Abstract
Recently, several reports have been published in support of the idea that protein synthesis occurs in both the nucleus and the cytoplasm. This proposal has generated a great deal of excitement because, if true, it would mean that our thinking about the compartmentalization of cell functions would have to be re-evaluated. The significance and broad implications of this phenomenon require that the experimental evidence used to support it be carefully evaluated. Here, we critique the published evidence in support of, or in opposition to, the question of whether translation occurs in the nucleus. Arguments in support of nuclear translation focus on three issues: (1) the presence of translation factors and ribosomal components in the nucleus, and their recruitment to sites of transcription; (2) amino acid incorporation in isolated nuclei and in nuclei under conditions that should not permit protein import; and (3) the fact that nuclear translation would account for observations that are otherwise difficult to explain. Arguments against nuclear translation emphasize the absence (or low abundance) from nuclei of many translation factors; the likely inactivity of nascent ribosomes; and the loss of translation activity as nuclei are purified from contaminating cytoplasm. In our opinion, all of the experiments on nuclear translation published to date lack critical controls and, therefore, are not compelling; also, traditional mechanisms can explain the observations for which nuclear translation has been invoked. Thus, while we cannot rule out nuclear translation, in the absence of better supporting data we are reluctant to believe it occurs.
INTRODUCTION
The nuclear envelope of eukaryotic cells separates the nucleus and cytoplasm. It is thought to partition transcription and processing of messenger RNAs (mRNAs), which occur in the nucleus, from protein synthesis (translation), which is observed in the cytoplasm. Several recent reports have challenged this widely accepted idea by asserting that translation can also occur in the nucleus. Indeed, nuclear translation is an alluring hypothesis, as it provides a seemingly simple explanation for observations suggesting that newly synthesized mRNAs are monitored by translation before they are released into the cytoplasm.
Here we analyze the arguments for and against the existence of nuclear translation, and discuss a variety of interpretations of the published data. Three types of recent evidence are germane: reports that elements of the translation machinery are in the nucleus, reports that amino acids can be incorporated into proteins in nuclear preparations, and models that invoke nuclear translation to explain how protein synthesis can affect nucleus-associated events in gene expression.
IS THE TRANSLATION MACHINERY PRESENT IN THE NUCLEUS?
For nuclear translation to occur, essential components such as ribosomes, tRNAs, and translation factors must be present in the nucleus, in addition to the mRNA. Ribosomes and tRNAs are synthesized and undergo maturation in the nucleus, and immunological studies have indicated that certain translation factors are present in the nucleoplasm.
Newly synthesized and processed nuclear tRNAs and mRNAs probably could function if they were exposed to the translational machinery, as they appear to undergo complete splicing and end-maturation prior to export. Indeed, tRNAs are aminoacylated within the nucleus, apparently as a means of quality control before export to the cytoplasm (Lund and Dahlberg 1998; Sarkar et al. 1999). Although mRNP remodeling during, or subsequent to, export alters the complement of proteins bound to an mRNA, the primary sequences of the transcripts do not appear to change after arrival in the cytoplasm, indicating that the nuclear mRNA could be translated.
In contrast to tRNAs and mRNAs, evidence from several organisms suggests that newly assembled ribosomal subunits are not functional until they are exported. Certain yeast shuttling proteins such as Tif6p exit the nucleus with the 60S subunits and are removed by cytoplasmic factors (Senger et al. 2001). Both Tif6p and its mammalian ortholog TIF6 inhibit the association of the 40S and 60S subunits (Si and Maitra 1999). Moreover, in Xenopus oocyte nuclei, no newly synthesized 80S ribosomes can be found (E. Lund and J.E. Dahlberg, unpubl.). In yeast, nascent 40S subunits are not found on polyribosomes until the small rRNA undergoes cytoplasmic maturation (from 20S to 18S rRNA; Venema and Tollervey 1999). Thus, if functional 80S ribosomes exist in nuclei, they are likely to be different from those destined for use in the cytoplasm. Alternatively, a small subpopulation of ribosomes might exist that supports very low levels of translation—say one initiation per mRNA—and go undetected.
Although several immunological studies have demonstrated the presence of initiation factors such as eIF4E and eIF4G in cell nuclei, it is unclear that all essential translation factors are there (Etchison-and-Etchison 1987; Lejbkowicz et al. 1992; Iborra et al. 2001; McKendrick et al. 2001). Recently, Görlich and coworkers addressed this question (Bohnsack et al. 2002), using carefully controlled expression of GFP-tagged proteins in stably transfected mammalian cells. They found that nuclei of these cells had 1% or less of cytoplasmic levels of the following translation initiation, elongation, and termination factors: eIF2, eIF2B, eIF4A1, eIF5, eEF1A, eEF1B, or eEF2 and eRF1. Moreover, this study and experiments on EF1A by Kutay and collaborators (Calado et al. 2002) showed that several very active export receptors function to keep the nuclear levels of these factors low. Both groups conclude that it is highly unlikely that nuclei contain sufficient amounts of all translation factors needed to sustain significant levels of nuclear translation. Two caveats of this interpretation are that most of these proteins were fused to GFP, which might have influenced their distribution, and that even modest levels of translation factors might support low levels of protein synthesis if they were highly concentrated at specific locations within nuclei.
Thus, the available data on the presence or absence of functional ribosomes and translation factors in nuclei cannot completely exclude the possibility that nuclear translation occurs. However, if nuclear translation does occur, it is likely to differ significantly from cytoplasmic translation with regard to both the identity and function of the components involved.
CAN AMINO ACID INCORPORATION BE OBSERVED IN ISOLATED NUCLEI OR NUCLEAR PREPARATIONS?
Evidence in support of nuclear translation was obtained by Cook and colleagues, who reported that amino acids could be incorporated in isolated, highly purified nuclei (Iborra et al. 2001). They calculated that about 10%–15% of cell translation was nuclear. They also showed that tagged amino acids (provided in the form of biotin-lysine-tRNALys and BODIPY-lysine-tRNALys) accumulated in nuclei of cells that had been permeabilized by a mild detergent treatment. This accumulation was stimulated by transcription conditions, indicating that translation and transcription were coupled in these nuclei. Some of the experimental preparations and protocols used in these experiments have been questioned. For example, the purity of isolated nuclei is critically important, as was discussed in a debate on nuclear translation almost 25 years ago by Goidl and Allen (1978).
In this issue of RNA, Deutscher and colleagues address the question of purity of nuclear preparations (Nathanson et al. 2003). When they prepared HeLa cell nuclei by the method of Iborra et al. (2001) they observed essentially the same extent of amino acid incorporation per 106 nuclei, thereby confirming the earlier published results. However, concern about the small amount of cytoplasmic contamination in their nuclear preparation led these authors to subject the nuclei to further purification, reducing the remaining cytoplasmic contamination several hundred-fold. Under these conditions, the level of amino acid incorporation fell by about the same factor, becoming essentially undetectable, thus making it likely that the amino acid incorporation observed with the less purified nuclei resulted from cytoplasmic contaminants. However, even though this purification did not affect the ability of nuclei to support transcription, it cannot be ruled out that key translation factors could have been lost, thereby causing the loss of translation.
Another concern with the experiments of Iborra et al. (2001) relates to the integrity of the nuclei in the permeabilized cells. If the nuclei were permeabilized during preparation of the cells, various translation factors might leak in or out. Although large particles of dextran were excluded from these nuclei, biotin-lysine-tRNALys (and BODIPY-lysine-tRNALys) must have been able to cross the nuclear envelope, as that step was required for introducing the tagged amino acids into the nuclei. tRNAs have not been shown to enter intact nuclei, so the ability of tRNALys to enter the nucleus indicates that the nuclei may have been permeabilized by the detergent treatment. The authors also observed incorporation of amino acids in the presence of thapsigargin, a compound thought to inhibit protein import but which is without effect on the import of several proteins (Marshall et al. 1997; Strubing and Clapham 1999); the ability of this reagent to block protein import under these experiments was not described. Nuclear entry of 40-kD dextran beads was reported to be inhibited (in data not presented), but, again, the presence of aminoacylated tRNAs in the nuclei shows that the entry of macromolecules into the nuclei was not blocked. Thus, before these experiments can be considered as compelling evidence for nuclear translation, both the purity of the isolated nuclei and the integrity of the nuclei in the permeabilized cells must be demonstrated.
Recently, Rosbash and colleagues (Brogna et al. 2002) presented additional evidence in support of the coupling of transcription and translation in the nucleus. They observed that ribosome components and some translation factors accumulate at the sites of RNA polymerase II transcription in the polytene chromosomes of Drosophila salivary glands. They also reported the incorporation of amino acids at the sites of transcription, even when protein import from the cytoplasm was inhibited. The implications of these results would be that splicing and translation occurred in a coupled fashion at the site of transcription, thereby demonstrating nuclear translation in a minimally perturbed system.
Unfortunately, the specificity and effectiveness of several crucial reagents were not described, making the significance of the results unclear. Perhaps the most serious omission is lack of information about the specificity toward Drosophila proteins of the antibodies, which had been generated against peptides of human ribosomal proteins (r-proteins). Several important antibodies used in this study were described as having nonspecific binding when they were originally isolated (Nadano et al. 2000); this raises the possibility that the proteins detected in the Drosophila preparations were not r-proteins. Also, the concentration (1.0%) of Triton X-100 present in the buffers used to dissect the salivary glands may have permeabilized the nuclear envelope, allowing artificial access of cytoplasmic materials. The rRNA sequences associated with the polytene chromosomes may well be precursors of rRNAs (Ringborg et al. 1970). In any case, it should be noted that the presence of r-proteins and rRNA sequences does not demonstrate the presence of functional ribosomes. Experiments designed to look directly for incorporation of 35S-labeled amino acids at sites of transcription suffer from the fact that the authors used thapsigargin to inhibit import of labeled proteins from the cytoplasm; like Iborra et al. (2001; see above), these authors did not report the efficacy of thapsigargin as an inhibitor of protein import in the cells used in their experiments.
Thus, although the experiments of Iborra et al. (2001) and Brogna et al. (2002) are consistent with nuclear translation, both studies lack important controls that would rule out alternative explanations such as contamination, disruption of nuclei, or ineffective reagents. Consequently, neither report can be taken as presenting compelling evidence in favor of nuclear translation.
IS NUCLEAR TRANSLATION NEEDED TO EXPLAIN HOW PROTEIN SYNTHESIS COULD AFFECT NUCLEAR EVENTS ASSOCIATED WITH GENE EXPRESSION?
Nuclear translation would provide a convenient way to explain how protein synthesis, which monitors mRNA codons in a reading-frame-sensitive manner, could affect the levels of nucleus-associated mRNA (for review, see Maquat 1995; Brogna 2001; Wilkinson and Shyu 2002). The appeal of such an explanation accounts for much of the excitement generated by the Iborra et al. paper discussed above. However, one must question if such a “nucleocentric” model is needed to explain effects of translation on the turnover of nucleus-associated mRNA.
Cytoplasmic monitoring of nucleus-associated mRNAs
Transcription errors, inappropriate splicing, and mutation can generate defective mRNAs containing premature termination codons (PTCs), which cause the termination of translation within the body of the message (Maquat and Carmichael 2001). The major mRNA surveillance process, called nonsense-mediated mRNA decay (NMD), can reduce the levels of PTC-containing mRNAs (PTC+ mRNAs) to 5%–30% of that of the wild-type mRNA in both the nucleus-associated and cytoplasmic fractions of cells. NMD is initiated when a PTC is recognized in frame by a ribosome; compensatory mutations that put the PTC out of frame result in stabilization of the transcript, as do nonsense-suppressor tRNAs. Inhibitors of protein synthesis such as cycloheximide also stabilize nucleus-associated PTC-containing mRNAs, confirming that their turnover is directly coupled to translation (for review, see Maquat 1995; Frischmeyer and Dietz 1999).
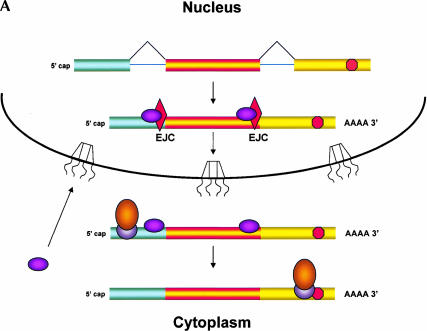
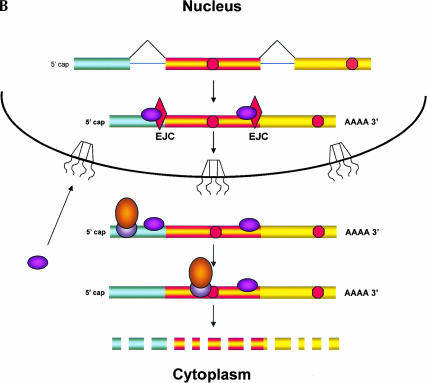
In spite of its effects on nucleus-associated mRNA, NMD need not occur within the nucleus per se; instead, it could take place in the perinuclear cytoplasm (Fig. 1 ▶). Current models of NMD propose that a ribosome initiates a “pioneer round of translation” on newly made mRNA bound to nuclear cap binding protein (Fortes et al. 2000; Ishigaki et al. 2001). During this initial round of translation, the ribosome is thought to remove the exon junction complex (EJC) from the mRNA (Dostie and Dreyfuss 2002; Lejeune et al. 2002); the EJC is a group of proteins that associate with the mRNA during splicing and that, among other things, promote mRNA export and provide a mark needed for execution of NMD (Le Hir et al. 2001). Because the normal termination codon is almost always in the 3′ terminal exon, the mRNA has no associated EJC proteins by the time the pioneering ribosome reaches this stop codon. However, if the ribosome encounters a termination codon in an mRNA that still has a bound EJC, this codon is recognized as a PTC and several additional proteins are recruited to the mRNA, initiating mRNA destruction by NMD (Page et al. 1999; Kim et al. 2001; Lykke-Andersen et al. 2001; Wagner and Lykke-Andersen 2002). All of these NMD-related events would occur in the perinuclear cytoplasm if the pioneering round of translation occurs during nuclear export of the mRNA (Maquat 2002; Schell et al. 2002). As discussed by Nathanson et al. (2003), the perinuclear cytoplasm would fractionate with nuclei in most purification protocols, so events that occur there would appear to be nuclear.
FIGURE 1.
Nonsense mediated decay of mRNAs. The monitoring of mRNAs for premature termination codons (PTCs) is mediated by ribosomes during the “pioneering round of translation.” Several proteins associate with sequences near exon/intron borders of the spliced mRNA, forming the exon junction complex (EJC). Some EJC proteins are removed during export whereas others are removed during translation and thus serve as indicators of whether a particular region of the mRNA has been translated. (A) If no EJC proteins are on the mRNA when the pioneering ribosome encounters a chain termination codon, this codon is read as a normal termination signal, at the 3′ end of the coding region. (B) If the ribosome has not traversed all exon/intron borders, EJC protein(s) remain on the mRNA and serve as an indication that any termination codon encountered is a PTC. As a consequence, nonsense-mediated decay (NMD) is initiated, resulting in destruction of the mRNA. The pioneering round of translation is shown here as happening in the cytoplasm (or the perinuclear cytoplasm), and with the released proteins being imported into the nucleus.
Recently, Mühlemann and colleagues (Bühler et al. 2002) used an innovative approach to determine whether NMD, and hence translation, takes place prior to the nuclear export of mRNAs. They observed NMD of a PTC+ mRNA in the nuclear fraction of HeLa cells, even when they inhibited mRNA export. To inhibit export, the authors used transiently transfected cells expressing the Matrix (M) protein of VSV, an inhibitor of nucleo-cytoplasmic transport (Her et al. 1997) that slows, but does not block, export. Although this result appears to give strong support for nuclear translation, several issues remain to be clarified before the data can be considered definitive. For example, all mRNA quantitations were normalized to the levels of 18S rRNA in the respective cell compartments, but the extent to which M protein affects the levels of this RNA standard is unknown. Thus it is unclear how much the M protein affected the distribution of mRNAs between the intranuclear and perinuclear compartments. Also, the authors could not rule out differential effects of M protein on the production of PTC+ versus PTC− mRNAs (see discussion of NAS, below).
Could NMD in the perinuclear cytoplasm explain the destabilization of the PTC+ mRNA in the presence of M protein? Because mRNAs continue to be exported in the presence of M protein (albeit slowly), they could be subjected to perinuclear monitoring for PTCs during export. NMD is likely to be the slowest step in the process of export and proofreading, because it involves several complicated steps not required for simple export and proofreading. If NMD is the rate-limiting step even in the presence of M protein, the reduced export would not affect the levels of PTC+ mRNA. That would be true regardless of whether NMD occurs on the nuclear or cytoplasmic side of the nuclear envelope. Consequently, although the results presented are consistent with NMD occurring in the nucleus, an interpretation based on cytoplasmic proofreading of the mRNA is also valid.
Effects of PTCs in mRNA on the splicing of pre-mRNA
In contrast to NMD, the splicing of pre-mRNAs is a decidedly intranuclear event; yet the presence of PTCs in certain spliced mRNAs can affect utilization of alternative splice sites, leading to the skipping of the PTC-containing exon and/or accumulation of the unspliced pre-mRNA at the site of transcription (Mendell and Dietz 2001; Mühlemann et al. 2001; Maquat 2002). At least two different processes have been shown to be responsible. Because both of them are initiated by mutations that often generate PTCs in the sequence of the mRNA, they can be difficult to distinguish from each other (Cartegni et al. 2002).
In one process, the mutation alters an exonic splicing enhancer (ESE), an RNA sequence in the pre-mRNA that acts as a binding site for one or more splicing factors; these, in turn, affect the efficiency of utilization of nearby splice sites (Liu et al. 2001). Thus, mutation of an ESE affects splicing directly and does not involve the monitoring of a PTC (or a missense codon) in the spliced product. A recent paper by Beemon and coworkers (Caputi et al. 2002) is an elegant illustration of this principle.
The other process, called nonsense-associated alternative splicing (NAS), is initiated by reading-frame-sensitive recognition of PTCs in certain mRNAs during translation; unlike NMD, NAS occurs infrequently and is observed only with specific exons of particular mRNAs. The best documented example of NAS, studied extensively by Wilkinson and colleagues, involves the exon encoding the hypervariable VDJ region of human T-cell receptor-β (TCR-β) mRNA; this exon is created by gene rearrangements likely to generate PTCs. In this case, manifestations of NAS include accumulation of the corresponding pre-mRNA, increased use of possible alternative latent splice sites, and presumably a decrease in the normal splicing of the PTC-containing mRNA (Mühlemann et al. 2001; Wang et al. 2002a; Qian et al. 1993).
The large VDJ exon is not optimal for splicing, so its inclusion in the normal TCR-β mRNA is likely to require one or more specific splicing-enhancer binding proteins. Recent experiments show that NAS requires translation of the PTC-containing TCR-β mRNA, but it does not depend on NMD per se (Mendell et al. 2002; Wang et al. 2002b). Moreover, a trans-acting signal must be transmitted from a translating ribosome (sensing the PTC) to the splicing apparatus because a PTC created by the (normal) splicing of one TCR-β pre-mRNA affects the splicing of another TCR-β pre-mRNA molecule (Wang et al. 2002a).
Wilkinson and collaborators (Wang et al. 2002b; Wilkinson and Shyu 2002) have proposed a model for NAS, in which translation occurs in the nucleus at—or near—the site of transcription and splicing. In this model, a PTC in the mature mRNA elicits NAS of related pre-mRNAs because a stalled ribosome affects the local concentration of splicing factors by some unknown mechanism (Wang et al. 2002a).
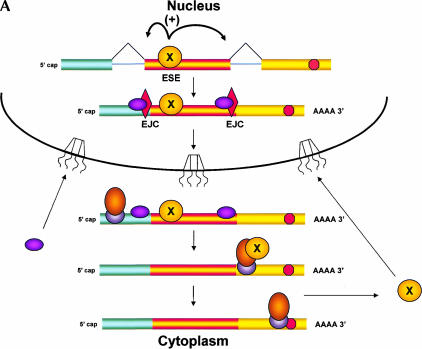
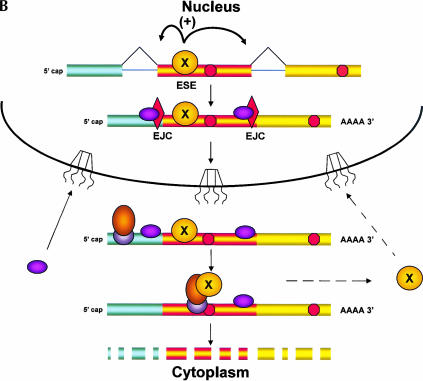
Here, we propose an alternative model, in which the PTC-containing mRNA is monitored only in the cytoplasm (Fig. 2 ▶). In this scenario, a splicing factor that binds to the VDJ exon exits the nucleus with the mRNA and, like other shuttling splicing factors (Caceres et al. 1998), needs to be recycled back for efficient splicing to continue. A delay in the return of this factor would decrease its concentration within the nucleus, thereby slowing the normal splicing of the TCR-β pre-mRNA. The slowing of splicing of the VDJ exon would account for the accumulation of pre-mRNA at the site of transcription (Mühlemann et al. 2001) and the use of alternative splice sites that leads to skipping of the VDJ exon (Wang et al. 2002a).
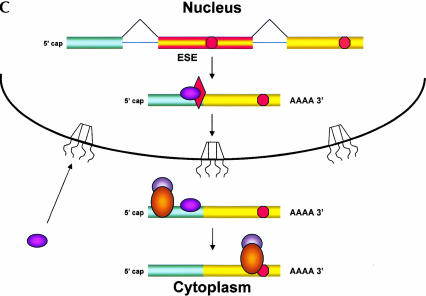
FIGURE 2.
A model for coupling nuclear splicing to cytoplasmic translation. The presence of PTCs in specific exons of particular mRNAs can alter the splicing of the pre-mRNA; like NMD, this nonsense-associated alternative splicing (NAS) uses translation to detect the PTC. If the pioneering round of translation occurs in the perinuclear cytoplasm rather than in the nucleus, a signal indicating the presence of the PTC must be transmitted from the ribosome back to the nuclear splicing machinery. (A) This model proposes that a splicing factor (denoted X) binds to an splicing enhancer sequence (ESE) in a nonoptimal exon of a pre-mRNA, allowing for inclusion of the exon in the mRNA, and that this factor remains bound to the ESE as the mRNA exits the nucleus. In the cytoplasm, the ESE binding factor is transferred to the pioneering ribosome and released only when this ribosome reaches the termination codon at the 3′ end of the coding region. Release of the factor allows it to recycle back to the nucleus, to support further rounds of splicing. (B) If the pioneering ribosome encounters a PTC (a termination codon to the 5′ side of an EJC), it is unable to release the factor, thereby retarding return of the factor to the nucleus. Recognition of the PTC also results in NMD; the presence of the factor on the pioneering ribosome may even enhance NMD, resulting in “Super NMD”, as is observed with TCR-β mRNA. (C) A reduced level of the ESE binding factor would result in skipping of the ESE-containing exon during splicing. This cytoplasmic feedback model predicts that in cells expressing both the wild-type and PTC-containing alleles of the same gene, splicing of both types of pre-mRNAs would be affected.
We postulate that the first ribosome binds the exon-specific splicing factor as it translates the VDJ exon of the mRNA and releases it only upon encountering the normal translation termination codon, in an mRNA devoid of EJC proteins. Thus, the efficient return of this factor to the nucleus would depend on translation of the entire coding region of the mRNA, an event that would not occur if the ribosome encountered a PTC during the pioneering round of translation. Comparable sequestration of other sequence-specific splicing factors might account for other forms of splice site selection that lead to skipping of specific PTC-containing sequences (Gersappe et al. 1999; Li et al. 2002). Association of the VDJ exon binding protein(s) with cytoplasmic ribosomes might also account for the special, highly efficient NMD (VDJ-exon dependent “Super NMD”) to which this TCR-β mRNA is subject (Gudikote and Wilkinson 2002).
A distinction between the nuclear and cytoplasmic translation models is the testable prediction that cytoplasmic sequestration of a factor would affect the splicing of the pre-mRNAs synthesized off both alleles of a TCR-β gene. To our knowledge, this prediction has not yet been tested in a controlled experiment. Thus, the possibility remains that PTC monitoring during both NMD or NAS occurs by translation in the perinuclear cytoplasm, obviating the need to propose nuclear translation at this time.
CONCLUSIONS
Does translation occur in the nuclei of cells? As discussed here, published papers on this subject are in conflict. The papers that appear to support the notion of nuclear translation lack critical information, leading us to question their conclusions until further data or more definitive experiments are presented. Also, a compelling case has not been made that it is needed to explain how the translation machinery influences the processing and turnover of mRNAs. However, papers that argue against nuclear translation, in trying to prove the absence of an event, are also open to criticisms, such as those concerning the activities of the preparations used or the sensitivities of the detection systems employed. Thus, the possibility remains that evidence for the existence of nuclear translation may some day be obtained. Although the idea of nuclear translation has a certain appeal, it unnecessarily posits the existence of an elaborate system for which there is no clear evidence. Because we are reluctant to invoke complex explanations when simpler ones suffice, at this time we favor the idea that translation does not occur in the nuclei of cells.
Acknowledgments
We thank Marvin P. Wickens and Phillip Anderson for critically reading the manuscript and M. Wilkinson, H. Dietz, D. Görlich, and U. Kutay for communication of unpublished data. We are supported by National Institutes of Health Grants GM30220 to J.E.D. and GM51836 to E.B.G. We apologize to many colleagues whose contributions were not cited due to limited space.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2121703.
REFERENCES
- Bohnsack, M., Regener, K., Schwappach, B., Saffrich, R., Paraskeva, E., Hartmann, E., and Gorlich, D. 2002. Exp5 exports eEF1A via tRNA from nuclei and synergises with other transport pathways to confine translation to the cytoplasm. EMBO J. 21: 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna, S. 2001. Pre-mRNA processing: Insights from nonsense. Curr. Biol. 11: R838–R841. [DOI] [PubMed] [Google Scholar]
- Brogna, S., Sato, T., and Rosbash, M. 2002. Ribosome components are associated with sites of transcription. Mol. Cell 10: 93–104. [PubMed] [Google Scholar]
- Bühler, M., Wilkinson, M., and Mühlemann, O. 2002. Intranuclear degradation of nonsense codon-containing mRNA. EMBO Rep. 3: 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, J.F., Screaton, G.R., and Krainer, A.R. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & Dev. 12: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado, A., Treichel, N., Müller, E.-C., Otto, A., and Kutay, U. 2002. Exportin 5 mediated nuclear export of elongation factor 1A and tRNA. EMBO J. 21: 6216–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi, M., Kendzior, R.J., and Beemon, K.L. 2002. A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes & Dev. 16: 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285–298. [DOI] [PubMed] [Google Scholar]
- Dostie, J. and Dreyfuss, G. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Etchison, D. and Etchison, J.R. 1987. Monoclonal antibody-aided characterization of cellular p220 in uninfected and poliovirus-infected HeLa cells: Subcellular distribution and identification of conformers. J. Virol. 61: 2702–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes, P., Inada, T., Preiss, T., Hentze, M.W., Mattaj, I.W., and Sachs, A.B. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6: 191–196. [PubMed] [Google Scholar]
- Frischmeyer, P.A. and Dietz, H.C. 1999. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8: 1893–1900. [DOI] [PubMed] [Google Scholar]
- Gersappe, A., Burger, L., and Pintel, D.J. 1999. A premature termination codon in either exon of minute virus of mice P4 promoter-generated pre-mRNA can inhibit nuclear splicing of the intervening intron in an open reading frame-dependent manner. J. Biol. Chem. 274: 22452–22458. [DOI] [PubMed] [Google Scholar]
- Goidl, J. and Allen, W. 1978. Does protein synthesis occur within the nucleus? Trends Biol. Sci. 3: N225–N228. [Google Scholar]
- Gudikote, J.P. and Wilkinson, M.F. 2002. T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J. 21: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her, L.S., Lund, E., and Dahlberg, J.E. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. [See comments.] Science 276: 1845–1848. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., Jackson, D.A., and Cook, P.R. 2001. Coupled transcription and translation within nuclei of mammalian cells. [See comments.] Science 293: 1139–1142. [DOI] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Kim, V.N., Kataoka, N., and Dreyfuss, G. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science 293: 1832–1836. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejbkowicz, F., Goyer, C., Darveau, A., Neron, S., Lemieux, R., and Sonenberg, N. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA 89: 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Ishigaki, Y., Li, X., and Maquat, L.E. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 21: 3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Wachtel, C., Miriami, E., Yahalom, G., Friedlander, G., Sharon, G., Sperling, R., and Sperling, J. 2002. Stop codons affect 5′ splice-site selection by surveillance of splicing. Proc. Natl. Acad. Sci. USA 99: 5277–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., Cartegni, L., Zhang, M.Q., and Krainer, A.R. 2001. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. [See comments.] Nat. Genet. 27: 55–58. [DOI] [PubMed] [Google Scholar]
- Lund, E. and Dahlberg, J.E. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. [See comments.] Science 282: 2082–2085. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., and Steitz, J.A. 2001. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 1995. When cells stop making sense: Effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1: 453–465. [PMC free article] [PubMed] [Google Scholar]
- ——— 2002. NASty effects on fibrillin pre-mRNA splicing: Another case of ESE does it, but proposals for translation-dependent splice site choice live on. Genes & Dev. 16: 1743–1753. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173–176. [DOI] [PubMed] [Google Scholar]
- Marshall, I.C., Gant, T.M., and Wilson, K.L. 1997. Ionophore-releasable lumenal Ca2+ stores are not required for nuclear envelope assembly or nuclear protein import in Xenopus egg extracts. Cell Calcium 21: 151–161. [DOI] [PubMed] [Google Scholar]
- McKendrick, L., Thompson, E., Ferreira, J., Morley, S.J., and Lewis, J.D. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 21: 3632–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell, J.T. and Dietz, H.C. 2001. When the message goes awry: Disease-producing mutations that influence mRNA content and performance. Cell 107: 411–414. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., ap Rhys, C.M.J., and Dietz, H.C. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419–422. [DOI] [PubMed] [Google Scholar]
- Mühlemann, O., Mock-Casagrande, C.S., Wang, J., Li, S., Custodio, N., Carmo-Fonseca, M., Wilkinson, M.F., and Moore, M.J. 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8: 33–43. [DOI] [PubMed] [Google Scholar]
- Nadano, D., Ishihara, G., Aoki, C., Yoshinaka, T., Irie, S., and Sato, T.A. 2000. Preparation and characterization of antibodies against human ribosomal proteins: Heterogeneous expression of S11 and S30 in a panel of human cancer cell lines. Jpn J. Cancer Res. 91: 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson, L., Xia, T., and Deutscher, M. 2003. Nuclear protein synthesis: A re-evaluation. RNA (this issue). [DOI] [PMC free article] [PubMed]
- Page, M.F., Carr, B., Anders, K.R., Grimson, A., and Anderson, P. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19: 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, L., Theodor, L., Carter, M., Vu, M.N., Sasaki, A.W., and Wilkinson, M.F. 1993. T cell receptor-β mRNA splicing: Regulation of unusual splicing intermediates. Mol. Cell. Biol 13: 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringborg, U., Daneholt, B., Edstrom, J.E., Egyhazi, E., and Rydlander, L. 1970. Evidence for transport of preribosomal RNA from the nucleolus to the chromosomes in Chironomus tentans salivary gland cells. J. Mol. Biol. 51: 679–686. [DOI] [PubMed] [Google Scholar]
- Sarkar, S., Azad, A.K., and Hopper, A.K. 1999. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96: 14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell, T., Kulozik, A.E., and Hentze, M.W. 2002. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol. Rev. 3: 1006.1–1006.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger, B., Lafontaine, D.L., Graindorge, J.S., Gadal, O., Camasses, A., Sanni, A., Garnier, J.M., Breitenbach, M., Hurt, E., and Fasiolo, F. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8: 1363–1373. [DOI] [PubMed] [Google Scholar]
- Si, K. and Maitra, U. 1999. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell. Biol. 19: 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubing, C. and Clapham, D.E. 1999. Active nuclear import and export is independent of lumenal Ca2+ stores in intact mammalian cells. J. Gen. Physiol. 113: 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 261–311. [DOI] [PubMed] [Google Scholar]
- Wagner, E. and Lykke-Andersen, J. 2002. mRNA surveillance: The perfect persist. J. Cell Sci. 115: 3033–3038. [DOI] [PubMed] [Google Scholar]
- Wang, J., Hamilton, J.I., Carter, M.S., Li, S., and Wilkinson, M. 2002a. Alternatively spliced TCR mRNA induced by disruption of reading frame. Science 297: 108–110. [DOI] [PubMed] [Google Scholar]
- Wang, J., Chang, Y., Hamilton, J.I., and Wilkinson, M.F. 2002b. Nonsense-associated altered splicing: A frame-dependent response distinct from nonsense-mediated decay. Mol. Cell. 10: 951–957. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M. and Shyu, A. 2002. RNA surveillance by nuclear scanning? Nat. Cell Biol. 4: E144–E147. [DOI] [PubMed] [Google Scholar]