to eEF1A (and its prokaryotic counterpart EF-Tu) with much lower affinity, suggesting that the aminoacyl group is a fundamental requirement for ternary complex formation (20, 43). Functionally equivalent to the aminoacylated tRNA (aa-tRNA) acceptor/T arms, the TLS of TMV consists of three coaxially stacked helical segments with a total length of 11 bp (Fig. 1), of which the last two helices are formed by PK structures (21, 54). Although the UPD domain involved in binding eEF1A/GTP is also composed of PKs, they are structurally different from those of the TLS. Our experiments clearly demonstrate that this eEF1A interaction requires specific structural features of PK2 and PK3 of the UPD that do not at all resemble either tRNA or TMV TLS. Furthermore, in competition experiments, aa-tRNA does not compete with the UPD/eEF1A complex (data not shown). The PK2-PK3 region and the aa-TLS region might interact with eEF1A independently or simultaneously; it is also possible that the eEF1A molecule complexed with the UPD might interact with the downstream aa-TLS and mediate interaction with both structural domains. This observation is consistent with evidence showing that eEF1A has at least two RNA binding sites and is able to bind aa-tRNA and high-molecular-weight rRNAs simultaneously (56). These results raise intriguing questions regarding the origin of eEF1A binding sites in the UPD. One possibility may be that the PKs of the UPD mimic another cellular ligand of eEF1A—rRNA—in their interaction with eEF1A. In line with this, rRNA but not tRNA was found to specifically inhibit the functional interaction of the viral replicase and RNAs of cucumber mosaic virus that, like TMV, has a PK-rich 3′-UTR with a TLS (51).
eEF1A requires GTP to form a stable complex with aa-tRNA. This GTP dependence is due to structural changes occurring upon GTP binding to eEF1A (43). Probably due to functional mimicry, the interaction of eEF1A with the aa-TLS of plant viruses, including TMV, requires GTP and is inhibited by GDP. In the present study, we found that the GTP-bound form of eEF1A is also specifically required for interaction with the UPD.
In addition to GTP, Mg2+ is also strictly required for the interaction of eEF1A with the TMV UPD. Mg2+ ions are an essential cofactor in the binding of GTP to eEF1A (7) and are also involved in the formation of PK structures (61). Interestingly, removal of Mg2+ in our UV cross-linking experiments stimulated binding of a wheat germ protein to the TMV 3′-UTR (p30) (Fig. 5C). Although the identity and specificity of p30 remain to be determined, it is tempting to speculate that the alternative hairpin-loop structure of the TMV 3′-UTR might be a prerequisite for its binding.
Possible functions of the eEF1A/TMV RNA 3′-UTR complex in translation.
eEF1A is an abundant and highly conserved protein in eukaryotic cells and is one of the most extensively characterized proteins of the translational machinery (13, 33, 46). The canonical role of eEF1A is to bring aa-tRNA to the A site of the ribosome during translation in a GTP-dependent mechanism (43). To fulfill its functional role in protein synthesis, eEF1A specifically interacts with a number of macromolecules, e.g., aa-tRNA, guanine nucleotides, components of the ribosome, and several other proteins of the translational apparatus.
The eEF1A binding site in the TMV 3′-UTR UPD colocalizes with the region known to be largely responsible for facilitating translation (22, 38). Moreover, results of other studies suggest that the phylogenetically conserved higher-order structure of the two coterminal PKs of the UPD, and particularly the conserved primary sequence within PK3, is essential for the translational regulation associated with the TMV RNA 3′-UTR (38). The latter study showed that stems 1 and 2 of PK3 are particularly important for high level expression of TMV U1 RNA in plant protoplasts. In our hands, mutants in this region (including some identical to those tested by Leathers et al. [38]) also interfered with eEF1A binding (Fig. 9), suggesting a role for eEF1A in the regulation of translation of TMV mRNAs.
How might eEF1A contribute to the translational regulation mediated by the TMV RNA 3′-UTR? The 3′-UTR/eEF1A complex may be involved in stimulation of translational elongation or some other cotranslational event. Specific association of eEF1A with the 3′-UTR might significantly increase the local concentration of eEF1A on virus mRNAs and promote translation elongation by preventing diffusion of factors from actively translated virus mRNA, thus giving viral translation a selective advantage. Plant cell heat shock protein HSP101 is involved in TMV RNA 5′-3′ communication via interaction with both the Ω leader and PKs 3 and 4 within the TMV 3′-UTR (38, 60). Since the sites of interaction for eEF1A and HSP101 on the TMV 3′-UTR partially overlap and both require the conserved PK3, the two proteins could either bind simultaneously or compete for binding and participate in different functions of the virus RNA. Further experiments will be required to distinguish between these possibilities.
An alternative possibility is that the eEF1A-UPD interaction negatively affects translation, possibly by assisting interaction of viral replicase proteins with the viral 3′-UTR, thereby participating in inhibition of TMV gRNA translation prior to replication.
There is also the possibility that the eEF1A/TMV 3′-UTR complex may be involved in the attachment of TMV RNAs to the cytoskeleton and endoplasmic reticulum (ER), thereby regulating efficiency of protein synthesis. In general, protein synthesis can be enhanced by a close spatial association of the ribosome and other factors involved in translation with the cytoskeleton (2, 31, 57). eEF1A interacts with actin filaments, tubulin, and the ER and apparently mediates the general attachment of cellular mRNA to the cytoskeleton (2, 3, 13, 31, 45). TMV genomic RNA is associated with the plant cytoskeleton and ER throughout the infection cycle (42). Moreover, TMV RNA is transported intracellularly, probably via microtubules and/or the ER (28), and eEF1A has been implicated as a cofactor of the viral transport process (17).
Possible functions of the eEF1A/TMV RNA 3′-UTR complex in replication.
TMV replicates via production of negative-sense RNA intermediates; the 3′-terminal portion of the genomic RNA contains a unique promoter that is specifically recognized by the viral replicase complex (49, 64). An essential part of this promoter is located within the UPD of the TMV 3′-UTR (Fig. 1). Deletion of this domain reduces both TMV amplification in protoplasts and minus-strand RNA synthesis in an in vitro assay to undetectable levels, suggesting an important role in viral replication (12, 49, 59). The PKs of the UPD have been suggested to play a role in the assembly of the TMV replicase complex (12). Similar roles for the 3′-UTRs of alfalfa mosaic virus and BMV RNAs have also been proposed (52, 63). Specific binding of eEF1A to elements of the TMV 3′-UTR UPD, required for both initiation of viral negative-strand RNA synthesis and enhancing viral replication in cis, suggests a role for eEF1A-binding in both template recognition for viral replication and the assembly of the TMV replication complex. However, mutations in the UPD or TLS, which in our hands eliminate eEF1A binding, are still able to support TMV RNA synthesis (12, 49, 59). Thus, eEF1A binding to the TMV 3′-UTR might simply enhance replication efficiency, as observed for aminoacylation of TYMV (25). Nevertheless, host proteins of 50, 54, and 56 kDa consistently copurify with the viral replicase complex (48); p56 is immunologically related to GCD10, the RNA-binding subunit of yeast eIF3. Whether the 50- or 54-kDa proteins are equivalent to eEF1A remains to be seen. There is increasing evidence of participation of eEF1A in replication of animal viruses (4, 14 27).
Possible requirement for eEF1A in other steps of the TMV infection cycle.
It is likely that eEF1A bound to the 3′-UTR of TMV RNA is also a component of the virus mRNP involved in localization of the TMV replication bodies. This view is further supported by the recent finding that levels of eEF1A, TMV replicase, and transport protein are specifically enriched in the virus bodies produced by TMV in tobacco leaves or protoplasts (17). The virus body was also shown to be enriched in ribosomes, indicating an enhanced level of translation. Such specific accumulation of eEF1A in the virus bodies may reflect a high requirement for this host factor during TMV infection (17).
In addition to its specific binding to aa-tRNA, eEF1A has a weak and noncooperative general affinity for RNA, which is independent of GTP and occurs via RNA-binding sites distinct from those for aa-tRNA (56). These RNA-binding sites may play a role in the attachment of eEF1A to the ribosome or to mRNA. Replication of TMV RNA takes place in close association with components of the cellular cytoskeleton and the ER (28). Recent experiments suggest that the virus MP participates in anchoring of the viral genomic RNA to the ER and microtubules during middle and late stages of TMV infection (8, 42). Given that eEF1A has been implicated in viral transport processes (17), other potential roles of the eEF1A interaction to TMV RNA 3′-UTR, such as anchoring the TMV RNA replication complex to a specific membrane or cytoskeleton in a host cell or participation in cell-to-cell spread of virus RNA, should also be examined in the future.
It remains an open question whether aminoacylation of TMV RNA and the consequent interaction with eEF1A is important for virus infectivity. Although efficient infectivity of TYMV depends on aminoacylation, aminoacylation-defective mutants of BMV RNA-3 give rise to infectious virus (41). Moreover, aminoacylation is not always observed in viral RNAs which possess a TLS (25, 41). In view of our results, this might be explained by a general requirement for eEF1A binding to the viral 3′-RNA, i.e., not necessarily specifically to aa-TLS, for virus viability.
Acknowledgments
We thank Manfred Heinlein and Johannes Fütterer for critical comments on the manuscript. We are very grateful to Mike Rothnie for his help in preparing some of the figures. We also thank Oleg Kurnasov (IPR, Pushchino, Russia) for construction of several plasmids, Diana Dominguez for helpful advice concerning UV cross-linking assays, and Yuri Dorochov and J. G. Atabekov (MSU, Moscow, Russia) for providing clones TMV U1 and cr-TMV 3′-UTR. We appreciate Herbert Angliker and Peter Müller for DNA sequencing and synthesis of the DNA oligonucleotides, respectively, Etienne Herzog and Orlene Guerra-Peraza for valuable advice, and members of T. Hohn's laboratory for continuous encouragement.
This research was supported in part by grants from INTAS (96-00857 and 99-00720).
REFERENCES
- 1.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassell, G. J., C. M. Powers, K. L. Taneja, and R. H. Singer. 1994. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol. 126:863-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassell, G. J., Y. Oleynikov, and R. H. Singer. 1999. The travels of mRNAs through all cells large and small. FASEB J. 13:3450-3454. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 71:6433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal, T., and G. G. Carmichael. 1979. RNA replication function and structure of Q-beta replicase. Methods Enzymol. 60:628-638.379538 [Google Scholar]
- 6.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348:125-132. [DOI] [PubMed] [Google Scholar]
- 8.Boyko, V., L. J. van Der, J. Ferralli, E. Suslova, M. O. Kwon, and M. Heinlein. 2000. Cellular targets of functional and dysfunctional mutants of tobacco mosaic virus movement protein fused to green fluorescent protein. J. Virol. 74:11339-11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, D., and L. Gold. 1996. RNA replication by Q beta replicase: a working model. Proc. Natl. Acad. Sci. USA 93:11558-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browning, K. S., J. Humphreys, W. Hobbs, G. B. Smith, and J. M. Ravel. 1990. Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J. Biol. Chem. 265:17967-17973. [PubMed] [Google Scholar]
- 11.Buck, K. W. 1999. Replication of tobacco mosaic virus RNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:613-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandrika, R., S. Rabindran, D. J. Lewandowski, K. L. Manjunath, and W. O. Dawson. 2000. Full-length tobacco mosaic virus RNAs and defective RNAs have different 3′ replication signals. Virology 273:198-209. [DOI] [PubMed] [Google Scholar]
- 13.Condeelis, J. 1995. Elongation factor 1-alpha, translation and the cytoskeleton. Trends Biochem. Sci. 20:169-170. [DOI] [PubMed] [Google Scholar]
- 14.Das, T., M. Mathur, A. K. Gupta, G. M. Janssen, and A. K. Banerjee. 1998. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 αβγ for its activity. Proc. Natl. Acad. Sci. USA 95:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deiman, B. A., A. K. Koenen, P. W. Verlaan, and C. W. Pleij. 1998. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J. Virol. 72:3965-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding, X. S., Y. Bao, S. A. Carter, and R. S. Nelson. 1998. Host factor involved in virus replication and cell-to-cell movement. Annu. Rep. Samuel Roberts Noble Found. 1998:94-96. [Google Scholar]
- 18.Dorokhov, Y., P. A. Ivanov, V. K. Novikov, A. A. Agranovsky, S. Y. Morozov, V. A. Efimov, R. Casper, and J. G. Atabekov. 1994. Complete nucleotide sequence and genome organization of a tobamovirus infecting cruciferae plants. FEBS Lett. 350:5-8. [DOI] [PubMed] [Google Scholar]
- 19.Dreher, T. W., C. H. Tsai, and J. M. Skuzeski. 1996. Aminoacylation identity switch of turnip yellow mosaic virus RNA from valine to methionine results in an infectious virus. Proc. Natl. Acad. Sci. USA 93:12212-12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreher, T. W., O. C. Uhlenbeck, and K. S. Browning. 1999. Quantitative assessment of EF-1α·GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 274:666-672. [DOI] [PubMed] [Google Scholar]
- 21.Felden, B., C. Florentz, R. Giege, and E. Westhof. 1996. A central pseudoknotted three-way junction imposes tRNA-like mimicry and the orientation of three 5′ upstream pseudoknots in the 3′ terminus of tobacco mosaic virus RNA. RNA 2:201-212. [PMC free article] [PubMed] [Google Scholar]
- 22.Gallie, D. R., and V. Walbot. 1990. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev. 4:1149-1157. [DOI] [PubMed] [Google Scholar]
- 23.Gallie, D. R., J. N. Feder, R. T. Schimke, and V. Walbot. 1991. Functional analysis of the tobacco mosaic virus tRNA-like structure in cytoplasmic gene regulation. Nucleic Acids Res. 19:5031-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallie, D. R. 1998. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Goodwin, J. B., J. M. Skuzeski, and T. W. Dreher. 1997. Characterization of chimeric turnip yellow mosaic virus genomes that are infectious in the absence of aminoacylation. Virology 230:113-124. [DOI] [PubMed] [Google Scholar]
- 26.Gultyaev, A. P., E. van Batenburg, and C. W. Pleij. 1994. Similarities between the secondary structure of satellite tobacco mosaic virus and tobamovirus RNAs. J. Gen. Virol. 75:2851-2856. [DOI] [PubMed] [Google Scholar]
- 27.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 28.Heinlein, M., H. S. Padgett, J. S. Gens, B. G. Pickard, S. J. Casper, B. L. Epel, and R. N. Beachy. 1998. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10:1107-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, T., S. M. Tahara, and M. C. Lai-Michel. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito, T., and M. M. Lai. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254:288-296. [DOI] [PubMed] [Google Scholar]
- 31.Jansen, R. P. 1999. RNA-cytoskeletal associations. FASEB J. 13:455-466. [PubMed] [Google Scholar]
- 32.Joshi, R. L., J. M. Ravel, and A. L. Haenni. 1986. Interaction of turnip yellow mosaic virus Val-RNA with eukaryotic elongation factor EF-1a. Search for a function. EMBO J. 5:1143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinzy, T. G., and E. Goldman. 2000. Nontranslational functions of components of the translational apparatus, p. 973-997. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Krieg, J., S. Hartmann, A. Vicentini, W. Glasner, D. Hess, and J. Hofsteenge. 1998. Recognition signal for C-mannosylation of Trp-7 in RNase 2 consists of sequence Trp-x-x-Trp. Mol. Biol. Cell 9:301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lai, M. M. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 37.Lax, S. R., S. J. Lauer, K. S. Browning, and J. M. Ravel. 1996. Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol. 118:109-128. [DOI] [PubMed] [Google Scholar]
- 38.Leathers, V., R. Tanguay, M. Kobayashi, and D. R. Gallie. 1993. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol. Cell. Biol. 13:5331-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewandowski, D. J., and W. O. Dawson. 1998. Deletion of internal sequences results in tobacco mosaic virus defective RNAs that accumulate to high levels without interfering with replication of the helper virus. Virology 251:427-437. [DOI] [PubMed] [Google Scholar]
- 40.Litvak, S., A. Tarrago, L. L. Tarrago, and J. E. Allende. 1973. Elongation factor viral genome interaction dependent on the aminoacylation of turnip yellow mosaic virus RNA and tobacco mosaic virus RNA. Nat. New Biol. 241:88-90. [DOI] [PubMed] [Google Scholar]
- 41.Mans, R.-M. W., C.-W. A. Pleij, and L. Bosch. 1991. tRNA-like structures. Function and evolutionary significance. Eur. J. Biochem. 201:303-324. [DOI] [PubMed] [Google Scholar]
- 42.Mas, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merrick, W. C., and J. Nyborg. 2000. The protein biosynthesis elongation cycle, p. 89-125. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Metz, A. M., R. T. Timmer, and K. S. Browning. 1992. Isolation and sequence of a cDNA encoding the cap binding protein of wheat eukaryotic protein synthesis initiation factor 4F. Nucleic Acids Res. 20:4096.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore, R. C., and R. J. Cyr. 2000. Association between elongation factor-1α and microtubules in vivo is domain dependent and conditional. Cell Motil. Cytoskeleton 45:279-292. [DOI] [PubMed] [Google Scholar]
- 46.Negrutskii, B. S., and A. V. El'skaya. 1998. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid Res. Mol. Biol. 60:47-78. [DOI] [PubMed] [Google Scholar]
- 47.O'Farrell, P. Z., H. M. Goodman, and P. H. O'Farrell. 1977. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 12:1133-1141. [DOI] [PubMed] [Google Scholar]
- 48.Osman, T. A., and K. W. Buck. 1997. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J. Virol. 71:6075-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osman, T. A., C. L. Hemenway, and K. W. Buck. 2000. Role of the 3′ tRNA-like structure in tobacco mosaic virus minus-strand RNA synthesis by the viral RNA-dependent RNA polymerase in vitro. J. Virol. 74:11671-11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pleij, C. W., and L. Bosch. 1989. RNA pseudoknots: structure, detection, and prediction. Methods Enzymol. 180:289-303. [DOI] [PubMed] [Google Scholar]
- 51.Quadt, R., and E. M. Jaspars. 1991. Characterization of cucumber mosaic virus RNA-dependent RNA polymerase. FEBS Lett. 279:273-276. [DOI] [PubMed] [Google Scholar]
- 52.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rietveld, K., R. Van Poelgeest, C. W. Pleij, J. H. van Boom, and L. Bosch. 1982. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 10:1929-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rietveld, K., W. A. Pleij, and L. Bosch. 1983. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 2:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slobin, L. I. 1980. The role of eucaryotic factor Tu in protein synthesis. The measurement of the elongation factor Tu content of rabbit reticulocytes and other mammalian cells by a sensitive radioimmunoassay. Eur. J. Biochem. 110:555-564. [DOI] [PubMed] [Google Scholar]
- 56.Slobin, L. I. 1983. Binding of eucaryotic elongation factor Tu to nucleic acids. J. Biol. Chem. 258:4895-4900. [PubMed] [Google Scholar]
- 57.Stapulionis, R., and M. P. Deutscher. 1995. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. USA 92:7158-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strauss, J. H., and E. G. Strauss. 1999. Viral RNA replication: with a little help from the host. Science 283:802-804. [DOI] [PubMed] [Google Scholar]
- 59.Takamatsu, N., Y. Watanabe, T. Meshi, and Y. Okada. 1990. Mutational analysis of the pseudoknot region in the 3′ noncoding region of tobacco mosaic virus RNA. J. Virol. 64:3686-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanguay, R. L., and D. R. Gallie. 1996. Isolation and characterization of the 102-kilodalton RNA-binding protein that binds to the 5′ and 3′ translational enhancers of tobacco mosaic virus RNA. J. Biol. Chem. 271:14316-14322. [DOI] [PubMed] [Google Scholar]
- 61.van Belkum, A., J. P. Abrahams, C. W. Pleij, and L. Bosch. 1985. Five pseudoknots are present at the 204 nucleotides long 3′ noncoding region of tobacco mosaic virus RNA. Nucleic Acids Res. 13:7673-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vende, P., M. Piron, N. Castagne, and D. Poncet. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 74:7064-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vlot, A. C., L. Neeleman, H. J. Linthorst, and J. F. Bol. 2001. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 75:6440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watanabe, T., A. Honda, A. Iwata, S. Ueda, T. Hibi, and A. Ishihama. 1999. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J. Virol. 73:2633-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiland, J. J., and T. W. Dreher. 1993. Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc. Natl. Acad. Sci. USA 90:6095-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeyenko, V. V., L. A. Ryabova, D. R. Gallie, and A. S. Spirin. 1994. Enhancing effect of the 3′-untranslated region of tobacco mosaic virus RNA on protein synthesis in vitro. FEBS Lett. 354:271-273. [DOI] [PubMed] [Google Scholar]