Abstract
Since porcine endogenous retroviruses (PERVs) can infect cultured human cells, they are a potential hazard to xenotransplantation. For this reason, endogenous retroviruses from the Westran (Westmead Hospital transplantation) inbred line of pigs were analyzed by using consensus primers for the type A and type B viruses to amplify 1.8-kb envelope gene fragments. After preliminary analysis with restriction enzymes KpnI and MboI, 31 clones were sequenced. Between types A and B, five recombinant clones were identified. Fifty-five percent of clones (17 of 31) had premature stop codons within the envelope protein-encoding region. Endogenous retroviruses in Westran pigs were physically mapped by fluorescence in situ hybridization (FISH) using PERV-A and PERV-B envelope clones as probes to identify at least 32 integration sites (19 PERV-A sites and 13 PERV-B sites). The chromosomal sites of integration in the Westran strain are quite different from those in the European Large White pig. The recombinant clones suggest that defective PERVs could become infective through recombination and further that PERVs might recombine with human endogenous retroviruses in xenotransplants.
Porcine endogenous retroviruses (PERVs) are proviruses, inherited in a stable Mendelian fashion (31), although they can also be acquired through infection. Type C virus particles released in two cell lines from pig kidney were first described by Breese (6), with particles being morphologically similar to those of the mouse type C leukemia viruses. Soon after, C-type viral particles in five different pig leukemia cell lines were reported (2). Todaro et al. (43) showed that porcine retroviruses were present in multiple copies in the porcine genome in DNA from different tissues and cells. Lieber et al. (23) described the biological and immunological properties of porcine type C viruses, finding cell lines from most mammalian species resistant to infection by these viruses, which are otherwise very similar to other mammalian C-type viruses by morphological, biochemical, and immunological criteria. Suzuka et al. (39) reported the isolation of a swine C-type retrovirus from a porcine malignant lymphoma, which was subsequently partially characterized by restriction digestion of the 8.8-kb viral clone (40). This Tsukuba-1 clone has subsequently been sequenced and found 99% identical with PERV from miniature swine lymphocytes (1).
PERVs have taken on a new significance with the advent of xenotransplantation, as they are thought to have the potential to cross the species barrier. Due to the acute shortage of human organs and tissues for transplantation, the use of nonhuman species for xenotransplantation is considered a possible solution. Success in modulating immunological rejection by transgenic modifications to animals has raised the possibility of clinical introduction of xenotransplantation (11, 16, 37). Pigs are regarded as a better source for xenotransplantation into humans than primates for safety, financial, ethical, and practical reasons. Clinical trials with pig xenografts have been carried out; the trials included perfusion with pig livers or porcine hepatocytes as a bridging strategy for hepatic failure, the use of pancreatic islet cells as a treatment for chronic diabetes, and the implantation of fetal neuronal tissue as a therapy for Parkinson's disease (8, 12, 17).
Recently porcine retroviruses have become a focus of concern, as they infect human cells in vitro (27, 34, 50), although there is no evidence that this occurs in vivo in baboons (26) or in humans (30, 32, 35, 41). Akiyoshi et al. (1) suggested that the risk of viral infection would be increased in xenotransplantation by the presence of factors commonly associated with viral infection, namely, immune suppression, graft versus host disease, graft rejection, viral coinfection, and cytotoxic therapies. Very recently, the transplantation of porcine pancreatic islets into SCID (severe combined immunodeficiency) mice led to in vivo expression of PERVs, reinforcing fear about the risk of PERV infection in immunosuppressed human patients (46).
The viral envelope is the major determinant of host range and is essential for infection. Two main types of pig retrovirus, PERV-A and PERV-B, which differ by 507 bases in their envelope (env) genes, are widely distributed in different pig breeds, as assessed by Southern hybridization (22). PERVs are present at approximately 50 copies in different breeds of pig (1, 22).
Host range analysis initially showed that PERVs are restricted in their species tropism, infecting only porcine cells and not cell lines derived from a range of species including chimpanzees, rhesus monkeys, horses, minks, bats, rabbits, cows, cats, dogs, and mice (43). The receptor specificities of PERV-A, PERV-B, and PERV-C were investigated with LacZ pseudotype vectors (42). The results showed no cross-interference, implying that the Env proteins recognize different cell surface receptors. Also, host range analyses by the vector transduction assay showed that PERV-A and PERV-B viruses have wider host ranges, including several human cell lines, than PERV-C viruses, which infected only two pig cell lines and one human cell line (42). Recently, the in vitro host range of PERV was studied in human primary cells and cell lines, as well as in cell lines from nonhuman primates and other species. The analysis revealed that there were three distinct patterns of susceptibility to infection among these host cells. First, some cells are resistant to infection in the assay. Second, other cells are infected by virus but the cells are not permissive to productive replication and spread. Third, the final category of cells is permissive for productive infection and spread (51). Very recently, four novel groups of gamma retroviruses (γ2 to γ5) and four novel groups of beta retroviruses (β1 to β4) (33) in addition to the previously recognized gamma 1 retroviruses (PERV-A, -B, and -C) were detected in pigs. Also a full-length novel PERV-E, a gamma retrovirus, has been identified by screening a porcine genomic library with distinct reverse transcriptase sequences as hybridization probes (25).
In 1994, the transplantation research group at Westmead Hospital in Sydney, Australia, initiated an inbreeding program in a stock of feral pigs as resources for transplantation research and in the long term as potential donors for xenotransplantation. The line is called Westran (Westmead transplantation), and pigs from it are the subject of the research reported here. Specifically, we describe a study of sequencing and mapping PERV env in the Westran pig line as a preliminary component of the assessment of these viruses as hazards for xenotransplantation.
MATERIALS AND METHODS
Animal resources.
The Westran line is believed to be descended from a pair of pigs released on Kangaroo Island, off the coast of South Australia, in 1803 by a French navigator and explorer, Nicholas Baudin (10). Captured feral pigs from Kangaroo Island were transferred to Adelaide University for biomedical research in 1976 (28). After being maintained as a very small colony for about 15 years, a pair of full sibs was transferred to Westmead Hospital in Sydney for transplantation research. Since then, the core breeding line has been maintained by deliberate full-sib mating up to the current eighth generation. Assessment of genetic and immunological composition has been performed by a Westmead/University of Sydney research team using ABO blood grouping and mixed lymphoctye reaction and lymphocytotoxity assay nonreactivity and monitoring hyperpolymorphic microsatellite markers. The highly inbred status of the line has been confirmed by finding very high levels of microsatellite homozygosity (94% [47 of 50] of microsatellites are monomorphic at the eighth generation of full-sib mating) (W. J. Hawthorne, J. S. Burgess, Y. Chen, S. Walters, T. Patel, J. Clarke, L. Weston, P. J. O'Connell, C. Moran, J. R. Chapman, and R. D. M. Allen, Abstr. 7th World Congr. Int. Pancreas Islet Transplant., p. 143). Crossover skin grafts also have been performed in Westran pigs. Littermate same-sex skin grafts are accepted long term without evidence of rejection. The sequences of endogenous retroviruses from a generation 5 inbred boar (no. 115) have been analyzed. PERV locations were mapped on chromosome spreads from this boar and its son from a sib mating no. 167.
Primers and PCR conditions.
Consensus primer pairs PERV-F (5′-CATGCATCCCACGTTAAGC-3′) and PERV-R (5′-ACCATCCTTCAAACCACCC-3′) (22), chosen from the highly conserved regions at either end of the PERV-A (GenBank accession no. Y12238) and -B (GenBank accession no. Y12239) envelope genes, were used to search for novel variants in the less conserved region and to amplify as many envelope fragments as possible from PERV proviruses in the pig genome. DNA polymerases Taq and Pfu were employed in this study. PCR with Taq DNA polymerase was carried out in a 50-μl volume with approximately 100 ng of template genomic DNA, 0.2 mM (each) primer, 600 μM (each) deoxynucleoside triphosphate (dNTP), 1 or 2 mM MgCl2, 5 μl of 10× PCR buffer (Invitrogen), and 2.5 U of Taq polymerase. This mixture was overlaid with 1 drop of mineral oil. Amplification was performed in a PTC-100 programmable thermal controller (MJ Research Inc.) for 45 cycles with denaturation for 1 min at 94°C, annealing for 1 min at 65°C (PERV-A and -B), and extension for 1 min at 72°C. The final extension was for 20 min at 72°C. The Pfu proofreading DNA polymerase was chosen for the second set of PERV PCRs because its error rate is known to be lower than that of Taq DNA polymerase. PCR was carried out in a 50-μl volume with approximately 100 ng of template genomic DNA, 0.2 mM (each) primer, 0.5 mM (each) dNTP, 10× PCR buffer (Stratagene), and 2.5 U of cloned Pfu polymerase (Stratagene). This mixture was overlaid with 1 drop of mineral oil. Amplification was performed in an MJ thermocycler with initial denaturation for 3 min at 95°C, followed by 35 cycles of denaturation for 1 min at 95°C, annealing for 1 min at 65°C, and extension for 5 min at 72°C. The final extension was for 10 min at 72°C.
Cloning of PERV PCR products and preliminary identification of clone types by restriction digestion.
The env PCR products were gel purified (BRESAclean DNA purification kit; Bresatec) and ligated into the pCR-Blunt plasmid vector (Invitrogen) for Pfu polymerase-generated PCR products and a pCR2.1-TOPO plasmid vector (Invitrogen) for Taq polymerase-generated PCR products and transformed into Escherichia coli TOP10 bacteria (Invitrogen) as suggested by the manufacturer.
Restriction enzymes KpnI and MboI (Promega) were used for preliminary screening of the clones for characteristic features of PERV-A and -B (22).
Sequencing of PERV clones.
A SequiTherm EXCEL long-read DNA sequencing kit (Epicentre) and Li-Cor sequencer (model 4200) were used to read approximately 1.8 kb of full-length insert sequences by using a pair of M13 vector primers (M13F, 5′-TTTCCCAGTCACGACGTTG-3′, and M13R, 5′-GGATAACAATTTCACACAGG-3′) labeled with different infrared-sensitive dyes suitable for the Li-Cor sequencing system. Sequences were analyzed with Base ImageIR software, version 4.1 (Li-Cor Inc.).
SeqEd software, version 1.0.3 (Perkin-Elmer, Applied Biosystems), was used to amalgamate and orient the sequences with respect to published PERV sequences. Full sequences of the PERV env PCR products were assembled by overlapping forward and reverse sequencing products. The overlapping of sequences at each end of the long reads may compensate for the less accurate reads at the ends.
Alignment of the PERV env sequences was performed with the Clustalw and Pileup programs of the multiple sequence alignment option in GCG by using the World Wide Web ANGIS interface (http://www.angis.org.au/). The putative amino acid sequences were determined with the Translate program in GCG.
Chromosomal localization of PERVs.
Westran porcine chromosomes were derived from blood lymphocytes cultured for 72 h in AminoMax (Life Technologies). To synchronize cells in mid-S phase, 300 μg of thymidine (Sigma)/ml was added 24 h before finishing the cell culture. At day 3, the cells were rinsed three times with Dulbecco's phosphate-buffered saline (Commonwealth Serum Labs). They were suspended once again in the culture medium containing 20 μg of 5-bromodeoxyuridine (Sigma)/ml and 0.5 μg of 5-fluorodeoxyuridine (Sigma)/ml and further incubated for 6[1/2] h, followed by a 1-h spindle fiber disruption with colchicine (Sigma). Harvest and slide preparation by air drying were by standard techniques.
PERV clones Taq-82 and Taq-9 were used as probes for PERV-A and PERV-B, respectively. To improve the efficiency and specificity of hybridization, the PERV inserts were excised from the vector by EcoRI restriction digestion. To label PERV probes with biotin, a BioNick labeling system (Life Technologies) was employed for nick translation. Parallel incorporation of a trace of [3H]dATP (Amersham) indicated that 50 and 47 pmol of biotin-14-dATP were incorporated into 1 μg of PERV-A and PERV-B probes, respectively. Immunochemical detection and staining by the PPD11 method (21) were as described previously (48). Cells showing positive signals on porcine chromosomes under blue-light excitation were photographed on color-positive film for analysis.
The fluorescence in situ hybridization (FISH) signal, appearing as yellow grains on R-banded chromosomes, was scored, and the data were plotted onto pig standard R-band ideograms of about 300 bands (18). Twenty cells from each animal (no. 115 and 167) were counted for each of the PERV-A and PERV-B probes.
Statistical analysis of hybridization signals.
The Zmax test (14) was used to analyze the cumulative FISH data from 20 metaphase cells to determine the significance of each hybridization location. This test was originally designed for analyzing grain counts from radioactive in situ hybridization but is ideally suited for the present situation, where there are multiple sites of relatively weak hybridization which must be distinguished from background labeling.
Nucleotide sequence accession numbers.
GenBank accession numbers for 31 Westran PERV envelope sequences are AF426916 through AF426946.
RESULTS
Restriction enzyme digestion for screening PERV clones.
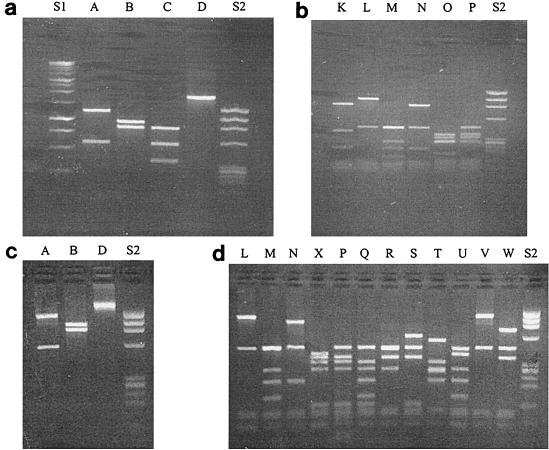
Cloned PERV-A and -B env PCR products, amplified by Taq DNA polymerase (64 clones) and Pfu proofreading DNA polymerase (346 clones), were screened by KpnI and MboI restriction enzymes. Four patterns of KpnI digestion (Fig. 1a) and six patterns of MboI digestion (Fig. 1b) were identified in the PCR product amplified by Taq DNA polymerase. Each of the KpnI/Pfu patterns corresponded to a KpnI/Taq pattern (A, B, and D). Four of the MboI/Pfu patterns corresponded to MboI/Taq patterns (L, M, N, and P). Three patterns of KpnI digestion (Fig. 1c) and 12 patterns of MboI digestion (Fig. 1d) were identified in a PCR product amplified by the Pfu proofreading DNA polymerase. Combined results for the two restriction enzymes were that 9 types of clone were recognizable for clones amplified by Taq DNA polymerase and 13 types were recognizable for clones amplified by Pfu proofreading DNA polymerase (Table 1).
FIG. 1.
Restriction digestion patterns of PERV clones. (a and b) Four patterns (A to D) of KpnI digestion and six patterns (K to P) of MboI digestion were identified in the PCR product amplified by Taq DNA polymerase. (c and d) Three patterns (A, B, and D) of KpnI digestion and 12 patterns (L, M, N, X, and P to W) of MboI digestion were identified in the PCR product amplified by Pfu proofreading DNA polymerase. Lanes S1 and S2, 1-kb ladder and φX174/HaeIII marker (Promega) size standards, respectively.
TABLE 1.
Restriction enzyme digestion and sequence analysis of PERV clones
| Polymerase used | Digestion pattern | No. of clones | No. of clones sequenced [ID(s)a] | Most likely type from sequence comparison | Presence of premature stop codon [ID(s)] |
|---|---|---|---|---|---|
| Taq | AK | 1 | 1 (1) | A+B | Yes (1) |
| AM | 10 | 3 (6, 10, 11) | A | Yes (6, 10, 11) | |
| AN | 4 | 2 (8, 82) | A | Yes (8, 82) | |
| AO | 1 | 1 (17) | A | Yes (17) | |
| AP | 4 | 2 (54, 56) | A | Yes (54) | |
| BL | 41 | 6 (3, 4, 9, 12, 14, 20) | B | Yes (3, 12) | |
| BN | 1 | 1 (24) | B | No | |
| CL | 1 | 1 (21) | A+B | No | |
| DL | 1 | 1 (29) | B | Yes (29) | |
| Pfu | AV | 2 | 1 (251) | A | No |
| AM | 15 | 1 (6) | A | No | |
| AN | 17 | 1 (11) | A | No | |
| AX | 1 | 1 (260) | A+B | Yes (260) | |
| AP | 48 | 1 (3) | A | No | |
| AQ | 5 | 1 (56) | A | Yes (56) | |
| AR | 2 | 1 (62) | A | No | |
| AS | 4 | 1 (112) | A | Yes (112) | |
| AT | 1 | 1 (115) | A | Yes (115) | |
| AU | 1 | 1 (232) | A+B | Yes (232) | |
| AW | 1 | 1 (295) | A | Yes (295) | |
| BL | 248 | 1 (1) | B | No | |
| DU | 1 | 1 (345) | A+B | No |
ID(s), clone identification number(s).
KpnI restriction digestion pattern A is characteristic of PERV-A and pattern B is characteristic of PERV-B, based on the published PERV-A and PERV-B sequences (22). Digestion patterns AM, AN, AP, and BL were found for clones generated with both Taq and Pfu polymerases. On the other hand, restriction digestion patterns AK, AO, BN, CL, and DL were found only in the clones generated by Taq DNA polymerase, and restriction enzyme patterns AV, AX, AQ, AR, AS, AT, AU, AW, and DU were found only in the clones generated by Pfu DNA polymerase. These unique clones constitute 8% (5 of 64) of the Taq DNA polymerase-amplified clones and 5% (18 of 346) of the Pfu DNA polymerase-amplified clones. Among both Taq- and Pfu-amplified clones, the BL type is predominant, constituting 64% (41 of 64) of the Taq polymerase-amplified clones and 72% (248 of 346) of the Pfu-amplified clones (Table 1).
Sequences of PERV clones.
By using the restriction digestion patterns to ensure inclusion of the widest possible range of clone types, 18 Taq-amplified clones and 13 Pfu-amplified clones were sequenced (Table 1). Table 2 summarizes the sequence differences among PERV classes including published PERV-A (GenBank accession no. Y12238), PERV-B (GenBank accession no. Y12239), and PERV-C (GenBank accession no. AF038600) sequences (1, 22).
TABLE 2.
Average numbers of nucleotide differences in pairwise comparisons of sequences within and between PERV classes
| PERV class | PERV class of clones for comparison | No. of sequence comparisons | No. of differences (bp)
|
|
|---|---|---|---|---|
| Mean ± SD | Range | |||
| PERV-A | PERV-A | 153 | 31.5 ± 11.53 | 5-54 |
| PERV-B | 180 | 397.9 ± 4.50 | 388-408 | |
| PERV-C | 18 | 241 ± 6.76 | 222-254 | |
| Recombinant PERV | 90 | 144.5 ± 104.71 | 60-356 | |
| PERV-B | PERV-B | 45 | 10.4 ± 5.62 | 1-24 |
| PERV-C | 10 | 444.3 ± 2.41 | 440-448 | |
| Recombinant PERV | 50 | 283.9 ± 94.45 | 97-351 | |
| PERV-C | Recombinant PERV | 5 | 301.0 ± 30.33 | 274-346 |
To determine the envelope types, each clone was initially aligned with the published PERV-A and PERV-B sequences. Seventeen clones had sequences very similar to the PERV-A sequence, differing from it by only 44 to 54 bases. They are designated PERV-A clones. Nine clones had sequences very similar to the PERV-B sequences, differing from it by only 1 to 15 bases. These are designated PERV-B clones. The remaining five clones differed from both PERV-A and PERV-B by at least 94 bases. Sequence comparison showed that these five clones are actually recombinants between PERV-A and PERV-B. Excluding recombinant clones, the absolute number of nucleotide differences among PERV-A clones is between 5 and 54 bp in 1,785 bp. For PERV-B clones, there were between 1 and 24 bp differences in 1,776 bp. There are about 400 bp different between the PERV-A and PERV-B groups (Table 2).
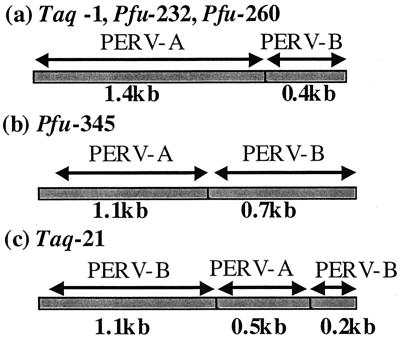
The recombinant clones were classified into three groups based on the patterns of breakpoints between PERV-A and -B sequences (Fig. 2). For Taq-1, Pfu-232, and Pfu-260, different polymerases gave the same breakpoints, suggesting that this recombinant sequence reflects genuine PERV sequence in the pig genome and that it is not an artifact of the PCR process. Clone Taq-21 shows double recombination between PERV-A and PERV-B.
FIG. 2.
Three types of recombinant envelope gene sequence between PERV-A and PERV-B.
Frameshift mutations and premature stop codons.
The nucleotide sequence alignments of the 17 PERV-A clones, 9 PERV-B clones, and 5 recombinant clones together with the PERV-A, PERV-B, and PERV-C published sequences show that 55% of the clones (17 of 31) sequenced have premature stop codons within the envelope protein-encoding region, which would prevent the retrovirus from making a full-length envelope protein recognizable by the cell surface receptor for the virus. Table 3 lists the positions of premature stop codons and their causes. Fourteen stops were caused by frameshift mutation, and only three were caused by base substitutions. A hot spot for frameshift mutations was found at nucleotide position 1134, with 10 of 14 found at this position. Three clones, namely, Taq-10, Taq-11, and Pfu-232, have two frameshift mutations.
TABLE 3.
Sequence analyses of 17 clones with premature stop codons
| Clone IDa | Stop codon positionb | Cause(s) of stop codonc |
|---|---|---|
| Taq-1 | 416 | ΔT nt 1134 (frameshift mutation) |
| Taq-3 | 578 | Base substitution C1733T |
| Taq-6 | 467 | ΔA nt 1250 (frameshift mutation) |
| Taq-8 | 416 | ΔT nt 1134 (frameshift mutation) |
| Taq-10 | 330 | ΔC nt 815 (frameshift mutation), ΔT nt 1134 (frameshift mutation) |
| Taq-11 | 428 | ΔT nt 1134, ΔA nt 1250 (frameshift mutation) |
| Taq-12 | 242 | Base substitution A725T |
| Taq-17 | 395 | Base substitution C1184T |
| Taq-29 | 373 | ΔA nt 1042 (frameshift mutation) |
| Taq-54 | 416 | ΔT nt 1134 (frameshift mutation) |
| Taq-82 | 416 | ΔT nt 1134 (frameshift mutation) |
| Pfu-56 | 416 | ΔT nt 1134 (frameshift mutation) |
| Pfu-112 | 416 | ΔT nt 1134 (frameshift mutation) |
| Pfu-115 | 511 | Base substitution C1532T |
| Pfu-232 | 428 | ΔT nt 1134, ΔA nt 1250 (frameshift mutation) |
| Pfu-260 | 416 | ΔT nt 1134 (frameshift mutation) |
| Pfu-295 | 416 | ΔT nt 1134 (frameshift mutation) |
ID, identification.\
Position of the stop codon in relation to the position of the codon for the first amino acid in the sequence.\
Nucleotide (nt) positions are positions of mutations in the nucleotide sequence. The nucleotide numbering starts from the 5′ forward primer sequence (nucleotide 1).
Chromosomal distributions of PERVs.
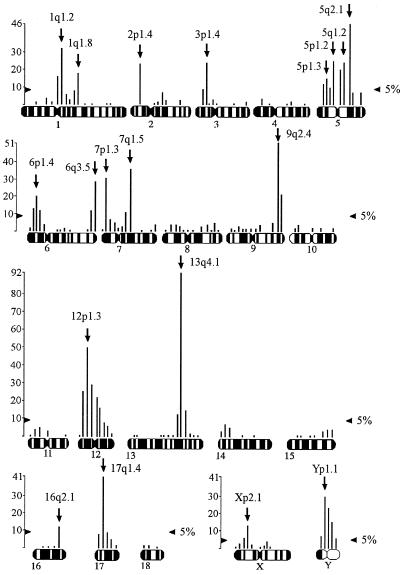
FISH signals for each PERV-A and PERV-B envelope probe were scored. For animal 115 with the PERV-A probe, 478 grains were found distributed over 106 chromosomal segments, giving an average density of 4.51 grains per segment. Over 131 positions from animal 167, 745 grains were found with the same PERV-A probe, giving a higher mean grain density of 5.69. The pooled PERV-A FISH signals for these two animals (115 and 167) are presented on a secondary plot with mean density of 7.84 (1,223 grains/156 chromosomal positions) (Fig. 3).
FIG. 3.
The results of PERV-A hybridization pooled across two animals (115 and 167). The vertical scale is number of grains. The 5% significance threshold for the Zmax test is indicated. Arrows, most likely band locations of PERVs.
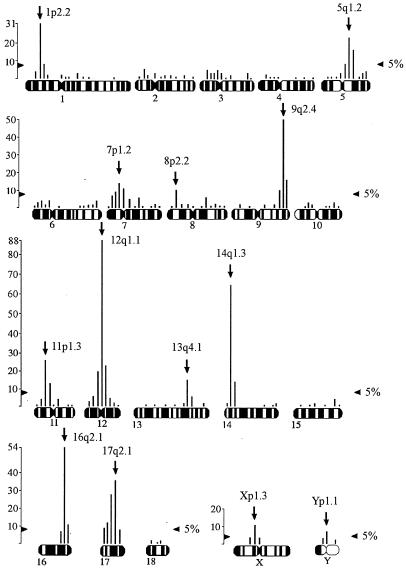
Similarly, hybridization of the PERV-B probe to metaphases from animal 115 and animal 167 were scored. Only pooled PERV-B FISH results are presented (Fig. 4). The mean grain density with this probe is 3.6 (342 grains/95 positions) for animal 115. Again animal 167 had a higher mean density of 4.44 (560 grains/126 positions). The pooled mean density is 6.05 (902 grains/149 positions) from the results for two animals (115 and 167).
FIG. 4.
The results of PERV-B hybridization pooled across two animals (115 and 167). The vertical scale is number of grains. The 5% significance threshold for the Zmax test is indicated. Arrows, most likely band locations of PERVs.
Chi-square homogeneity analysis (29) shows no interaction between animal and probe, thus validating tests of the marginal totals separately for the effects of animal and probe against the null hypothesis of equal numbers of grains (signal intensity). For the comparison of PERV-A versus PERV-B, the chi-square value of 48.79 with one degree of freedom indicates a very highly significant deviation from expectation: the PERV-A probe hybridized much more strongly than PERV-B, presumably due to a larger number of PERV-A inserts. For the comparison of hybridization intensity between animals 115 and 167, the chi-square test (χ12 = 110.69) indicates a highly significant deviation from the expected equal signal intensity. The most likely explanation for this difference is the superior quality of the chromosome cultures and spreads from animal 167.
Zmax test statistics have been calculated for the hybridization data from animals 115 and 167 and for the pooled results for the two animals. In these analyses, each site of hybridization was treated as a different segment. It was assumed as a first approximation that each segment is the same size. It should be noted that, since both animals were males, segments on the sex chromosomes (X and Y chromosomes) occur only half as often as autosomal segments, and this has been taken into account in applying the Zmax statistic.
There are 20 significant sites in animal 115 and 26 in animal 167 with the PERV-A probe (data not shown). For the pooled data, there are 41 significant PERV-A sites (Fig. 3). However, it is highly unlikely that there are so many sites of PERV-A insertion. In many cases, significant sites are in contiguous bands and are almost certainly the results of “spillover” of signal between bands. For example, the site at 9q2.5 in Fig. 3 is almost certainly spillover from the major peak at 9q2.4.
After making allowances for contiguous significant sites, there were 14 major PERV-A peaks identified in animal 115 and 17 major PERV-A peaks identified in animal 167 (Table 4). Thirteen of the significant PERV-A major peaks identified above (1q1.2, 2p1.4, 3p1.4, 5q2.1, 6q3.8, 7p1.3, 7q1.5, 9q2.4, 12p1.3, 13q4.1, 17q1.4, Xp2.1, Yq) are identical in animals 115 and 167, confirming that these locations have genuine PERV-A insertions. There are another five strongly suggestive PERV-A locations (1q1.8, 5p1.3, 5p1.2, 5q1.2, 6p1.4) which are significant in one animal but which are present below the significance threshold in the other animal. The site at 16q2.1 is also a suggestive PERV-A location, although it is significant only in pooled results. The patterns of peaks for both animals are identical (data not shown). Therefore there are 19 PERV-A locations, including suggestive locations, identified in the Westran line. The results for the two Westran animals are quite consistent, as expected. However, the Westran PERV-A locations are quite different from those of Large White pigs (Table 4).
TABLE 4.
Sites of significant major PERV-A peaks in Westran animals 115 and 167 and pooled results for these two animals
| Chromosome | Site of peak for animal(s):
|
Published PERV-A locationa | ||
|---|---|---|---|---|
| 115 (20 cells) | 167 (20 cells) | 115 + 167 (40 cells) | ||
| 1 | 1q1.2 | 1q1.2 | 1q1.2 | |
| 1q1.8 | 1q1.8 | |||
| 1q2.1 | ||||
| 1q2.3 | ||||
| 1q2.4 | ||||
| 2 | 2p1.4 | 2p1.4 | 2p1.4 | |
| 3 | 3p1.4 | 3p1.4 | 3p1.4 | |
| 5 | 5p1.3 | 5p1.3 | ||
| 5p1.2 | 5p1.2 | |||
| 5q1.2 | 5q1.2 | |||
| 5q2.1 | 5q2.1 | 5q2.1 | ||
| 6 | 6p1.4 | 6p1.4 | ||
| 6q3.5 | 6q3.5 | 6q3.5 | ||
| 7 | 7p1.3 | 7p1.3 | 7p1.3 | |
| 7q1.5 | 7q1.5 | 7q1.5 | ||
| 8 | 8p1.2 | |||
| 9 | 9q2.4 | 9q2.4 | 9q2.4 | |
| 12 | 12p1.3 | 12p1.3 | 12p1.3 | |
| 13 | 13q4.1 | 13q4.1 | 13q4.1 | |
| 13q4.2 | ||||
| 13q4.3 | ||||
| 13q4.9 | ||||
| 16 | 16q2.1 | |||
| 17 | 17q1.4 | 17q1.4 | 17q1.4 | |
| X | Xp2.1 | Xp2.1 | Xp2.1 | |
| Y | Yq | Yp1.1 | Yp1.1 | Yp1.2 |
Published locations in a Large White pig from reference 36.
The Zmax test indicates 10 significant sites of hybridization in animal 115 and 17 significant sites in animal 167 with the PERV-B probe (data not shown). However, after allowing for the contiguous sites apparently affected by spillover of hybridization signal, it is reasonable to conclude that there are 9 significant major peaks in animal 115 and 13 in animal 167 (Table 5). The differences between animals 115 and 167 are the peaks on chromosomes 8, 13, X, and Y. All these different PERV-B locations are significant in animal 167 but do not exceed the significance threshold in animal 115 (data not shown). However, they are significant in the pooled results for the two animals (Fig. 4).
TABLE 5.
Significant PERV-B locations in animals 115 and 167 and pooled results for these two animals
| Chromosome | Site of peak for animal(s):
|
Published PERV-B locationa | ||
|---|---|---|---|---|
| 115 (20 cells) | 167 (20 cells) | 115 + 167 (40 cells) | ||
| 1 | 1p2.2 | 1p2.2 | 1p2.2 | |
| 4 | 4p1.1 | |||
| 5 | 5q2.1 | 5q1.2 | 5q1.2 | |
| 7 | 7p1.2 | 7p1.1 | 7p1.2 | 7p1.2→p1.1 |
| 8 | 8p2.2 | 8p2.2 | ||
| 9 | 9q2.4 | 9q2.4 | 9q2.4 | |
| 9q2.6 | ||||
| 10 | 10p1.2 | |||
| 11 | 11p1.3 | 11p1.3 | 11p1.3 | |
| 11q1.4 | ||||
| 12 | 12q1.1 | 12q1.1 | 12q1.1 | |
| 13 | 13q4.1 | 13q4.1 | ||
| 13q4.2 | ||||
| 14 | 14q1.3 | 14q1.3 | 14q1.3 | |
| 14q2.8 | ||||
| 16 | 16q2.1 | 16q2.1 | 16q2.1 | |
| 17 | 17q2.1 | 17q2.1 | 17q2.1 | 17q2.1 |
| X | Xp1.3 | Xp1.3 | ||
| Y | Yq | Yq | ||
Published locations in a Large White pig from reference 36.
Nine significant PERV-B major peaks (1p2.2, 5q2.1, 7p1.2, 9q2.4, 11p1.3, 12q1.1, 14q1.3, 16q2.1, 17q2.1) for animals 115 and 167 are identical, confirming these nine as unequivocal PERV-B locations in the Westran line. There are four more suggestive locations (8p2.2, 13q4.1, Xp1.3, Yq) in Westran pigs. A comparison of these PERV-B locations with the published PERV-B locations in Large White pigs (36) shows that four PERV-B sites, located on chromosomes 7, 9, 13, and 17, are identical or very close, indicating that these are probably common PERV-B insertions in these two breeds (Table 5 and Fig. 4).
DISCUSSION
About 30% of the 31 envelope sequences described here are classified as PERV-A, whether from Taq or Pfu polymerase-amplified clones. However, the clones sequenced in this study are a nonrandom sample because selection of clones for sequencing was based on restriction enzyme digestion results. The restriction digestion results for 410 randomly chosen clones (Table 1) show that 27.8% (114 of 410) are PERV-A, that 71% (291 of 410) are PERV-B, and that 1.2% (5 of 410) can be classified as recombinant clones between PERV-A and PERV-B, providing a more accurate representation of the relative occurrence of these three classes.
Initially Taq DNA polymerase was used to generate PCR products for cloning. However due to a substantial level of minor sequence differences between clones, which might have been artifacts of the inability of Taq to correct errors during DNA replication, proofreading polymerase Pfu was tried. Use of the formula derived by Cantor and Smith (7) to calculate the predicted misincorporation rate for the PERV amplification showed that the misincorporation rate for a product of the size analyzed here was 0.65 nucleotide for the Taq DNA polymerase and 0.08 nucleotide for Pfu DNA polymerase. Thus two Taq clones on average would be expected to differ by 1.3 nucleotides, a Taq clone sequence versus a Pfu clone sequence would be expected to differ by 0.73 nucleotide, and a pair of Pfu clones would be expected to differ by 0.16 nucleotide if misincorporation during PCR was the only source of difference between them. We can conclude that comparisons of Pfu clone sequences with other Pfu sequences or Taq clone sequences are very unlikely to show artifactual differences due to PCR. Even comparisons of Taq sequences with other Taq sequences are unlikely to be seriously affected by amplification artifacts, as only about one such nucleotide difference is expected on average.
The consensus primers used in this study did not amplify any new types of PERVs, only PERV-A and PERV-B. It was later revealed that the forward consensus primer had good homology with the PERV-C sequence but the reverse primer did not. Therefore PERV-C-specific primers were tested in Westran pigs and amplified PCR products of the expected size in several trials (data not shown). These results indicate that the Westran line is PERV-C positive. The copy number of PERV-C has been estimated as 8 to 15 copies per genome for specific strains of inbred and outbred miniature pigs (1). There is evidence of variable PERV-C copy number in other pigs. Some pigs have even been found to be negative for PERV-C (5, 25). Even though PERV-C has a narrower host range than PERV-A and PERV-B (42), it will be important to further characterize PERV-C in Westran pigs if they are ever to be used for xenotransplantation.
The sequencing results suggest that a large proportion of PERVs (17 of 31) are defective due to premature stop codons in the envelope gene. Indeed, 11 clones have a frameshift mutation caused by a deletion of the same nucleotide at position 1135. While some of these clones may simply reflect resampling of the same PERV insertion from the PCR product, it is unlikely that all 11 do. Thus this site may be a hot spot for deletion or may reflect independent insertions of PERVs with the same deletion during the evolutionary history of the pig. Bebenek et al. (3) indicated that the reverse transcriptase of HIV-1 is relatively error prone and that errors are nonrandomly distributed. They found base substitution and one-base frameshift mutational hot spots mainly due to the template-primer slippage. Subject to further verification, these results are encouraging as they indicate that a substantial proportion of the PERVs in the Westran line constitute little potential hazard in xenotransplantation as they are nonfunctional.
The three types of recombinant retroviruses that showed recombination between PERV-A and PERV-B were an unexpected and interesting finding. There is ample precedent for this type of recombination among retroviruses in other species. Retroviruses package two complete viral genomic RNAs in each virion, and this specific configuration facilitates recombination by template swapping during reverse transcription. Recombination between retrovirus genomes has been demonstrated during mixed infection with genetically marked avian tumor viruses (4, 19, 47, 53, 54), murine leukemia viruses (15, 52), and human retroviruses (9). Also, exogenous viruses can recombine with endogenous retrovirus sequences (13, 38, 49). Recently, new recombinants between species have been discovered. For example, baboon endogenous retrovirus (BaEV) is a recombinant retrovirus containing type C gag-pol genes and a type D env gene probably arising by recombination of two primate viruses (24, 45). Similarly, RD-114 of cats is a recombinant between env of BaEV and gag-pol of Papio cynocephalus endogenous retrovirus (44).
Although the possibility that the recombinant clones are PCR artifacts cannot be excluded completely, at least one of the apparently recombinant PERVs is likely to be a genuine recombinant because three clones showing the same pattern of recombination were amplified with two DNA polymerases (Fig. 2).
The FISH results show that there are at least 13 definite PERV-A locations and 9 definite PERV-B locations and a possible further 6 PERV-A and 4 PERV-B locations. The higher number of locations for PERV-A than for PERV-B is more like the pattern that Le Tissier et al. (22) found for European pigs than the pattern they found for Asian pigs, even though Westran pigs have a mitochondrial DNA sequence typical of Asian breeds (20). However the distributions of PERV-A and PERV-B in European versus Asian pigs were not accurately estimated by Le Tissier et al. (22), and some Asian breeds could possibly have more PERV-A than PERV-B. Also Asian breed mitochondrial DNA has introgressed into several European breeds. Le Tissier et al. (22) showed that PERV-A proviruses are present at between 10 and 23 copies and that PERV-B proviruses are present at between 7 and 12 copies in different pig breeds. On average, inbreeding is as likely to cause loss of a PERV site as it is to cause fixation. Thus the inbred Westran line might be expected to have fewer sites than outbred animals. However, there is a possibility that some PERV locations could not be detected by the method used in the present studies because of mismatch between the probe and target causing weak hybridization signals.
Comparison of the FISH results between animals 115 and 167 indicates consistent hybridization patterns except for four locations detected with the PERV-B probe (8p2.2, 13q4.1, Xp1.3, Yq) (Table 5). Based on the high level of inbreeding and close relationship of these animals, these differences are unlikely to be due to differences in the presence of hybridization targets. In each case, the sites were detected in animal 167, which has an overall higher signal intensity, and were absent from 115. Thus it appears that they slipped below the threshold for detection in animal 115, due to the lower efficiency of hybridization and/or signal detection in this animal.
Hybridization of PERV-A and PERV-B probes to the same site is also possible. There are five possible sites (5q2.1, 9q2.4, 13q4.1, 16q2.1, Yp1.1) showing hybridization peaks in the same chromosomal locations with PERV-A and PERV-B probes (Fig. 3 and 4). There are three possible explanations. First, distinct PERV-A and PERV-B insertions may lie close to each other. Second, small regions of highly conserved sequence between PERV-A and PERV-B probes, where the forward and reverse primers are located, could contribute to some cross-hybridization although the cross-hybridization signal would be expected to be very small. Third, recombinant PERVs (Fig. 2) could generate hybridization signals with both probes, likely to be more equal in intensity. Sequence analysis of PERV clones has shown the existence of three possible types of recombinant PERV clones. These three sites might correspond to the three different recombinant PERVs.
The PERV-A locations in Westran pigs are quite different from those in Large White pigs (Table 4). Rogel-Gaillard et al. (36) reported eight PERV-A locations on four different chromosomes (1, 8, 13, and Y) of their Large White pig. On the other hand, consistent significant PERV-A sites on 13 different chromosomes in Westran pigs are observed. The PERV-A locations on 10 chromosomes (2, 3, 5, 6, 7, 9, 12, 16, 17, and X) are so far unique to Westran pigs. However, a PERV-A site on the Y chromosome seems to be the same in the Large White and the Westran lines. On chromosome 1, there are two significant PERV-A sites (1q1.2 and 1q1.8) in Westran pigs and three sites (1q2.1, 1q2.3, 1q2.4) in the Large White pig. Of these, only the sites at 1q1.8 in Westran pigs and 1q2.1 in the Large White pig could possibly be the same, although misallocation is highly unlikely given that these sites are well separated. It is probable that the significant PERV-A site on 13q4.1 in Westran pigs could be the same as that in the Large White pig allocated to the adjacent location at 13q4.2.
About one-half of the PERV-B locations appear to be located in the same or adjacent chromosomal bands in Westran and Large White pigs (Table 5). They are the sites on chromosomes 7, 9, 13, and 17. A PERV-B site was mapped to 7p1.2 (Fig. 4) in this study, very close to the known location of the swine major histocompatibility complex (SLA complex) class I region. Two distinct PERV-B integration loci, at 7p1.1 and 7p1.2-1.1, very close to the SLA complex have been identified by using BAC clones as FISH probes by Rogel-Gaillard et al. (36), with one corresponding to the Westran 7p1.2 site (Table 5). Seven different chromosomes (chromosomes 1, 5, 8, 12, 16, X, and Y) have PERV-B sites in Westran pigs absent in Large White pigs. PERV-B sites on chromosomes 4 and 10 in Large White pigs are absent from the Westran pig. For chromosomes 11 and 14, a single PERV-B site has been identified in Westran and Large White pigs. However, the locations are so far apart (14q1.3 in Westran pigs and 14q2.8 in Large White pigs), being located on different arms for chromosome 11 (11p1.3 in Westran pigs and 11q1.4 in Large White pigs), that they clearly represent independent insertions in these breeds. Thus, as expected, different pig breeds share some PERV insertions in their genomes but the breeds also have other unique locations.
In conclusion, this study has characterized and mapped PERVs in the Westran pig strain, in the process identifying numerous novel sites of insertion, establishing that a substantial proportion of the retrovirus inserts have defective envelope genes, and recognizing recombinants between PERV-A and PERV-B. The occurrence of recombinant PERVs also provides a salutary reminder of the potential for recombinational repair of defective retroviruses and for recombination of human and porcine retroviruses during xenotransplantation. Defective PERVs could possibly regain infectious potential through recombination. Furthermore PERVs could recombine with human endogenous retroviruses to generate totally novel retroviruses. Thus these results corroborate the recombinogenic potential of retroviruses and highlight the potential danger of intra- and/or interspecies recombination of PERVs in xenotransplantation.
Acknowledgments
We gratefully acknowledge Herman Raadsma, Marylin Jones, and Gina Attard for access to and help with their Li-Cor sequencer at the Camden Campus of the University of Sydney and Cindy D. K. Bottema for use of her laboratory facilities at the Waite Campus of the University of Adelaide. We also thank Jeremy Chapman, Phil O'Connell, Wayne Hawthorne, and Jane Burgess at Westmead Hospital for assistance, access to unpublished results, and discussions.
The award of a part-time research fellowship from the Research Foundation of The Queen Elizabeth Hospital and the support of Repromed are gratefully acknowledged by G. C. Webb. This work was supported by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. S. Porterfield, and A. T. de Madrid. 1971. C-type virus particles in pig kidney cell lines. J. Gen. Virol. 10:195-198. [DOI] [PubMed] [Google Scholar]
- 3.Bebenek, K., J. Abbotts, J. D. Roberts, S. H. Wilson, and T. A. Kunkel. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948-16956. [PubMed] [Google Scholar]
- 4.Blair, D. G., W. S. Mason, E. Hunter, and P. K. Vogt. 1976. Temperature-sensitive mutants of avian sarcoma viruses: genetic recombination between multiple or coordinate mutants and avian leukosis viruses. Virology 75:48-59. [DOI] [PubMed] [Google Scholar]
- 5.Bösch, S., C. Arnauld, and A. Jestin. 2000. Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J. Virol. 74:8575-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breese, S. S. 1970. Virus-like particles occurring in culture of stable pig kidney cell line. Arch. Gesamte Virusforsch. 30:401-404. [DOI] [PubMed] [Google Scholar]
- 7.Cantor, C. R., and C. L. Smith. 1999. Genomics: the science and technology behind the human genome project. John Wiley & Sons, New York, N.Y.
- 8.Chari, R. S., B. H. Collins, J. C. Magee, J. M. DiMaio, A. D. Kirk, R. C. Harland, R. L. McCann, J. L. Platt, and W. C. Meyers. 1994. Brief report: treatment of hepatic failure with ex vivo pig-liver perfusion followed by liver transplantation. N. Engl. J. Med. 331:234-237. [DOI] [PubMed] [Google Scholar]
- 9.Clavel, F., M. D. Hoggan, R. L. Willey, K. Strebel, M. A. Martin, and R. Repaske. 1989. Genetic recombination of human immunodeficiency virus. J. Virol. 63:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, H. M. 1954. Kangaroo Island's wild pigs. S. Aust. Nat. 28:57-61. [Google Scholar]
- 11.Cozzi, E., and D. J. White. 1995. The generation of transgenic pigs as potential organ donors for humans. Nat. Med. 1:964-966. [DOI] [PubMed] [Google Scholar]
- 12.Deacon, T., J. Schumacher, J. Dinsmore, C. Thomas, P. Palmer, S. Kott, A. Edge, D. Penney, S. Kassissieh, P. Dempsey, and O. Isacson. 1997. Histological evidence of fetal pig neural cell survival after transplantation into a patient with Parkinson's disease. Nat. Med. 3:350-353. [DOI] [PubMed] [Google Scholar]
- 13.Elder, J. H., J. W. Gautsch, F. C. Jensen, R. A. Lerner, J. W. Hartley, and W. P. Rowe. 1977. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc. Natl. Acad. Sci. USA 74:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewens, W. J., R. C. Griffiths, S. N. Ethier, S. A. Wilcox, and J. A. Graves. 1992. Statistical analysis of in situ hybridization data: derivation and use of the Zmax test. Genomics 12:675-682. [DOI] [PubMed] [Google Scholar]
- 15.Faller, D. V., and N. Hopkins. 1978. T1 oligonucleotides that segregate with tropism and with properties of gp70 in recombinants between N- and B-tropic murine leukemia viruses. J. Virol. 26:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fodor, W. L., B. L. Williams, L. A. Matis, J. A. Madri, S. A. Rollins, J. W. Knight, W. Velander, and S. P. Squinto. 1994. Expression of a functional human complement inhibitor in a transgenic pig as a model for the prevention of xenogeneic hyperacute organ rejection. Proc. Natl. Acad. Sci. USA 91:11153-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groth, C. G., O. Korsgren, A. Tibell, J. Tollemar, E. Moller, J. Bolinder, J. Ostman, F. P. Reinholt, C. Hellerstrom, and A. Andersson. 1994. Transplantation of porcine fetal pancreas to diabetic patients. Lancet 344:1402-1404. [DOI] [PubMed] [Google Scholar]
- 18.Gustavsson, I. 1988. Standard karyotype of the domestic pig. Hereditas 109:151-157. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, S., and H. Hanafusa. 1972. Genetic recombination with avian tumor virus. Virology 49:37-44. [DOI] [PubMed] [Google Scholar]
- 20.Kim, K.-I., J.-H. Lee, K. Li, Y.-P. Zhang, S.-S. Lee, J. Gongora, and C. Moran. 2002. Phylogenetic relationships of Asian and European pig breeds determined by mitochondrial DNA D-loop sequence polymorphism. Anim. Genet. 33:19-25. [DOI] [PubMed] [Google Scholar]
- 21.Lemieux, N., B. Dutrillaux, and P. E. Viegas. 1992. A simple method for simultaneous R or G banding and fluorescence in situ hybridization of small single-copy genes. Cytogenet. Cell Genet. 59:311-312. [DOI] [PubMed] [Google Scholar]
- 22.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 23.Lieber, M. M., C. J. Sherr, R. E. Benveniste, and G. J. Todaro. 1975. Biological and immunological properties of porcine type C viruses. Virology 66:616-619. [DOI] [PubMed] [Google Scholar]
- 24.Mang, R., J. Goudsmit, and, A. C. van der Kuyl. 1999. Novel endogenous type C retrovirus in baboons: complete sequence, providing evidence for baboon endogenous virus gag-pol ancestry. J. Virol. 73:7021-7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mang, R., J. Maas, X., Chen, J. Goudsmit, and A. C. van der Kuyl. 2001. Identification of a novel type C porcine endogenous retroviruses: evidence that copy number of retroviruses increases during host inbreeding. J. Gen. Virol. 82:1829-1834. [DOI] [PubMed] [Google Scholar]
- 26.Martin, U., G. Steinhoff, V. Kiessig, M. Chikobava, M. Anssar, T. Morschheuser, B. Lapin, and A. Haverich. 1998. Porcine endogenous retrovirus (PERV) was not transmitted from transplanted porcine endothelial cells to baboons in vivo. Transplant Int. 11:247-251. [DOI] [PubMed] [Google Scholar]
- 27.Martin, U., V. Kiessig, J. H. Blusch, A. Haverich, K. von der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692-964. [DOI] [PubMed] [Google Scholar]
- 28.McIntosh, G. M., and A. Pointon. 1981. The Kangaroo Island strain of pig in biomedical research. Aust. Vet. J. 57:182-185. [DOI] [PubMed] [Google Scholar]
- 29.Mead, R., R. N. Curnow, and A. M. Hasted. 1993. Statistical methods in agriculture and experimental biology. Chapman and Hall, London, United Kingdom.
- 30.Paradis, K., G. Langford, Z. Long, W. Heneine, P. Sandstrom, W. M. Switzer, L. E. Chapman, C. Lockey, D. Onions, and E. Otto. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. Science 285:1236-1241. [DOI] [PubMed] [Google Scholar]
- 31.Patience, C., D. A. Wilkinson, and R. A. Weiss. 1997. Our retroviral heritage. Trends Genet. 13:116-120. [DOI] [PubMed] [Google Scholar]
- 32.Patience, C., G. S. Patton, Y. Takeuchi, R. A. Weiss, M. O. McClure, L. Rydberg, and M. E. Breimer. 1998. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet 352:699-701. [DOI] [PubMed] [Google Scholar]
- 33.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 35.Pitkin, Z., and C. Mullon. 1999. Evidence of absence of porcine endogenous retrovirus (PERV) infection in patients treated with a bioartificial liver support system. Artif. Organs 23:829-833. [DOI] [PubMed] [Google Scholar]
- 36.Rogel-Gaillard, C., N. Bourgeaux, A. Billault, M. Vaiman, and P. Chardon. 1999. Construction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements. Cytogenet. Cell Genet. 85:205-211. [DOI] [PubMed] [Google Scholar]
- 37.Sharma, A., J. Okabe, P. Birch, S. B. McClellan, M. J. Martin, J. L. Platt, and J. S. Logan. 1996. Reduction in the level of Gal(α1,3)Gal in transgenic mice and pigs by the expression of an α(1,2)fucosyltransferase. Proc. Natl. Acad. Sci. USA 93:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephenson, J. R., G. R. Anderson, S. R. Tronick, and S. A. Aaronson. 1974. Evidence for genetic recombination between endogenous and exogenous mouse RNA type C viruses. Cell 2:87-94. [DOI] [PubMed] [Google Scholar]
- 39.Suzuka, I., K. Sekiguchi, and M. Kodama. 1985. Some characteristics of a porcine retrovirus from a cell line derived from swine malignant lymphomas. FEBS Lett. 183:124-128. [DOI] [PubMed] [Google Scholar]
- 40.Suzuka, I., N. Shimizu, K. Sekiguchi, H. Hoshino, M. Kodama, and K. Shimotohno. 1986. Molecular cloning of unintegrated closed circular DNA of porcine retrovirus. FEBS Lett. 198:339-343. [DOI] [PubMed] [Google Scholar]
- 41.Switzer, W. M., V. Shanmugam, L. Chapman, and W. Heneine. 1999. Polymerase chain reaction assays for the diagnosis of infection with the porcine endogenous retrovirus and the detection of pig cells in human and nonhuman recipients of pig xenografts. Transplantation 68:183-188. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todaro, G. J., R. E. Benveniste, M. M. Lieber, and C. J. Sherr. 1974. Characterization of a type C virus released from the porcine cell line PK (15). Virology 58:65-74. [DOI] [PubMed] [Google Scholar]
- 44.van der Kuyl, A. C., J. T. Dekker, and J. Goudsmit. 1999. Discovery of a new endogenous type C retrovirus (FcEV) in cats: evidence for RD-114 being an FcEVGag-Pol/baboon endogenous virus BaEVEnv recombinant. J. Virol. 73:7994-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Kuyl, A. C., R. Mang, J. T. Dekker, and J. Goudsmit. 1997. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J. Virol. 71:3666-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Laan, L. J. W., C. Lockey, B. C. Griffeth, F. S. Frasier, C. A. Wilson, D. E. Onions, B. J. Hering, Z. Long, E. Otto, B. E. Torbett, and D. R. Salomon. 2000. Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature 407:501-504. [DOI] [PubMed] [Google Scholar]
- 47.Vogt, P. K. 1971. Genetically stable reassortment of markers during mixed infection with avian tumor viruses. Virology 46:947-952. [DOI] [PubMed] [Google Scholar]
- 48.Webb, G. C., S. Jitrapakdee, C. D. K. Bottema, and J. Wallace. 1997. Assignment of the rat pyruvate carboxylase gene to band 1q43 by in situ hybridisation. Cytogenet. Cell Genet. 79:151-152. [DOI] [PubMed] [Google Scholar]
- 49.Weiss, R. A., W. S. Mason, and P. K. Vogt. 1973. Genetic recombinants and heterozygotes derived from endogenous and exogenous avian RNA tumor viruses. Virology 52:535-552. [DOI] [PubMed] [Google Scholar]
- 50.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, C. A., S. Wong, M. Van Brocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, P. K., and J. A. McCarter. 1973. Genetic studies of temperature-sensitive mutants of Moloney murine leukemia virus. Virology 53:319-326. [DOI] [PubMed] [Google Scholar]
- 53.Wyke, J. A., and J. A. Beamand. 1979. Genetic recombination in Rous sarcoma virus: the genesis of recombinants and lack of evidence for linkage between pol, env and src genes in three factor crosses. J. Gen. Virol. 43:349-364. [DOI] [PubMed] [Google Scholar]
- 54.Wyke, J. A., J. G. Bell, and J. A. Beamand. 1975. Genetic recombination among temperature-sensitive mutants of Rous sarcoma virus. Cold Spring Harbor Symp. Quant. Biol. 39:897-905. [DOI] [PubMed] [Google Scholar]